Abstract
Three-dimensional (3D) cardiac magnetic resonance (CMR) perfusion imaging is promising for the precise sizing of defects and for providing high perfusion contrast, but remains an experimental approach primarily due to the need for large-dimensional encoding, which, for traditional 3DFT imaging, requires either impractical acceleration factors or sacrifices in spatial resolution. We demonstrated the feasibility of rapid 3D CMR perfusion imaging using a stack-of-spirals acquisition accelerated by non-Cartesian k-t SENSE, which enables entire myocardial coverage with an in-plane resolution of 2.4 mm. The optimal under-sampling pattern was employed to achieve the largest separation between true and aliased signals, which is a pre-requisite for k-t SENSE reconstruction. Flip angle and saturation recovery time were chosen to ensure negligible magnetization variation during the transient data acquisition. We compared the proposed 3D perfusion method with the standard 2DFT approach by consecutively acquiring both data during each R-R interval in cardiac patients. The mean and standard deviation of the correlation coefficients between time intensity curves (TICs) of 3D vs 2DFT were 0.94 and 0.06 across seven subjects. The linear correlation between the two sets of upslope values was significant (r=0.78, P<0.05).
Keywords: 3D CMR perfusion, stack-of-spirals acquisition, k-t SENSE, respiratory motion correction
Introduction
For robust and accurate identification of myocardial ischemia, cardiac magnetic resonance (CMR) perfusion sequences need to meet several technical requirements, including high spatial resolution, large spatial coverage, high contrast-to-noise ratio (CNR), and high temporal resolution (1). Two-dimensional (2D) multi-slice saturation-prepared acquisition with parallel imaging has been the most established method. With this approach, 3–4 short-axis slices with 2–3 mm in-plane spatial resolution can be obtained per R-R interval on clinical scanners (2). Recently, the spatial coverage and spatial resolution have been improved by using advanced acceleration techniques such as k-t SENSE, compressed sensing, and highly constrained backprojection (3–5).
Three-dimensional (3D) CMR perfusion imaging has been sought as an alternative to the 2D multi-slice approach due to its potential advantages (6–8). Three-dimensional volumetric imaging of the entire myocardium enables more accurate quantification of the extent of myocardial ischemia, which has been shown to be the most important prognostic factor (9,10). Three-dimensional CMR also inherently provides higher signal-to-noise ratio (SNR) and CNR, as well as a greater potential for high rates of acceleration due to increased sensitivity encoding capacity and spatio-temporal sparsity.
Despite the aforementioned potential advantages, 3D CMR perfusion imaging has remained as an experimental method mainly due to the requirement of large-dimensional encoding during a limited acquisition time. The feasibility of 3D perfusion imaging was demonstrated using parallel-imaging-accelerated 3DFT acquisition, but only moderate spatial resolution (2.8×4.2×10 mm3) was achievable as a tradeoff for complete left ventricular (LV) coverage (6,7,11). Recently, Vitanis et al. reported high-resolution (2.3×2.3×10 mm3) 3D CMR perfusion imaging using compartment-based k-t principal component analysis, which improved reconstruction accuracy by further exploiting signal correlation within each of the right ventricle, LV and myocardium (8).
While Cartesian readout has been used predominantly in CMR perfusion sequences, a spiral trajectory is a promising alternative, particularly for 3D, due to its high readout efficiency. Potential issues in using spiral readout for CMR perfusion include data inconsistency artifacts caused by spiral k-space modulation during the transient data acquisition, image blurring caused by field inhomogeneity, and susceptibility artifacts. Recently, Salerno et al. demonstrated that these potential artifacts could be abated through careful choice of imaging parameters (12) by acquiring high-quality three-slice perfusion images in patients. In our study, stack-of-spirals readouts combined with non-Cartesian k-t SENSE were employed for rapid acquisition of 3D perfusion data. We developed a whole-heart 3D spiral CMR perfusion imaging method with a spatial resolution of 2.4×2.4×9 mm3 and evaluated its performance in comparison with standard 2D multi-slice perfusion imaging.
Methods
Pulse sequence
The pulse sequence consisted of two saturation recovery acquisition modules for consecutive 3D spiral and single-slice 2DFT perfusion imaging per each R-R interval (Fig. 1). The 3D spiral imaging consisted of a saturation preparation, saturation recovery time (TSR,spr), a fat saturation pulse, and a 3D stack-of-spirals data acquisition. Immediately after the 3D spiral acquisition, a saturation recovery 2DFT acquisition was carried out in an analogous manner. Four-chamber-view cine images were used to identify the start of quiescent diastolic phase relative to the R wave. The 3D spiral acquisition was synchronized to the stable diastolic phase by detecting the R wave at every TR of the 2DFT acquisition and adjusting the subsequent delay to the saturation pulse for the next spiral acquisition (TD in Fig. 1). During the first cardiac cycle, low-resolution images were acquired at two different echo times with the saturation pulse turned off and used to compute a field map.
Figure 1.
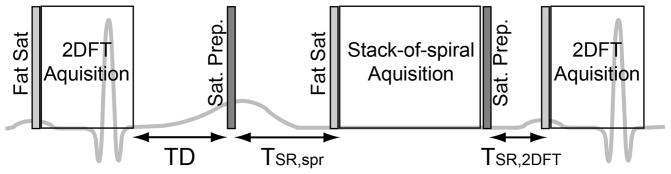
Schematic of the pulse sequence for whole-heart 3D spiral and single-slice 2DFT perfusion imaging. The pulse sequence consists of two consecutive modules of a saturation RF pulse, a saturation recovery time, a fat saturation RF pulse, and a GRE acquisition. The 3D spiral acquisition was synchronized to mid-diastole by using ECG gating and adjusting the delay time between the 2DFT acquisition and the saturation preparation for the next spiral imaging (TD).
k-t SENSE sampling and reconstruction
A set of ten dual-density spiral trajectories was designed for a given in-plane field-of-view (FOVr) and spatial resolution such that inner k-space was 5-fold over-sampled and outer k-space was fully (Nyquist) sampled with a transition centered at 18% of the k-space radius (Fig. 2a). Two interleaves were acquired out of the ten interleaves per kz encoding step, with the net effect being full sampling of the inner k-space and 5-fold under-sampling of the outer k-space. Images were reconstructed from the under-sampled k-space data using non-Cartesian k-t SENSE reconstruction (13).
Figure 2.
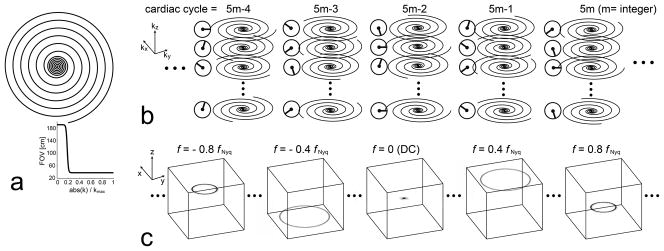
(a): Dual density spiral interleaf. Inner k-space was 5-fold over-sampled and outer k-space was fully (Nyquist) sampled, with a Fermi function-based transition centered at 18% of the k-space radius. (b): Temporal sampling scheme of 5-fold under-sampled stack-of-spirals acquisition in kx-ky-kz-t domain. With respect to ten interleaves per kz for full FOV, two acquired interleaves were rotated by two interleaves in the kz direction (72°), and were rotated by four interleaves over cardiac cycles (144°). (c): Illustration of the resultant point spread function (PSF) in x-y-z-f domain. The PSF is non-zero at only five temporal frequencies (DC, ±0.4 fNyq, ±0.8 fNyq) due to the lattice structure of the sampling pattern. Four ring-shaped side-lobes are maximally displaced from the main-lobe, which significantly reduces potential overlap between a true object and its aliases.
In time-resolved 3D under-sampling, aliased signals can be described by a convolution between a fully sampled signal and a point spread function in x-y-z-f space. A large separation between the main lobe and side lobes is generally favored to minimize the overlap between the true signal and its aliases (14). In searching for an optimal sampling function that achieves the largest separation, we restricted paired sampling indices of spiral interleaves and kz encoding to conform to a lattice structure, as in Ref. (14) with Cartesian sampling. Figure 2 represents the selected under-sampling strategy in kx-ky-kz-t domain (b), and the corresponding PSF in x-y-z-f domain (c). The sampled spiral interleaves were rotated by two interleaves (72°) in the kz direction, and were also rotated by four interleaves (144°) over cardiac cycles. For instance, at the (5m-4)th cardiac cycle (m=integer), sampled spiral indices were [1,6], [2,7], [3,8], and [4,9] at kz = 1,2,3, and 4, respectively. The distance between the main-lobe and the first side-lobe was 0.4×FOVr on the x-y plane, 0.4×FOVz in the z-direction, and 0.4 fNyq in the frequency direction. The distance between the main-lobe and the second side-lobe was 0.2×FOVr on the x-y plane, 0.2×FOVz in the z-direction, and 0.8 fNyq in the frequency direction.
k-t SENSE reconstruction with linear off-resonance correction was performed by solving the following system equation iteratively using the conjugate gradient (CG) method (15).
| (1) |
where ρxf and dkt are an unknown 4D image in x-y-z-f space and stack-of-spirals data measured by multiple receiver coils, respectively. The encoding matrix A is defined as A = [ΦFx→kS Ff→t] where Ff→t, S, and Fx→k denote 1D inverse Fourier transform from f to t, multiplication by coil sensitivities, and 3D Fourier transform from x to k, respectively, and Φ denotes phase modulation of k-space samples by the estimated DC value of field inhomogeneity, f0. Mxf represents approximated signal distribution in x-y-z-f space learned from the low resolution training data. To speed up convergence of the CG algorithm, a diagonal preconditioner D was used whose entries are the inverses of the diagonal elements of the system matrix .
The reference single-slice 2DFT perfusion imaging was accelerated by Cartesian k-t SENSE. Fifteen phase encoding lines at k-space center were always sampled to be used for training data, and the remaining outer k-space was under-sampled sequentially in kx-ky-t space by a factor of three.
Selection of sequence parameters
Perfusion imaging with a non-Cartesian acquisition is prone to k-space amplitude modulation during the transient data acquisition, which may cause severe distortion of PSF and thus noticeable image artifacts (12). Sequence parameters were selected to ensure a negligible magnetization variation while maintaining high perfusion contrast in the following way.
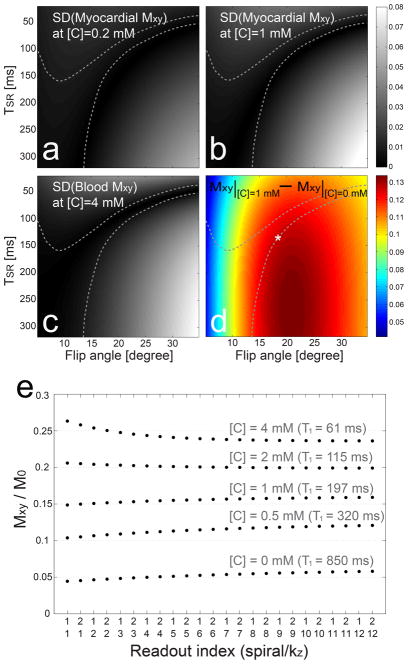
Transverse magnetizations during a saturation recovery acquisition were simulated as a function of saturation recovery time TSR, readout flip angle α, and T1 values of the myocardium and blood pool. The concentration of the contrast agent [C] was assumed to range from 0 to 1 mmole/L in the myocardium and from 0 to 4 mmol/L in blood pool, and was converted to T1 values using a relaxivity of 3.9 L/mmol/s (16). Figure 3 shows the standard deviation of magnetization as a function of TSR and α when [C] is 0.2 mM (a), 1 mM (b) in the myocardium, and 4 mM (c) in blood pool, respectively. Despite large T1 difference among the three cases, the region of small magnetization variations (dark areas) does not significantly change. Candidates of paired parameters (TSR, α) were restricted to ones that produce standard deviations smaller than 0.01 M0 for all tested T1 values. Among these candidates, (TSR, α) was selected to produce the largest perfusion contrast defined as the difference between myocardial magnetization at [C]=1 mM and myocardial magnetization at [C]=0. Figure 3d shows the region of small magnetization variation (enclosed by dotted contours), and the selected pair of parameters (white asterisk), on top of the color map of the simulated perfusion contrast. The magnetization response simulated using the selected (TSR, α) confirms only slight variation in a wide range of T1 values (Fig. 3e).
Figure 3.
Selection of flip angle α and saturation recovery time TSR. Transverse magnetizations from saturation recovery imaging were simulated over a range of α, TSR, and the concentration of the contrast agent [C]. The standard deviation (SD) of the magnetization response exhibits weak dependency on contrast concentration as shown in a–c with [C] = 0.2 mM, 1 mM and 4 mM, respectively. Among candidate sets of (α, TSR) that generate SDs smaller than 0.01 M0 (enclosed by dotted contours), we selected one that produced the largest perfusion contrast defined as differential transverse magnetizations between [C] = 1 mM and [C] = 0 mM. The selected pair is denoted by white asterisk on top of color visualization of the perfusion contrast in d. The resultant magnetization responses using the selected parameters show only marginal variations over a range of contrast concentration (e). Furthermore, only two spiral interleaves are acquired per kz encode and thus, the in-plane modulation along spiral trajectories will be negligible.
Imaging protocols
The dual imaging of 3D spiral and 2DFT perfusion was performed on a GE 1.5 Tesla scanner, using an eight-channel cardiac coil array for signal reception. Rest perfusion scans were performed in 7 patients (2 males, age=54±15) who were undergoing clinically ordered CMR examinations. Written informed consent as approved by the Institutional Review Board was obtained from all participants.
The 3D spiral imaging parameters included saturation recovery time = 140 ms, flip angle = 17°, spatial resolution = 2.4×2.4×9 mm3, FOV = 38×38×90 cm3, 10 section slices (after discarding 2 edge slices), TR = 9.8 ms, spiral readout time = 7.1 ms, readout bandwidth = 125 kHz, acquisition time = 230 ms, and the number of time frames = 40. The 2DFT imaging parameters were chosen for a standard perfusion sequence based on Ref (2): saturation recovery time = 40 ms, flip angle = 12°, spatial resolution = 2.4×2.4 mm2, FOV = 33×33 cm2, slice thickness = 9 mm, TR = 2.8 ms, readout bandwidth = 62.5 kHz, acquisition time = 157 ms. 2DFT data were acquired at the location of the center slice of 3D spiral images. Contrast media (0.1 mmol/kg, Multi-Hance) was injected at a rate of 3–4 ml/s followed by 20 ml saline. Subjects were instructed to hold their breath as long as possible.
Image registration and analysis
The time series of 3D perfusion images was registered to compensate for respiratory drift. A 2D polygonal ROI was specified on a mid-short-axis slice, and was copied to the center eight slices to generate a 3D cylindrical ROI. 3D translations were iteratively found that produced the largest correlation between consecutive cardiac cycles. Due to signal changes in blood pools and the myocardium over time, mutual information was used as a correlation measure, which calculates a degree of similarity based on image contrast (17).
Region-based time intensity curve (TIC) analyses were performed for a comparison between 2DFT perfusion image and the center slice of 3D spiral image. Prior to analyses, raw perfusion images were divided by pre-contrast images obtained by averaging data from the 4th through 7th cardiac cycles, to compensate for the different sequence parameters between the two acquisitions. First, correlation coefficients between the TICs of 3D spiral and 2DFT images were calculated to investigate similarity of overall temporal dynamics captured by the two acquisitions. This calculation excluded data at the first two and the last two time frames, which tend to suffer from filtering effects of k-t SENSE reconstruction. Second, upslope values were computed by linear fitting of the TIC during signal enhancement to investigate similarity of a semi-quantitative perfusion index. Peak SNR and perfusion CNR were computed for 3D spiral perfusion (18,19).
Results
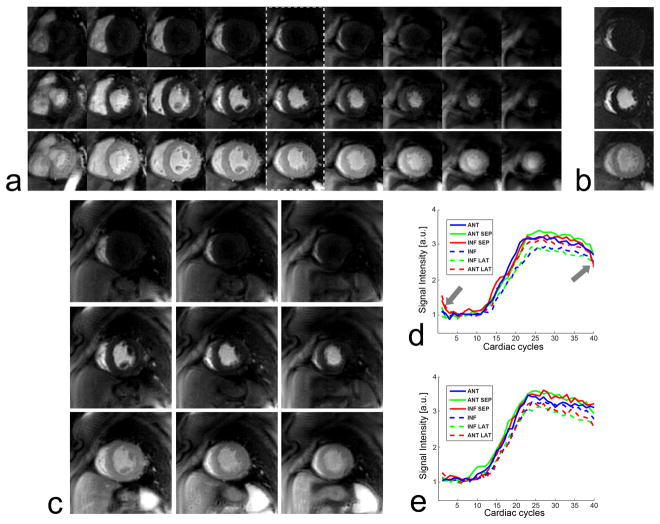
Figure 4 contains representative 3D spiral and 2DFT perfusion images from a patient without perfusion defects. Images at peak RV enhancement, peak LV enhancement, and peak myocardial enhancement are shown from the top to the bottom rows in Figs. 4a and 4b. Contrast enhancement is homogeneous over the entire myocardium, consistent with normal perfusion. k-t SENSE reconstruction effectively suppressed aliasing artifacts of the 5-fold spiral under-sampling. The 2DFT images and the corresponding center slice of the 3D data (denoted by the dotted rectangle) exhibit nearly the same contrast uptake and wash-out through the RV, LV, and myocardium. This is confirmed by the TICs from the two data sets (Figs. 4d and 4e). The number of CG iterations in the image reconstruction varied from 7 to 15 over seven data sets, which resulted in reconstruction time of 16 – 34 min on a desktop computer equipped with a 2.67 GHz Intel processor and 8 GB RAM.
Figure 4.
Representative 3D spiral perfusion images (a) and single-slice 2DFT perfusion images (b) in a patient without perfusion defects. Peak RV enhancement, peak LV enhancement, and peak myocardial enhancement are shown from the top to the bottom rows. Less cropped versions of 3D spiral images are also shown for the 4th, 5th and 6th slices (c). Due to the normal perfusion, contrast enhancement is homogeneous over the entire myocardium. The 2DFT perfusion images and the corresponding center slice of 3D perfusion images (denoted by dotted rectangle) show nearly the same dynamics of contrast uptake, as validated by their time intensity curves (c, d). The temporal filtering effects of k-t SENSE reconstruction caused errors in the first few and last few time frames (denoted by arrows), yet their effects on the depiction of contrast enhancement appear to be negligible.
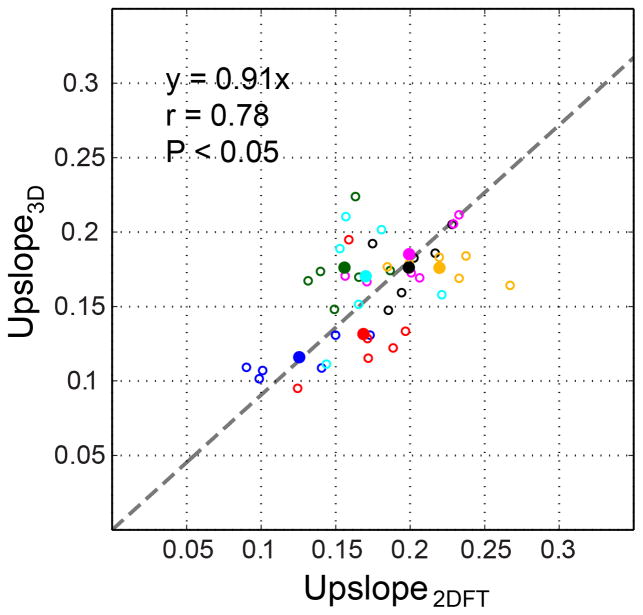
The mean and standard deviation of the correlation coefficients between TICs of 2DFT images and TICs of the center slice of 3D images were 0.94 and 0.06 across 42 myocardial segments from 7 subjects. This demonstrates excellent correlation between the temporal dynamics of the contrast enhancement captured by 2DFT and 3D spiral perfusion imaging. Figure 5 is a scatter plot of the TIC upslopes of 3D spiral and 2DFT data. All 42 upslope pairs across 7 subjects (6 per subject, open circle) as well as 7 pairs of per-subject average upslope (solid circles) are shown. The linear correlation of the 7 pairs of average upslopes was significant (r=0.78, P < 0.05). The slope of the linear fit of the 7 pairs was slightly smaller than 1.0, presumably due to the non-linear relationship between the saturation recovery time and signal intensity, which cannot be completely compensated by the pre-contrast normalization. Peak SNR and CNR of 3D spiral images were 66.7±22.8 and 20.2±8.7.
Figure 5.
Scatter plot of normalized TIC upslope values of 2DFT perfusion images and the center slice of 3D perfusion images. 42 upslope pairs across 7 subjects (6 pairs per subject) are displayed by open circles, and 7 average upslope pairs are displayed by solid circles, using a unique color per subject. Linear fitting of the 7 average upslope pairs results in y = 0.91x.
Discussion
We have demonstrated the feasibility of k-t SENSE accelerated 3D spiral cardiac perfusion MRI with a comparison to 2DFT perfusion MRI. With the interleaved under-sampling scheme that maximizes the distance between the main-lobe and side-lobes, aliases from 5-fold under-sampled data were effectively suppressed using the iterative non-Cartesian k-t SENSE reconstruction, which resulted in high-quality perfusion images covering the entire LV. The reference 2DFT perfusion images and 3D spiral perfusion images at the same slice location showed high correlation in overall temporal dynamics as well as TIC upslopes.
The contiguous 3D coverage may be advantageous for the registration of perfusion images, which can expedite subsequent quantitative perfusion analyses. We observed that the 3D translation-based registration significantly improved the continuity of TICs in a few data sets that involved small to moderate respiratory drifts. However, the accuracy of the motion compensation needs to be investigated further though systematic studies in a large number of data sets containing different levels of respiratory motion. Another practical issue of the registration used in this study is the need for manual ROI specification, which is a fundamental drawback of rigid registration methods. Application of fully automatic registration methods based on non-rigid models may improve the consistency of registration results and reduce the processing time (20,21).
The sensitivity to respiratory motion is a practical issue of the presented 3D perfusion imaging. Respiratory motion reduces spatio-temporal correlation of perfusion data and thus increases k-t SENSE reconstruction error, which may appear as spatio-temporal blurring and ghosting artifact (or whirling artifact in spiral imaging) (22,23). All patients in the present study were able to maintain a good breath-hold at least until the peak myocardial enhancement (about 25 cardiac cycles). In a few patients, significant respiratory motion occurred after the peak enhancement, but images during contrast enhancement in the LV and the myocardium were not severely affected (mild blurring), allowing qualitative and TIC-based assessment of perfusion. Temporally constrained k-t reconstruction using principal component analysis was recently shown to improve robustness to respiratory motion, and its application to 3D spiral perfusion may be promising to reduce the respiratory motion sensitivity (23).
A limitation of this study is that only rest perfusion scans were performed as subjects were recruited from those who were undergoing routine clinical CMR examinations. All seven patients who participated in this study showed normal first-pass contrast enhancement in both 3D and 2DFT perfusion images. To validate the clinical utility of the proposed 3D perfusion imaging technique, stress perfusion studies in patients with ischemic defects would be necessary. The main goal of this study was to demonstrate the performance of k-t SENSE reconstruction of under-sampled stack-of-spirals perfusion data by a comparison with the reference 2DFT perfusion.
A relaxivity of 3.9 L/mmol/s was assumed in the Bloch simulation for determining the saturation recovery time and flip angle as Gd-DTPA (Magnevist) was routinely used at the time of preliminary studies. It is acknowledged, however, that perfusion scans on patients were performed with Gd-BOPTA (Multi-Hance), which meant the sequence parameters were less optimal for the higher relaxivity (8.1 L/mmol/s). However, a Bloch simulation showed that the standard deviation of magnetization and perfusion contrast were minimally affected by the less optimal parameters, due to weak dependency of Gadolinium concentration on transient magnetization response. The standard deviation was still smaller than 0.01 M0 for all tested Gadolinium concentrations when simulated with a relaxivity of 8.1 L/mmol/s and the parameters used in this study. Also, simulated perfusion contrast was smaller than the optimal case by only 6% (0.17 M0 vs 0.18 M0).
Conclusions
We have demonstrated the feasibility of a high-resolution whole-heart first-pass 3D perfusion imaging method using a k-t SENSE accelerated stack-of-spirals acquisition. An optimal under-sampling scheme was employed for the maximal separation of true and aliased signals, and sequence parameters were carefully chosen to minimize transient data inconsistency artifacts. As a result, high-quality images were acquired across the entire LV with a 2.4 mm in-plane spatial resolution, as validated by excellent correlation with the reference 2DFT perfusion images in terms of overall temporal dynamics and upslope values of TICs.
Acknowledgments
This work was supported by an American Heart Association postdoctoral fellowship to T. Shin (09POST2150025).
Grant Sponsors:
National Institutes of Health (R01 HL039297)
References
- 1.Gerber BL, Raman SV, Nayak KS, Epstein FH, Ferreira P, Axel L, Kraitchman DL. Myocardial first-pass perfusion cardiovascular magnetic resonance: history, theory, and current state of the art. J Cardiovasc Magn Reson. 2008;10:18. doi: 10.1186/1532-429X-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kellman P, Arai AE. Imaging sequences for first pass perfusion - a review. J Cardiovasc Magn Reson. 2007;9(3):525–537. doi: 10.1080/10976640601187604. [DOI] [PubMed] [Google Scholar]
- 3.Plein S, Ryf S, Schwitter J, Radjenovic A, Boesiger P, Kozerke S. Dynamic contrast-enhanced myocardial perfusion MRI accelerated with k-t SENSE. Magn Reson Med. 2007;58(4):777–785. doi: 10.1002/mrm.21381. [DOI] [PubMed] [Google Scholar]
- 4.Otazo R, Kim D, Axel L, Sodickson DK. Combination of compressed sensing and parallel imaging for highly accelerated first-pass cardiac perfusion MRI. Magn Reson Med. 2010;64(3):767–776. doi: 10.1002/mrm.22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ge L, Kino A, Griswold M, Mistretta C, Carr JC, Li D. Myocardial perfusion MRI with sliding-window conjugate-gradient HYPR. Magn Reson Med. 2009;62(4):835–839. doi: 10.1002/mrm.22059. [DOI] [PubMed] [Google Scholar]
- 6.Kellman P, Zhang Q, Larson AC, Simonetti OP, McVeigh ER, Arai AE. Cardiac first-pass perfusion MRI using 3D true FISP parallel imaging using TSENSE. Proceeding of ISMRM, 12th Annual Meeting; Kyoto. 2004. p. 310. [Google Scholar]
- 7.Shin T, Hu HH, Pohost GM, Nayak KS. Three-dimensional first-pass myocardial perfusion imaging at 3T: Feasibility study. J Cardiovasc Magn Reson. 2008;10:57. doi: 10.1186/1532-429X-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vitanis V, Manka R, Giese D, Pedersen H, Plein S, Boesiger P, Kozerke S. High resolution three-dimensional cardiac perfusion imaging using compartment-based k-t principal component analysis. Magn Reson Med. 2011;65(2):575–587. doi: 10.1002/mrm.22620. [DOI] [PubMed] [Google Scholar]
- 9.Brown KA, Boucher CA, Okada RD, Guiney TE, Newell JB, Strauss HW, Pohost GM. Prognostic value of exercise thallium-201 imaging in patinets presenting for evaluation of chest pain. J Am Coll Cardiol. 1983;1:994–1001. doi: 10.1016/s0735-1097(83)80100-4. [DOI] [PubMed] [Google Scholar]
- 10.Hachamovitch R, Berman DS, Shaw LJ, Kiat H, Cohen I, Cabico JA, Friedman J, Diamond GA. Incremental prognostic value of myocardial perfusion single photon emission computed tomography for the prediction of cardiac death. Circ. 1998;97(6):535–543. doi: 10.1161/01.cir.97.6.535. [DOI] [PubMed] [Google Scholar]
- 11.Shin T, Pohost GM, Nayak KS. Systolic 3D first-pass myocardial perfusion MRI: Comparison with diastolic imaging in healthy subjects. Magn Reson Med. 2010;63(4):858–864. doi: 10.1002/mrm.22315. [DOI] [PubMed] [Google Scholar]
- 12.Salerno M, Sica CT, Kramer CM, Meyer CH. Optimization of spiral-based pulse sequences for first-pass myocardial perfusion imaging. Magn Reson Med. 2011;65(6):1602–1610. doi: 10.1002/mrm.22746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsao J, Boesiger P, Pruessmann KP. k-t BLAST and k-t SENSE: dynamic MRI with high frame rate exploiting spatiotemporal correlations. Magn Reson Med. 2003;50(5):1031–1042. doi: 10.1002/mrm.10611. [DOI] [PubMed] [Google Scholar]
- 14.Tsao J, Kozerke S, Boesiger P, Pruessmann KP. Optimizing spatiotemporal sampling for k-t BLAST and k-t SENSE: application to high-resolution real-time cardiac steady-state free precession. Magn Reson Med. 2005;53(6):1372–1382. doi: 10.1002/mrm.20483. [DOI] [PubMed] [Google Scholar]
- 15.Hansen MS, Baltes C, Tsao J, Kozerke S, Pruessmann KP, Eggers H. k-t BLAST reconstruction from non-Cartesian k-t space sampling. Magn Reson Med. 2006;55(1):85–91. doi: 10.1002/mrm.20734. [DOI] [PubMed] [Google Scholar]
- 16.Pintaske J, Martirosian P, Graf H, Erb G, Lodemann KP, Claussen CD, Schick F. Relaxivity of Gadopentetate Dimeglumine (Magnevist), Gadobutrol (Gadovist), and Gadobenate Dimeglumine (MultiHance) in human blood plasma at 0.2, 1. 5, and 3. Tesla. Invest Radiol. 2006;41(3):213–221. doi: 10.1097/01.rli.0000197668.44926.f7. [DOI] [PubMed] [Google Scholar]
- 17.Pluim JPW, Maintz JBA, Viergever MA. Mutual-information-based registration of medical images: a survey. IEEE Trans Med Imaging. 2003;22(8):986–1004. doi: 10.1109/TMI.2003.815867. [DOI] [PubMed] [Google Scholar]
- 18.Reeder SB, Wintersperger BJ, Dietrich O, Lanz T, Greiser A, Reiser MF, Glazer GM, Schoenberg SO. Practical approaches to the evaluation of signal-to-noise ratio performance with parallel imaging: application with cardiac imaging and a 32-channel cardiac coil. Magn Reson Med. 2005;54(3):748–754. doi: 10.1002/mrm.20636. [DOI] [PubMed] [Google Scholar]
- 19.Fenchel M, Helber U, Simonetti OP, Stauder NI, Kramer U, Nguyen CN, Finn JP, Claussen CD, Miller S. Multislice first-pass myocardial perfusion imaging: Comparison of saturation recovery (SR)-TrueFISP-two-dimensional (2D) and SR-TurboFLASH-2D pulse sequences. J Magn Reson Imaging. 2004;19(5):555–563. doi: 10.1002/jmri.20050. [DOI] [PubMed] [Google Scholar]
- 20.Milles J, van der Geest RJ, Jerosch-Herold M, Reiber JH, Lelieveldt BP. Fully automated motion correction in first-pass myocardial perfusion MR image sequences. IEEE Trans Med Imaging. 2008;27(11):1611–1621. doi: 10.1109/TMI.2008.928918. [DOI] [PubMed] [Google Scholar]
- 21.Li C, Sun Y. Nonrigid registration of myocardial perfusion MRI using pseudo ground truth. Med Image Comput Comput Assist Interv. 2009;12(Pt 1):165–172. doi: 10.1007/978-3-642-04268-3_21. [DOI] [PubMed] [Google Scholar]
- 22.Plein S, Kozerke S, Suerder D, Luescher TF, Greenwood JP, Boesiger P, Schwitter J. High spatial resolution myocardial perfusion cardiac magnetic resonance for the detection of coronary artery disease. Eur Heart J. 2008;29(17):2148–2155. doi: 10.1093/eurheartj/ehn297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedersen H, Kozerke S, Ringgaard S, Nehrke K, Kim WY. k-t PCA: temporally constrained k-t BLAST reconstruction using principal component analysis. Magn Reson Med. 2009;62(3):706–716. doi: 10.1002/mrm.22052. [DOI] [PubMed] [Google Scholar]