Abstract
Antisense transcript, long non-coding RNA HOTAIR is a key player in gene silencing and breast cancer and is transcriptionally regulated by estradiol. Here, we have investigated if HOTAIR expression is misregulated by bisphenol-A (BPA) and diethylstilbestrol (DES). Our findings demonstrate BPA and DES induce HOTAIR expression in cultured human breast cancer cells (MCF7) as well as in vivo in the mammary glands of rat. Luciferase assay showed that HOTAIR promoter estrogen-response-elements (EREs) are induced by BPA and DES. Estrogen-receptors (ERs) and ER-coregulators such as MLL-histone methylases (MLL1 and MLL3) bind to the HOTAIR promoter EREs in the presence of BPA and DES, modify chromatin (histone methylation and acetylation) and lead to gene activation. Knockdown of ERs down-regulated the BPA and DES induced expression of HOTAIR. In summary, our results demonstrate that BPA and DES exposure alters the epigenetic programming of the HOTAIR promoters leading to its endocrine disruption in vitro and in vivo.
Introduction
Non-coding RNAs (ncRNA) are a group of regulatory RNAs that are not translated into protein (Mattick and Makunin 2006). NcRNAs that are longer than 200 nucleotides (nt) are classified as long non-coding RNAs (lncRNA) (Carninci et al. 2005). LncRNA consists of more than 50% of transcript in human cells (Khalil et al. 2009; Niland et al. 2012). LncRNAs are major players in diverse biological processes that include genome packaging, chromatin organization, dosage compensation, genomic imprinting, and gene regulation (Brown et al. 1991; Sleutels et al. 2002; Carninci et al. 2005; Rinn et al. 2007; Gupta et al. 2010). Similar to protein coding mRNA, many lncRNA are transcribed by RNA polymerase II (RNA pol II), capped, spliced, and polyadenylated (Rinn et al. 2007; Cabili et al. 2011). In addition, lncRNA may be transcribed from both sense and antisense strands of the genome (Ponting et al. 2009). HOTAIR (HOX Antisense Intergenic RNA) is a recently discovered 2.2 kb long lncRNA that is located at the antisense strand of the HOXC gene locus in chromosome 12, flanked by HOXC11 and HOXC12 (Rinn et al. 2007). HOTAIR silences the HOXD locus genes present at chromosome 2 in humans, in trans, via recruitment of various gene silencing machineries (Rinn et al. 2007). For example, HOTAIR interacts with PRC2 (polycomb repressive complex 2) and histone demethylases LSD1/CoREST/REST complexes through it 5′- and 3′-end, respectively (Rinn et al. 2007; Tsai et al. 2010). Notably, PRC2 complex that contains histone H3-lysine 27 (H3K27)-specific histone methylases EZH2, along with several other polycomb group (PcG) of proteins (Wang et al. 2010). LSD1 is a histone H3K4-specific demethylase (Wang et al. 2011). Notably, H2K27-methylation and H3K4-demthylation are critical for gene silencing. In summary, HOTAIR recruits the machinery required for gene silencing (PRC2 and LSD1-complexes) to the promoters of target gene that modify chromatins which, in turn, lead to gene silencing (Tsai et al. 2010).
A growing body of literature has revealed that HOTAIR is highly expressed in variety of cancers including breast tumor (Gupta et al. 2010), hepatocellular carcinoma (Geng et al. 2011; Yang et al. 2011; Ishibashi et al. 2013), pancreatic cancer (Kim et al. 2012), non-small cell lung cancer (Nakagawa et al. 2013; Zhuang et al. 2013), colorectal cancer (Kogo et al. 2011), gastrointestinal stromal tumor (Niinuma et al. 2012), esophageal squamous cell carcinoma (Lv et al. 2013), sarcoma (Milhem et al. 2011) and others. HOTAIR expression levels are correlated with tumor metastases and loss of HOTAIR has been linked with decrease in cancer invasiveness (Gupta et al. 2010; Geng et al. 2011). However, although HOTAIR has been shown to play a fundamental role in cancer, little is known about its transcriptional regulation. Recent studies from our laboratory demonstrated that HOTAIR is a critical player in survival and maintenance of breast cancer cells (Bhan et al. 2013). We also found that HOTAIR is transcriptionally regulated by estradiol in breast cancer cells, which is mediated via the co-ordination of estrogen-receptors and various ER-coregulators including mixed lineage leukemia (MLL) family of histone methyl-transferases (Bhan et al. 2013).
Since HOTAIR is a critical player in the maintenance and viability of breast cancer cells and is transcriptionally regulated by estrogen (Bhan et al. 2013), we hypothesized that its transcription may be influenced by estrogenic endocrine disrupting chemicals (EDCs) and synthetic estrogens. An endocrine disruptor is an exogenous substance or mixture that alters function(s) of the endocrine system and consequently causes adverse health effects in an intact organism, or its progeny, or (sub) populations. EDCs interact with hormone receptors even at very low concentrations and interfere with hormone signaling affecting various hormonally regulated processes including reproduction and development (Brown and Lamartiniere 1995; Calle et al. 1996; BLOCK et al. 2000; Munoz-de-Toro et al. 2005; Susiarjo et al. 2007; Adamsson et al. 2008; Baba et al. 2009; Doherty et al. 2010a; Mahoney and Padmanabhan 2010; Gibert et al. 2011; Wolstenholme et al. 2012; Xu et al. 2012). Chronic or acute exposure to EDCs results in harmful health effects including birth defects, diabetes, cancers, reproductive problems, early puberty, and obesity (Hwang et al. 2008; Jacobs et al. 2008; Nakanishi 2008; Navas and Segner 2008; Phillips and Foster 2008; Schoeters et al. 2008; Wohlfahrt-Veje et al. 2009; Grun 2010; Casals-Casas and Desvergne 2011; Lee et al. 2012; Fenichel et al. 2013; Vuorinen et al. 2013). EDCs may induce aberrant and altered gene expression. For example, exposure to estrogenic EDCs such as bisphenol-A (BPA) and synthetic estrogens such as diethylstilbestrol (DES) alters uterine HOX gene expression and induces developmental changes in the female reproductive tract (Ben-Jonathan and Steinmetz 1998; Akbas et al. 2004; Bromer et al. 2010b; Doherty et al. 2010b). Perinatal exposure to BPA alters mammary gland development in mice (Krishnan et al. 1993; Saunders et al. 1997). BPA is commonly found in plastics, metallic storage containers, and other routinely used consumables. DES is an active synthetic nonsteroidal estrogen drug that has been used for various hormonal disorders (Titus-Ernstoff et al. 2001), induction of fertility, and others (Levi et al. 1996). Women exposed to DES while pregnant have been identified as having increased incidence of breast cancer (Greenberg et al. 1984; Calle et al. 1996). Daughters born after in utero exposure to DES are also at a higher risk for breast cancer (Troisi et al. 2007). Thus, BPA and DES exposure are of serious health concern.
In the present study, we investigated the impact of the BPA and DES on transcriptional regulation of breast cancer associated lncRNA HOTAIR both in vitro and in vivo. Our results demonstrate that the HOTAIR gene is dysregulated upon exposure to nanomolar concentrations of BPA and DES in breast cancer cells as well as in vivo, in the mammary glands of ovariectomized rats. BPA and DES exposure recruits ERs and ER-coregulators at the HOTAIR promoter, alters the epigenetic modifications leading to HOTAIR dysregulation.
Experimental Procedure
Cell culture and treatment with estradiol, BPA and DES
The MCF7 cell line (human breast cancer, ER-positive) was obtained from American Type Cell Culture Collection (ATCC) and cultured in our laboratory as described by us previously (Ansari et al. 2013a; Ansari et al. 2013b; Bhan et al. 2013). Cells were grown and maintained in Dulbecco’s modified Eagle’s media (DMEM; Sigma-Aldrich, St. Louis, MO) supplemented with 10% heat inactivated fetal bovine serum, 2 mM L-glutamine, 100 units/mL penicillin and 0.1 mg streptomycin/mL. Cells were maintained in a humidified incubator with 5 % CO2 and 95 % air at 37 °C.
For the treatment with estradiol, DES, and BPA (Sigma-Aldrich), cells were grown and maintained for at least 3 generations in phenol-red free DMEM-F12 media (Sigma-Aldrich) supplemented with 10% charcoal stripped serum (FBS), 2 mM L-glutamine and 100 units/mL penicillin and 0.1 mg/mL streptomycin. Cells were grown up to 60 % confluency in 60 mm culture plates and treated with varying concentrations of estradiol, BPA and DES for 4 h. Then the cells were harvested for RNA/protein extraction or for ChIP assays (Ansari et al. 2009a; Ansari et al. 2013a; Ansari et al. 2013b; Bhan et al. 2013).
RNA extraction, cDNA synthesis, RT-PCR and real-time PCR (qPCR)
For the RNA extraction, we followed similar protocols as described by us earlier (Ansari et al. 2009a; Ansari et al. 2013a; Ansari et al. 2013b; Bhan et al. 2013). Briefly, control and treated cells (BPA or DES) were harvested, centrifuged at 500 g for 5 min at 4 °C, and then resuspended in diethyl pyrocarbonate (DEPC) treated buffer A (20 mM Tris-HCl (pH 7.9); 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol (DTT), and 0.2 mM phenylmethylsulfonyl fluoride (PMSF, Sigma-Aldrich)), for 10 min on ice. Cells were centrifuged at 3500 g for 5 min at 4 °C, and the supernatant was subjected to phenol-chloroform (1:1 phenol and chloroform saturated with 1 x TE). The aqueous layer was mixed with 3 volume of ethanol, incubated at −80 °C for 4 h and then centrifuged at 13000 g for 30 min at 4 °C. The RNA pellets were air dried, dissolved in DEPC treated water containing 0.5 mM EDTA, and quantified using a nanodrop spectrophotometer.
For cDNA synthesis, 2.4 μM of oligo dT (Promega, Madison, WI) was mixed with 500 ng of the RNA in a 12 μL total volume and incubated at 70 °C for 10 min. The mix was then added to a cocktail of 100 units of MMLV reverse transcriptase (Promega, Madison, WI), 1X first strand buffer (Promega, Madison, WI), 100 μM dNTPs (each), 1 mM DTT, and 20 units of RNaseOut (Invitrogen, Carlsbad, CA) and the volume was made up to 25 μL (using DEPC treated water). This mixture was incubated at 37 °C for 1 h for the reverse transcription. Each cDNA product was diluted to 100 μL, and 5 μL of the diluted cDNA was subjected to PCR amplification using specific primer pairs as described in Table 1.
Table 1.
Nucleotide sequence of antisense and primers Table 1 Nucleotide sequence of antisense and primers
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| PCR primers | ||
| MLL1 | GAGGACCCCGGATTAAACAT | GGAGCAAGAGGTTCAGCATC |
| MLL2 | AGGAGCTGCAGAAGAAGCAG | CAGCCAAACTGGGAGAAGAG |
| MLL3 | CATATGCACGACCCTTGTTG | ACTGCTGGATGTGGGGTAAG |
| MLL4 | CCCTCCTACCTCAGTCGTCA | CAGCGGCTACAATCTCTTCC |
| GAPDH | CAATGACCCCTTCATTGACC | GACAAGCTTCCCGTTCTCAG |
| hHOTAIR | GGTAGAAAAAGCAACCACGAAGC | ACATAAACCTCTGTCTGTGAGTGCC |
| rHOTAIR | GTTAACATGACCAGCGATCTGA | AATTAATTAGTGCCTCCCAGTCC |
| ERα | AGCACCCTGAAGTCTCTGGA | GATGTGGGAGAGGATGAGGA |
| ERβ | AAGAAGATTCCCGGCTTTGT | TCTACGCATTTCCCCTCATC |
| HOTAIR ERE1 | TGCCTATATTTCTCTCCCTTAC | AGAAGAGGTGGAAGCCAGGAAC |
| HOTAIR ERE2 | CTCCAGGTGGCTTATTTGTATCTT | CTGCTGCAGAGAATTTCAGGT |
| HOTAIR ERE3 | TCAAGGAAAGAAGGCCCTGGC | AGGTATTGATGCTGTGGCCAG |
| HOTAIR ERE4 | TATGGCTTAGTTTTTCAACAA | TCAGTGGCCAGGGCCTTCTTT |
| GAPDH promoter | CAATGACCCCTTCATTGACC | GACAAGCTTCCCGTTCTCAG |
| Cloning primers | ||
| HOTAIR Promoter | GGTACCCTGTGAGGAGACAGCTGCTGGACa | AAGCTTAGGCTGGAACAGATCCCAAACAAAa |
| HOTAIR ERE1 | GGTACCCTTGCCTATATTTCTCTCCCTTACAGa | CTCGAGGCTCGTAAAATAGGGCTTTTATGGa |
| HOTAIR ERE2 | GGTACCCTCCAGGTGGCTTATTTGTATCTTAa | CTCGAGCTGCCTTAACTTTGGTCCAGCTACa |
| HOTAIR ERE3 | GGTACCTCAAGGAAAGAAGGCCCTGGCa | CTCGAGAGGTATTGATGCTGTGGCCAGa |
| HOTAIR ERE4 | GGTACCTATGGCTTAGTTTTTCAACAAa | CTCGAGTCAGTGGCCAGGGCCTTCTTTa |
| Antisense | ||
| MLL1 | TGCCAGTCGTTCCTCTCCACb | |
| MLL2 | ACTCTGCCACTTCCCGCTCAb | |
| MLL3 | CCATCTGTTCCTTCCACTCCCb | |
| MLL4 | CCTTCTCTTCTCCCTCCTTGTb | |
| ERα | CATGGTCATGGTCAGb | |
| ERβ | GAATGTCATAGCTGAb | |
| Scramble | CGTTTGTCCCTCCAGCATCTb | |
cloning primers flanked by appropriate restriction sites
Phosphodiester linkages replaced by phosphorothioate linkages.
In general, PCR reaction reactions were carried out for 31 cycles (30 s at 94 °C for denaturation, 30 s at 60 °C for annealing, 45 s at 72 °C for elongation) and final PCR products were analyzed in 1.5 % agarose gel electrophoresis. For the qPCR reactions, 5 μL of diluted cDNA were mixed with 5 μL Sso EvaGreen supermix (Bio-Rad, Hercules, CA) and 2 μM each primer and final volume was made up to 12 μL. PCR reaction reactions were carried out in CFX96 real-time detection system (Bio-Rad, Hercules, CA) for 40 cycles (5 s at 95 °C for denaturation and 10 s at 60 °C for both annealing and elongation). Data analyses were performed using CFX manager software (Bio-Rad, Hercules, CA). Each experiment was repeated three times with three replicates each time.
ERs knockdown via antisense transfection
Cells were grown up to 60% confluency in 60 mm culture plates and transfected with ERs (ERα and ERβ) and scramble (no homology to ERs) antisense oligonucleotides independently (Table 1) using iFect transfection reagent (K.D. Medical, Columbia, MD) as previously described (Ansari et al. 2011b; Ansari et al. 2012a; Shrestha et al. 2012). Prior to transfection, a cocktail of transfection reagent and antisense oligonucleotide was made as follows. Initially, 12 μL (12 μg) of iFect reagent was mixed with 200 μL DMEM (without FBS and antibiotics) in an Eppendorf tube. In a separate Eppendorf, antisense oligonucleotide was mixed with 100 μL DMEM (without supplements). Then the diluted antisense solution was mixed with diluted iFect reagents and allowed to stand for 45 min in dark. In the meantime, cells were washed twice with supplement free DMEM and then 1.7 mL of supplement free DMEM was added to each cell culture plate. Finally, antisense-transfection reagents cocktail was applied to the cell plates and incubated for 24 h. Then 2 mL of DMEM with all supplements including 20% FBS was added in each well, incubated for additional 24 h and then harvested for RNA/protein extraction or for ChIP assays (Ansari et al. 2013a).
Chromatin immunoprecipitation assay (ChIP)
ChIP assays were performed using MCF7 cells and an EZ Chip™ chromatin immuno-precipitation kit (Millipore, Temecula, CA) as described previously (Ansari et al. 2013b). In brief, MCF7 cells were treated with 100 nM BPA and 10 nM DES separately for 4 h, fixed with 4% formaldehyde, and then sonication to shear the chromatins (~ 300 bp long DNA fragments).
The fragmented chromatin was pre-cleaned with protein-G agarose beads and subjected to immuno-precipitation with antibodies specific to ERα, ERβ, MLL1, MLL2, MLL3, MLL4, H3K4-trimethyl, acetylated histone, CBP, p300, TAF250, β-actin (control) and RNA polymerase II (RNAPII) overnight. Immuno-precipitated chromatins were washed and de-proteinized to obtain purified DNA fragments that were PCR amplified using various primers corresponding to different ERE regions of HOTAIR promoter (Table 1). Antibodies were purchased from commercial sources that are as follows: MLL1 (Abgent, San Diego, CA, AP6182a, MLL2 (Abgent, San Diego, CA, AP6183a), MLL3 (Abgent, AP6184a), MLL4 (Sigma, St. Louis, MO, AV33704), ERα (D-12, sc-8005, Santa Cruz Biotechnology, Dallas, TX), ERβ (H-150, Santa Cruz biotechnology, sc-8974), H3K4-trimethyl (EMD-millipore, Temecula, CA, 07-473), RNA pol II (RNAPII, Abcam, Cambridge, MA, 8WG16), β-actin (Sigma, A2066), CBP (A22, Santa Cruz Biotechnology, Sc369) and p300 (N15, Santa Cruz Biotechnology, Sc584) (Ansari et al. 2012a) (Bhan et al. 2013).
Dual luciferase reporter assay
HOTAIR full length promoter (−2050 to + 5 nt) and individual estrogen response elements (EREs) such as ERE1 (−536 to −924 nt), ERE2 (−1315 to −1531 nt), ERE3 (−1623 to −1755 nt), and ERE4 (−1731 to −1833 nt) regions were cloned and inserted upstream of the luciferase gene in a pGL3 vector (Promega, Madison, WI) (cloning primers in Table 1) (Bhan et al. 2013). MCF7 cells (50% confluency) were grown in 6 well plate in DMEM-F12 media, and co-transfected with 1500 ng of these ERE-pGL3 constructs along with 150 ng of a reporter plasmid containing renilla luciferase (pRLTk, Promega, Madison, WI) as an internal transfection control using FuGENE6 transfection reagent. Control transfections were done using empty pGL3 vector without any ERE insertion. At 24 h post transfection, cells were treated with 100 nM BPA and 10 nM DES separately, and incubated for additional 4 h and then subjected to luciferase assay, using Dual luciferase reporter assay kit (Promega, Madison, WI) as instructed. Luciferase activities were normalized to the renilla and plotted. Each treatment was performed in four parallel replicates and the experiment was repeated at least twice (Ansari et al. 2011b; Bhan et al. 2013).
Animal Experiments
Subjects
Twelve 90 day old, experimentally naïve, adult, female, Sprague-Dawley rats were triple housed in a temperature and humidity-controlled environment under a 12h reversed light/dark cycle with lights on at 7 p.m. and off at 7 a.m. All animals had free access to food and water throughout the study and were maintained and cared for in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Ovariectomy
Rats were anesthetized with a 2–3%isoflurane-oxygen vapor mixture and ovariectomized (OVX) (Maclusky 2005; Betancourt AM 2010; Eilam-Stock et al. 2012) Following a 4–5 day surgical recovery period, all rats underwent vaginal lavage testing daily for 8 consecutive days to confirm cessation of estrous cycling. All OVXs performed were confirmed as complete and thus, no animals were eliminated on the basis of an incomplete procedure (Inagaki et al. 2012).
Drugs and drug treatments
BPA: 10 mg of BPA was dissolved in 1 mL of ethanol to create a stock solution that was stored at −4 °C. DES and estradiol were dissolved in peanut oil to yield final concentrations of 5 μg/mL. BPA was dissolved in ethanol and brought up to a final concentration of 50 μg/mL with saline. Rats were given subcutaneous injections of either BPA (25 μg/kg), estradiol (5 μg), or DES (5 μg/kg), (n=4) 24 and 4 hours prior to sacrifice. Animals were sacrificed via rapid decapitation and mammary gland tissue was collected from each rat, flash frozen on dry ice, and then stored at −80 °C until RNA extraction (Maclusky 2005; Betancourt AM 2010; Eilam-Stock et al. 2012).
RNA extraction
RNA extraction was carried out using ZyGEM kit (Hamilton, New Zealand) according to the manufacturer’s protocol. The RNA was reverse transcribed to cDNA and subjected to qPCR using rat specific primers (Table 1) (Ansari et al. 2013b).
Statistical analysis
Each experiment was done in two to three replicates and then cells were pooled (and treated as one sample), subjected to RNA extraction, RT-PCR and ChIP analysis and each experiment was repeated at least three times (n = 3). The real time PCR analysis of such samples were done in three parallel replicate reactions and repeated thrice (n = 3). For luciferase assay each treatment was carried out in four replicates and the experiment was repeated at least twice (n = 2). Normally distributed data were analyzed by ANOVA and non-normally distributed data were analyzed using student-t tests (SPSS) to determine the level of significance between individual treatments. The treatments were considered significantly different at p ≤ 0.05.
Results
HOTAIR expression is induced by endocrine disruptors BPA and DES in breast cancer cells in vitro and in the mammary gland of rats in vivo
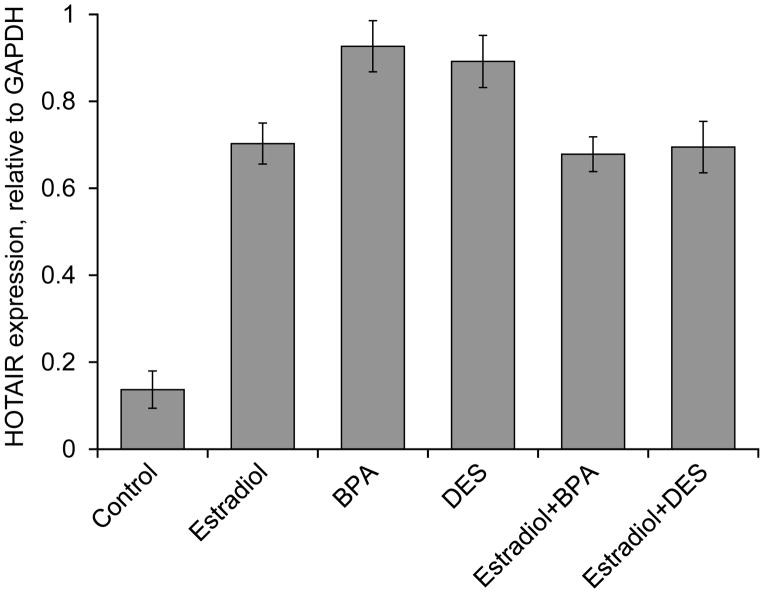
As HOTAIR is known to be overexpressed in breast carcinoma (Gupta et al. 2010) and transcriptionally regulated by estradiol (Bhan et al. 2013), we hypothesized that its expression may be dysrupted by synthetic estrogens and estrogenic endocrine disrupting chemicals (EDCs). To test this, we treated human breast cancer cells, MCF7, with BPA and DES. In parallel, we also treated the cells with estradiol separately as a positive control. RNA from the control and treated cells was examined for the levels of HOTAIR expression. The qPCR analyses showed BPA (100 nM) and DES (10 nM) treatment induced (~ 6 fold) the expression of HOTAIR, in comparison to the control (untreated cells) (Figure 1). As expected, HOTAIR level was also induced (~5 fold) upon treatment with estradiol (0.1 nM) (Figure 1). The level of induction of HOTAIR by BPA and DES was slightly reduced in presence of estradiol, likely due to competition (Figure 1). The dose-dependent expression of HOTAIR by BPA, DES and estradiol is shown in Supplementary Figure S1. These results showed that HOTAIR is optimally expressed when cells are treated with 0.1 nM estradiol, 10 nM BPA and 100 nM DES, respectively. Therefore these were the concentrations chosen for further mechanistic studies in this manuscript. Notably, our analyses showed that higher concentrations of BPA (100 nM) and DES (10 nM) than estradiol (0.1 nM) were required to attain optimal level of HOTAIR expression and this might be likely due to their differential affinity towards to estrogen-receptors (ERs).
Figure 1.
Effect of 17β-estradiol (estradiol), bisphenol-A (BPA) and diethylstilbestrol (DES) on HOTAIR gene expression in MCF7 cells. (A) MCF7 cells were grown in phenol red free DMEM-F-12 media and treated with 0.1 nM estradiol, 100 nM BPA and 10 nM DES separately and in combination of 0.1 nM estradiol + 100 nM BPA and 0.1 nM estradiol + 10 nM DES. RNA from the control and treated cells were analyzed by qPCR using primers specific to HOTAIR. GAPDH was used as the control. Bars indicate standard errors. Each experiment was repeated for thrice with three parallel replicates. P values < 0.05 were considered to be significant.
HOTAIR expression is upregulated in vivo, in the mammary glands of rats upon exposure to BPA, DES as well as estradiol
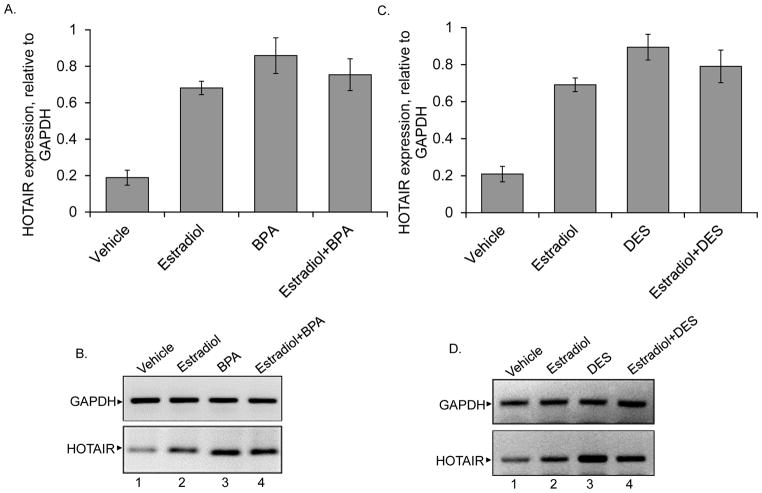
To examine if the HOTAIR gene is regulated by estradiol or if it is misregulated upon exposure to BPA and DES in vivo, we exposed ovariectomized (OVX) adult female rats to estradiol, BPA, or DES, and examined HOTAIR expression in mammary gland tissues. In brief, OVX Sprague Dawley rats were subjected to acute levels of estradiol (5μg), and BPA (25μg/kg) or DES (5μg/kg) at 24 h and 4 h prior to sacrifice. These doses of estradiol, BPA and DES were chosen because these concentrations were used previously by other laboratories and were found to be effective in in vivo analyses of gene expression and to influence behaviors and other neural functions (Stangl et al. 2002; Maclusky 2005; Adamsson et al. 2008; Li et al. 2009; Betancourt AM 2010; Casals-Casas and Desvergne 2011; Eilam-Stock et al. 2012; Nanjappa et al. 2012). RNA was isolated from the mammary glands from the control and treated animals using ZyGEM kit, reverse transcribed, and subjeted to qPCR analyses for the expression of HOTAIR using rat specific HOTAIR primers (He et al. 2011) (Figures 2A–D). GAPDH was used as control. The qPCR products were also analyzed in agarose gel and these data were shown in Figure 2B and D. Our results demonstrate that HOTAIR gene is up regulated 3.3 and 4.1 folds in the rat mammary glands by estradiol and BPA, respectively (Figures 2A–B). Mammary glands of animals that were given combination treatments of estradiol + BPA also upregulated HOTAIR by 3.6 fold compared to the untreated control animals (Figure 2A–B). However, the levels of HOTAIR upregulation were slightly suppressed in cases of estradiol + BPA combination treatments, in comparison to the BPA alone treatment (Figure 2A–B). This could be due to the competitive mode of regulation by estradiol and BPA. Similar to BPA and estradiol, treatment with DES or DES + estradiol, also resulted in upregulation of HOTAIR by 4.3 and 3.8 folds, respectively (Figures 2C–D). These results demonstrated that HOTAIR is misregulated upon exposure to the estrogenic EDCs and synthetic estrogens like BPA and DES, even in the absence of estrogen, in vivo.
Figure 2.
In vivo effect of estradiol, BPA and DES on HOTAIR expression. Ovariectomized adult female rats were administered with acute doses of estradiol (5 μg), BPA (25 μg/kg), and DES (5 μg/kg), for 24 h, either separately or in combination. RNA from the control, estradiol, BPA and estradiol/BPA treated mammary glands were analyzed by qPCR (panel A). The agarose gel analysis of the qPCR products is shown in panel B. GAPDH was used as a loading control. Panel C shows the effects of estradiol, DES and estradiol + DES on HOTAIR expression, analyzed via qPCR and agarose gene analysis of the qPCR products is shown in panel D. Each experiments were performed with three replicate (n = 3). Bars indicate standard errors. P values < 0.05 were considered to be significant.
HOTAIR promoter EREs are responsive to BPA and DES
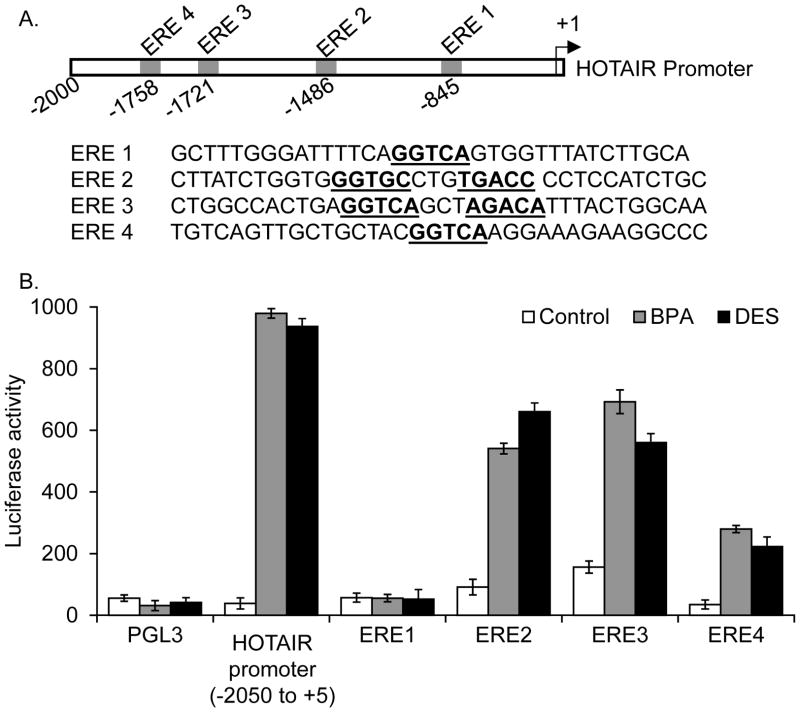
Since HOTAIR is found to be an estradiol-responsive gene and is induced by BPA and DES, we investigated the potential mechanism of BPA and DES induced expression of HOTAIR. The HOTAIR promoter contains multiple putative EREs close to the transcription start site (within −2000 nt upstream) (Bhan et al. 2013). The ERE2 (GGTGCnnnTGACC) and ERE3 (GGTCAnnnAGACA) appear to be imperfect full EREs with two base mismatches in comparison to the consensus full ERE (TGACCnnnGGTCA) (Figure 3A) (Bhan et al. 2013). To examine the potential roles of these EREs in transcriptional dysregulation of HOTAIR by BPA and DES, we cloned (Bhan et al. 2013) each ERE in a luciferase based reporter construct, pGL3 and analyzed their response to BPA and DES exposure using luciferase based reporter assay. In addtion, we also cloned the full length promoter (−2050 to +5 nt region) in the pGL3 construct and analyzed its BPA and DES response. In brief, we transfected each ERE-pGL3 constructs into MCF7 cells and then exposed to BPA (100 nM) or DES (10 nM), seperately. Notably, along with ERE-pGL3 constructs, a renilla luciferase vector was co-transfected as an interenal transfection control. An empty pGL3 vector was also transfected as a negative control. The control and treated cell extracts were analyzed by dual luciferase kit and the luciferase induction (normalized to renilla expression) was plotted for various ERE-pGL3 constructs. These analyses demonstrated that HOTAIR full length promoter is significantly induced (32 and 20.5 fold induction by BPA and DES, respectively) upon exposure to BPA and DES (Figure 3B). Individual ERE clones such as ERE2 and ERE3 containing pGL3 constructs also showed high levels of luciferase induction in the presence of BPA and DES (Figure 3B). In case of ERE2, the levels of luciferase activities are induced by about 16.8 and 16 folds for BPA, and DES, respectively (Figure 3B). In case of ERE3, the levels of luciferase activity were 18.4 and 16.8 folds for BPA and DES, respectively (Figure 3B). ERE4 showed moderate level of BPA and DES response (Figure 3B). On the contrary, ERE1 do not show any luciferase activity as compared to the empty pGL3 (control) upon treatment with BPA and DES, suggesting it to be a non-functional ERE. These observations indicate HOTAIR promoter is transcriptionally induced upon exposure to BPA and DES and primarily ERE2 and ERE3, which are potential imperfect full EREs, appear to coordinate the BPA and DES induced HOTAIR expression.
Figure 3.
HOTAIR promoter EREs are responsive to BPA and DES treatments. (A) HOTAIR gene promoter EREs (termed as ERE1, ERE2, ERE3 and ERE4 locations and the neighboring sequences are shown). HOTAIR promoter regions spanning ERE1, ERE2 ERE3 and ERE4 as well as the full-length promoter (−2050 to +5 nt region) were cloned individually (clones 1–4) into a luciferase based reporter construct, pGL3, used for transfection and reporter assay. (B) Luciferase based reporter assay. ERE-pGL3, full-length promoter-pGL3 or empty pGL3 (vector control) constructs were transfected into MCF7 cells separately for 24 h. A renilla luciferase construct was also co-transfected along with ERE-pGL3 constructs as an internal transfection control. Cells were then treated with 100 nM BPA and 10 nM DES and subjected to luciferase assay by using dual-Glo Luciferase Assay kit. The luciferase activities (normalized to renilla activity) were plotted. The experiment was repeated thrice with four parallel replicate (n =3). Bars indicate standard errors. P values < 0.05 were considered to be significant.
Estrogen-receptors (ERs) are essential for BPA and DES induced HOTAIR expression
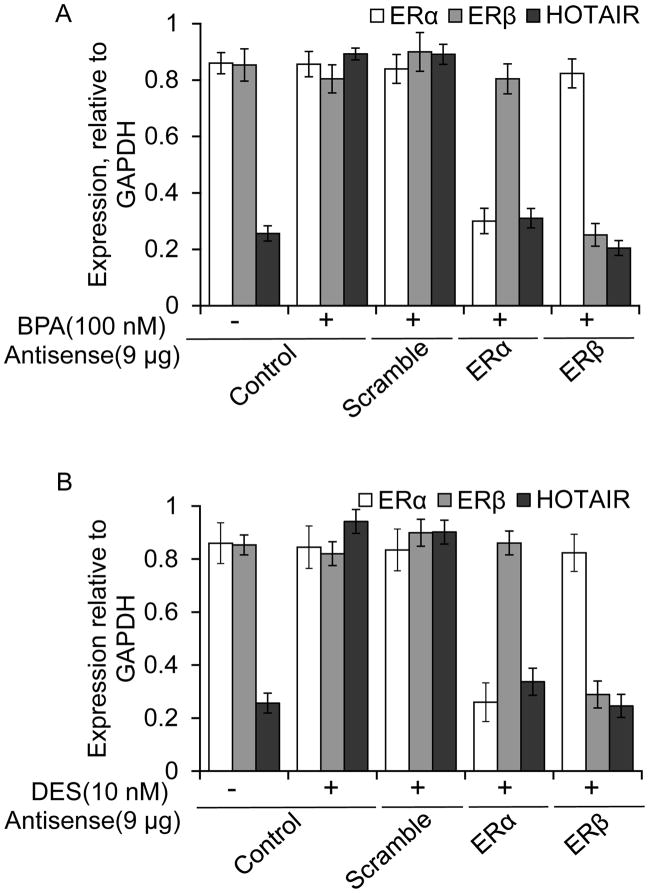
Estradiol mediated gene regulation is well known to be coordinated via involvement of estrogen-receptors such as ERα and ERβ (Mangelsdorf et al. 1995; Horwitz et al. 1996; Nilsson and Gustafsson 2002; Lonard and O’Malley 2005; Chen et al. 2006). To examine, if ERs are associated with the BPA and DES induced HOTAIR expression, we knocked down ERα and ERβ independently using respective antisense oligonucleotide (Ansari et al. 2011a; Ansari et al. 2011b; Ansari et al. 2012a; Ansari et al. 2013a; Bhan et al. 2013), in MCF7 cells, exposed the cells to BPA and DES and examined their impacts on HOTAIR expression. In brief, MCF7 cells were transfected with ERα and ERβ and scramble (as control) antisense separately and then treated with 100 nM BPA and 10 nM DES, independently for 4 h. RNA was analyzed by qPCR for the expression of HOTAIR (relative to GAPDH). The levels of respective ER-knockdown are examined both in RNA levels (Figures 4A–B) and protein levels (data not shown) (Ansari et al. 2011a; Ansari et al. 2012a; Bhan et al. 2013). For examining the specificity of ERα-knockdown, the expression of ERβ (as well as GAPDH) was examined as specificity control and vice versa (Figures 4A–B and supplementary figure S2). As seen in Figure 4A, the application of the scramble antisense does not affect the expression of ERα and ERβ, or the BPA-induced HOTAIR expression. However, application of ERα-antisense specifically knocked down ERα (but not ERβ) and that resulted in down-regulation of BPA-induced HOTAIR expression (Figure 4A, agarose gel of RT-PCR products is shown the supplementary Figure S2). Similarly, application of ERβ-antisense specifically knocked down ERβ (but not ERα) and down-regulated BPA-induced HOTAIR expression (Figure 4A and supplementary figure S2). These analyses demonstrate that ERα and ERβ are key players in BPA-induced HOTAIR expression. Similarly, independent knockdown of ERα and ERβ, also down-regulated the DES-induced expression of HOTAIR, indicating key roles for both ERα and ERβ, in the DES induced HOTAIR misregulation (Figure 4B and supplementary Figure S2)
Figure 4.
Roles of ERs on BPA and DES-induced expression of HOTAIR. (A–B) MCF7 cells were transfected with ERα, ERβ or scramble antisense (9 μg each) separately for 48 h and treated with BPA (100 nM) (Panel A) and DES (10 nM) (Panel B) for additional 4 h. RNA was isolated and subjected to real-time quantification (qPCR) of HOTAIR expression (relative to GAPDH). In case of ERα-knockdown, ERβ-PCR analysis was performed as a specificity control, in addition to GAPDH and vice versa. Each experiment was repeated at least thrice (n = 3). Bars indicate standard errors. P values < 0.05 were considered to be significant.
ERs and ER-coactivators such as MLL-histone methylases bind to the HOTAIR promoter and modify chromatins in presence of BPA and DES
In addition to ERs, ER-coactivators are integral components of estrogen-dependent gene activation (Lee et al. 2000; Lee et al. 2001). During activation of estradiol-responsive genes, ERs interact with various ER-coregulators that bridge ERs to chromatin modifying proteins and basal transcription machinery and that ultimately lead to gene activation (Mangelsdorf et al. 1995; Horwitz et al. 1996; Nilsson and Gustafsson 2002; Lonard and O’Malley 2005; Chen et al. 2006). Many ER coactivators have been discovered, including SRC1 family protein, CREB-binding protein (CBP/p300), p/CAF, and ASCOM [activating signal cointegrator-2 (ASC2) complexes] (Bulynko and O’Malley 2011; Ansari et al. 2013a). Recent studies from our laboratory and others demonstrate that mixed lineage leukemia (MLLs) histone methylases (Dreijerink et al. 2006; Lee et al. 2008a; Lee et al. 2008b; Bulynko and O’Malley 2011) act as ER co-activators and regulate estrogen-responsive genes (Jeong et al. 2011; Umezawa et al. 2009; Valekunja et al. 2013). MLLs are well recognized histone 3 lysine (H3K4) specific histone methyltransferases that aid in gene activation (Hess 2004; Ansari et al. 2009b; Ansari et al. 2012b). MLLs possess multiple LXXLL domains (also called nuclear receptor box or NR box) through which they may interact with NRs and thus, participate in NR-mediated gene activation (Lee et al. 2000; Lee et al. 2001; Dreijerink et al. 2006; Ansari and Mandal 2010).
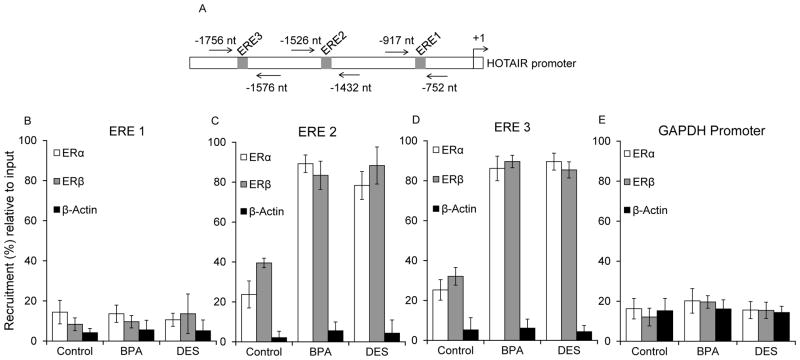
To further investigate the mechanism of BPA and DES induced HOTAIR expression, we examined the bindings of ERs and several ER-coactivators such as histone methylases MLL1-MLL4, CBP and p300 in the HOTAIR promoter in the absence and presence of BPA and DES using chromatin immunoprecipitation (ChIP) assay (Shrestha et al. 2012; Ansari et al. 2013a). Briefly, MCF7 cells were treated with BPA (100 nM) and DES (10 nM) for 4 h, fixed with formaldehyde, sonicated to shear the chromatin, and subjected to chromatin immuno-precipitation using ERα, ERβ, MLLs (MLL1–4), CBP, p300, and β-actin (control) antibodies. ChIP DNA fragments were PCR (qPCR)-amplified using primers specific to various ERE regions of the HOTAIR promoter (Figures 5–6 and supplementary Figure S3–4). Figures 5A–D (and supplementary figure S3) showed the binding of both ERα and ERβ in ERE1-ERE4 regions of HOTAIR promoter in the absence and presence of BPA and DES. These analyses showed that there was no recruitment of ERs in the ERE1 region irrespective of BPA and DES (Figures 5B and supplementary figure S3). Interestingly, ERα and ERβ were enriched in both ERE2 and ERE3 regions in a BPA- and DES-dependent manner (Figures 5C–D and supplementary figure S3). ERE4 was not so much sensitive to BPA and DES induced binding of ERs (data not shown, supplementary figure S3). There was no significant binding of ERs in the GAPDH promoter (a non-specific DNA region control) (Figure 5E). These analyses further demonstrate that ERs are key players in the BPA and DES induced HOTAIR expression and furthermore ERE2 and ERE3 regions of HOTAIR promoter are primarily associated with BPA- and DES -induced HOTAIR expression.
Figure 5.
BPA and DES-induced binding of ERs in the HOTAIR promoter EREs. (A–D) MCF7 cells were treated with 100 nM BPA and 10 nM DES, separately for 4 h and subjected to ChIP assay using antibodies specific to ERα and ERβ. ChIP DNA fragments were PCR-amplified (qPCR) using primers specific to different ERE regions in the HOTAIR promoter. Panel A shows the position of the primers in the HOTAIR promoter. Panel B, C and D show the ChIP analysis with ERα and ERβ antibodies, respectively, at different ERE regions (ERE1–3). Panel E demonstrates ChIP analysis of the GAPDH promoter (non-specific DNA control) with ERα and ERβ antibodies. Each experiment was repeated at least thrice. Bars indicate standard errors.
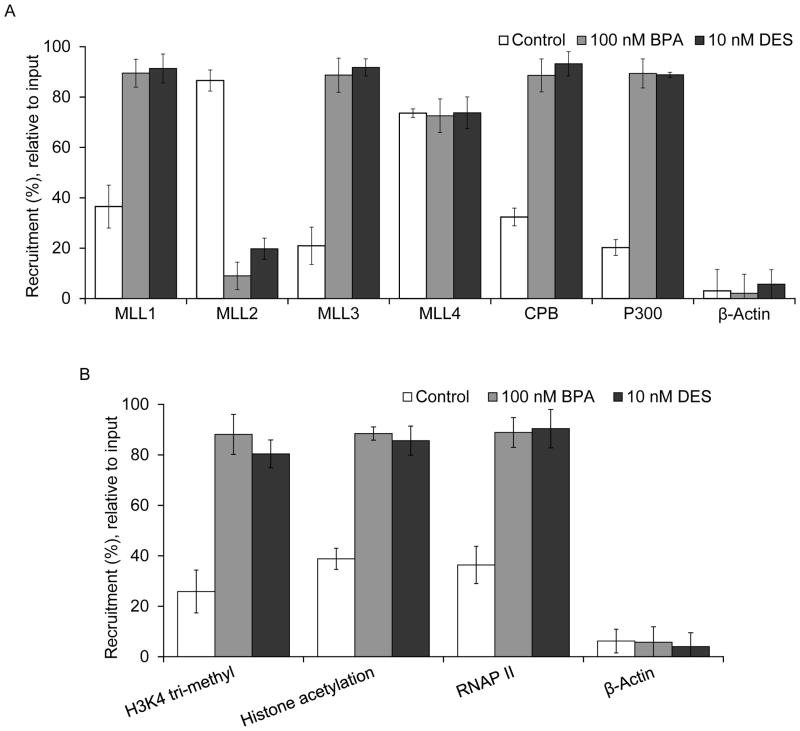
Figure 6.
Roles of MLLs, ER-coregulators, and histone modification. (A–B) MCF7 cells were treated with 100 nM BPA and 10 nM DES, separately for 4 h and subjected to ChIP assay using antibodies specific to MLLs (MLL1–4), CBP, p300, H3K4-trimethyl, histone acetyl, RNA polymerase II (RNAP II) and β-actin (control). ChIP DNA fragments were PCR-amplified (qPCR) using primers specific to ERE2 region in the HOTAIR promoter. Panel A shows the ChIP analysis of MLLs (MLL1–4), CBP, p300 and β-actin on the ERE2 of the HOTAIR promoter. Panel B shows the ChIP analysis of H3K4-trimethyl, histone acetyl, RNA polymerase II (RNAP II) and β-actin (control) on the ERE2 region of the HOTAIR promoter. Each experiment was repeated at least thrice. Bars indicate standard errors. P values < 0.05 were considered to be significant
Because ERs are recruited to ERE2 and ERE3 regions of the HOTAIR promoter, we examined the recruitment of various ER-coregulators such as MLLs (MLL1–4), CBP and p300 into the ERE2 and ERE3 regions in the absence and presence of BPA and DES, using ChIP assay (Figures 6A and supplementary figure S4). These analyses showed that CBP, p300, and histone methylases, MLL1 and MLL3 were enriched in the ERE2 and ERE3 regions in the presence of BPA and DES (Figure 6A and supplementary Figure S4). In contrast, we observed the constitutive binding of MLL2 in the HOTAIR promoter (ERE2), MLL2 was delocalized from the HOTAIR promoter upon exposure of BPA and DES (Figure 6A and supplementary figure S4). MLL4 binding was not significantly affected by BPA and DES exposure, though some amount of constitutive binding of MLL4 were observed at the HOTAIR promoter. No binding of MLLs was observed in the GAPDH promoter (non-specific DNA control) (supplementary figure S4). These observations demonstrated that, along with ERs, ER-coregulators such as MLL1 and MLL3 histone methylases and CBP and p300 are involved in the BPA and DES induced HOTAIR expression.
As histone acetylases and methyl-transferases are involved in DES- and BPA- mediated HOTAIR gene activation, we examined the levels of histone acetylation and histone H3K4-tri-methylation status, and the recruitment of RNA Polymerase II (RNAP II) on to the HOTAIR promoter in the absence and presence of DES and BPA. Notably, H3K4-tri-methylation and histone acetylation are essential post-translational histone modifications that are critical for gene expression activation (Ansari and Mandal 2010). MLLs are well known to be histone H3K4-specific methyl-transferases that are critical players in gene activation (Ansari and Mandal 2010). ChIP analyses demonstrated that the levels of histone H3K4-trimethylation, histone acetylation and the levels of RNA polymerase II were increased at the HOTAIR promoter (ERE2) in the presence of BPA, and DES (Figure 6B and supplementary figure S4). Overall, these analyses demonstrated that ERs, ER-coregulators such as histone methylases MLL1 and MLL3, CBP, p300 (histone acetylase) are associated with the HOTAIR promoter in presence of BPA and DES, and introduce the histone H3K4-trimethylation and acetylations in the promoter histones leading to HOTAIR gene activation.
Discussion
Non-coding RNA constitutes a major portion of transcripts present in eukaryotic cells (Khalil et al. 2009). Most of them play regulatory roles in controlling gene expression, silencing, and chromatin organization (Qureshi et al. 2010; Pan et al. 2011; Harries 2012; Niland et al. 2012; Sierra-Miranda et al. 2012; Xia et al. 2013). The long non-coding RNA HOTAIR, which is located in chromosome 12 within HOXC locus, controls the expression of genes in the HOXD cluster, in a trans fashion (Rinn et al. 2007; Tsai et al. 2010). HOTAIR RNA interacts with gene silencing machinery, such as PRC2 and LSD1 complexes, and recruits them into the target gene promoters which leads to target gene silencing (Tsai et al. 2010). Increasing numbers of studies indicate that HOTAIR is up regulated in various types of human cancers including breast cancer (Gupta et al. 2010). HOTAIR overexpression is linked with cancer invasiveness and metastasis (Gupta et al. 2010; Geng et al. 2011). Recent studies from our laboratory demonstrate that HOTAIR is transcriptionally regulated by estradiol in breast cancer cells (MCF7), which is coordinated via ERs and various ER-coregulators (Bhan et al. 2013). SiSENSE (small interfering SENSE oligonucleotide) mediated knockdown of HOTAIR induced apoptosis in breast cancer cells indicating the crucial importance of HOTAIR in the survival and maintenance of breast cancer cells (Bhan et al. 2013). HOTAIR regulates various apoptotic genes such as BCL2, BID (BH3 interacting domain death agonist) etc. and therefore knockdown of HOTAIR resulted in apoptotic cell death (Bhan et al. 2013). HOTAIR also regulates various tumor suppressor genes including PCDH10 (Protocadherin-10), PCDHB5 (Protocadherin β-5), JAM2 (Junction adhesion molecule 2) etc. (Gupta et al. 2010). Overexpression or knockdown of HOTAIR misregulated the expression of PCDH10, PCDHB5, JAM2 etc. and is thus closely associated with the initiation and progression of tumors (Gupta et al. 2010; Tsai et al. 2010).
In this manuscript, we investigated if the HOTAIR gene may be misregulated by estrogenic endocrine disrupting chemicals (EDCs) and synthetic estrogens that are associated with major health risks. Specifically, we have investigated the epigenetic mechanism of endocrine disruption of HOTAIR upon exposure to BPA and DES in vitro and in vivo. BPA is a well-known estrogenic endocrine disruptor and DES is a synthetic estrogen. Both BPA and DES contains the multiple phenolic moieties in their structures through which they may interact with estrogen-receptors by mimicking estradiol phenolic moiety and therefore interfere with normal estrogen signaling (Blair et al. 2000; Yearley et al. 2008). BPA and DES exposure has been implicated with variety of health disorders in humans and other organism (Smith and Taylor 2007). In utero exposure to high levels of BPA causes alterations in estrous cyclicity and results in abnormal mammary gland developments in rodents (Markey et al. 2001; Muñoz-de-Toro et al. 2005; Fenton 2006). BPA exposure induces significant developmental consequences in the female reproductive tract such as early onset of puberty, alterations in hormone production, and histological changes in the vagina and mammary glands (Smith and Taylor 2007). Low-dose gestational BPA exposure increased ERα and progesterone receptor (PR) expression in the adult murine uterine endometrium (Markey et al. 2005; Bromer et al. 2010a). In humans, exposure to BPA has been associated with recurrent miscarriage (Sugiura-Ogasawara et al. 2005). Similarly, DES, which hast been extensively used as a synthetic estrogen for the treatment hormonal disorder including miscarriage, has been linked with an increased risk of cancer in exposed women and children (Smith and Taylor 2007). Despite the high health risk associated with BPA and DES exposure, understanding about their molecular mechanism of action is still limited. Here we have investigated the impact of BPA and DES exposure on the transcriptional regulation of a breast cancer associated lncRNA, HOTAIR and investigated the molecular mechanism.
Our studies demonstrated that, HOTAIR is induced significantly upon exposure to low concentrations of BPA and DES in vitro, in cultured breast cancer cells MCF7. Most importantly, we also observed that HOTAIR expression is upregulated in the rat mammary glands upon treatment with BPA and DES as well as estradiol, indicating estradiol-mediated transcriptional regulation of HOTAIR and its potential endocrine disruption by BPA and DES in vivo. Mechanistic studies demonstrated that estrogen-receptors (ERα and ERβ) coordinate the BPA and DES induced HOTAIR expression in MCF7 cells. Knockdown of ERs resulted in down-regulation of BPA and DES-induced HOTAIR expression. HOTAIR promoter EREs (especially ERE2 and ERE3) are induced upon exposure to BPA and DES (luciferase assay), indicating their potential involvement in endocrine disruption mediated via BPA and DES. ChIP analysis showed that ERs (ERα and ERβ) binds to the HOTAIR promoter EREs (ERE2 and ERE3) in the presence of BPA and DES. Notably, based on the ER-knockdown experiments (Figure 4) and ChIP analyses (Figure 5), we observed that both ERα and ERβ are involved and necessary for the BPA- and DES-induced HOTAIR expression. These observations suggest that ERα and ERβ may form heterodimer and bind to the HOTAIR promoter during BPA- and DES-induced gene expression. Similar to ERs, ER-coregulators such as the MLL-family of histone methylases (MLL1 and MLL3), and CBP and p300 (histone acetylases) were bound to the HOTAIR promoter ERE regions in upon exposure to BPA and DES. BPA and DES treatment also altered the epigenetic marks such as histone H3K4-trimethylation, histone acetylation etc. that are crucial for gene activation. Thus, exposure to BPA and DES, alters the epigenetic programming of the HOTAIR promoter, which resulted in turning on HOTAIR gene expression, even in the absence of physiological estrogen.
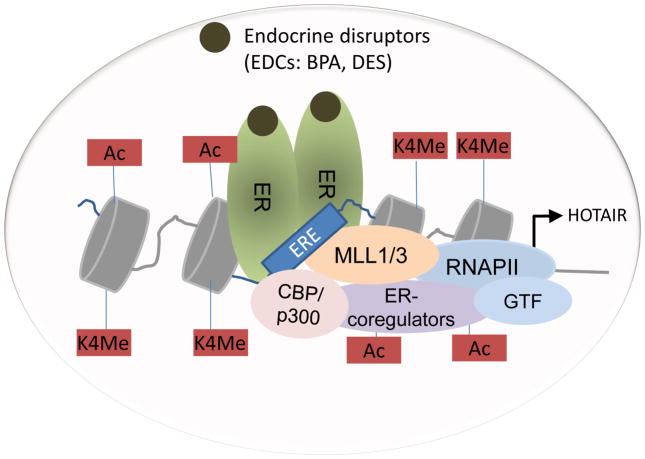
Notably, previous studies from our laboratory demonstrated that HOTAIR is an estrogen responsive gene (Bhan et al. 2013). Estrogen-receptors (ERα and ERβ) along with ER-coregulators such as histone methylases, MLL1 and MLL3, CBP and p300, were bound to the HOTAIR promoter in an estradiol dependent manner (Bhan et al. 2013). These observations indicate that the epigenetic mechanism of transcriptional activation of HOTAIR by BPA and DES is similar to that of estradiol mediated HOTAIR activation. These observations also further indicate that HOTAIR gene expression may be disrupted upon exposure to BPA and DES both in vitro and in vivo, even in the absence of estradiol, which may contribute towards various human pathogenesis including breast cancer. A model showing the roles of MLLs and ERs during the BPA and DES induced disruption of HOTAIR is shown in Figure 7.
Figure 7.
Models showing the roles of ERs, MLLs and other ER-coregulators during BPA and DES mediated endocrine disruption of HOTAIR. Steroidogenic EDCs like BPA and DES binds to ERs (ERα and ERβ), in a similar fashion to estradiol. Activated ERs (dimerized) bind to the functional EREs of the HOTAIR promoter. ER-coregulators such as MLL1 and MLL3, CBP/p300, and other ER-coregulators are recruited to the HOTAIR promoter EREs. Promoter histones are methylated (H3K4-trimethylated) and acetylated followed by recruitment of general transcription factors (GTFs) and RNA polymerase II (RNAP II), transcription initiation and gene activation.
It is important to note that, HOTAIR is an antisense transcript and lncRNA. Therefore, our studies also demonstrate that endocrine disruptors can disrupt the noncoding RNAs and can induce antisense transcripts, in a similar fashion as protein coding genes from the sense strands. Our studies revealed novel epigenetic mechanism of endocrine disruption, novel roles of MLL histone methylases and their coordination with ERs and various ER-coregulators during endocrine disruption, both in vitro and in vivo. Although further in vivo analyses are required to understand the detailed mechanism of HOTAIR gene expression and misregulation by EDCs, synthetics estrogens, and other environmental toxins/chemical, our observations indicate that exposure to BPA or DES may turn on the expression of HOTAIR in vivo, in a very similar fashion to estrogen even in the absence of estrogen, and that may result in adverse health effects including cancer and other hormonally regulated disorders.
Supplementary Material
Research Highlights.
Long non-coding RNA HOTAIR is a key player in breast cancer
HOTAIR is induced by endocrine disruptors BPA and DES in vitro and in vivo
MLL-histone methylases and ERs coordinate the BPA/DES-induced HOTAIR expression
BPA/DES alter the histone methylation/acetylation status at the HOTAIR promoter
Revealed novel epigenetic mechanism of endocrine disruption of lncRNA HOTAIR
Acknowledgments
We thank all the Mandal laboratory members for helpful discussions. Research in the Mandal laboratory is supported in part by grants from the National Institutes of Health (1R15 ES019129-01 and 2R15CA113747-02), and the American Heart Association (0765160Y).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamsson NA, Brokken LJ, Paranko J, Toppari J. In vivo and in vitro effects of flutamide and diethylstilbestrol on fetal testicular steroidogenesis in the rat. Reprod Toxicol. 2008;25:76–83. doi: 10.1016/j.reprotox.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Akbas GE, Song J, Taylor HS. A HOXA10 estrogen response element (ERE) is differentially regulated by 17 beta-estradiol and diethylstilbestrol (DES) Journal of molecular biology. 2004;340:1013–1023. doi: 10.1016/j.jmb.2004.05.052. [DOI] [PubMed] [Google Scholar]
- Ansari KI, Hussain I, Kasiri S, Mandal SS. HOXC10 is overexpressed in breast cancer and transcriptionally regulated by estrogen via involvement of histone methylases MLL3 and MLL4. J Mol Endocrinol. 2012a;48:61–75. doi: 10.1530/JME-11-0078. [DOI] [PubMed] [Google Scholar]
- Ansari KI, Hussain I, Shrestha B, Kasiri S, Mandal SS. HOXC6 Is transcriptionally regulated via coordination of MLL histone methylase and estrogen receptor in an estrogen environment. J Mol Biol. 2011a;411:334–349. doi: 10.1016/j.jmb.2011.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari KI, Kasiri S, Hussain I, Bobzean SA, Perrotti LI, Mandal SS. MLL histone methylases regulate expression of HDLR-SR-B1 in presence of estrogen and control plasma cholesterol in vivo. Mol Endocrinol. 2013a;27:92–105. doi: 10.1210/me.2012-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari KI, Kasiri S, Hussain I, Mandal SS. Mixed lineage leukemia histone methylases play critical roles in estrogen-mediated regulation of HOXC13. The FEBS journal. 2009a;276:7400–7411. doi: 10.1111/j.1742-4658.2009.07453.x. [DOI] [PubMed] [Google Scholar]
- Ansari KI, Kasiri S, Mandal SS. Histone methylase MLL1 has critical roles in tumor growth and angiogenesis and its knockdown suppresses tumor growth in vivo. Oncogene. 2013b;32:3359–3370. doi: 10.1038/onc.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari KI, Kasiri S, Mishra BP, Mandal SS. Mixed lineage leukaemia-4 regulates cell-cycle progression and cell viability and its depletion suppresses growth of xenografted tumour in vivo. British journal of cancer. 2012b;107:315–324. doi: 10.1038/bjc.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari KI, Mandal SS. Mixed lineage leukemia: roles in gene expression, hormone signaling and mRNA processing. FEBS J. 2010;277:1790–1804. doi: 10.1111/j.1742-4658.2010.07606.x. [DOI] [PubMed] [Google Scholar]
- Ansari KI, Mishra BP, Mandal SS. MLL histone methylases in gene expression, hormone signaling and cell cycle. Front Biosci (Landmark Ed) 2009b;14:3483–3495. doi: 10.2741/3466. [DOI] [PubMed] [Google Scholar]
- Ansari KI, Shrestha B, Hussain I, Kasiri S, Mandal SS. Histone methylases MLL1 and MLL3 coordinate with estrogen receptors in estrogen-mediated HOXB9 expression. Biochemistry. 2011b;50:3517–3527. doi: 10.1021/bi102037t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba K, Okada K, Kinoshita T, Imaoka S. Bisphenol A disrupts Notch signaling by inhibiting gamma-secretase activity and causes eye dysplasia of Xenopus laevis. Toxicol Sci. 2009;108:344–355. doi: 10.1093/toxsci/kfp025. [DOI] [PubMed] [Google Scholar]
- Ben-Jonathan N, Steinmetz R. Xenoestrogens: the emerging story of bisphenol a. Trends Endocrinol Metab. 1998;9:124–128. doi: 10.1016/s1043-2760(98)00029-0. [DOI] [PubMed] [Google Scholar]
- Betancourt AMEI, Desmont RA, Russo J, Lamartiniere CA. In-utero exposure to bisphenol A shifts the window of susceptibility for mammary carcinogenesis in the rat. Environ Health Perspect. 2010;118:1614–1619. doi: 10.1289/ehp.1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense Transcript Long Noncoding RNA (lncRNA) HOTAIR is Transcriptionally Induced by Estradiol. J Mol Biol. 2013 doi: 10.1016/j.jmb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair RM, Fang H, Branham WS, Hass BS, Dial SL, Moland CL, Tong W, Shi L, Perkins R, Sheehan DM. The Estrogen Receptor Relative Binding Affinities of 188 Natural and Xenochemicals: Structural Diversity of Ligands. Toxicological Sciences. 2000;54:138–153. doi: 10.1093/toxsci/54.1.138. [DOI] [PubMed] [Google Scholar]
- BLOCK K, KARDANA A, IGARASHI P, TAYLOR HS. In utero diethylstilbestrol (DES) exposure alters Hox gene expression in the developing müllerian system. The FASEB Journal. 2000;14:1101–1108. doi: 10.1096/fasebj.14.9.1101. [DOI] [PubMed] [Google Scholar]
- Bromer JG, Zhou Y, Taylor MB, Doherty L, Taylor HS. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. The FASEB Journal. 2010a;24:2273–2280. doi: 10.1096/fj.09-140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromer JG, Zhou Y, Taylor MB, Doherty L, Taylor HS. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. FASEB J. 2010b;24:2273–2280. doi: 10.1096/fj.09-140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Brown NM, Lamartiniere CA. Xenoestrogens alter mammary gland differentiation and cell proliferation in the rat. Environ Health Perspect. 1995;103:708–713. doi: 10.1289/ehp.95103708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulynko YA, O’Malley BW. Nuclear receptor coactivators: structural and functional biochemistry. Biochemistry. 2011;50:313–328. doi: 10.1021/bi101762x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle EE, Mervis CA, Thun MJ, Rodriguez C, Wingo PA, Heath CW. Diethylstilbestrol and Risk of Fatal Breast Cancer in a Prospective Cohort of US Women. American journal of epidemiology. 1996;144:645–652. doi: 10.1093/oxfordjournals.aje.a008976. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Casals-Casas C, Desvergne B. Endocrine disruptors: from endocrine to metabolic disruption. Annu Rev Physiol. 2011;73:135–162. doi: 10.1146/annurev-physiol-012110-142200. [DOI] [PubMed] [Google Scholar]
- Chen J, Kinyamu HK, Archer TK. Changes in Attitude, Changes in Latitude: Nuclear Receptors Remodeling Chromatin to Regulate Transcription. Molecular Endocrinology. 2006;20:1–13. doi: 10.1210/me.2005-0192. [DOI] [PubMed] [Google Scholar]
- Doherty L, Bromer J, Zhou Y, Aldad T, Taylor H. In Utero Exposure to Diethylstilbestrol (DES) or Bisphenol-A (BPA) Increases EZH2 Expression in the Mammary Gland: An Epigenetic Mechanism Linking Endocrine Disruptors to Breast Cancer. Hormones and Cancer. 2010a;1:146–155. doi: 10.1007/s12672-010-0015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty LF, Bromer JG, Zhou Y, Aldad TS, Taylor HS. In utero exposure to diethylstilbestrol (DES) or bisphenol-A (BPA) increases EZH2 expression in the mammary gland: an epigenetic mechanism linking endocrine disruptors to breast cancer. Horm Cancer. 2010b;1:146–155. doi: 10.1007/s12672-010-0015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreijerink KM, Mulder KW, Winkler GS, Hoppener JW, Lips CJ, Timmers HT. Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Res. 2006;66:4929–4935. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
- Eilam-Stock T, Serrano P, Frankfurt M, Luine V. Bisphenol-A impairs memory and reduces dendritic spine density in adult male rats. Behav Neurosci. 2012;126:175–185. doi: 10.1037/a0025959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenichel P, Chevalier N, Brucker-Davis F. Bisphenol A: An endocrine and metabolic disruptor. Ann Endocrinol (Paris) 2013;74:211–220. doi: 10.1016/j.ando.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Fenton SE. Endocrine-Disrupting Compounds and Mammary Gland Development: Early Exposure and Later Life Consequences. Endocrinology. 2006;147:s18–s24. doi: 10.1210/en.2005-1131. [DOI] [PubMed] [Google Scholar]
- Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39:2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- Gibert Y, Sassi-Messai S, Fini JB, Bernard L, Zalko D, Cravedi JP, Balaguer P, Andersson-Lendahl M, Demeneix B, Laudet V. Bisphenol A induces otolith malformations during vertebrate embryogenesis. BMC Dev Biol. 2011;11:4. doi: 10.1186/1471-213X-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ER, Barnes AB, Resseguie L, Barrett JA, Burnside S, Lanza LL, Neff RK, Stevens M, Young RH, Colton T. Breast Cancer in Mothers Given Diethylstilbestrol in Pregnancy. New England Journal of Medicine. 1984;311:1393–1398. doi: 10.1056/NEJM198411293112201. [DOI] [PubMed] [Google Scholar]
- Grun F. Obesogens. Curr Opin Endocrinol Diabetes Obes. 2010;17:453–459. doi: 10.1097/MED.0b013e32833ddea0. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries LW. Long non-coding RNAs and human disease. Biochem Soc Trans. 2012;40:902–906. doi: 10.1042/BST20120020. [DOI] [PubMed] [Google Scholar]
- He S, Liu S, Zhu H. The sequence, structure and evolutionary features of HOTAIR in mammals. BMC Evolutionary Biology. 2011;11:102. doi: 10.1186/1471-2148-11-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10:500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Horwitz KB, Jackson TA, Bain DL, Richer JK, Takimoto GS, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- Hwang HM, Park EK, Young TM, Hammock BD. Occurrence of endocrine-disrupting chemicals in indoor dust. Sci Total Environ. 2008;404:26–35. doi: 10.1016/j.scitotenv.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T, Frankfurt M, Luine V. Estrogen-Induced Memory Enhancements Are Blocked by Acute Bisphenol A in Adult Female Rats: Role of Dendritic Spines. Endocrinology. 2012;153:3357–3367. doi: 10.1210/en.2012-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 2013;29:946–950. doi: 10.3892/or.2012.2219. [DOI] [PubMed] [Google Scholar]
- Jacobs MN, Janssens W, Bernauer U, Brandon E, Coecke S, Combes R, Edwards P, Freidig A, Freyberger A, Kolanczyk R, et al. The use of metabolising systems for in vitro testing of endocrine disruptors. Curr Drug Metab. 2008;9:796–826. doi: 10.2174/138920008786049294. [DOI] [PubMed] [Google Scholar]
- Jeong KW, Kim K, Situ AJ, Ulmer TS, An W, Stallcup MR. Recognition of enhancer element-specific histone methylation by TIP60 in transcriptional activation. Nat Struct Mol Biol. 2011;18:1358–1365. doi: 10.1038/nsmb.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2012;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- Krishnan AV, Stathis P, Permuth SF, Tokes L, Feldman D. Bisphenol-A: an estrogenic substance is released from polycarbonate flasks during autoclaving. Endocrinology. 1993;132:2279–2286. doi: 10.1210/endo.132.6.8504731. [DOI] [PubMed] [Google Scholar]
- Lee HR, Hwang KA, Park MA, Yi BR, Jeung EB, Choi KC. Treatment with bisphenol A and methoxychlor results in the growth of human breast cancer cells and alteration of the expression of cell cycle-related genes, cyclin D1 and p21, via an estrogen receptor-dependent signaling pathway. Int J Mol Med. 2012;29:883–890. doi: 10.3892/ijmm.2012.903. [DOI] [PubMed] [Google Scholar]
- Lee JS, Kim KI, Baek SH. Nuclear receptors and coregulators in inflammation and cancer. Cancer letters. 2008a;267:189–196. doi: 10.1016/j.canlet.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Lee JW, Cheong JH, Lee YC, Na SY, Lee SK. Dissecting the molecular mechanism of nuclear receptor action: transcription coactivators and corepressors. Exp Mol Med. 2000;32:53–60. doi: 10.1038/emm.2000.10. [DOI] [PubMed] [Google Scholar]
- Lee JW, Lee YC, Na SY, Jung DJ, Lee SK. Transcriptional coregulators of the nuclear receptor superfamily: coactivators and corepressors. Cell Mol Life Sci. 2001;58:289–297. doi: 10.1007/PL00000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee J, Lee S-K, Lee JW. Activating Signal Cointegrator-2 Is an Essential Adaptor to Recruit Histone H3 Lysine 4 Methyltransferases MLL3 and MLL4 to the Liver X Receptors. Molecular Endocrinology. 2008b;22:1312–1319. doi: 10.1210/me.2008-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi F, Lucchini F, Pasche C, La Vecchia C. Oral contraceptives, menopausal hormone replacement treatment and breast cancer risk. Eur J Cancer Prev. 1996;5:259–266. doi: 10.1097/00008469-199608000-00006. [DOI] [PubMed] [Google Scholar]
- Li MWM, Mruk DD, Lee WM, Cheng CY. Disruption of the blood-testis barrier integrity by bisphenol A in vitro: Is this a suitable model for studying blood-testis barrier dynamics? The International Journal of Biochemistry & Cell Biology. 2009;41:2302–2314. doi: 10.1016/j.biocel.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard DM, O’Malley BW. Expanding functional diversity of the coactivators. Trends Biochem Sci. 2005;30:126–132. doi: 10.1016/j.tibs.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Lv XB, Lian GY, Wang HR, Song E, Yao H, Wang MH. Long noncoding RNA HOTAIR is a prognostic marker for esophageal squamous cell carcinoma progression and survival. PLoS One. 2013;8:e63516. doi: 10.1371/journal.pone.0063516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclusky H, Leranth The environmental estrogen bisphenol A inhibits estradiol-induced hippocampal synaptogenesis. 2005. Environ Health Perspect. 2005;11:675–679. doi: 10.1289/ehp.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney MM, Padmanabhan V. Developmental programming: impact of fetal exposure to endocrine-disrupting chemicals on gonadotropin-releasing hormone and estrogen receptor mRNA in sheep hypothalamus. Toxicol Appl Pharmacol. 2010;247:98–104. doi: 10.1016/j.taap.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markey CM, Luque EH, Munoz de Toro M, Sonnenschein C, Soto AM. In Utero Exposure to Bisphenol A Alters the Development and Tissue Organization of the Mouse Mammary Gland. Biology of Reproduction. 2001;65:1215–1223. doi: 10.1093/biolreprod/65.4.1215. [DOI] [PubMed] [Google Scholar]
- Markey CM, Wadia PR, Rubin BS, Sonnenschein C, Soto AM. Long-Term Effects of Fetal Exposure to Low Doses of the Xenoestrogen Bisphenol-A in the Female Mouse Genital Tract. Biology of Reproduction. 2005;72:1344–1351. doi: 10.1095/biolreprod.104.036301. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Human Molecular Genetics. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Milhem MM, Knutson T, Yang S, Zhu D, Wang X, Leslie KK, Meng X. Correlation of MTDH/AEG-1 and HOTAIR Expression with Metastasis and Response to Treatment in Sarcoma Patients. J Cancer Sci Ther. 2011:S5. [PMC free article] [PubMed] [Google Scholar]
- Munoz-de-Toro M, Markey CM, Wadia PR, Luque EH, Rubin BS, Sonnenschein C, Soto AM. Perinatal exposure to bisphenol-A alters peripubertal mammary gland development in mice. Endocrinology. 2005;146:4138–4147. doi: 10.1210/en.2005-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-de-Toro M, Markey CM, Wadia PR, Luque EH, Rubin BS, Sonnenschein C, Soto AM. Perinatal Exposure to Bisphenol-A Alters Peripubertal Mammary Gland Development in Mice. Endocrinology. 2005;146:4138–4147. doi: 10.1210/en.2005-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N, Sato I, Takahashi S, Kondo T, Satoh K. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 2013;436:319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- Nakanishi T. Endocrine disruption induced by organotin compounds; organotins function as a powerful agonist for nuclear receptors rather than an aromatase inhibitor. J Toxicol Sci. 2008;33:269–276. doi: 10.2131/jts.33.269. [DOI] [PubMed] [Google Scholar]
- Nanjappa MK, Simon L, Akingbemi BT. The industrial chemical bisphenol A (BPA) interferes with proliferative activity and development of steroidogenic capacity in rat Leydig cells. Biol Reprod. 2012;86:135, 131–112. doi: 10.1095/biolreprod.111.095349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas JM, Segner H. In-vitro screening of the antiestrogenic activity of chemicals. Expert Opin Drug Metab Toxicol. 2008;4:605–617. doi: 10.1517/17425255.4.5.605. [DOI] [PubMed] [Google Scholar]
- Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki Y, et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72:1126–1136. doi: 10.1158/0008-5472.CAN-11-1803. [DOI] [PubMed] [Google Scholar]
- Niland CN, Merry CR, Khalil AM. Emerging Roles for Long Non-Coding RNAs in Cancer and Neurological Disorders. Front Genet. 2012;3:25. doi: 10.3389/fgene.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson S, Gustafsson JA. Estrogen receptor action. Crit Rev Eukaryot Gene Expr. 2002;12:237–257. doi: 10.1615/critreveukaryotgeneexpr.v12.i4.10. [DOI] [PubMed] [Google Scholar]
- Pan YF, Feng L, Zhang XQ, Song LJ, Liang HX, Li ZQ, Tao FB. Role of long non-coding RNAs in gene regulation and oncogenesis. Chin Med J (Engl) 2011;124:2378–2383. [PubMed] [Google Scholar]
- Phillips KP, Foster WG. Endocrine toxicants with emphasis on human health risks. J Toxicol Environ Health B Crit Rev. 2008;11:149–151. doi: 10.1080/00927870701873115. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders PT, Majdic G, Parte P, Millar MR, Fisher JS, Turner KJ, Sharpe RM. Fetal and perinatal influence of xenoestrogens on testis gene expression. Adv Exp Med Biol. 1997;424:99–110. doi: 10.1007/978-1-4615-5913-9_19. [DOI] [PubMed] [Google Scholar]
- Schoeters G, Den Hond E, Dhooge W, van Larebeke N, Leijs M. Endocrine disruptors and abnormalities of pubertal development. Basic Clin Pharmacol Toxicol. 2008;102:168–175. doi: 10.1111/j.1742-7843.2007.00180.x. [DOI] [PubMed] [Google Scholar]
- Shrestha B, Ansari KI, Bhan A, Kasiri S, Hussain I, Mandal SS. Homeodomain-containing protein HOXB9 regulates expression of growth and angiogenic factors, facilitates tumor growth in vitro and is overexpressed in breast cancer tissue. FEBS J. 2012;279:3715–3726. doi: 10.1111/j.1742-4658.2012.08733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra-Miranda M, Delgadillo DM, Mancio-Silva L, Vargas M, Villegas-Sepulveda N, Martinez-Calvillo S, Scherf A, Hernandez-Rivas R. Two long non-coding RNAs generated from subtelomeric regions accumulate in a novel perinuclear compartment in Plasmodium falciparum. Mol Biochem Parasitol. 2012;185:36–47. doi: 10.1016/j.molbiopara.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Smith CC, Taylor HS. Xenoestrogen exposure imprints expression of genes (Hoxa10) required for normal uterine development. The FASEB Journal. 2007;21:239–246. doi: 10.1096/fj.06-6635com. [DOI] [PubMed] [Google Scholar]
- Stangl H, Graf G, Yu L, Cao G, Wyne K. Effect of estrogen on scavenger receptor BI expression in the rat. Journal of Endocrinology. 2002;175:663–672. doi: 10.1677/joe.0.1750663. [DOI] [PubMed] [Google Scholar]
- Sugiura-Ogasawara M, Ozaki Y, Sonta S-i, Makino T, Suzumori K. Exposure to bisphenol A is associated with recurrent miscarriage. Human Reproduction. 2005;20:2325–2329. doi: 10.1093/humrep/deh888. [DOI] [PubMed] [Google Scholar]
- Susiarjo M, Hassold TJ, Freeman E, Hunt PA. Bisphenol A exposure in utero disrupts early oogenesis in the mouse. PLoS Genet. 2007;3:e5. doi: 10.1371/journal.pgen.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Sugihara A, Uchida K, Sato T, Ohta Y, Katsu Y, Watanabe H, Iguchi T. Developmental effects of perinatal exposure to bisphenol-A and diethylstilbestrol on reproductive organs in female mice. Reproductive Toxicology. 2002;16:107–116. doi: 10.1016/s0890-6238(02)00005-9. [DOI] [PubMed] [Google Scholar]
- Titus-Ernstoff L, Hatch EE, Hoover RN, Palmer J, Greenberg ER, Ricker W, Kaufman R, Noller K, Herbst AL, Colton T, et al. Long-term cancer risk in women given diethylstilbestrol (DES) during pregnancy. Br J Cancer. 2001;84:126–133. doi: 10.1054/bjoc.2000.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troisi R, Hatch EE, Titus-Ernstoff L, Hyer M, Palmer JR, Robboy SJ, Strohsnitter WC, Kaufman R, Herbst AL, Hoover RN. Cancer risk in women prenatally exposed to diethylstilbestrol. Int J Cancer. 2007;121:356–360. doi: 10.1002/ijc.22631. [DOI] [PubMed] [Google Scholar]
- Tsai M-C, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa R, Yamada M, Horiguchi K, Ishii S, Hashimoto K, Okada S, Satoh T, Mori M. Aberrant histone modifications at the thyrotropin-releasing hormone gene in resistance to thyroid hormone: analysis of F455S mutant thyroid hormone receptor. Endocrinology. 2009;150:3425–3432. doi: 10.1210/en.2008-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valekunja UK, Edgar RS, Oklejewicz M, van der Horst GT, O’Neill JS, Tamanini F, Turner DJ, Reddy AB. Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1554–1559. doi: 10.1073/pnas.1214168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuorinen A, Odermatt A, Schuster D. In silico methods in the discovery of endocrine disrupting chemicals. J Steroid Biochem Mol Biol. 2013 doi: 10.1016/j.jsbmb.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Wang J, Lu F, Ren Q, Sun H, Xu Z, Lan R, Liu Y, Ward D, Quan J, Ye T, et al. Novel Histone Demethylase LSD1 Inhibitors Selectively Target Cancer Cells with Pluripotent Stem Cell Properties. Cancer Research. 2011;71:7238–7249. doi: 10.1158/0008-5472.CAN-11-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Jin Q, Lee J-E, Su I-h, Ge K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proceedings of the National Academy of Sciences. 2010;107:7317–7322. doi: 10.1073/pnas.1000031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlfahrt-Veje C, Main KM, Skakkebaek NE. Testicular dysgenesis syndrome: foetal origin of adult reproductive problems. Clin Endocrinol (Oxf) 2009;71:459–465. doi: 10.1111/j.1365-2265.2009.03545.x. [DOI] [PubMed] [Google Scholar]
- Wolstenholme JT, Edwards M, Shetty SR, Gatewood JD, Taylor JA, Rissman EF, Connelly JJ. Gestational exposure to bisphenol a produces transgenerational changes in behaviors and gene expression. Endocrinology. 2012;153:3828–3838. doi: 10.1210/en.2012-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, Xiao BX, Guo JM. Acting mechanisms and research methods of long noncoding RNAs. Yi Chuan. 2013;35:269–280. doi: 10.3724/sp.j.1005.2013.00269. [DOI] [PubMed] [Google Scholar]
- Xu X, Hong X, Xie L, Li T, Yang Y, Zhang Q, Zhang G, Liu X. Gestational and lactational exposure to bisphenol-A affects anxiety- and depression-like behaviors in mice. Horm Behav. 2012;62:480–490. doi: 10.1016/j.yhbeh.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- Yearley EJ, Zhurova EA, Zhurov VV, Alan Pinkerton A. Experimental electron density studies of non-steroidal synthetic estrogens: Diethylstilbestrol and dienestrol. Journal of Molecular Structure. 2008;890:240–248. [Google Scholar]
- Zhuang Y, Wang X, Nguyen HT, Zhuo Y, Cui X, Fewell C, Flemington EK, Shan B. Induction of long intergenic non-coding RNA HOTAIR in lung cancer cells by type I collagen. J Hematol Oncol. 2013;6:35. doi: 10.1186/1756-8722-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.