Abstract
Myeloproliferative neoplasms (MPNs) such as chronic myelogenous (CML) and chronic myelomonocytic leukemias (CMML) are frequently induced by tyrosine kinase oncogenes. Although these MPNs are sensitive to tyrosine kinase inhibitors such as imatinib, patients often relapse upon withdrawal of therapy. We used a model of MPN, which is induced by co-expression of the oncoproteins HIP1/PDGFβR (H/P) and AML1/ETO (A/E) from their endogenous loci, to examine the mechanisms of disease development and recurrence following imatinib withdrawal. Although the MPN displayed a full hematologic response to imatinib, 100% of the diseased mice relapsed upon drug withdrawal. MPN persistence was not due to imatinib resistance mutations in the H/P oncogene or massive gene expression changes. Within one week of imatinib treatment, more than 98% of gene expression changes induced by the oncogenes in isolated hematopoietic stem and progenitor cells (LSKs) normalized. Supplementation of imatinib with G-CSF or arsenic trioxide reduced MPN-initiating cell frequencies and the combination of imatinib with arsenic trioxide cured a large fraction of mice with MPNs. In contrast, no mice in the imatinib-treated control cohorts were cured. These data suggest that treatment with a combination of arsenic trioxide and imatinib can eliminate refractory MPN-initiating cells and reduce disease relapse.
INTRODUCTION
Despite the clinical response of BCR/ABL and HIP1/PDGFβR (H/P) induced myeloproliferative neoplasms (MPNs), such as chronic myelogenous (CML) and chronic myelomonocytic leukemias (CMML), to the tyrosine kinase inhibitors (TKIs) specific to the ABL, PDGFR and c-KIT kinases (e.g. imatinib, nilotinib and desatinib), disease persistence in patients on these drugs is a significant problem [1-4]. While oncogenic tyrosine kinase inhibition with imatinib has led to reduced mortality rates for patients with BCR-ABL associated CML [5] and PDGFβR mutant associated CMML [6, 7], a majority of treated patients still have malignant cells that expand into frank disease when drug is discontinued [5, 8]. TKI resistance mutations, amplification of kinase transcripts, reduction of intracellular TKI concentrations or lack of addiction to the oncogenic kinase are all possible mechanisms that enable persistence of TKI treated MPN-initiating cells.
Although the characteristics of MPN-initiating cells in CML and CMML have not been fully elucidated, these cells are thought to share many phenotypic characteristics with hematopoietic stem cells (HSCs), including self-renewal, multi-potency and quiescence [9]. Studies of the CD34+ fraction of CML samples in culture have found that quiescent cells are insensitive to imatinib [10] and become sensitive upon addition of high concentrations of growth factors that promote hematopoietic proliferation and mobilization [11, 12]. The molecular mechanisms underlying the enhancement of a cell’s sensitivity to imatinib by cell cycle entry are not known. There is only suggestive in vivo data with interferon-alpha indicating that the imatinib therapeutic index may be improved with hematopoietic mobilization in individuals with MPNs. Interferon-alpha has pleiotropic cellular effects, including mobilizing activity, and it has been tested as a TKI supplement in humans with CML with favorable outcomes [13]. These limited in vivo data indicate that further studies in mice and humans of HSC mobilizers as additives to continuous imatinib therapy are warranted [13].
We have used a tyrosine kinase oncogene-induced MPN mouse model, which expresses the combination of H/P and AML1/ETO (A/E) oncogenes from conditional knock-in alleles to probe the mechanism(s) of disease persistence in the presence of imatinib [14]. The H/P oncogene is expressed as a result of a t(5;7) chromosomal translocation and is a member of a large family of mutations that involve translocations with the PDGFβR gene, which lead to CMML. There are at least 20 different chromosomal translocation partners for the PDGFβR tyrosine kinase found in CMML [15-17]. Recently, a recurrent PDGFβR mutation, EBF1-PDGFβR, was also identified in Philadelphia chromosome-negative acute lymphoblastic leukemias [18]. The A/E oncogene is expressed as a result of the t(8:21) chromosomal translocation and is frequently found in M2 type acute myeloid leukemias [19]. A/E has not only been reported in a patient with a PDGFβR mutation [20], but and is also frequently present in neoplasms that co-express other tyrosine kinase mutations such as aberrant c-Kit, JAK2 or Flt-3 [18, 21]. Furthermore, AML1 (aka RUNX1) mutations are the most frequent aberrations found in patients with progressed CML [22]. The H/P;A/E combination knock-in model is useful because the knock-in alleles express single copies of the human oncogenes from their endogenous loci resulting in a rapid onset, uniform MPN allowing for therapeutic studies of large numbers of mice with neoplasia in an immune-competent syngeneic background [14]. We used this knock-in model of MPN to investigate cellular and molecular changes contributing to disease persistence in the presence of TKIs and to discover if alternative treatment options are more effective than imatinib therapy alone.
RESULTS
MPN-initiating cells in the H/P;A/E MPN model survive imatinib treatment
We have previously reported that 100% of Mx1-Cre; Hip1LSL-HP;Aml1LSL-AE (H/P;A/E) mice rapidly develop an aggressive, imatinib-responsive MPN following activation of the “knocked-in” human oncogenes [14]. However, we found that the bone marrow in imatinib-treated H/P;A/E mice still contained MPN-initiating cells as assayed by successful bone marrow transplantation of imatinib treated disease into lethally irradiated syngeneic recipients. Whether the remaining MPN-initiating cells were sufficient to cause relapse post drug withdrawal was not known. To answer this, we maintained a cohort of H/P;A/E mice in disease remission with imatinib for four weeks and then a group were taken off therapy. All mice were sacrificed one week after imatinib withdrawal and found to have splenomegaly (Figure 1A, left) and decreased long-term HSC (LT-HSCs) frequencies, a previously characterized hallmark of the H/P;A/E MPN [14] (Figure 1A, right). Irrespective of how long the mice were maintained on imatinib (longest continuous treatment to date has been six months), relapse was observed post drug withdrawal, indicating that enough MPN-initiating cells survived imatinib therapy to cause relapse. This same relapse phenomenon was observed with the use of the more potent cousin of imatinib, nilotinib (Figure 1B). These data further support the observation that TKI treatment suppresses the H/P;A/E MPN but does not cure the disease.
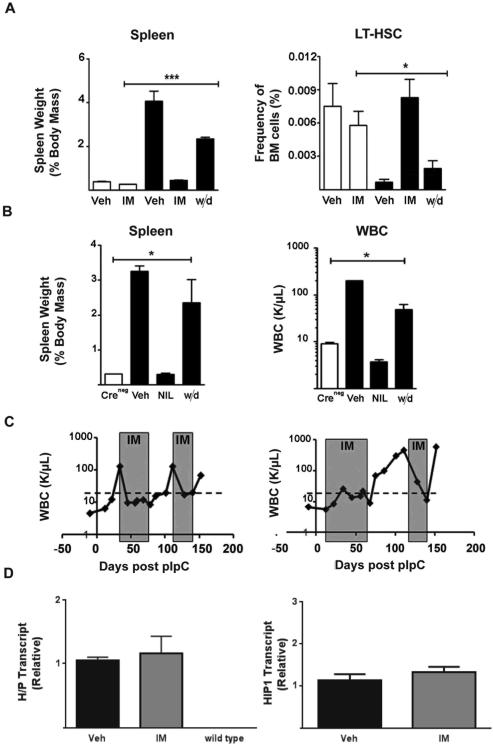
Figure 1. MPN-initiating cells in the H/P;A/E MPN model survive imatinib treatment.
(A) Control Creneg;H/P;A/E (open bars) mice were treated with vehicle (Veh, n=12) or imatinib (IM, n=3) for 28 days. Mx-1Cre;H/P;A/E (black bars) diseased mice were also treated with vehicle for 28 days (Veh, n= 8) or imatinib (IM, n=8). One group of H/P;A/E mice was treated with imatinib for 28 days, drug was discontinued and mice were evaluated seven days later (withdrawal (w/d), n=8). Left, Spleen weight was normalized to body mass. Right, Frequency of LT-HSCs (CD150+CD48−LSK) from H/P;A/E diseased mice and control Creneg mice treated with vehicle, imatinib or imatinib plus withdrawal. In all cases, the vehicle and withdrawal mice were afflicted with hepatosplenomegaly, leukocytosis and diminished LT-HSC frequency (p<0.005 compared to Creneg controls).
(B) Diseased H/P;A/E (black bars) mice were treated for 21 days with vehicle (Veh, n=3) or nilotinib (NIL, n=3). One group was treated with nilotinib for 14 days followed by discontinuation of treatment. The mice were analyzed seven days later (w/d, n=3). Control mice (Creneg, n=3; open bars) were not treated and were used as a reference for normal or baseline numbers. Left, Spleen weight was normalized to body mass. Right, WBC from H/P;A/E diseased mice and control Creneg mice treated with vehicle, nilotinib or nilotinib plus withdrawal (in all cases the different between vehicle or withdrawal and nilotinib were significant; p<0.005 ).
(C) Left, WBC from a representative mouse upon discontinuation of imatinib treatment and re-starting imatinib therapy 48 days later. Right, WBC from a representative mouse in which imatinib therapy was initiated as “prevention” therapy shortly after (10 days) pIpC treatment. A total of 10 H/P;A/E mice were put on a two-month course of imatinib therapy given twice daily immediately after pIpC induction. Drug was then discontinued. Disease was indicated by elevated levels of WBCs in the peripheral blood. All 10 mice given “preventative treatment” presented with disease within two weeks of drug withdrawal (Table S1). Gray boxes indicate periods of imatinib treatment in both cases. Dashed line is drawn at a WBC of 30K/μl; counts above the line indicate disease.
(D) H/P and Hip1 message levels were not elevated in bone marrow after imatinib therapy. Mice were treated with vehicle (n=4) or twice daily imatinib (n=4) for 14 days. Bone marrow was harvested from both femurs for H/P expression analysis (left). Wild type murine Hip1 levels were tested to determine if imatinib led to general upregulation of the Hip1 locus (right). Error bars represent standard deviation. There were no significant differences between the two conditions (vehicle v. imatinib).
Analysis of peripheral blood upon re-treatment with the same dose of imatinib in previously relapsed H/P;A/E mice indicated that the MPN-initiating cells remained sensitive to the same dose of therapy following relapse (Figure 1C, left). The drug sensitivity was also present in diseased mice that were started on imatinib prior to disease onset, in an original attempt to prevent disease altogether (Figure 1C, right, Table S1). Upon discontinuation of imatinib therapy in this “prevention” cohort of mice, disease incidence remained 100%, indicating that imatinib does not prevent the disease. In fact, we have sequenced the knockin allele from two different Hip1+/LSL-H/P spleens after a month of imatinib therapy and have not identified de novo mutations in the H/P sequence. H/P transcript levels in bone marrow from imatinib treated mice did not show increased H/P expression compared to vehicle treated mice (Figure 1D). These data suggest that de novo H/P TKI resistance mutations or oncogene amplifications conferring resistance to TKIs were not the cause of imatinib refractoriness.
Hematopoietic stem and progenitor cell alterations during H/P;A/E-induced MPN normalize with imatinib therapy
Next, we sought to characterize the MPN-initiating cells responsible for disease relapse and persistence. We initially used MRP8-Cre transgenic mice to drive oncogene expression solely in late granulocyte macrophage progenitors (GMP) [23] and observed that H/P;A/E induced MPNs did not develop in mice that express the oncogenes even in those mice that were aged for 2 years (data not shown and Figure S1). This suggested that the MPN-initiating cells originate from the LT-HSCs or the earliest progenitors, such as short-term HSCs (ST-HSCs), multipotent progenitors (MPPs), or common myeloid progenitors (CMPs). Additionally, we performed transplantation of Lineage−Kit+ (LK) (contains CMP, GMP, MEP) and LSK (containing LT-HSC, ST-HSC and MPP) bone marrow fractions into lethally irradiated mice and found that only the LSK population transferred disease suggesting that CMPs are unlikely to contain MPN-initiating cells (Table 1). To investigate whether an abnormal sub-population of progenitors persisted within the LSK in the presence of imatinib as a candidate MPN-initiating cell, we analyzed the frequency of early progenitors in the LSK population with and without imatinib treatment and found that all MPN-associated abnormalities such as decreased MPPs and LT-HSCs and relative increase in ST-HSCs resolved with imatinib (Figure 2A). This complete resolution of early progenitor abnormalities in the presence of imatinib prevented identification of a single persistent MPN-initiating cell population using this analysis.
Table 1.
Transplantation of bone marrow cells from mice with imatinib treated Mx1-Cre;H/P;A/E-induced MPNs
| Donor Cell No. |
Cell Population* |
Donor (n) | Fraction of Recipients with MPN (%) |
|---|---|---|---|
| 5 | LT-HSC | 2 | 12/23 (52%) |
| 50 | LT-HSC | 2 | 12/43 (83%) |
| 50 | LSK** | 2 | 0/15 (0%) |
| 5,000 | LSK | 2 | 2/14 (14%) |
|
| |||
| 50 | LK*** | 3 | 0/15 (0%) |
| 5,000 | LK | 3 | 0/15 (0%) |
|
| |||
| 2,000,000 | WBM | 2 | 10/30 (33%) |
H/P;A/E diseased mice were treated for 14 days with imatinib prior to transplant of their bone marrow cells into lethally irradiated recipient mice. This was necessary because when the mice were not treated, the majority of diseased cells were terminally differentiated neutrophils that diluted out any transplantable cells in the bone marrow. In fact, LT-HSCs are difficult to detect in diseased bone marrow. WBM or FACS isolated populations were co-transplanted with 200,000 wild type bone marrow protective cells.
LSK=Lin-Sca+Kit+ is a population in wild type mice known to contain LT-HSC, ST-HSC and MPP early progenitors.
LK=Lin-Sca-Kit+ is a population of cells in the wild type mice known to contain CMP, MEP, and GMP committed progenitors.
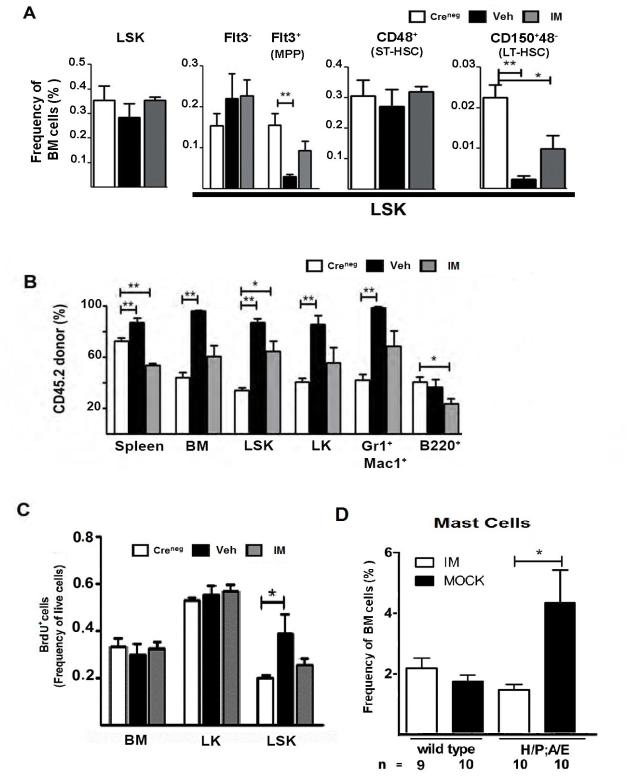
Figure 2. Hematopoietic stem and progenitor cell frequencies normalize with imatinib therapy during H/P;A/E-induced MPN but are not eliminated.
(A) Creneg controls (white bars) or H/P;A/E diseased mice were treated with vehicle (black bars) or imatinib (grey bars) for 7-14 days (Creneg n=7, Veh n=7, IM n=4). The overall frequencies of various stem and progenitor cells within the LSK population (Flt3−LSK and Flt3+LSK=MPP; CD48+=ST-HSC; CD150+4−=LT-HSC) are shown. LSK=Lin−Sca+cKit+. *p<0.05, **p<0.005 compared to Creneg controls.
(B) CD45.2 bone marrow cells from un-induced Mx1-Cre;H/P;A/E (H/P;A/E) or control Creneg;H/P;A/E (Creneg) mice (C57Bl/6J strain) were transplanted with equal numbers of CD45.1 wild type bone marrow cells (SJL strain) into lethally irradiated SJL/J (CD45.1) recipients. Each bar on the graph represents cells from either Creneg control (open bars), vehicle-treated (black bars), or imatinib (grey bars) treated animals (Creneg n=5, Veh n=7, IM n=6). *p<0.05, **p<0.005 compared to Creneg controls.
(C) Mice with H/P;A/E-induced MPN were treated with vehicle (black bars) or imatinib (grey bars) for 14-35 days. Twenty-four hours prior to sacrifice, mice were injected with 1 mg BrdU. BrdU incorporation was measured as frequency of live cells from whole bone marrow, LK and LSK cells.
(D) Frequency of bone marrow cells from wild type (n=10) and diseased H/P;A/E mice that were treated with imatinib (open bars) or mock (black bars) therapy (n=10 for each group). CD117+,Fc?R1+,GP49+ cells were scored as mast cells. Error bars represent the standard error of the mean. Significance was determined using a two-tailed t-test. *p<0.05.
We next attempted to determine which cells persisted in the presence of imatinib by using a chimeric CD45.1/CD45.2 transplant system that permits the analysis of mice with the H/P and A/E oncogenes expressed in only a fraction of the hematopoietic cell compartment. With the expression of these two different isotypes of the pan-hematopoietic marker CD45 (CD45.1 and CD45.2), we were able to differentiate between the recipient and donor populations. After engraftment of a 1:1 mix of un-induced Mx1-Cre;H/P;A/E CD45.2 to wild type CD45.1 cells, the degree of chimerism was stable at 1:1 (CD45.2:CD45.1), and no differences in blood counts or spleen weights between Creneg and Mx1-Cre;H/P;A/E mice were observed. The MPN was then induced with pIpC activation of Mx1-Cre and, as expected, CD45.2 donor cell chimerism approached 100% as the MPN developed (see sample plots in Figure S2A, Figure 2B). Unexpectedly, after one month of imatinib therapy, the fraction of donor cells (CD45.2) versus recipient cells (CD45.1) in the bone marrow returned to starting ratios of approximately 1:1 with the exception of the donor LSK population, which persisted at a statistically significantly higher frequency than starting frequencies no matter how long mice were maintained on imatinib (Figure 2B). There was a trend for the other myeloid populations (whole bone marrow, LK and Gr1+Mac1+) to persist, as well, but none of these differences from the control were statistically significant. Even after longer treatment courses of two mice with 150 days of imatinib therapy, a 1:1 ratio of donor and recipient cells in most bone marrow progenitor compartments remained steady without significant differences from each other, with exception of the slight increase in the LSK population (data not shown). These data are consistent with the assertion that without tyrosine kinase signaling, H/P;A/E MPN-initiating cells are inhibited from generating a full blown MPN but are not eliminated, and in fact differentiate into terminally differentiated hematopoietic cells similar to the differentiation of surrounding wild type LT-HSCs.
Imatinib inhibits hyperproliferation of progenitors from H/P;A/E mouse bone marrow
Because the donor (CD45.2) percentage of the LSK population was elevated compared to other progenitors in the chimera assay (Figure 2B), we examined if these cells demonstrated hyperproliferation during imatinib therapy. Control or diseased mice were treated with vehicle or imatinib before intraperitoneal injection with bromodeoxyuridine (BrdU), which labels 6% of HSCs in normal bone marrow in 24 hours [24]. The mice were sacrificed at 24 hours and bone marrow cells were analyzed for BrdU incorporation. LSK cells from H/P;A/E mice showed a two-fold increase in BrdU incorporation (p<0.04) compared to controls (Figure 2C). However, the frequency of BrdU incorporation in LSKs from H/P;A/E mice returned to control frequencies in the presence of imatinib (Figure 2C). These data indicate that diseased cells within the LSK compartment are hyperproliferative as a result of H/P;A/E signaling but that these cells return to baseline rates of proliferation in the presence of imatinib.
Gene expression changes in LSKs from H/P;A/E mice lead to the identification of a mast-cell specific signature
To identify whether molecular factors contributed to the persistence of MPN-initiating cells, we sought to determine disease-related molecular changes in LSKs in the presence of imatinib. To do this, we compared the gene expression profiles of the Flt3−Lin−Sca+Kit+ (Flt3−LSK) cells in the presence or absence of imatinib. We isolated Flt3−LSK cells from the bone marrow of healthy (Creneg) control mice, diseased (H/P;A/E) mice, or imatinib-treated diseased mice (Imatinib treated–H/P;A/E) and performed RNA expression analysis using Affymetrix arrays. We selected the LSK population of stem and progenitor cells to test in the presence of imatinib because they remained slightly elevated in the imatinib treated chimeric mice (Figure 2B) and were able to transfer the disease into lethally irradiated recipients whereas the more mature progenitor populations did not (Table 1). Mx1-Cre induced H/P;A/E-expression in Flt3−LSK cells led to significant (p<0.01; 2-tailed t-test) changes (2-fold or greater compared to Creneg control) in 355 genes, 82% of which were down-regulated. Further, of these 355 genes, 98% were completely restored to their basal levels upon treatment with a one-week imatinib course (Table S2). This indicated that the majority of disease-associated changes could be rapidly resolved with imatinib. The 355 altered genes did not group into significant pathways including those predicted to be downstream of tyrosine kinases. We used a variety of pathway analysis tools that included Ingenuity Pathway Analysis and Wiki-pathways as well as manual curation and inspection. There were nine gene expression changes that were shared with a recently published 95 gene MPN-initiating cell signature obtained from JunB knockout mice with MPNs [25]. Again, these nine genes did not fall into a single pathway. Because these shared gene changes (highlighted in green in Table S2) resolved with imatinib therapy, we did not investigate them further.
Out of the two percent of altered messages that did not show complete resolution in imatinib-treated diseased mice, we found four genes that exhibited a greater than 10-fold increase in expression compared to control Creneg LSK cells (Table 2; Pla2g7, Cpa3, Ms4a2 and Gp49a). All of these genes are expressed specifically in mast cells [26-29]. After longer term treatment (four weeks) with imatinib, the expression levels of the mast cell genes in LSK cells normalized to Creneg control levels, suggesting that these mast cell-like diseased progenitor cells are eventually cleared (data not shown).
Table 2.
Expression analysis of Flt3-LSK cells from Mx1-Cre;H/P;A/E MPN mice identify a mast cell signature
| Gene Name | Gene Symbol |
Untreated H/P;A/E/cntrl |
IM Treated H/P;A/E/cntrl |
|---|---|---|---|
| tryptase beta 2 | Tpsb2 | 2.70 | 3.79 |
|
| |||
| phospholipase A2, group VII | Pla2g7 | 12.50 | 2.60 |
|
| |||
| carboxypeptidase A3, mast cell | Cpa3 | 49.64 | 23.02 |
|
| |||
| membrane-spanning 4-domains, sf A, member 2 |
Ms4a2 | 62.10 | 7.40 |
|
| |||
| glycoprotein 49 A | Gp49a | 202.10 | 44.22 |
The relative expression profiles of Flt3-LSKs from untreated, diseased mice (n=6) and imatinib-treated (IM) mice (n=6) were compared to LSKs from Creneg H/P;A/E control (cntrl) mice (n=6). Gene expression is depicted as a ratio of H/P;A/E MPN cells over control cells. Genes were selected if ratios were greater than two with limited variability between mice (p < 0.01 using a two-tailed t-test). Of the 355 genes that were differentially affected by disease progression, five out of seven remained perturbed in presence of short-term imatinib were the mast cell specific genes listed here.
Because we observed elevated expression of mast cell specific genes in the LSK population after short-term imatinib treatment, we assessed the frequency of mature mast cells in diseased H/P;A/E and control Creneg mice. Indeed, the mast cell frequencies, as measured by elevated numbers of CD117(c-kit)+,FcεRI+,GP49+ cells, increased 10-fold in the peripheral blood (Figure 2D). Following a two-week course of imatinib or mock treatments, peripheral blood was analyzed for mast cell frequencies and the percentage of CD117(c-kit)+,FcεRI+,GP49+ mast cells in the peripheral blood of imatinib-treated diseased mice resolved. These findings suggest that these newly identified malignant mast cells and their progenitors do not persist in the presence of imatinib. Therefore, the expression array analysis using the LSK population did not identify persistent gene expression patterns that contributed to imatinib refractoriness.
HSCs and Other Progenitors Do Not Display CD47 Upregulation during Imatinib Treatment
A potential mechanism for imatinib refractoriness is MPN-initiating cell evasion of normal immunosurveillance. Normal HSCs upregulate surface expression of the phagocytosis resistance marker CD47 in response to hematopoietic mobilizing stimuli (e.g., lipopolysaccharide [LPS] and granulocyte colony-stimulating factor [G-CSF]), and this expression protects HSCs from macrophage-mediated phagocytosis during peripheral migration [30]. The authors of the latter study hypothesized that LICs may use this normal physiologic process to evade therapy. In their study, CD47 was upregulated on stem, progenitor, and Mac1+ cells in an Mrp8-BcrAbl/Bcl2 transgenic mouse model of CML [31, 32]; however, those mice progressed to blast crisis and it is not known when during disease progression CD47 expression increased. To test this hypothesis in the H/P;A/E model of CML, we measured the CD47-FITC fluorescence intensity of various cell populations of Creneg control mice as well as of cells from vehicle- and imatinib-treated H/P;A/E mice. The largest increase of CD47 staining was detected in the LSK compartment (Figure 3A, black bars, p<0.05); however, imatinib treatment reduced CD47 expression to Creneg control levels in this population (grey bars). In fact, no consistent elevation of CD47 in the LT-HSC population that contains the most leukemogenic cells was observed even in the absence of imatinib treatment. These data suggest that no unique leukemogenic cell population is marked by CD47 expression for protection from peripheral phagocytosis as a mechanism of MPN-initiating cell persistence. Immune evasion by increasing expression of CD47 does not underlie MPN-initiating cell persistence in this mouse model of MPN.
Figure 3. Analysis of Molecular Signals of Immune Evasion (CD47) and Cellular Survival or Proliferation as Possible Mediators of MPN-initiating cell Persistence.
(A) Creneg control or H/P;A/E diseased mice were treated with vehicle or imatinib for 14 days, and bone marrow cells were stained with CD47-FITC. Left, Representative histogram plots of CD47 fluorescence intensity of different bone marrow cell populations. The dotted line depicts the Creneg control median. Right, Quantification of mean fluorescence intensity for multiple cell populations. All vehicle values are significantly greater than Creneg values (*p<0.005), but none of the imatinib values are significantly different than the controls. Creneg, n=3; vehicle-treated (Veh), n=3; and imatinib-treated (IM), n=3. WBM=whole bone marrow, Lin−=lineage negative, LK=Lin−c-Kit+, LSK=Lin−c-Kit+Sca1+, LTHSC=CD150+CD48−CD41−LSK.
(B) Representative PTEN western blot analysis of extracts (75 μg per lane) from whole spleens (wild type [n=6], H/P;A/E + vehicle-treated [n=5], and H/P;A/E + imatinib (IM)-treated [n=5] diseased mice). Spleen histology from each condition is represented below the western blot. See also Figure S3.
Signal Transduction Molecules in Spleens from Imatinib-Treated Diseased Mice Displayed a Delayed Recovery Compared to Histology
Because constitutive signaling by the H/P oncogene was expected due to its constitutive activity in BaF3 cells [33], we tested extracts of spleens from diseased vehicle and imatinib treated mice for altered downstream signaling events as represented by PTEN, AKT, STAT5 or MAPK phosphorylation. Distinct changes in expression of several of these proteins in the diseased spleens compared to wild-type spleens were observed and were initially assumed to result from a change in the cellular composition of the diseased spleens to that of primarily terminally differentiated neutrophils; however, 10 days of imatinib treatment restored the gross phenotype of the spleens, but the molecular alterations did not resolve. For example, AKT and STAT5 were present in wild type spleens but not detected in spleen extracts from mice with MPNs treated with imatinib for 10 days (Figure S3). PTEN was present in the diseased spleens but a new faster migrating protein band was also observed (Figure 3B, Vehicle). It is quite possible that this band represents the cleaved caspase-3 PTEN product identified previously [34]. In contrast, MAPK and actin were not consistently altered. Mouse HIP1 and HIP1-related, recently found to be substrates of PDGFRβ fusion oncogenes [35] were also deficient in the diseased spleens (Figure S3).
Upon longer treatment with imatinib (25 days), the spleen histology (Figure 3B, bottom), weight and WBC parameters as well as the AKT and STAT5 levels were no different than those found in wild type mice (Figure S3). Despite these apparent signs of physical and molecular normalcy, western blot analysis continued to demonstrate the PTEN alteration. However, the amount of this “cleavage” product was much reduced. These data indicate that the faster migrating PTEN band may be a reliable marker of residual disease in imatinib-treated mice. It is tempting to speculate that this PTEN product may have dominant negative activity leading to deficient PTEN inhibition of the PtdIns3-kinase pathway. However, this hypothetical mechanism for promotion of MPN-initiating cell survival in the presence of tyrosine kinase inhibition is untested. Investigating this hypothesis further by sorting out what cells in the spleen express this PTEN product, defining what this product is, and determining its enzymatic activity will be important future steps.
G-CSF and arsenic trioxide synergize with imatinib to reduce MPN-initiating cells
Because imatinib did not cure the H/P;A/E mice, we screened drugs that were readily available from the clinic pharmacy for therapeutic effects in our mice. In addition to availability, we chose drugs that, based on previously reported activity in leukemias, may be alternatives to imatinib or have synergy with imatinib in the H/P;A/E mice. We initially screened the histone deacetylase inhibitor, suberoylanilide hydroxamic acid (SAHA) (recently shown to synergize with imatinib in a BCR/ABL transgenic model of leukemia [36]), Rapamycin (mTor inhibitor), GM-CSF (promotes differentiation of myeloid progenitors [37]), G-CSF (promotes hematopoietic mobilization [38]) and arsenic trioxide (promotes both differentiation and mobilization of hematopoietic cells [39]). None of these agents demonstrated anti-MPN activity when used as test treatments for the H/P;A/E MPN. We then tested their ability to potentiate imatinib therapy by determining whether or not they prevented disease transfer from imatinib treated mice into lethally irradiated recipient mice. This assay is considered a surrogate of MPN-initiating cell activity. From this drug screen, G-CSF and arsenic trioxide showed promise.
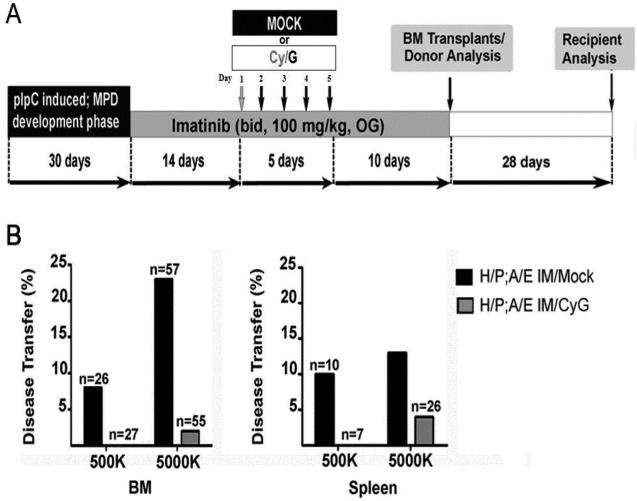
We initially used a Cyclophosphamide/G-CSF (Cy/G) HSC “mobilization” regimen to supplement imatinib [38]. As mentioned earlier, one potential mechanism by which LICs evade kinase inhibitor therapy is by “hiding” in a specific cellular state, such as quiescence [1, 40]; analogous to the mechanism by which normal HSCs avoid chemotherapeutic toxicity. Mice were divided into two groups: one group received mock injections and the other received injections of Cy/G in the presence of imatinib. Both groups were treated with 14-days of imatinib pre-treatment for a hematologic response and then imatinib plus mock or Cy/G injections occurred (“mock” and “Cy/G;” Figure 4A). All mice were continuously treated with another 10 days of imatinib following mobilization. After the five-day imatinib supplementation, shifts in peripheral blood myeloid populations were observed in all Cy/G treated mice; consistent with the fact that G-CSF induces myeloid differentiation and mobilization from the bone marrow. No significant hematological differences were detected between the mobilized and mock groups of donor animals at the end of the 29-day treatment course (Figure S4). After therapy, bone marrow and spleen cells were then transplanted into the lethally irradiated recipients as described previously [14]. As expected, a significant number (25%) of transplant recipient mice that received five million bone marrow cells from imatinib-only treated H/P;A/E donor mice developed an MPN (Table 3 and Figure 4B). In contrast, the addition of Cy/G to imatinib therapy led to an almost complete abrogation of disease transfer even at the highest transferred dose (Table 3 and Figure 4B, five million cells, left hand panel). Similar results were also obtained using spleen cells as transplant material (Table 3 and Figure 4B, right hand panel). Further, when tested alone, G-CSF, but not Cytoxan, eliminated transplantable MPN-initiating cells (Table 3). These findings suggest that G-CSF reduces MPN-initiating cell frequencies when used in combination with imatinib.
Figure 4. G-CSF synergizes with imatinib to reduce MPN-initiating cell frequency.
(A)G-CSF (G) treatment protocol for cohorts of mice with H/P;A/E-induced MPN (Mock, n=6; Cy/G n=6). H/P;A/E diseased mice were injected with the Cy/G mobilizing agent or vehicle alone for five days. Both treatments occurred in the continuous presence of imatinib as depicted schematically in Figure 3A. After a total of 29 days of imatinib therapy, bone marrow or spleen cells were transplanted into lethally irradiated syngeneic recipient mice. Wild type bone marrow cells (500,000 cells) were included in each transplant for radioprotection. Hematopoietic analysis of the recipients for the disease occurred 28 days later.
(B) Mice treated with imatinib plus Cy/G (grey bars) had reduced ability to transfer disease from bone marrow or spleen into secondary lethally irradiated bone marrow recipients compared to mice treated with imatinib alone (black bars).
Table 3.
Supplementation of imatinib (IM) with G-CSF or arsenic trioxide decreases leukemogenic cell frequency in H/P;A/E mice
| Donor Cell No. | Treatment* | Donor (n) | Fraction of Recipients with MPN (%)** |
|---|---|---|---|
| Bone Marrow Transplants | |||
|
| |||
| 500,000 | Mock/IM | 6 | 2/26 (8%) |
| 5,000,000 | Mock/IM | 8 | 12/43 (28%) |
| 500,000 | CyG/IM | 6 | 0/27 (0%) |
| 5,000,000 | CyG/IM | 8 | 1/40 (2.5%) |
|
| |||
| 5,000,000 | G/IM | 3 | 0/43 (0%) |
| 5,000,000 | ASO/IM | 3 | 2/45 (4.4%) |
|
| |||
| Spleen Transplants | |||
|
| |||
| 500,000 | Mock/IM | 2 | 1/10 (10%) |
| 5,000,000 | Mock/IM | 2 | 2/10 (20%) |
| 500,000 | CyG/IM | 2 | 0/7 (0%) |
| 5,000,000 | CyG/IM | 2 | 0/8 (0%) |
H/P;A/E diseased mice were either injected with the Cy/G, G-CSF (G), arsenic trioxide (ASO), or vehicle alone. All treatments occurred in the continuous presence of imatinib as depicted schematically in Figure 3A. Twenty-nine days after imatinib therapy, bone marrow or spleen cells were transplanted into lethally irradiated syngeneic recipient mice. Control bone marrow cells (500,000 cells) were included in each transplant for radioprotection.
MPN-initiating cell frequency was diminished by G-CSF and ASO as determined by the inability of mice treated with these drugs and imatinib to transfer disease from bone marrow or spleen into secondary recipients. See also Figure 3.
In addition to measuring the effects of supplemental G-CSF on imatinib activity in MPN-initiating cells, we also treated a cohort of mice with imatinib and 14 days of supplemental arsenic trioxide, a drug with complex effects. One effect of arsenic trioxide treatment is bone marrow mobilization and it has been reported that arsenic supplementation of cytosine arabinoside (ara-C) chemotherapy improved elimination of MPN-initiating cells [39]. Similar to what was observed with G-CSF supplementation of imatinib, we observed disease did not effectively transplant when arsenic trioxide was added to imatinib (Table 3). Limiting dilution calculations indicate that imatinib treated mice have an MPN-initiating cell frequency of 1/14,000,000 bone marrow cells. The groups treated with either imatinib plus CyG or imatinib plus arsenic had significantly lower frequencies, resulting in over a 15-fold and 7-fold decrease in frequency of MPN-initiating cells in the bone marrow, respectively. These data indicate that a reduction of MPN-initiating cells, as assayed by disease transfer, can be achieved by the supplementation of continuous imatinib therapy with Cy/G-CSF, G-CSF or arsenic trioxide.
Addition of arsenic trioxide to imatinib results in reduction of disease relapse
Although these transplantation studies support the idea that G-CSF and arsenic trioxide can reduce MPN-initiating cell frequency, it is not clear from these studies whether or not these combined therapies eliminate enough MPN-initiating cells to be of clinical use. We generated large cohorts of mice with MPNs for these relapse experiments by transplanting a 50:50 mix of wild type and un-induced Mx1-Cre;H/P;A/E cells into lethally irradiated recipient mice. Mice were then treated with pIpC and developed an MPN within 30 days of induction. MPN-bearing mice were then treated for 14 days with imatinib to obtain a hematologic response and then imatinib was supplemented with Cy/G-CSF or arsenic trioxide. Mice were evaluated for relapse 28 days after all treatment was discontinued (Figure 5A). The addition of Cy/G-CSF to imatinib resulted in no effect on relapse rate or frequency (Figure 5B) despite imatinib’s ability to reduce transplantable MPN-initiating cells (Figure 4B). In contrast, addition of arsenic trioxide to imatinib did prevent relapse. The initial cohort of mice treated with imatinib or imatinib plus arsenic trioxide resulted in a cure of 5 out of the 14 imatinib plus arsenic-treated mice (35.7% vs. 0% imatinib alone; p<0.001) as measured by no evidence of MPN three months after drug withdrawal (Figure 5C). Necropsies showed normal spleens, peripheral blood and bone marrow progenitor cell frequencies. These results were observed again in a second validation cohort of diseased mice. In this case, the combined treatment of imatinib and arsenic trioxide led to disease eradication in an even larger fraction of the mice, an effect that was again not seen in any of the mice treated with imatinib alone (Figure 5D). At the end of the second experiment, all imatinib/mock treated mice showed evidence of MPN. Conversely, only one-fourth (n=3/12) of the imatinib/arsenic trioxide treated mice showed evidence of an MPN. Collectively, data from these two assays for MPN-initiating cell persistence, transplant and disease relapse, indicate that these two compounds, G-CSF and even more effective, arsenic, enhance the ability of imatinib to eradicate MPN-initiating cells.
Figure 5. The addition of arsenic trioxide to imatinib results in the elimination of MPN-initiating cells and leads to decreased relapse rates.
(A) Treatment protocol for mice with H/P;A/E-induced MPN. H/P;A/E diseased mice were treated with imatinib for 14 days and subsequently injected with either the Cy/G mobilizing agent for five days, or arsenic trioxide for 14 days. Both treatments were given in the continuous presence of imatinib, as depicted. Treatments were stopped and relapse analysis was performed 28 days later.
(B) WBC counts at disease onset, after imatinib treatment, or after imatinib and Cy/G combination therapy. Weeks one through three on the x-axis indicates time after withdrawal of treatment.
(C) WBC counts at disease onset, after imatinib treatment, or after imatinib with additional daily arsenic trioxide injections for two weeks. Weeks 1-3 on the x-axis indicates time after withdrawal of treatment. *p<0.05 as determined using Pearson Chi-square analysis.
(D) H/P;A/E induced MPN mice (n=28) were tested for the therapeutic effects of supplemental arsenic. Mice were randomly assigned to imatinib or imatinib supplemented with arsenic trioxide and observed over an eight week period and then sacrificed for hematopoietic analysis. (*p< 0.05 as determined by the Mantel-Cox and Gehan-Breslow-Wilcoxon tests). Nine of the 12 (75%) imatinib plus arsenic treated mice were with normal size livers and spleens vs. all of the imatinib treated mice relapsed with hepatosplenomegaly.
DISCUSSION
Previous studies indicate that patients diagnosed with both BCR/ABL and PDGFβR mutant chronic myeloid leukemias and maintained on imatinib are not cured of their disease [1, 6-8, 41]. Very few patients achieve major molecular remissions on imatinib or nilotinib, and most experience relapse upon drug discontinuation [5, 8]. Upon therapy re-initiation, disease is usually suppressed, indicating that de novo TKI resistance mutations in tyrosine kinase oncogenes do not contribute to the majority of drug refractoriness. These observations suggest that while TKI therapies result in the eradication of a large number of cancerous cells, they do not eliminate MPN-initiating cells that maintain the disease. It would be a relief for patients currently maintained on chronic TKI therapy if new therapies that abolish MPN-initiating cells to yield more cures and allow for drug discontinuation were developed.
With multiple lines of evidence demonstrating that imatinib therapy suppresses but does not cure the MPN in the H/P;A/E mice (Figure 1), we investigated potential mechanisms of MPN-initiating cell persistence. Tyrosine kinase resistance mutations were not a likely cause, as disease relapse was successfully re-treated with the same dose of imatinib or nilotinib (Figure 1A-D). We originally postulated that imatinib leads to the eradication of MPN-initiating cell progeny but not the MPN-initiating cell itself (Figure 5, Situation I). However, we observed that the oncogene expressing cells were present in all progenitors during imatinib therapy, indicating normal hematopoiesis was not inhibited (Figure 2B, Situation II). These data are consistent with a report by Corbin and colleagues where cultured human Bcr/Abl-expressing CD34+ cells also displayed normal hematopoiesis in the presence of imatinib [42]. The ultimate goal is therefore to convert the cytostatic effect of imatinib (Figure 5, Situation II) into a cytotoxic effect (Figure 5, Situation III).
In another attempt to understand why MPN-initiating cells in our H/P;A/E model of MPN persist in the presence of TKIs, we studied gene expression in untreated and imatinib treated LSK cells from H/P; A/E mice. LSKs were tested because they were able to transplant the MPNs into lethally irradiated mice (Table 1). The majority of the 355 genes (82%) with altered expression in the H/P;A/E expressing LSK cells were down-regulated. Ninety-eight percent of these changes resolved with one week of imatinib therapy and there was complete resolution within four weeks of imatinib therapy. We also observed decreased protein expression of AKT, PI3K, STAT5 and the HIP1 family in diseased spleen extracts. These RNA and protein data led us to speculate that the constitutively active H/P tyrosine kinase by using ATP continuously may serve as an energy sink for the cell. This would lead the cell to conserve its limited ATP and decrease overall protein and RNA synthesis to the bare necessities. These data suggest that the disease transferring cells in the LSK population do not express a hallmark gene signature that helps explain the mechanism(s) of MPN-initiating cell persistence.
Another possible mechanism of MPN-initiating cell persistence involves evasion from immunosurveillance. Based on previous reports, the hematopoietic surface marker CD47 is upregulated in myeloid leukemias, and by binding its receptor SIRPα on macrophages, this surface protein provides protection from phagocytosis [30]. In the H/P;A/E-induced MPN, CD47 was indeed upregulated on LSK stem/early progenitor-like cells. However, surface CD47 levels returned to wild type levels upon a response to imatinib. These data suggest that immune evasion via persistence of upregulated CD47 expression is not the principal mechanism of MPN-initiating cell persistence in this model of MPN.
Because there was no distinct mechanism for disease persistence identified, we chose to screen a selection of drugs readily available in the clinic for 1) intrinsic anti-MPN activity or 2) synergistic activity with imatinib. We screened SAHA (HDAC inhibitor), Rapamycin (MTor inhibitor), GM-CSF, G-CSF and arsenic trioxide and did not observe anti-MPN activity. However, we found that G-CSF and arsenic trioxide enhanced imatinib’s ability to reduce MPN-initiating cells (Figure 6, situation III). Both of these agents have HSC mobilization activities suggesting that other HSC mobilizers such as AMD3100 [43] could also synergize with imatinib warranting investigation into such therapeutic activity in the future. However, a recent report of AMD3100 and CNS toxicity by Agarwal and colleagues is a concern [44]. In this study of a model of CML with retroviral expression of BCR/ABL, the combination of BCR/ABL kinase inhibition and plerixaflor (AMD3100) failed to be synergistic against leukemia and was, in fact, neurotoxic due to CNS invasion by CML cells [44].
Figure 6. Models of MPN-initiating cell persistence.
Schematic of normal myeloid development with dark blue HSCs (cycling between quiescent and activated states) differentiating into myeloid progenitors (light blue) which divide into terminally differentiated mature myeloid cells (lightest blue). The persistence of an MPN-initiating cell (depicted as a dark orange) leads to hyper-production of myeloid progenitors (light orange) and mature myeloid cells (lightest orange). Three possibilities are presented for response to tyrosine kinase inhibition by imatinib.
Situation I assumes that myeloid progenitors with the oncogenic kinase are the only cells sensitive to elimination by imatinib. In this scenario, MPN-initiating cells circumvent kinase inhibition and continue to aberrantly differentiate and self-renew. However, the continued presence of imatinib rapidly dispatches the overproduced myeloid progenitors, leaving behind a relatively rare (but active) population of MPN-initiating cells.
Situation II shows that the MPN-initiating cells and mutant progenitors depend on oncogenic kinase signaling for leukemogenesis, but not survival. In this scenario, kinase inhibition by imatinib causes reversion to normal HSC differentiation. The end result is a hematopoietic compartment with a mixed population of wild type and mutant cells that are phenotypically similar.
Situation III indicates what is desired to achieve a cure where MPN-initiating cells are converted to imatinib sensitivity as a result of additional drugs such as arsenic trioxide.
The G-CSF data are consistent with previous data indicating that cultured CD34+ cells from CML patients have a cytostatic response to imatinib that becomes cytotoxic when proliferative cytokines, such as G-CSF, are added to the culture media [10-12]. A recent “negative” clinical trial of combined imatinib and G-CSF in CML was distinct from our suggested protocol as it did not test treatment of patients with continuous imatinib during the G-CSF phase of therapy [45]. It is possible that this treatment schedule may not be optimal. However, even in our mouse model, relapse rate was not altered by G-CSF supplementation of imatinib but was altered by arsenic trioxide supplementation of imatinib.
Further analysis of arsenic and other mobilizers in the clinic is quite feasible as oncologists are already familiar with its FDA approved use for patients with acute promyelocytic leukemia [46]. Ito and colleagues have reported results supporting the assertion that the mobilizing activity of arsenic trioxide is what is responsible for their observed improved elimination of CML-initiating cells by ara-C when supplemented with arsenic trioxide [39]. Also, a phase I/II trial for TKI-resistant accelerated or acute phase CML, which compared imatinib in the presence or absence of arsenic, is listed on the NCI trial web site (clinicaltrials.gov Identifier NCT00053248) but the rationale/results for this trial have not been reported. Future studies that investigate the efficacy of supplementing imatinib or its cousins with arsenic trioxide to cure rather than suppress disease in individuals with MPNs are warranted. The characterization and drug targeting of MPN-initiating cells by using such models as the H/P;A/E mice treated with imatinib and arsenic trioxide have the potential to contribute to the identification and preclinical testing of new therapies for patients with MPNs such as CML and CMML.
METHODS
Mice and transplant model
The pIpC induction of disease in the Mx1-Cre;Hip1+/LSL-H/P;Aml1+/LSL-A/E mouse has been described previously [14]. The Hip1+/LSL-H/P mice have been donated to the Jackson Laboratory. MRP8-CreIRESgfp (Dr. Irv Weissman, Stanford) mice were crossed with Hip1+/LSL-H/P;Aml1+/LSL-A/E mutant mice. Mice were genotyped as described previously [14].
For transplant experiments, donor mice were on a C57Bl/6 (CD45.2) genetic background. For bone marrow transplant, B6.SJL-Ptprc<a>Pepc<b>/BoyJ (SJL) recipient mice (CD45.1, Jackson Labs #002014) were irradiated twice (520 rads each time) separated by 3 hours. Donor cells (CD45.2) and radioprotective or recipient cells (CD45.2) were co-injected retro-orbitally within 24 h of the second dose of irradiation. Creneg;H/P;A/E donors were used as a control for the heterozygous state of the Hip1 and Aml1 loci. The proportion of each cell population that was donor-derived was represented as a percentage calculated as ([CD45.2]/[CD45.1+CD45.2])x100. Mice were housed in the Unit for Laboratory Animal Medicine at the University of Texas Southwestern Medical Center and were monitored regularly for evidence of morbidity and/or abnormal peripheral blood cell counts.
Flow cytometry
Flow cytometric analyses of progenitor frequencies (BD FACS-Canto) and sorting of Flt3−LSKs (BD FACS-AriaII) for array analysis were performed as described previously [47]. BrdU incorporation studies and Annexin V staining were performed according to the manufacturer’s instructions (BD Biosciences). Peripheral blood, bone marrow and spleen cells from imatinib or mock treated Mx1-Cre;H/P;A/E MPN mice and controls were analyzed for CD117(c-kit)+, FcεRI +, GP49+ mast cells. Mast cell specific antibodies (CD117 clone 2B8, FcεR1 clone MAR-1 and GP49 clone H1.1) were acquired from eBioscience.
For analysis of engraftment and chimerism, peripheral blood was collected from the submandibular plexus and analyzed for CD45.1 and CD45.2 as previously described [47]. Bone marrow samples from wild type C57BL/6 (CD45.2) and wild type SJL (CD45.1) mice were analyzed in parallel to set the appropriate flow cytometry gates for CD45 expression. The degree of chimerism was calculated by dividing the number of CD45.2-positive donor cells by the total number of cells expressing either CD45 marker ([CD45.2]/[CD45.1+CD45.2]) to exclude non-hematopoietic CD45 negative cells from the analysis.
Western blot analysis
Spleen extracts were prepared and subjected to western analysis as described previously [48]. For each lane, 75 μg of extract was subjected to SDS-6% or 10% polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose. The antibodies were as follows: polyclonal anti-HIP1 UM354 (1:5000, Assay Designs, Inc.), monoclonal anti-HIP1r (IC5), polyclonal anti-PTEN (1:1000, Cell Signaling), polyclonal anti- AKT (1:1000, Cell Signaling), polyclonal anti-Stat5 (1:200, Santa Cruz), polyclonal anti- MAPK (1:1000, Cell Signaling), polyclonal anti-phospho-MAPK (p42/p44, 1:1000, Cell Signaling) and polyclonal anti-actin (1:1000, Sigma).
RNA samples and isolation
For bone marrow expression arrays 10,000 Flt3−LSKs from bone marrow of the various experimental mice (Creneg;H/P;A/E, Mx1-Cre;H/P;A/E untreated; Mx1-Cre;H/P;A/E treated with one week of twice daily imatinib and a second cohort with four weeks of imatinib) were sorted directly into TRIzol (Ambion) or RNAlater (Life technologies) and RNA was isolated using TRIzol according to the manufacturer’s instructions.
Quantitative PCR
Mice with H/P;A/E MPNs were treated with imatinib or mock for one month, twice daily and bone marrow was harvested from both femurs or from spleens. Splenic and bone marrow RNA was extracted by TRIzol (Invitrogen) and treated with DNase I according to the manufacturer’s instructions. RNA concentrations were quantified in triplicate with the Nanodrop 2000 Spectrophotometer (Thermo Scientific). Two μg total RNA was used to synthesize the first cDNA strand with the SuperScriptTM III First-Strand Synthesis System for RT-PCR (Invitrogen). The Sybr Green quantitative reaction was carried out according to the manufactures instructions with primers specific for H/P, murine HIP1, GAPDH and HPRT.
cDNA synthesis and labeling
For each chip, RNA from 30,000 Flt3−LSKs (bone marrow cells sorted from three mice were combined) was amplified, converted to cDNA and fragmented and labeled using the Ovation RNA Amplification V2 and Encore Biotin Module kits according to the manufacturer’s instruction (NuGEN, San Carlos CA).
Oligonucleotide array hybridization and analysis
Labeled fragmented cDNA (4.4 ug) was hybridized for 18 hr at 45oC to Affymetrix Mouse Genome 430 2.0 short oligomer arrays, which detect approximately 44,000 mouse transcripts representing over 34,000 well-characterized mouse genes and ests (Affymetrix, Santa Clara, CA). Arrays were washed and stained using a Fluidics Station 450 (Affymetrix) according to the manufacturer’s recommended procedures. The arrays were stained with phycoerythrein-conjugated streptavidin (Invitrogen, Carlsbad, CA) and the fluorescence intensities were determined using a GCS 3000 7G high-resolution confocal laser scanner and AGCC software (Affymetrix). The scanned images were analyzed using Expression Console v2.0 software (Affymetrix). Quality control metrics for cRNA integrity, sample loading and variations in staining were determined after background correction and signal summarization by MAS5.0 statistical algorithms resident in GCOS and standardization of each array by global scaling the average of the fluorescent intensities of all genes on an array to a constant mean target intensity of 250.
Array data analysis
Expression data were analyzed following background correction, probe set signal summarization and normalization by MAS5.0 with global scaling (TGT=250), RMA and/or PLIER [49]. Pairwise comparisons of single arrays were performed by MAS 5.0 in GCOS. Probe sets exhibiting significant differential expression were identified using GeneMaths XT (Applied Maths, Austin TX), based on the following criteria: 1) MAS5.0 detection p-values ,0.5 for all samples in at least one experimental group, 2) ANOVA p-value < 0.05, 3) Absolute Signal log ratio > 1.0 and independent t-test p-value < 0.05 for at least one pairwise comparison versus the control cre negative disease-free group. Gene annotation was obtained from the NCBI (www.ncbi.nlm.nih.gov), NetAffx (ww.affymetrix.com), the Gene Ontology Consortium (http://amigo.geneontology.org), the Kyoto Encyclopedia of Genes and Genomes (www.genome.jp/kegg), and WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt).
Drug preparation
Imatinib pills were dissolved in water and filtered through a 0.45-μm syringe filter and brought to a final concentration of 20 mg/ml (100 μl doses). Nilotinib (Tasigna, Novartis) was dissolved in 10% NMP/90% PEG-300 and administered once daily at a dose of 100 mg/kg by oral gavage. The cyclophosphamide/G-CSF (Cy/G) protocol for HSC mobilization has been previously described [38]. Briefly, during imatinib therapy, mice were injected intraperitoneally with 4 mg Cyclophosphamide (Cy;Bristol-Myers Squibb). G-CSF was then administered for 4 consecutive days by subcutaneous injection (5 μg/mouse; 100 μl of a 5 μg/100μl solution; approx. 250 μg/Kg). Arsenic Trioxide was administered with daily i.p. injections for 14 consecutive days (10 μg in 100 μl per 20 g mouse). Imatinib (Gleevec), Cy (Cytoxan), G-CSF (Neupogen), and arsenic (Trisenox) were purchased or graciously donated to the laboratory from the hospital pharmacy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bob Rooney, Luke Peterson, Phil (Zhe) Guan, Ivan Maillard, Yipin Wu and Alice Gauvin for their intellectual input and technical assistance. This work was supported by the following grants: CBTG CA009676 (STP), R01 CA82363-03 (TSR), R01 CA098730-01 (TSR), and a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research (TSR). TSR holds the Jeanne Ann Plitt Professorship in Breast Cancer Research and the H. Ben and Isabelle T. Decherd Chair in Internal Medicine at UT Southwestern Medical Center. TSR was supported as a Leukemia and Lymphoma Society Scholar during the time this work was completed.
ABBREVIATIONS
- A/E
AML1/ETO
- CBC
complete blood count
- CML
chronic myelogenous leukemia
- CMML
chronic myelomonocytic leukemia
- Cy/G
cyclophosphamide/G-CSF
- Cytarabine
ara-C
- GMP
granulocyte/monocyte progenitor
- H/P
HIP1/PDGFβR
- H/P;A/E
genotype of Mx1-Cre;Hip1LSL-hp/+;Aml1LSL-AE/+
- HSC
hematopoietic stem cell
- HSPC
hematopoietic stem and progenitor cell
- LIC
leukemia initiating cell
- LK
lineage−c-Kit+ immunophenotype
- LSK
lineage−Sca-1+c-Kit+ immunophenotype
- LS
lineage-Sca-1+c-Kit+/− immunophenotype
- LT-HSC
long-term hematopoietic stem cell
- MPN
myeloproliferative neoplasm
- MPP
multipotent progenitor
- pIpC
polyinosinic-polycytidylic acid
- ST-HSC
short-term hematopoietic stem cell
- TKI
tyrosine kinase inhibitor
- WBC
white blood cell count
Footnotes
CONFLICT OF INTREST
The authors declare no conflicts of interest.
Author Contributions: STP, ZLH, KIOW and SBF performed, designed, edited and interpreted the research. TSR designed and interpreted the research. TSR, STP, and VEMM wrote the paper.
References
- 1.Savona M, Talpaz M. Getting to the stem of chronic myeloid leukaemia. Nat Rev Cancer. 2008;8(5):341–50. doi: 10.1038/nrc2368. [DOI] [PubMed] [Google Scholar]
- 2.Cortes J, O'Brien S, Kantarjian H. Discontinuation of imatinib therapy after achieving a molecular response. Blood. 2004;104(7):2204–5. doi: 10.1182/blood-2004-04-1335. [DOI] [PubMed] [Google Scholar]
- 3.Higashi T, et al. Imatinib mesylate-sensitive blast crisis immediately after discontinuation of imatinib mesylate therapy in chronic myelogenous leukemia: report of two cases. Am J Hematol. 2004;76(3):275–8. doi: 10.1002/ajh.20096. [DOI] [PubMed] [Google Scholar]
- 4.Mauro MJ, Druker BJ, Maziarz RT. Divergent clinical outcome in two CML patients who discontinued imatinib therapy after achieving a molecular remission. Leuk Res. 2004;28 Suppl 1:S71–3. doi: 10.1016/j.leukres.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 5.Druker BJ, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355(23):2408–17. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 6.David M, et al. Durable responses to imatinib in patients with PDGFRB fusion gene-positive and BCR-ABL-negative chronic myeloproliferative disorders. Blood. 2007;109(1):61–4. doi: 10.1182/blood-2006-05-024828. [DOI] [PubMed] [Google Scholar]
- 7.Apperley JF, et al. Response to imatinib mesylate in patients with chronic myeloproliferative diseases with rearrangements of the platelet-derived growth factor receptor beta. N Engl J Med. 2002;347(7):481–7. doi: 10.1056/NEJMoa020150. [DOI] [PubMed] [Google Scholar]
- 8.Provenzano JD, Kuebler JP. Novel t(5;19) Translocation in a Patient with PDGFRB Associated Chronic Leukemia: Implications for Treatment Strategy. Case Rep Hematol. 2013;2013:709164. doi: 10.1155/2013/709164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang JC, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 2005;15(9):494–501. doi: 10.1016/j.tcb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Graham SM, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99(1):319–25. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- 11.Jorgensen HG, et al. Intermittent exposure of primitive quiescent chronic myeloid leukemia cells to granulocyte-colony stimulating factor in vitro promotes their elimination by imatinib mesylate. Clin Cancer Res. 2006;12(2):626–33. doi: 10.1158/1078-0432.CCR-05-0429. [DOI] [PubMed] [Google Scholar]
- 12.Holtz M, Forman SJ, Bhatia R. Growth factor stimulation reduces residual quiescent chronic myelogenous leukemia progenitors remaining after imatinib treatment. Cancer Res. 2007;67(3):1113–20. doi: 10.1158/0008-5472.CAN-06-2014. [DOI] [PubMed] [Google Scholar]
- 13.Preudhomme C, et al. Imatinib plus peginterferon alfa-2a in chronic myeloid leukemia. N Engl J Med. 2010;363(26):2511–21. doi: 10.1056/NEJMoa1004095. [DOI] [PubMed] [Google Scholar]
- 14.Oravecz-Wilson KI, et al. Persistence of leukemia-initiating cells in a conditional knockin model of an imatinib-responsive myeloproliferative disorder. Cancer Cell. 2009;16(2):137–48. doi: 10.1016/j.ccr.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grand FH, et al. p53-Binding protein 1 is fused to the platelet-derived growth factor receptor beta in a patient with a t(5;15)(q33;q22) and an imatinib-responsive eosinophilic myeloproliferative disorder. Cancer Res. 2004;64(20):7216–9. doi: 10.1158/0008-5472.CAN-04-2005. [DOI] [PubMed] [Google Scholar]
- 16.Jones AV, Cross NC. Oncogenic derivatives of platelet-derived growth factor receptors. Cell Mol Life Sci. 2004;61(23):2912–23. doi: 10.1007/s00018-004-4272-z. [DOI] [PubMed] [Google Scholar]
- 17.Tefferi A, Vardiman JW. Classification and diagnosis of myeloproliferative neoplasms: the 2008 World Health Organization criteria and point-of-care diagnostic algorithms. Leukemia. 2008;22(1):14–22. doi: 10.1038/sj.leu.2404955. [DOI] [PubMed] [Google Scholar]
- 18.Roberts KG, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22(2):153–66. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strout MP, et al. Core-binding factor (CBF) and MLL-associated primary acute myeloid leukemia: biology and clinical implications. Ann Hematol. 1999;78(6):251–64. doi: 10.1007/s002770050511. [DOI] [PubMed] [Google Scholar]
- 20.Golub TR, et al. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell. 1994;77(2):307–16. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]
- 21.Boissel N, et al. Incidence and prognostic impact of c-Kit, FLT3, and Ras gene mutations in core binding factor acute myeloid leukemia (CBF-AML) Leukemia. 2006;20(6):965–70. doi: 10.1038/sj.leu.2404188. [DOI] [PubMed] [Google Scholar]
- 22.Grossmann V, et al. A deep-sequencing study of chronic myeloid leukemia patients in blast crisis (BC-CML) detects mutations in 76.9% of cases. Leukemia. 2011;25(3):557–60. doi: 10.1038/leu.2010.298. [DOI] [PubMed] [Google Scholar]
- 23.Lagasse E, Weissman IL. bcl-2 inhibits apoptosis of neutrophils but not their engulfment by macrophages. J Exp Med. 1994;179(3):1047–52. doi: 10.1084/jem.179.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiel MJ, et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449(7159):238–42. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forsberg EC, et al. Molecular signatures of quiescent, mobilized and leukemia-initiating hematopoietic stem cells. PLoS One. 2010;5(1):e8785. doi: 10.1371/journal.pone.0008785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vainio P, et al. Phospholipase PLA2G7, associated with aggressive prostate cancer, promotes prostate cancer cell migration and invasion and is inhibited by statins. Oncotarget. 2011;2(12):1176–90. doi: 10.18632/oncotarget.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H, et al. Carboxypeptidase A3 (CPA3): a novel gene highly induced by histone deacetylase inhibitors during differentiation of prostate epithelial cancer cells. Cancer Res. 1999;59(12):2981–8. [PubMed] [Google Scholar]
- 28.Zuccolo J, et al. Phylogenetic analysis of the MS4A and TMEM176 gene families. PLoS One. 2010;5(2):e9369. doi: 10.1371/journal.pone.0009369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee KH, et al. Stimulatory function of gp49A, a murine Ig-like receptor, in rat basophilic leukemia cells. J Immunol. 2000;165(9):4970–7. doi: 10.4049/jimmunol.165.9.4970. [DOI] [PubMed] [Google Scholar]
- 30.Jaiswal S, et al. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138(2):271–85. doi: 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaiswal S, et al. Expression of BCR/ABL and BCL-2 in myeloid progenitors leads to myeloid leukemias. Proc Natl Acad Sci U S A. 2003;100(17):10002–7. doi: 10.1073/pnas.1633833100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majeti R, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138(2):286–99. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross TS, et al. Fusion of Huntingtin interacting protein 1 to platelet-derived growth factor beta receptor (PDGFbetaR) in chronic myelomonocytic leukemia with t(5;7)(q33;q11.2) Blood. 1998;91(12):4419–26. [PubMed] [Google Scholar]
- 34.Torres J, et al. Phosphorylation-regulated cleavage of the tumor suppressor PTEN by caspase-3: implications for the control of protein stability and PTEN-protein interactions. J Biol Chem. 2003;278(33):30652–60. doi: 10.1074/jbc.M212610200. [DOI] [PubMed] [Google Scholar]
- 35.Ames HM, et al. HIP1 Phosphorylation by Receptor Tyrosine Kinases. Mol Cell Biol. 2013 doi: 10.1128/MCB.00473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B, et al. Effective targeting of quiescent chronic myelogenous leukemia stem cells by histone deacetylase inhibitors in combination with imatinib mesylate. Cancer Cell. 2010;17(5):427–42. doi: 10.1016/j.ccr.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Connor RF, et al. Addition of sargramostim (GM-CSF) to imatinib results in major cytogenetic response in a patient with chronic myeloid leukemia. Leuk Res. 2006;30(10):1249–52. doi: 10.1016/j.leukres.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 38.Morrison SJ, Wright DE, Weissman IL. Cyclophosphamide/granulocyte colony-stimulating factor induces hematopoietic stem cells to proliferate prior to mobilization. Proc Natl Acad Sci U S A. 1997;94(5):1908–13. doi: 10.1073/pnas.94.5.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito K, et al. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453(7198):1072–8. doi: 10.1038/nature07016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Essers MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458(7240):904–8. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 41.Kavalerchik E, Goff D, Jamieson CH. Chronic myeloid leukemia stem cells. J Clin Oncol. 2008;26(17):2911–5. doi: 10.1200/JCO.2008.17.5745. [DOI] [PubMed] [Google Scholar]
- 42.Corbin AS, et al. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121(1):396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DiPersio JF, et al. Plerixafor and G-CSF versus placebo and G-CSF to mobilize hematopoietic stem cells for autologous stem cell transplantation in patients with multiple myeloma. Blood. 2009;113(23):5720–6. doi: 10.1182/blood-2008-08-174946. [DOI] [PubMed] [Google Scholar]
- 44.Agarwal A, et al. Effects of plerixafor in combination with BCR-ABL kinase inhibition in a murine model of CML. Blood. 2012;120(13):2658–68. doi: 10.1182/blood-2011-05-355396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drummond MW, et al. A pilot study of continuous imatinib vs pulsed imatinib with or without G-CSF in CML patients who have achieved a complete cytogenetic response. Leukemia. 2009;23(6):1199–201. doi: 10.1038/leu.2009.43. [DOI] [PubMed] [Google Scholar]
- 46.Antman KH. Introduction: the history of arsenic trioxide in cancer therapy. Oncologist. 2001;6 Suppl 2 doi: 10.1634/theoncologist.6-suppl_2-1. [DOI] [PubMed] [Google Scholar]
- 47.Yilmaz OH, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441(7092):475–82. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 48.Graves CW, et al. Use of a cryptic splice site for the expression of huntingtin interacting protein 1 in select normal and neoplastic tissues. Cancer Res. 2008;68(4):1064–73. doi: 10.1158/0008-5472.CAN-07-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irizarry RA, Wu Z, Jaffee HA. Comparison of Affymetrix GeneChip expression measures. Bioinformatics. 2006;22(7):789–94. doi: 10.1093/bioinformatics/btk046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.