Abstract
Aims
Active targeted transport of the nanoformulated redox enzyme, catalase, in macrophages attenuates oxidative stress and as such increases survival of dopaminergic neurons in animal models of Parkinson’s disease. Optimization of the drug formulation is crucial for the successful delivery in living cells. We demonstrated earlier that packaging of catalase into a polyion complex micelle (‘nanozyme’) with a synthetic polyelectrolyte block copolymer protected the enzyme against degradation in macrophages and improved therapeutic outcomes. We now report the manufacture of nanozymes with superior structure and therapeutic indices.
Methods
Synthesis, characterization and therapeutic efficacy of optimal cell-based nanoformulations are evaluated.
Results
A formulation design for drug carriers typically works to avoid entrapment in monocytes and macrophages focusing on small-sized nanoparticles with a polyethylene glycol corona (to provide a stealth effect). By contrast, the best nanozymes for delivery in macrophages reported in this study have a relatively large size (~200 nm), which resulted in improved loading capacity and release from macrophages. Furthermore, the cross-linking of nanozymes with the excess of a nonbiodegradable linker ensured their low cytotoxicity, and efficient catalase protection in cell carriers. Finally, the ‘alternatively activated’ macrophage phenotype (M2) utilized in these studies did not promote further inflammation in the brain, resulting in a subtle but statistically significant effect on neuronal regeneration and repair in vivo.
Conclusion
The optimized cross-linked nanozyme loaded into macrophages reduced neuroinflammatory responses and increased neuronal survival in mice. Importantly, the approach for nanoformulation design for cell-mediated delivery is different from the common requirements for injectable formulations.
Keywords: lood–brain barrier, catalase, cell-mediated drug delivery, neuroinflammation
The restriction of macromolecules entry to the brain is a major impediment to the development of new treatment strategies for neurodegenerative and neuroinfectious diseases, which include Parkinson’s (PD) and Alzheimer’s diseases, meningitis, encephalitis, prion disease, HIV-associated neurocognitive disorders [1,2] and stroke [3,4]. The pathobiology of these disorders have an inflammatory component in common [5], with production of reactive oxygen species (ROS) and proinflammatory factors, and consequential neurodegeneration [6–9]. The means to effectively treat such disorders by attenuating ROS production has met with limited success based, in part, on inefficient enzyme delivery to affected brain regions and across the blood–brain barrier (BBB) [10]. To this end, our own works have sought to improve brain delivery of one of the most potent in nature redox enzymes, catalase [11–1 4]. This has been based on the concept that immunocytes readily home to the sites of inflammation and can be used as vehicles for CNS drug delivery of antioxidant enzymes [15]. Active targeted drug transport to disease sites by the cell carriers offers several advantages, which include: improved drug efficacy, prolonged half-lives, time-controlled drug release, and diminished immunogenicity and cytotoxicity. In addition to drug transport across the BBB, the efficient delivery of antioxidants to the brain endothelium, a substantive ROS source [16], could also be beneficial for a range of neurologic disease-combating therapies [17–19].
Our laboratory sought to optimize the structure of catalase nanoparticles for cell-mediated delivery and therapeutic gain. We proposed to incorporate catalase in the block ionomer complexes (BICs) with a cationic block copolymer, poly(ethyleneimine) (PEI)–polyethylene glycol (PEG; PEI–PEG), termed ‘nanozyme’, which protected the enzyme within macrophages [13]. Preservation of catalase enzymatic activity in macrophages by such complexes was, in part, due to the decreasing of acidification of the cell’s endocytic compartments by the amino groups on the surface of the nanoparticles. This inhibited the protease activity and decreased the drug degradation in the host cells [20]. Here, we report further developments of drug nanoformulations for delivery in living cells. Contrary to a common approach for injectable drug formulations, the optimal particles were greater in size and stabilized by a nonbiodegradable cross-linker (Figure 1). This resulted in their higher loading capacity for macrophages, greater release and superior preservation of catalase enzymatic activity. Such nanozymes displayed significant therapeutic efficacy in in vitro and in vivo models of neuroinflammation. These results are in accordance with our prior findings regarding the cross-linking of BICs with another antioxidant enzyme, superoxide dismutase 1, which resulted in improved enzyme stability in the blood and brain [21]. Our ultimate goal is to obtain injectable catalase nanoformulations that may be loaded into cell carriers directly into the bloodstream and delivered to disease sites by active targeted transport in macrophages.
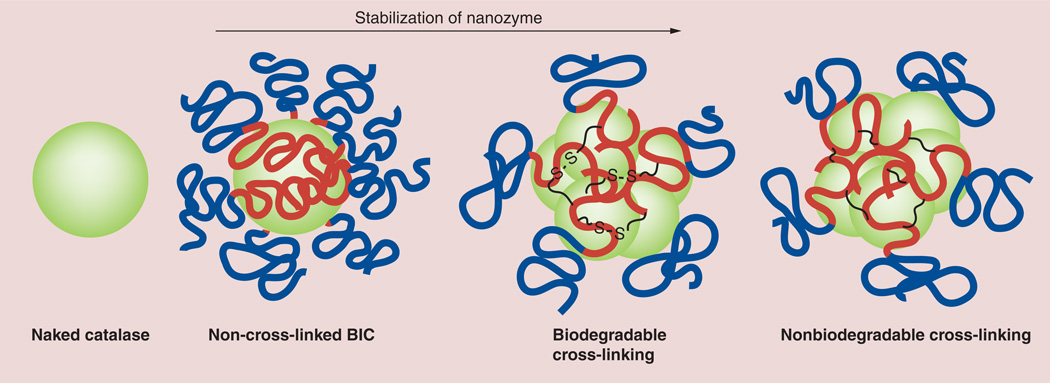
Figure 1. Nanozyme’s structure stabilization.
BICs spontaneously form in aqueous solution as a result of electrostatic coupling of catalase and a cationic block copolymer, poly(ethyleneimine)–polyethylene glycol. The obtained polyion complex, nanozyme, was further stabilized by cross-linking with an excess of biodegradable and nonbiodegradable linkers. BIC: Block ionomer complex.
Material & methods
Reagents
Catalase from bovine liver was purchased from Calbiochem (CA, USA). Bis-(sulfosuccinimidyl)suberate sodium salt (BS3), 3,3’-dithiobis-(sulfosuccinimidyl propionate) (DTSSP), 6-hydroxydopamine (6-OHDA), lipopolysaccharides (LPS), PEI (2K, branched, 50% aqueous solution), rhodamine isothiocyanate, and Triton™ X-100 were from Sigma-Aldrich (MO, USA). Methoxypolyethylene glycol epoxy was from Shearwater Polymer Inc. (AL, USA). PEI (2K)–PEG (10K) was synthesized as described previously by conjugation of PEI and methoxypolyethylene glycol epoxy [11]. 3H-labeled catalase was custom synthesized by PerkinElmer Life (MA, USA). IFN-γ was purchased from Peprotech Inc. (NJ, USA). A XenoLight™ RediJect D-Luciferin Ultra inflammation probe was purchased from Caliper LifeSciences (MA, USA). A description of the different antibodies (Abs) for catalase intracellular localization studies can be found in the Supplementary Material (see online at www.futuremedicine.com/doi/suppl/10.2217/NNM.13.115).
Assembling & cross-linking catalase nanozymes
A description of nanozymes used in this study is presented in Table 1. The BICs were produced by mixing catalase and a block copolymer (PEI–PEG) that bind electrostatically to each other and form nanoparticles with an enzyme–polyion complex core and PEG corona. Catalase and the block copolymer were separately dissolved in phosphate-buffered saline (PBS) at room temperature. Precalculated volumes of the block copolymer solution (0.25 mg/ml) were added drop-by-drop to the enzyme solution (0.5 mg/ml) to achieve the desired charge ratio (charge ratio [Z] = 2–9). The Z was calculated by dividing the number of amino groups in PEI–PEG protonated at pH 7.4 [22] by the total amount of glutamate and aspartate in the catalase. The mixture was supplemented with a six-times excess of either a biodegradable linker (DTSSP) or nonbiodegradable linker (BS3; with respect to catalase charges), incubated overnight at 4°C, and unreacted cross-linker was removed by desalting using NAP-10 Columns (GE Healthcare Bio-Sciences Corp., NJ, USA). Obtained nanozymes were characterized by electrophoretic retention, dynamic light scattering (DLS), nanoparticle tracking analysis and atomic force microscopy (AFM). A description of these methods can be found in the Supplementary Material.
Table 1.
Description of nanozymes.
| Sample ID | Nanoparticle | Z | Cross-linker |
|---|---|---|---|
| Catalase | Catalase alone | 0 | None |
| Non-Cl-2 | Non-Cl BIC | 2 | None |
| Non-Cl-6 | Non-Cl BIC | 6 | None |
| Cl-2-BS3 | Cl BIC | 2 | BS3 |
| Cl-4-BS3 | Cl BIC | 4 | BS3 |
| Cl-6-BS3 | Cl BIC | 6 | BS3 |
| Cl-9-BS3 | Cl BIC | 9 | BS3 |
| Cl-9-DTSSP | Cl BIC | 6 | DTSSP |
BIC: Block ionomer complex; BS3: Bis-(sulfosuccinimidyl)suberate sodium salt; Cl: Cross-linked; DTSSP: 3,3’-Dithiobis-(sulfosuccinimidyl propionate); ID: Identification; Z: Charge ratio.
Macrophage isolation, differentiation & cultivation
Bone marrow-derived macrophages (BMMs) were extracted from the femurs of 6–7-week-old C57Bl/6 male mice according to previously published protocols [23] and cultured for 12–14 days in DMEM medium (Invitrogen, CA, USA) supplemented with 1000 U/ml macrophage colony-stimulating factor (MCSF), a generous gift from Wyeth Pharmaceutical, MA, USA. RAW 264.7, a mouse macrophage cell line, was purchased from the American Type Culture Collection (VA, USA; catalog # TIB-71), and cultured in the DMEM supplemented with 10% fetal bovine serum. Human monocyte-derived macrophages (HMDMs) were obtained from the leukopaks of healthy donors, purified by countercurrent centrifugal elutriation [24] and cultured with MCSF [23].
To promote specific differentiation of RAW 264.7 macrophages, the cells were cultured in the presence of: IL-4 (to promote the M2 anti-inflammatory subtype), or IFN-γ and LPS (to obtain the M1 proinflammatory subtype). For M2 subset differentiation, macrophages were supplemented with IL-4 (40 ng/ml) for 48 h. For M1 subset differentiation, the cells were cultured in a mixture of IFN-γ (20 ng/ml) and LPS (100 ng/ml) for 48 h. Following incubation, the media was replaced with a mixture of: Abs to the mannose receptor (M2 type marker, anti-CD 206; 1 µg/ml; BD Biosciences, CA, USA), and Abs to CD 86 (M1 type marker, anti-CD 86; 2 µg/ml; BD Biosciences). The cells were incubated with the Abs for 1 h, washed, fixed and examined by confocal microscopy as described below (CD 86: λ = 405 nm; and CD 206: λ = 647 nm). An overexpression of specific markers related to the M1 or M2 subset of macrophages upon cell differentiation was confirmed by FACS.
Measures of nanozyme activity
The catalase activity of the obtained nanozymes in a cell-free system was measured using hydrogen peroxide decomposition [13]. The initial catalase activity was 38,000 U/mg protein. Next, the effect of cross-linking on the stability of the enzyme in BIC was examined upon incubation of different nanozymes (0.5 mg/ml catalase) with trypsin (10–5 M) or pronase (2 × 10−1 mg/ml) for 3 h at 37°C. Following incubation, the aliquots were subjected to catalytic activity assessment as described before. The stability of catalase was expressed in residual activity versus initial activity of nanozyme.
For evaluation of the activity of catalase released from the cell carriers, RAW 264.7 macrophages were loaded with different nanozyme formulations for 2 h, then the loading solutions were washed with PBS, and fresh complete media with serum was added to the cells. Following different periods of time, the media was collected and the antioxidant activity of the enzymes released from macrophages was assayed as described before. The concomitant media collected from nonloaded macrophages was examined in the control experiments, and the activity of released endogenous catalase was subtracted from the samples with nanozyme-preloaded macrophages.
Nanozyme accumulation & release
First, cytotoxicity of different nanozyme formulations in BMMs or RAW 267.4 macrophages were evaluated using a standard 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide assay [25], as described in [13]. A tracer dose of 3H-labeled catalase (4 µCi, 2 µl) was mixed with nonlabeled catalase solution (0.03 mg/ml), supplemented with PEI–PEG block copolymer and cross-linked (Cl) to obtain nanozyme, as described before. For accumulation studies, RAW 264.7 macrophages grown on 24-well plates (2.5 × 106 cells/plate) were treated with the 3H-labeled catalase alone or various nanozyme formulations in the assay buffer for different periods of time in the absence of serum [26, 27]. The concentration of catalase was kept the same in all samples (4 µCi/ml). After incubation, the cells were washed with PBS and solubilized in Triton X-100 (1%). For release studies, the cells were loaded with nanozymes for 2 h, washed with PBS and then incubated in fresh media for various times. A total of 100 µl of the cell concomitant media was then placed into 4 ml of a liquid scintillation cocktail, and the radioactivity levels were determined using a Tri-Carb® 4000 (Packard Bioscience; now PerkinElmer) scintillation counter. Assuming that radioactivity levels corresponded to the amount of 3H-labeled nanozyme, the obtained values were normalized for total cell protein content and expressed in ng of catalase per mg of total protein for loading experiments, and ng of catalase per ml of media.
Confocal microscopy
To study intracellular localization, the catalase was labeled with the Alexa Fluor® 680 or Alexa Fluor 488 Protein Labeling Kit (Invitrogen); the PEI or PEI–PEG were labeled with rhodamine isothiocyanate [11]. The intracellular localization studies were performed in HMDMs due to the specificity of the Abs used for different intracellular compartments staining. HMDMs grown in the chamber were treated with labeled Cl-6-BS3 (0.2 mg/ml) in assay buffer for various periods of time (5–60 min) at 37°C [28]. A detailed description of these evaluations can be found in the Supplementary Material.
To evaluate the transport of the macrophages into the brain with inflammation, RAW 264.7 macrophages were transduced with green fluorescent protein (GFP) firefly luciferase viral vector, as described in [29]. To eliminate nontransduced cells, macrophages were selected against puromycin (0.5 µg/ml). GFP-overexpressing macrophages were injected into 6-OHDA-intoxicated BALB/C mice on day 21 after intracranial (ic.) injection of the toxin. A total of 24 h later, mice were sacrificed and perfused with PBS and 4% paraformaldehyde. Brains were frozen, sectioned with a cryostate (10 µm thick) and examined by confocal microscopy (60× magnification). Healthy mice without brain inflammation (with PBS ic. injections) were used as a control group. The glass slides (five slides per brain) were sequentially treated with primary monoclonal mouse anti-CD11b Abs (BD Pharmingen, NJ, USA), the Abs defining macrophages; and secondary fluorescently labeled anti-Mouse-IgG-atto 647N (red; Sigma-Aldrich; 1:200 dilution), and then analyzed by confocal fluorescence microscopy.
For evaluation of catalase activity in the mouse brain, tissues from control animals were ic. injected with PBS or 6-OHDA, as described later, or PD mice were injected with 6-OHDA following intrajugular vein (intravenous; iv.) injection of BMM loaded with nanozyme (5 × 106 cells/mouse in 100 µl PBS) 48 h later. A total of 7 days after BMM treatment, animals were anesthetized with a ketamine/xylazine anesthetic cocktail and perfused transcardially with ice-cold PBS for 5 min. Ventral midbrain areas were dissected and placed in an ice-cold assay buffer (50 mM potassium phosphate buffer; pH 7.0), homogenized and centrifuged at 4000 rpm. Collected supernatant samples were analyzed for catalase activity using the Catalase Assay Kit (Cayman Chemical Company, MI, USA). Enzyme activity was expressed in nmol/min/ml.
Amplex® Red dye fluorescence assay
The decomposition of ROS in Cath.a neurons stimulated with 6-OHDA by different nanozymes released from preloaded macrophages was measured by fluorescence, as described before [11]. The effect of the supernatants collected from nonloaded macrophage ROS decomposition was evaluated with the control experiments. Specific details of this experiment can be found in the Supplementary Material.
Bioimaging & infrared spectroscopy of neuroinflammation
C57Bl/6 male mice were used in the in vivo image visualization and infrared spectroscopy (IVIS) experiments (five mice per group). The animals were treated in accordance with the Principles of Animal Care outlined by the US NIH and approved by the Institutional Animal Care and Use Committee of the University of Nebraska Medical Center (USA). For induction of brain inflammation, mice were ic. injected with LPS solution, as described in [14]. Specific details of these procedures can be found in the Supplementary Material. A total of 24 h after LP intoxication, animals were injected iv. with macrophages (5 × 106 cells/mouse in 100 µl PBS) preloaded with Cl nanozyme (Cl-6-BS3). Saline-treated animals, which were intoxicated with LPS 24 h before the treatment, were used in the control group. A total of 10 min before luminescent imaging (exposure time: 10 min), each animal received an intraperitoneal injection of XenoLight RediJect Inflammation probe. Animals were imaged at 1–30 days following the treatment using an IVIS® 200 Series imaging system (Xenogen Co., now Caliper LifeSciences). The chemoluminescent signal was quantified by Living Image® 2.50 software (Caliper LifeSciences) and presented as a radiance ratio of treated animal versus 24 h after LPS injection.
Immunohistochemical & stereological analyses
6-OHDA-intoxicated mice (six animals per group) were iv. injected either with PBS, nanozyme-preloaded macrophages (5 × 106 cells/mouse/100 µl), macrophages preloaded with naked catalase, macrophages alone, or Cl nanozyme alone (Cl-6-BS3; 0.5 mg/ml catalase) 48 h after intoxication. Healthy, nonintoxicated animals were ic. injected with PBS instead of 6-OHDA and used in control groups. A total of 5 weeks later, animals were sacrificed, perfused according to standard perfusion protocol, brains were removed, and immunohistochemical analysis was performed, as described in [29]. A detailed description can be found in the Supplementary Material.
Statistical analysis
For all experiments, data are presented as the mean ± the standard error of the mean. Tests for significant differences between the groups were performed using a one-way analysis of variance with multiple comparisons (Fisher’s pairwise comparisons) using GraphPad Prism® 6.0 (GraphPad software, CA, USA). A minimum p-value of 0.05 was chosen as the significance level for all tests.
Results
Nanozyme manufacture & characterization
Catalase has a net negative charge at neutral pH (phosphate buffer, pH 7.4). The BICs at various charge ratios (Z = 2–9) were produced by mixing the enzyme and cationic block copolymer PEI–PEG solutions, which bind electrostatically to each other, forming nanoparticles with the enzyme–polyion complex core and PEG corona. The cross-linking of the polyion complexes using nonbiodegradable (BS3) or biodegradable (DTSSP) linkers was confirmed by electrophoresis under denaturizing conditions and they were then transferred to nitrocellulose membranes for western blot (Supplementary Figure S1). Non-Cl BIC was unstable under denaturing conditions, resulting in protein release and subunit dissociation, similar to catalase alone. Cross-linking of BIC with both BS3 and DTSSP linkers retarded catalase electrophoretic mobility, suggesting that cross-linking afforded resistance to protein denaturation. It is worth noting that no free catalase was present in either Cl nanozyme preparation, suggesting that all catalase was incorporated into BIC and conjugated with PEI–PEG.
The hydrodynamic size and zeta-potential of nanozymes were determined by DLS (Table 2). Particles of naked catalase were approximately 13 nm in size with a low polydispersity index. This was close to the theoretical diameter (12.5 nm) of a single protein globule calculated from the molecular mass of the enzyme [30]. The formation of BICs (non-Cl nanozymes) resulted in slightly increased particle size, especially at Z = 2. Significantly larger aggregates were recorded for the Cl nanozymes, especially for BICs obtained with nonbiodegradable linkers (BS3). This was probably due to the cross-linking of several particles together, including bridging them through PEI–PEG that would not otherwise incorporate into BICs. It is worth noting that maximal particle size was recorded at Z = 6, with a mild, but statistically significant, decrease at Z = 9. The zeta-potential values decreased after the cross-linking, which may be indicative of consumption of the protein and/or PEI amino groups that reacted with the cross-linking reagents. Finally, a low polydispersity index for both Cl nanozymes indicates absence of small particles corresponding with the size of free catalase.
Table 2.
Hydrodynamic diameter of nanozymes measured by dynamic light scattering.
| Nanozyme | Z | Deff (nm) | PDI | Zeta-potential (mV) |
|---|---|---|---|---|
| Catalase alone | 0 | 12.5 ± 0.1 | 0.10 | −10.7 |
| Non-Cl-2 | 2 | 13.9 ± 0.1 | 0.20 | −0.8 |
| Non-Cl-6 | 6 | 24.2 ± 0.1 | 0.20 | −0.1 |
| Cl-2-BS3 | 2 | 79.0 ± 0.2 | 0.20 | −7. 0 |
| Cl-4-BS3 | 4 | 118.0 ± 0.2 | 0.20 | −4.9 |
| Cl-6-BS3 | 6 | 175.5 ± 0.2 | 0.20 | −1. 3 |
| Cl-9-BS3 | 9 | 161.3 ± 0.2 | 0.09 | −1. 8 |
| Cl-6-DTSSP | 6 | 152.7 ± 0.2 | 0.20 | −4.3 |
The Deff values represent the mean ± standard error of the mean of three separate measurements. BS3: Bis-(sulfosuccinimidyl)suberate sodium salt; Cl: Cross-linked; Deff: Hydrodynamic diameter; DTSSP: 3,3’-Dithiobis-(sulfosuccinimidyl propionate); PDI: Polydispersity index; Z: Charge ratio.
The size and polydispersity of the obtained nanoparticles were confirmed by a NS500 Nanoparticles Tracking Analysis system (NanoSight, Wiltshire, UK). This method allows the detection of a fraction of small particles in the presence of considerably greater particles, which is not detectable by DLS. The mean particle size for Cl-6-BS3 was 170 nm (Supplementary Figure S2), which correlated with the DLS results (Table 2).
The molecular weight of the obtained nanoparticles was determined using static light scattering analysis. For this purpose, solutions of Cl-6-BS3 were prepared at a range of concentrations (0.022–0.35 mg/ml), and the values of apparent molecular weight (inverse of molecular weight) obtained by light scattering were plotted over the range of nanoparticle concentrations (Supplementary Figure S3). The calculated molecular weight of the particles that was determined by the extrapolation of the linear dependence to zero concentration was 16,000 kDa, suggesting approximately 15 molecules of catalase in the particle.
AFM images confirmed the presence of small spherical particles for non-Cl nanozyme (non-Cl-6) and larger aggregates for the Cl nanozymes (Cl-2-BS3 and Cl-6-BS3) (Supplementary Figure S4). Consistent with DLS studies, the increase of PEI–PEG amount in BIC resulted in bigger particles. Therefore, AFM images provide average diameters and heights for non-Cl-6 (diameter: 11.5 ± 0.03 nm; height: 2.4 ± 0.08 nm), Cl-2-BS3 (diameter: 61.7 ± 0.07 nm; height: 29.8 ± 0.02 nm) and Cl-6-BS3 (diameter: 82.1.7 ± 0.08 nm; height: 36.8 ± 0.02 nm). Notably, as commonly observed for BICs, the sizes determined by AFM were smaller than those determined by DLS. This is typical for such systems, since AFM images are obtained in the absence of the solvent, while DLS determines the hydrodynamic diameter (i.e., the size of the swollen particles) [31]. AFM suggested that the particles of non-Cl-6 and Cl-2-BS3 have a common spherical morphology, while Cl-6-BS3 displayed a higher fraction of nonspherical formations (Supplementary Figure S4). The latter may be due to the cross-linking of multiple BIC particles, with the excess of PEI–PEG polymer present at Z = 6 serving as a particle ‘bridge’.
Effect of cross-linking on nanozyme cytotoxicity, loading & release
Covalent binding of PEI–PEG to catalase considerably diminished the toxicity of nanozymes in RAW 264.7 macrophages and BMMs (Supplementary Table S1). Although non-Cl nanozymes showed significant toxicity in RAW 264.7 macrophages (IC50: 0.3 ± 0.1 and 0.1 ± 0.1 mg/ml protein for non-Cl-2 and non-Cl-6, respectively), all Cl nanozymes were not toxic over all ranges of concentrations (up to 1 mg/ml). It is worth noting that BMMs were more resilient to the cytotoxic effects of nanozymes and did not show any decreases in survival for both Cl and non-Cl nanozymes (Supplementary Table S1).
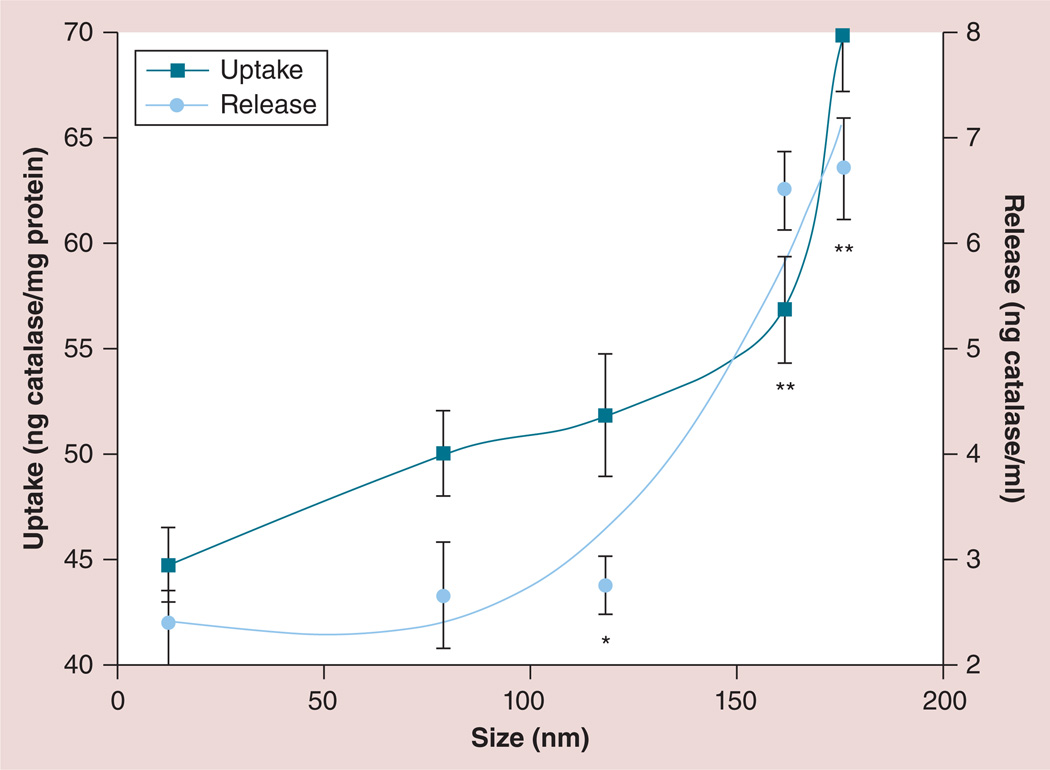
Next, for optimal for cell-mediated delivery, nanozymes should have a high loading capacity in macrophages and a sustained release at the disease site. Herein, we examined the effects of cross-linking on nanozyme accumulation and release in RAW 264.7 macrophages. First, the accumulation kinetics of some representative nanozymes, shown in Supplementary Figure S5a, suggested a rapid uptake. Consistent with our previous work [13], formation of BICs significantly reduced the amount of non-Cl nanozymes taken up by the cells compared with ‘naked’ catalase (Supplementary Figure S5A), possibly due to the effect of the PEG corona. However, the cross-linking of nanozymes significantly improved uptake in macrophages, especially for larger particles (Cl-6-BS3; Supplementary Figure S5B). Similar dependences were found in BMMs. Therefore, the cross-linking of nanozymes with both biodegradable and nonbiodegradable linkers (Cl-6-DTSSP and Cl-6-BS3, 30-min time point) increased their accumulation in the cells 1.5- and 2.3-times, respectively. Overall, the nanozyme loading capacity in macrophages exponentially increased with the particle size (Figure 2; squares). Preloaded RAW 264.7 macrophages released catalase into the media for at least 7 days (Supplementary Figure S6A). Consistent with these studies, cross-linking of nanozymes resulted in significant increases in catalase release from RAW 264.7 macrophages (Supplementary Figure S6B), which also correlated with nanozyme size (Figure 2; circles).
Figure 2. Effect of nanozyme size on loading and release from RAW 264.7 macrophages.
Uptake: the cells were treated for 2 h with different 3H-labeled cross-linked nanozymes (charge ratio = 2–9). Macrophages were then washed, solubilized in Triton™ X-100 (Sigma-Aldrich, MO, USA; 1%) and the amount of accumulated catalase was measured by radioactivity counter. Release: the cells were loaded with different 3H-labeled nanozymes as described before, washed with phosphate-buffered saline and incubated with catalase-free media for 2 h. Concomitant media was then collected and the amount of released catalase was accounted for by radioactivity counter. Results from eight wells (± standard error of the mean) demonstrated an exponential increase in accumulation (squares) and release (circles) of nanozyme with the increased size.
*p < 0.05.
**p < 0.005.
Effect of cross-linking on the preservation of catalase activity in the cell carriers
Based on these results, we selected nanozymes with a greater size (Cl-6-DTSSP and Cl-6-BS3) for further experiments. First, we examined catalase activity for each formulation in a cell-free system. It was demonstrated earlier that the incorporation of catalase into BIC with PEI–PEG at pH 7.4 did not significantly affect the enzyme activity. Here, we confirmed that no changes in the enzymatic activity of catalase were detected upon complex formation, as well as cross-linking with BS3 or DTSSP (Supplementary Table S2). Catalase in the obtained BICs was stable in water solutions; no decreases in the enzymatic activity were recorded for up to 7 days. Furthermore, the cross-linking of catalase with a block copolymer increased stability of catalase against a mixture of proteinases (trypsin and pronase) (Supplementary Figure S7). Therefore, 90–98% of the enzyme was preserved in nanozyme formulations, Cl-6-DTSSP and Cl-6-BS3, when only 10% of naked catalase and 50–80% of non-Cl nanozyme was active after incubation with proteinases.
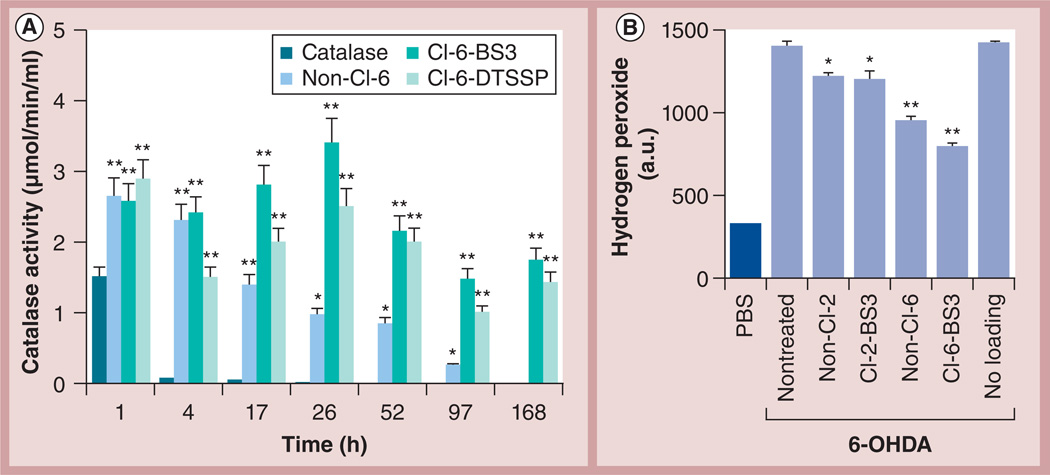
Finally, preservation of enzymatic activity in different catalase formulations as released from RAW 264.7 macrophages was evaluated by spectrophotometry (Figure 3A). Contrary to the free catalase that was completely inactivated inside macrophages after 4 h, macrophages preloaded with non-Cl-6 released active enzyme for at least 52 h. The cross-linking of nanozymes further increased the stability of catalase against degradation in the cells, especially at later time points (52–168 h), resulting in prolonged and sustained release of active catalase from macrophages. Stabilization with nonbiodegradable linker (Cl-6-BS3) protected catalase activity slightly better than with biodegradable linker (Cl-6-DTSSP). A similar pattern was demonstrated for Cl nanozymes released from BMMs. Therefore, enzymatic activity of Cl-6-BS3 released from preloaded BMMs over 24 h was 2.5-times greater than the activity of the corresponding non-Cl nanozyme, non-Cl-6.
Figure 3. Nanozyme cross-linking preserves catalase enzymatic activity in RAW 264.7 macrophages.
(A) ‘Naked’ catalase, or different catalase formulations: non-Cl nanozyme (non-Cl-6), Cl nanozyme with biodegradable linker (Cl-6-DTSSP), or Cl nanozyme with nonbiodegradable linker (Cl-6-BS3) were loaded into RAW 264.7 macrophages for 2 h. The cells were then washed and incubated with catalase-free media for various periods of time. Following incubation, the concomitant media was collected, and the activity of catalase released from preloaded macrophages was accounted for by spectrophotometry. Cross-linking of nanozymes considerably increased the stability of catalase against degradation in the cells, especially at longer time intervals. (B) Different catalase nanozymes (charge ratio = 2 and 6) were loaded into RAW 264.7 macrophages for 2 h. The cells were then washed and incubated in Kreb’s-Ringer buffer for 24 h. In parallel, Cath.A neurons were either incubated with assay buffer (PBS), or stimulated with 200 µM 6-OHDA to produce reactive oxygen species for 24 h. The supernatants collected from macrophages with the released enzyme were supplemented with Amplex® Red (Life Technologies, NY, USA) and horseradish peroxidase solutions and added to the activated neuronal cells. Control activated neurons were incubated with concomitant media from nonloaded macrophages. The amount of hydrogen peroxide produced by neuronal cells and decomposed by catalase released from macrophages was detected by fluorescence. The catalase activity was best protected by Cl nanozyme with a high excess of the block copolymer (Cl-6-BS3) released from macrophages. (A & B) Data represent the mean ± standard error of the mean (n = 4). Statistical significance of nanozyme activity compared with (A) catalase alone or (B) media collected from empty macrophages is shown by asterisks: *p < 0.05, **p < 0.005. 6-OHDA: 6-Hydroxydopamine; BS3: Bis-(sulfosuccinimidyl)suberate sodium salt; Cl: Cross-linked; DTSSP: 3,3’-Dithiobis-(sulfosuccinimidyl propionate).
To assess the antioxidant capacity of the catalase nanoformulations in an in vitro model of PD, RAW 264.7 macrophages preloaded with different nanozymes (Cl-2-BS3 and Cl-6-BS3) were incubated for 24 h in Krebs-Ringer buffer, and the concomitant media was collected and added to neurons treated with 6-OHDA. The amount of hydrogen peroxide produced by activated neurons and decomposed by catalase released from the cell carriers was assessed using Amplex® Red assay (Life Technologies, NY, USA) (Figure 3B). A greater effect was observed by Cl nanozymes that was consistent with their ability to preserve enzyme activity in the carrier cells (Figure 3A). Furthermore, catalase activities in nanozymes obtained at a higher Z (Cl-6-BS3) were protected in macrophages to a greater extent than at a lower Z (Cl-2-BS3). The supernatants collected from nonloaded macrophages had no effect on the hydrogen peroxide level. Together, this study suggests that the Cl nanozyme at Z = 6 was the most efficient in attenuating oxidative stress in an in vitro PD model. This is probably a result of both ROS inactivation in the neuronal media, as well as nanozyme efficient transport from cell carriers to the target cells.
Using ‘alternately activated’ macrophages
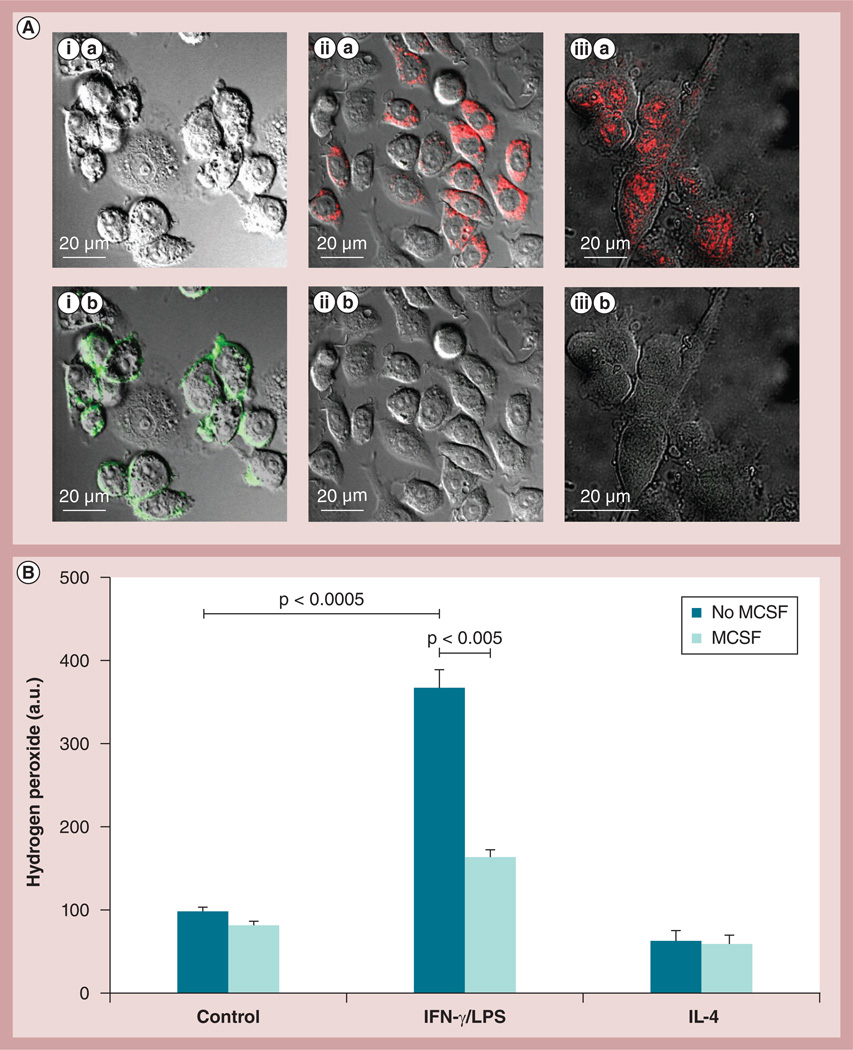
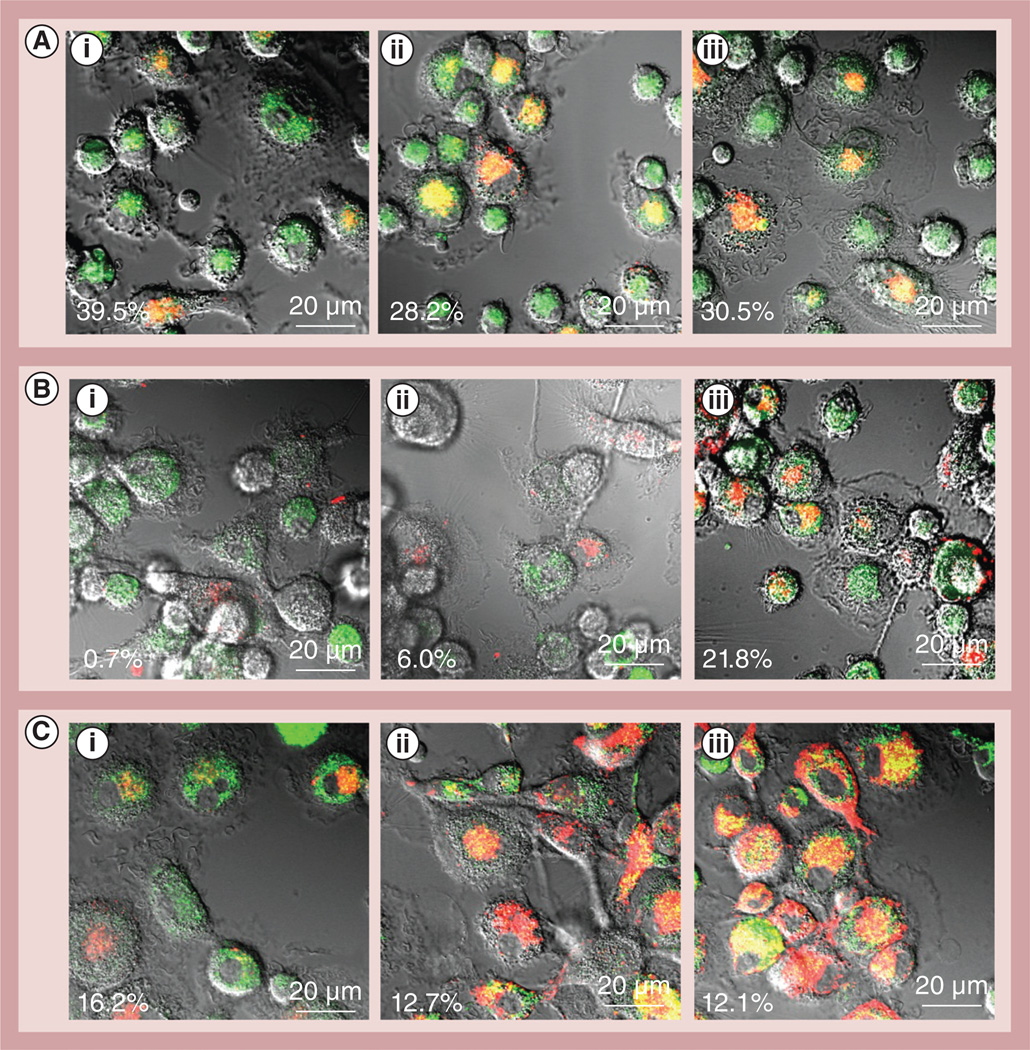
The possibility of using a particular subset of macrophages that would not cause additional inflammation at the disease site (as it is known for classically activated macrophages [32]), but would rather have neuroregenerative properties was investigated. For this purpose, RAW 264.7 macrophages were cultured in the presence of: IL-4 (for the M2 anti-inflammatory subtype); IFN-γ and LPS (for the M1 proinflammatory subtype); or MCSF as described in the ‘Materials & Methods’ section. The obtained subsets of macrophages were characterized by confocal microscopy (Figure 4A). Macrophages differentiated in the presence of IFN-γ/LPS demonstrated considerable expression of CD 86, and low, if any, levels of the mannose receptor (Figure 4A,i), which is indicative for the classically activated proinflammatory M1 subtype. By contrast, RAW 264.7 macrophages differentiated in the presence of IL-4 or MCSF showed high expression levels of the mannose receptor, and low, if any, expression of CD 86 (Figure 4A,iii), which is indicative for ‘alternately activated’ anti-inflammatory macrophages. The obtained results were confirmed by FACS. Indeed, treatment of macrophages with IFN-γ/LPS mixture resulted in upregulation of CD 86 expression, while treatment with IL-4 increased the expression levels of the mannose receptor (Supplementary Table S3). Next, R AW 264.7 macrophage responses to inflammatory signals representing the microenvironment at the disease tissues was examined in vitro. The macrophages differentiated with or without MCSF were supplemented with IFN-γ and LPS (to stimulate proinflammatory responses), and the amount of ROS in the concomitant media was measured by Amplex Red assay (Figure 4B). Cells cultured without MCSF released approximately four-times more ROS in the presence of IFN-γ and LPS. By contrast, macrophages grown in the presence of MCSF produced significantly lower levels of ROS. This indicates that a particular subset of alternatevly activated macrophages (M2) can sustain their anti-inflammatory functions even at the inflammation site and be utilized for targeted drug delivery, while limiting secondary inflammatory-mediated injury.
Figure 4. Macrophage differentiation toward alternatively activated (anti-inflammatory) macrophages.
(A) RAW 264.7 macrophages were cultured in the presence of (i) IFN-γ and LPS for the M1 proinflammatory subtype or (ii) IL-4 for the M2 anti-inflammatory subtype, (iii) or 100 µg/ml MCSF for 2 days as described in the ‘Materials & Methods’ section. The cells were then stained with a mixture of (A,i,a–A,iii,a) antibodies to CD 86 and (A,i,b–A,iii,b) mannose receptors for the M1 and M2 phenotype, respectively, and examined by confocal microscopy. (A,i) Macrophages differentiated in the presence of IFN-γ/LPS showed high expression of CD 86, but low, if any, expression of the mannose receptor, indicating the classically activated M1 subtype. (A,ii & A,iii) By contrast, cells differentiated in the presence of IL-4 or MCSF showed high expression of the mannose receptor, and low expression of CD 86, which is attributed to M2 macrophages. (B) A response of different subtypes of macrophages to proinflammatory conditions. RAW 264.7 macrophages cultured in the absence of MCSF demonstrated significant increases in ROS production in response to stimulation of IFN-γ/LPS. By contrast, macrophages cultured in the presence of MCSF did not demonstrate increased reactive oxygen species production, suggesting that a particular subset of the cell carriers (M2 subtype) would not cause addition inflammation in the brain upon arrival. Data represent the mean ± standard error of the mean (n = 6).
LPS: Lipopolysaccharide; MCSF: Macrophage colony-stimulating factor.
Intracellular localization of nanozyme in macrophages
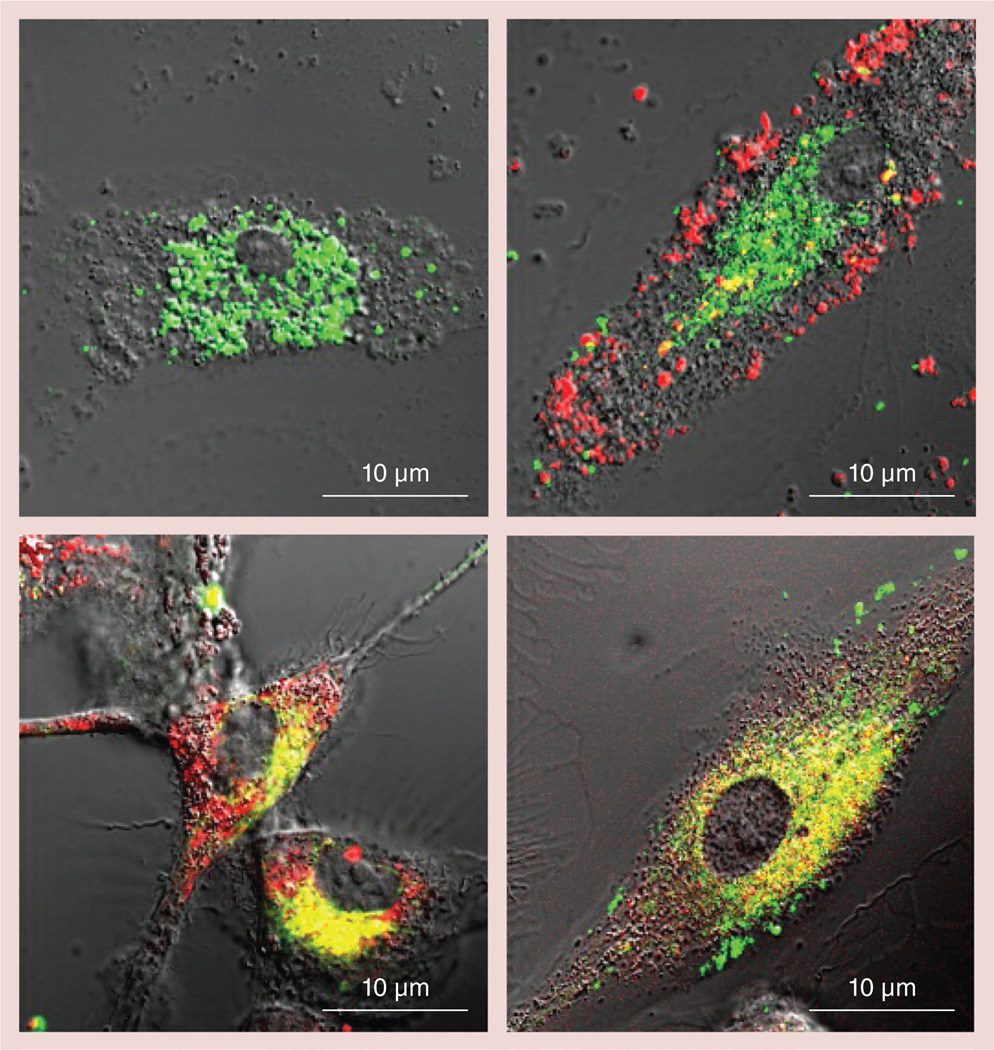
It has long been demonstrated that incorporation of DNA molecules into BICs with PEI–PEG results in the protection of DNA from nuclease degradation in biological milieu [22]. Similarly, we posit that the preservation of the active catalase by incorporation into BIC may be, at least in part, due to steric protection (masking) of the enzyme from proteolytic enzymes by polycation, as well as through PEI–PEG interactions with membranes, and change in ion currents, membrane potential and proton gradients, resulting in the decreases in acidification of endosomal compartments. The nanozyme cross-linking may further enhance this protective effect. To prove this hypothesis, we labeled catalase with Alexa 488 and PEI–PEG with rhodamine isothiocyanate, prepared non-Cl (non-Cl-6) or Cl (Cl-6-BS3) nanozymes, and studied their intracellular localization in BMMs by confocal microscopy (Figure 5). The cells were grown in the presence of MCSF as described in the ‘Materials & Methods’ section. As expected, the cross-linking of BIC (Cl-6-BS3) resulted in practically total colocalization of the polymer and catalase inside macrophages (Figure 5D). The block copolymer from non-Cl-6 was partway dissociated, resulting in the appearance of PEI–PEG fluorescence separate from that of catalase (Figure 5C). However, in this case, considerable colocalization of the labeled catalase and block copolymer still remained, which was consistent with the partial stabilization effect of non-Cl BICs. Control cells loaded with catalase alone are shown in Figure 5A. In this study, we also used BIC with PEI homopolymer (Z = 6) to assess whether the absence of a PEG corona could affect localization of the catalase and polycation in macrophages. Interestingly, the non-Cl polyion complex of homopolymer PEI was completely dissociated in macrophages, with PEI localizing close to the cell surface and catalase in vesicular compartments and/or cytoplasm (Figure 5B). This resulted in acidification of endosomal compartments, which was shown with pH-sensitive dye, LysoSensor™ Yellow/ Blue (Life Technologies), activation of proteases and fast catalase degradation [13]. Therefore, the presence of PEG chains on the surface on the nanozyme plays a stabilization role of the polyion complex inside the cells, possibly by preventing interactions of the polycation with negatively charged cell membranes that may lead to dissociation of non-Cl BIC.
Figure 5. Intracellular distribution of catalase and block copolymer in bone marrow-derived macrophages.
Different nanozymes were prepared with Alexa Fluor® 488-labeled catalase (Invitrogen, CA, USA; green) and rhodamine isothiocyanate-labeled polymers (poly(ethyleneimine) [PEI] or PEI–polyethylene glycol [PEG]; red). Bone marrow-derived macrophages were incubated for 2 h with (A) catalase-alone, or different nanozyme formulations: (B) non-cross-linked (non-Cl) PEI/catalase; (C) non-Cl PEI–PEG/catalase (non-Cl-6); or (D) Cl PEI–PEG/ catalase (Cl-6-bis-(sulfosuccinimidyl)suberate sodium salt), washed with phosphate-buffered saline, and then visualized by confocal microscopy. (D) Stabilization of nanozyme structure by cross-linking resulted in practically total colocalization of the polymer and catalase inside macrophages. (C) PEG corona in non-Cl nanozyme partially prevented block ionomer complex dissociation. (B) The non-Cl block ionomer complexes with PEI polymer were completely dissociated.
Nanozyme cross-linking affects intracellular trafficking in macrophages
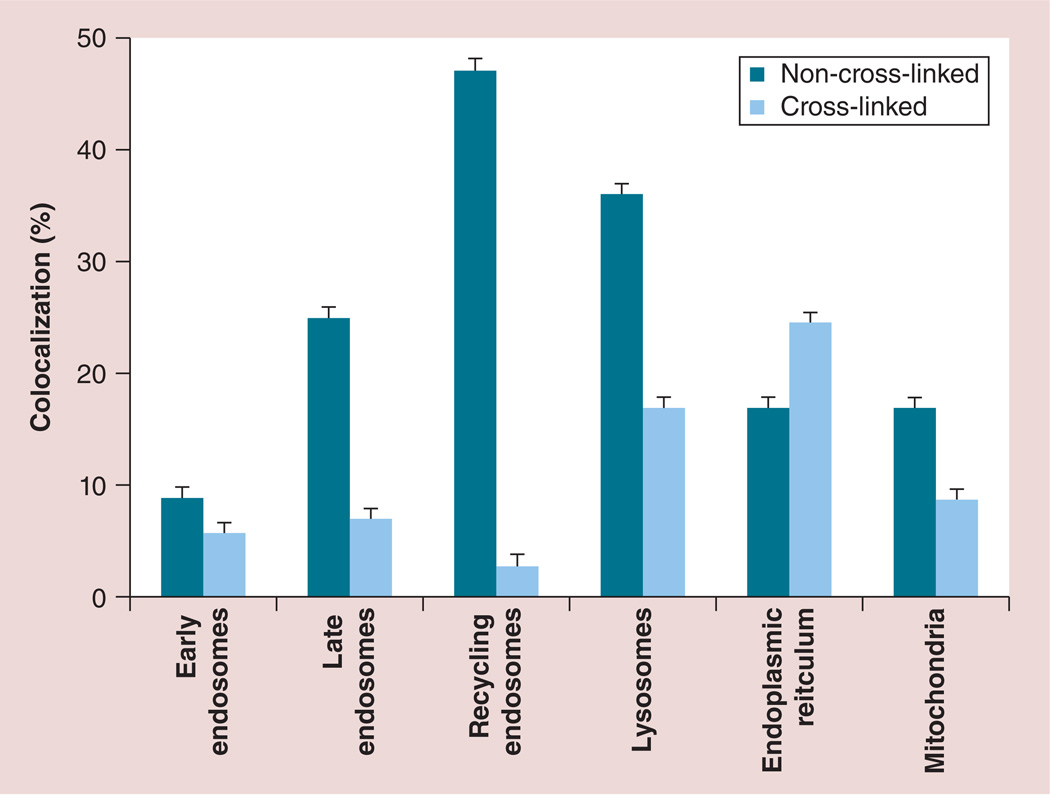
To examine nanozyme intracellular localization in the living cells, colocalization with main cellular compartments was evaluated by staining intracytoplasmic organelles, which included acidified endosomes, endoplasmic reticulum (ER) and mitochondria (Figure 6). Fluorescently labeled catalase was used in Cl nanozyme Cl-6-BS3. The HMDMs grown in the presence of MCSF were incubated with Cl-6-BS3 for various periods of time, washed and stained with markers for different intracellular compartments. Microphotographs demonstrated substantial colocalization of the nanozyme with LysoTracker® (Life Technologies; 28–39% colocalization; Figure 6 & Supplementary Table S3), and to a lesser degree with ER (1–20% colocalization; Figure 6 & Supplementary Table S3) and MitoTracker® (Life Technologies; 12–16% colocalization; Figure 6 & Supplementary Table S4). It is worth noting that Cl-6-BS3 was relocated from acidic endosomes to other cellular compartments at later time points (30–60 min; Supplementary Table S4). LysoTracker allows the examination of nanoparticle intracellular localization in live cells [33]. However, the marker stains all acidic compartments, including lysosomes, late endosomes and, to some extent, recycling endosomes. To study nanozyme localization in greater detail, specific staining with Abs to early, lat or recycling endosomes and lysosomes was performed in fixed human macrophages (Supplementary Figure S8 & Table S4). The Cl nanozyme Cl-6-BS3 first accumulated largely in early and late endosomes as determined by fluorescein isothiocyanate-conjugated mouse antibodies to EEA1 to early endosomes (BD Biosciences; 5–15% colocalization; Supplementary Figure S8 & Table S4) and mouse monoclonal antibodies to RAB 7 to late endosomes (Abcam, MA, USA; 7–17% colocalization; Supplementary Figure S8 & Table S4) immunostaining, respectively, and then was reshuffled to lysosomes (6.5–22% colocalization with LAMP1; to late endosomes Supplementary Figure S8 & Table S4). In contrast to a preferential compartmentalization of non-Cl BIC within recycling endosomes reported earlier [34], there was very little colocalization of Cl nanozyme with recycling endosomes as determined by Rab 11 Abs (3–10% colocalization; Supplementary Figure S8 & Table S4). It indicates that the cross-linking of catalase nanozymes altered their trafficking and intracellular localization in macrophages, resulting in nanozyme escape from endosomal compartments (in particular, recycling endosomes) into cytoplasm, and accumulation in ER and mitochondria with partial localization in lysosomes (Figure 7). It is worth nothing that a portion of nanozyme transferred to cytosol may be released inside exosomes, extracellular organelles secreted by macrophages, as was demonstrated in [34].
Figure 6. Nanozyme intracellular trafficking in macrophages.
Alexa Fluor® 680-labeled catalase (Invitrogen, CA, USA; red) was used to obtain cross-linked 6-bis-(sulfosuccinimidyl)suberate sodium salt. Live human monocyte-derived macrophages were incubated with cross-linked nanozyme for different times, and stained with: (A) LysoTracker® Green (Life Technologies, NY, USA; 150 nM), (B) ER-Tracker™ Green (Life Technologies; 1 µM), or (C) MitoTracker® Green (Life Technologies; 150 nM). The macrophages were incubated for (A,i, B,i & C,i) 5 min, (A,ii, B,ii & C,ii) 15 min and (A,iii, B,iii & C,iii) 30 min. Colocalization of nanozyme (red) and compartment staining (green) is manifested as yellow. Nanozyme accumulated first in acidified compartments and then reached mitochondria and endoplasmic reticulum. Refer also to Supplementary Table S3 for quantitative colocalization. The percentages in the bottom left hand of every image are the percentages of colocalization of nanozyme (red) and the compartment staining (green) specific for each slide, which is manifested in yellow colour.
Figure 7. Effect of nanozyme cross-linking on macrophage trafficking.
Human monocyte-derived macrophages were incubated with fluorescently labeled cross-linked and non-cross-linked nanozymes for 1 h, stained with different markers to specific intracellular compartments, and a percentage of nanozyme colocalization with these markers was evaluated using confocal microscopy images. Data represent the mean ± standard error of the mean (n = 40). Stabilization of nanozyme structure by cross-linking of nanoparticles drastically altered trafficking and intracellular localization in macrophages, shifting distribution from recycling and late endosomes to endoplasmic reticulum, mitochondria and, in part, lysosomes. Data for non-cross-linked nanozymes were reported in [34].
Macrophages accumulate in an inflamed mouse brain
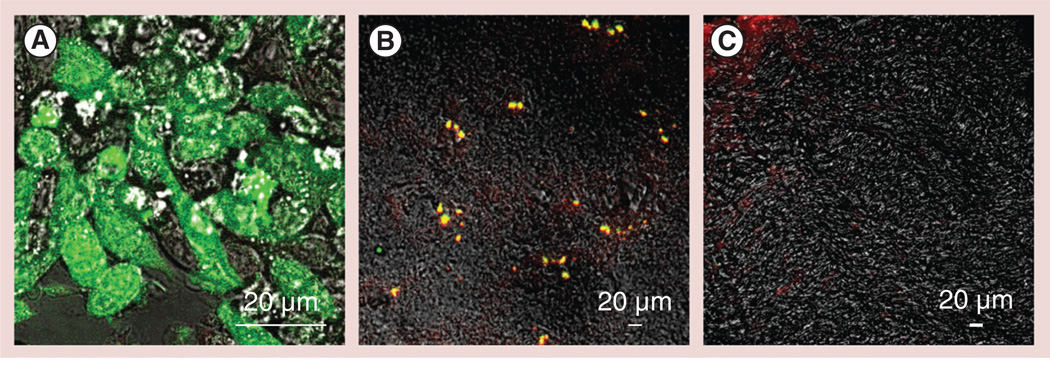
To examine whether macrophages reach the brain tissues, 6-OHDA-intoxicated mice were iv. injected with GFP-expressing RAW 264.7 macrophages. To track the cells in the mouse brain, macrophages were transduced with GFP firefly luciferase viral vector to overexpress the chimeric protein consisting of GFP and firefly luciferase (Figure 8A). Transduced macrophages were cultured for 5 days in the presence of MCSF to produce M2 ‘alternately activated’ macrophages, and then systemically administered into PD mice at the peak of inflammation (21 days after 6-OHDA intoxication). A total of 24 h later, mice were sacrificed and perfused as described in the ‘Materials & Methods’ section. Confocal images of brain slides taken from 6-OHDA-intoxicated mice demonstrate significant amounts of GFP-expressing macrophages in the midbrain (Figure 8B). Colocalization of GFP-expressing macrophages (green) and Abs to CD11b (red) manifested in yellow staining confirmed that the accumulated cells were in fact macrophages. It is worth nothing that macrophages were not detected in the brain of healthy mice (Figure 8C). Furthermore, catalase activity in the ventral midbrain of mice with brain inflammation was evaluated. Significant increases in the enzyme activity (p < 0.05) were found in PD mice treated systemically with macrophages loaded with nanoformulated catalase (15.8 ± 0.6 nmol/min/ml) compared with PD mice treated with PBS (7.8 ± 0.5 nmol/min/ml), or control healthy mice (7.9 ± 0.9 nmol/min/ml). Taken together, this indicated that catalase-loaded macrophages were able to cross the BBB and deliver their payload to the inflamed tissues.
Figure 8. Recruitment of green fluorescent protein-expressing RAW 264.7 macrophages to the substantia nigra pars compacta in 6-hydroxydopamine-intoxicated mice.
(A)Macrophages were transduced with green fluorescent protein (GFP) firefly luciferase virus and selected against katamycin. BALB/c mice were intracranially injected with 6-hydroxydopamine into the substantia nigra pars compacta, as described in the ‘Materials & Methods’ section. A total of 21 days later, the animals were injected into the intrajugular vein with GFP-expressing RAW 264.7 macrophages (green; 5 × 106 cells/ mouse in 100 µl). A total of 24 h later, the mice were sacrificed, and perfused with phosphate-buffered saline and 4% paraformaldehyde. (B) The brains were frozen, sectioned with a cryostat (10 µm thick) and examined by confocal microscopy (60× magnification). (C) Healthy mice without brain inflammation (with phosphate-buffered saline intracranial injections) were used as a control group. Slides were stained with primary antibodies to CD11b, a marker for macrophages, and secondary fluorescently labeled anti-mouse-IgG-atto 647N (red). Colocalization of GFP-expressing macrophages and CD11b antibodies, manifested in yellow staining, confirmed the presence of significant amounts of the genetically modified cells in the intoxicated brain. (C) No fluorescence in the healthy brain was found, indicating that systemically administered RAW 264.7 macrophages did not cross the blood–brain barrier in the absence of brain inflammation.
Nanozyme-loaded BMMs reduce neuroinflammation
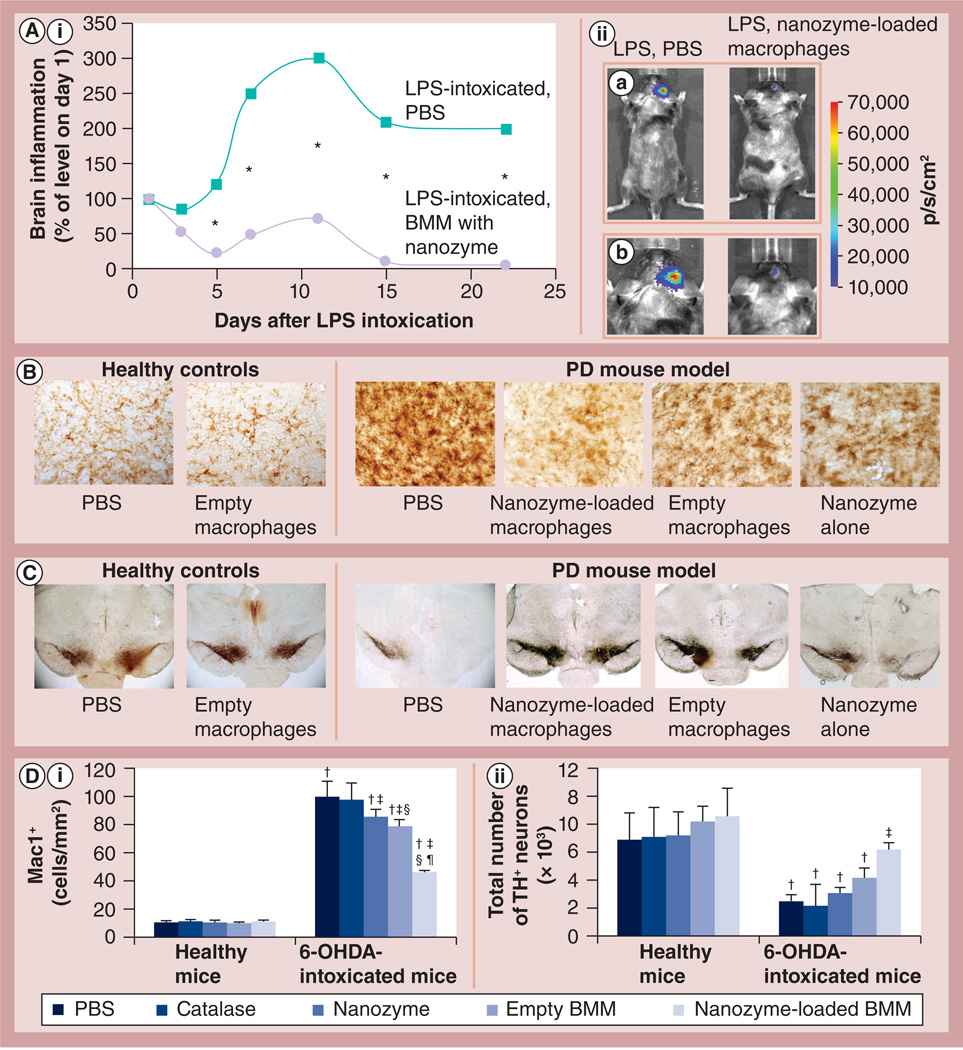
Linkages between neuroinflammation and nigrostriatal degeneration have been reported [35]. Therefore, attenuation of inflammation could serve to protect dopaminergic neurons in disease [8,9]. To examine whether macrophages preloaded with Cl nanozyme can reduce brain inflammation, IVIS studies were conducted. For this purpose, C57BL/6 mice were stereotactically injected with LPS into the substantia nigra pars compacta (SNpc) to produce inflammation, and then 24 h later one group was iv. injected with BMMs preloaded with the selected Cl-6-BS3 (Figure 9A,iia: whole body, Figure 9A,iib: head). Another group of LPS-intoxicated mice was iv. injected with PBS and used as a control. IVIS images of both groups were taken over 22 days. The chemiluminescent signal was quantified and presented as a radiance ratio of treated animal versus 24 h after LPS injection at various time points (Figure 8A). LPS ic. injections caused more than a threefold increase in brain inflammation levels of the control group. The Cl nanozyme administered in macrophages considerably decreased neuroinflammation in LPS-intoxicated mice. The prolonged anti-inflammatory effect was recorded more than 3 weeks after LPS intoxication. This signified the ability of nanozyme-loaded macrophages to reduce oxidative stress.
Figure 9. Effect of cross-linked nanozyme loaded into macrophages on the reduction of neuroinflammation and increase in neuronal survival in mice with brain inflammation.
(A,i)C57Bl/6 mice with LPS-induced encephalitis were injected intravenously with macrophages (5 × 106/mouse in 100 µl PBS) preloaded with cross-linked nanozyme (bis-(sulfosuccinimidyl)suberate sodium salt, charge ratio = 6; 2 mg/ml catalase) or PBS. (A,ii) Image visualization and infrared spectroscopy images over 20 days were taken 10 min after intraperitoneal injection of a XenoLight™ RediJect probe (Caliper LifeSciences, MA, USA). (A,ii,a) Whole body images and (A,ii,b) images of the mouse head for the corresponding time. The chemiluminescent signal was quantified and presented as radiance ratios of treated animal versus 24 h after LPS injection at various times. Macrophages preloaded with the cross-linked nanozyme caused prolonged decreases of neuroinflammation in LPS-intoxicated mice. Data represent mean ± standard error of the mean (n = 5). The statistical significance of inflammation levels compared with control LPS-intoxicated mice is shown by asterisk: *p < 0.05. (B–D) C57Bl/6 mice were intracranial injected with 6-OHDA. A total of 48 h later, animals were intravenously injected with macrophages preloaded with catalase alone, cross-linked nanozyme alone, empty macrophages or macrophages preloaded with cross-linked nanozyme, and 5 weeks later they were sacrificed and midbrain slides were stained for expression of (B) Mac1+, a marker for activated microglia or (C) tyrosine hydroxylase, a marker for dopaminergic neurons. (D) Quantification of microglial activation and neuronal survival. Values represent mean ± standard error of the mean (n = 6), and p < 0.05 compared with †: PBS (healthy controls); †: 6-ODHA (PD mouse model); §: catalase alone (in 6-OHDA intoxicated mice); and ¶: empty macrophages (in 6-OHDA-intoxicated mice). Selected images of several additional control groups are presented on Supplementary Figure S9. Whereas 6-OHDA treatment caused significant microglia activation and neuronal loss, administration of nanozyme-preloaded macrophages significantly decreased oxidative stress and increased neuronal survival. The administration of empty M2 macrophages slightly decreased microglia activation, and increased the number of dopaminergic neurons in mice with brain inflammation. 6-OHDA: 6-Hydroxydopamine; BMM: Bone marrow-derived macrophage; LPS: Lipopolysaccharide; PBS: Phosphate-buffered saline; PD: Parkinson’s disease
Next, potent anti-inflammatory and neuroprotective effects by macrophages loaded with Cl nanozyme were demonstrated in the mouse model of PD (Figure 9B–9D). 6-OHDA stereotactically injected into the SNpc produced considerable brain inflammation signified by upregulated expression of CD11b by microglia within the SNpc, which exhibited a more amoeboid morphology compared with ramified microglia in PBS-treated mice. By contrast, treatment of intoxicated mice with BMMs preloaded with Cl-6-BS3 resulted in decreased levels of CD11b and 50% fewer activated microglia cells compared with 6-OHDA-intoxicated control animals (Figure 9B & 9D). Furthermore, systemic administration of nanozyme-preloaded macrophages increased neuronal survival in 6-OHDA-intoxicated mice (Figure 9C & 9D). The numbers of tyrosine hydroxylase-expressing neurons in the SNpc of 6-OHDA animals treated with nanozyme-preloaded macrophages were significantly greater (p < 0.05) than those animals intoxicated with 6-OHDA and then injected with PBS. It is worth noting that empty macrophages as well as nanozyme alone had a subtle but statistically significant neuroprotective effect in mice with 6-OHDA ic. injections (Figure 9B–9D). We hypothesized that the particular subset of macrophages used in these studies (differentiated in the presence of MCSF) has a regeneration effect, as reported in [32]. No significant effect of catalase alone, nanozyme alone, BMMs preincubated with PBS or BMMs preloaded with naked catalase was found on microglia activation or neuronal survival in healthy mice (Figure 9B–9D & Supplementary Figure S9).
Discussion & conclusion
The inability of most potent therapeutics to cross the BBB following systemic administration necessitates the need to develop unconventional, clinically applicable drug-delivery systems to treat CNS disorders. Smart, biologically active vehicles are crucial to accomplish this challenging task. Using inflammatory response cells enables targeted drug transport and prolonged circulation times, along with reductions in cell and tissue toxicities. Here, we report the manufacture of new nanoformulations of nanozymes with chemically stabilized structure and improved loading capacity for the delivery of antioxidants in macrophages to disease inflamed brain areas.
It is worth noting that the approach for nanoformulation design for cell-mediated delivery should differ from the injectable formulations. In particular, we demonstrated that the increasing size of nanoparticles by cross-linking of several protein globules resulted in the exponentially increased loading capacity in macrophages as well as release from the cell carriers. This is consistent with the reported earlier studies that particles with approximately 1-µm diameter accumulated at a greater extent (2.6-fold) than smaller drug carriers of 500-nm diameter in macrophages [36]. Moreover, phagosomes may have different sizes depending on the size of the particles, which can range from as little as a few hundred nanometers [37, 38] to dozens of microns [39]. Next, a chemical cross-linking provided significant protection for the encapsulated enzyme, which remains capable of ROS decomposition without the need for particle enzyme release. Therefore, Cl nanozyme demonstrated significantly greater (approximately six-times) enzymatic activity than the non-Cl formulation. In addition, Cl nanozyme at Z = 6 provided the best protection of catalase activity against degradation inside the host cells in an in vitro PD model. Furthermore, the optimal nanozymes released from preloaded macrophages have potent antioxidant effects in neurons activated with 6-OHDA. This is probably a result of two separate processes: high catalytic activity of nanozyme released from the cell carriers, and efficient nanozyme transfer from preloaded macrophages into neurons. This suggests that macrophages preloaded with optimal structure nanozymes can mitigate oxidative stress associated with the neurodegenerative process.
The improved protection of Cl nanozymes against degradation in the cell carriers may be attributed to its intracellular distribution. The confocal images confirmed that a substantial portion of the block copolymer was dissociated from the non-Cl BIC upon entering macrophages, and remained in the peripheral areas of the cells. By contrast, all polymer molecules were colocalized with catalase when Cl nanozyme was used. The role of PEI–PEG is crucial in the preservation of catalase enzymatic activity in macrophages. According to our earlier studies, increasing the amount of the block copolymer in the BIC led to an increase in the endosomal pH, and resulted in better protection of the enzyme [13]. Indeed, an excess of free amino groups in the BIC nanoparticles at high Z values might buffer acidification of the endocytic compartments of the cells, thereby inhibiting protease activity with consequent decreases in catalase degradation [20]. In other words, a positively charged block copolymer might delay phagosomal maturation and as a result prevent degradation of the enzyme in macrophages. Alternatively, and perhaps supplementary to the polycation, PEG chains in the BIC can sterically protect catalase molecules against degradation by intracellular proteases. This effect should be more pronounced in the Cl nanozymes that do not dissociate in the cell compartments. Indeed, a nonbiodegradable linker (BS3) was more efficient in the preservation of nanozyme activity than a biodegradable one (DTSSP). Furthermore, the intracellular distribution studies indicate that Cl nanozymes enter fast through early and late endosomes, and then escape from endosomal compartments, probably due to the ‘proton sponge effect’, reaching largely the ER and mitochondria. Very few, if any, Cl nanozyme particles were found in recycling endosomes. By contrast, the non-Cl nanozyme was localized predominantly in recycled endosomes, as reported earlier [34].
Since macrophages are known to release different neurotoxic agents, including oxidative metabolites and proinflammatory cytokines, there is a significant concern as to whether the cell carriers can bring additional inflammation to the disease site. In fact, the BBB breakdown and entrance of inflammatory response cells to the brain under pathological conditions was traditionally suggested to have a negative role in secondary tissue degeneration. A number of therapeutic strategies for CNS neurodegenerative disorders were based on the prevention of monocyte–macrophage infiltration due to this reason [40]. However, developments in the last few decades began to change this dogma [41]. It has been demonstrated that macrophages may promote both injury and repair in the sites of CNS injury [32]. Here, we evaluated whether the cell carriers cause inflammation and neuronal damage in in vitro and in vivo studies. Our earlier studies and the present work show no cytotoxic effects in mice upon injection of a particular subset of macrophages differentiated in the presence of MCSF [12, 29]. We demonstrated here a subtle, but statistically significant, effect on improved neuronal survival in mice following iv. injection of these macrophages even without nanozyme. We hypothesized that differentiation of BMMs in the presence of MCSF resulted in a specific subset of anti-inflammatory (M2) macrophages. This was confirmed by confocal microscopy showing overexpression of the mannose receptor in these cells, which is indicative for a M2 subtype. Alternatively activated macrophages are known to release various factors that promote CNS repair, inducing neural tissue regeneration and renewal, matrix remodeling, debris clearance and angiogenesis [32]. Therefore, properly differentiated immune cells accumulating in traumatic, degenerative, ischemic, infectious and autoimmune lesions of the nervous system might provide a neuroprotective effect, which may further boost the therapeutic effect of cell-mediated drug-delivery systems.
Finally, we evaluated the potential of optimal Cl nanozymes to attenuate oxidative stress in a PD murine model. Cl catalase nanozyme administered in macrophages caused prolonged decreases of neuroinflammation in LPS-intoxicated mice, as demonstrated by IVIS. Immunohistochemical analysis confirmed systemic administration of macrophages with nanozyme resulted in a substantial and prolonged attenuation of neuroinflammation (over 5 weeks), and increased neuronal survival in 6-OHDA-intoxicated mice.
The mechanisms of the neuroprotective effects of cell-based catalase nanoformulations are still not well understood. In fact, immunocytes, mononuclear phagocytes (monocytes, macrophages and dendritic cells), lymphocytes and neutrophils, as well as stem cells exhibit an intrinsic homing property, enabling them to migrate to sites of injury, inflammation and tumor across the BBB in response to the release of cytokines/chemokines and upregulation of certain cell surface proteins in the diseased tissues and nearby blood vessels. Even in the healthy brain, perivascular macrophages, which reside on the parenchymal side of endothelial cells, originally come from circulating phagocytes, monocytes and macrophages, and have shown a remarkable capability to cross an intact BBB with an 80% turnover in 3 months [42,43]. It was reported that leukocytes traffic primarily between adjacent endothelial cells through the junctional complexes (paracellular migration) [44,45] or, in some cases, even through the endothelial cell itself (transcellular migration) [46]. Under pathological conditions, the rate of immunocyte transport to the inflamed brain tissues is further elevated [23,47–52]. Such frequent migration offers a unique opportunity to design novel cell-based drug formulations, when living cells act as Trojan horses carrying concealed drug cargoes while migrating across impermeable barriers (the BBB or blood–tumor barrier) to the sites of disease.
We demonstrated here that GFP-expressing macrophages accumulate in the brain in PD mice, which is consistent with our previous investigations that reported transport of fluorescently labeled [12] and luciferase-expressing [29] BMMs in the inflamed mouse brain. We also showed in this study increases in catalase enzymatic activity in the SNpc following adoptive transfer of nanozyme-loaded BMMs in PD mice. This confirmed our previous reports that BMMs preloaded with 125I-labeled [11], 3H-labeled [14] and fluorescently labeled [12] catalase nanozyme facilitated delivery of catalase to the inflamed brain. Nevertheless, this Trojan horse effect may not be the one and only mechanism of macrophage-mediated therapeutic effects. Therefore, several independent processes may serve to improve nanozyme-loaded macrophage therapeutics. In addition to the targeted tissue-specific delivery of therapeutics in BMMs [12,14], nanozyme-loaded macrophages were shown to prolong circulation in the blood, and therefore permit sustained release of catalase, allowing the enzyme to enter the brain, independent of carrier cells (‘depot effect’) [12,14]. Furthermore, catalase released from the macrophages in the liver and spleen can suppress peripheral leukocyte activation and protect nigrostratial neurons at a distance [12]. Finally, drug-loaded BMMs can transfer catalase nanoparticles to brain microvessel endothelial cells and increase BBB penetration and ROS decomposition [34]. We reported that such events occur through endocytosis-independent mechanisms, including fusion of cellular membranes, formation of macrophage bridging conduits, (i.e., filopodia and lamellipodia) that stretch to the neighbouring cells, and nanozyme incorporation into exosomes [28]. These extracellular vesicles released from preloaded macrophages may facilitate nanozyme transfer from carrier cells to target recipient cells [34]. Unfortunately, at this stage of understanding, we cannot discriminate which mechanism is the most important for the therapeutic effects of cell-based nanoformulations. A comprehensive examination and detailed understanding of these mechanisms would be of great value in designing strategies for cell-mediated drug delivery.
Overall, these studies demonstrated the feasibility of the therapeutic approach using cell-mediated catalase delivery for neuroprotection. Macrophages are attractive drug carriers due to their natural mobility, ability to efficiently engulf particles, penetrate the BBB and reach the site of neuropathology. The ultimate goal is to create such nanozymes that can be loaded to the cell carriers in the patient bloodstream and delivered by active targeted transport to the brain with inflammation.
Future perspective
Using cells as delivery vehicles enables targeted drug transport and prolonged circulation times along with reductions in cell and tissue toxicities. Immunocytes are highly mobile and capable of cell-to-cell transmission of drug-laden nanoparticles. Therefore, such systems represent a novel disease-combating strategy being applied to a spectrum of human disorders. However, this is not a simple task. The formulation design should provide high drug loading in cell carriers, efficient drug protection against degradation inside the cell carriers and sustained release at the disease site. In addition, the cell carriers should migrate in substantial quantities and be safe for the organism. Nevertheless, using optimal drug nanoformulations along with a specific subtype of ‘alternately activated’ anti-inflammatory macrophages opens the opportunity to exploit immune cells as Trojan horses for drug delivery.
Supplementary Material
Executive summary.
Cell-based drug delivery platform
-
▪
Macrophages preloaded with nanoformulated catalase, ‘nanozyme’, offer a new delivery system for active targeted transport of the antioxidant to the inflamed brain.
Nanozyme manufacture
-
▪
A formulation design of optimal particles delivered in macrophages differs from a common approach for injectable drug nanoformulations. They should have a relatively large size, be stabilized by excess of nonbiodegradable linker and be vectorized to the cell carriers.
Using a specific subtype of cell carriers
-
▪
Anti-inflammatory ‘alternatively activated’ macrophages that promote CNS repair should be used for drug delivery.
Macrophages accumulate in mouse inflamed brain
-
▪
Systemically administered macrophages were able to cross the blood–brain barrier and reach the brain tissues with inflammation.
Therapeutic effects of nanozyme-loaded bone marrow-derived macrophages
-
▪
Nanozyme loaded in macrophages reduced neuroinflammatory responses and increased neuronal survival in mice, providing a translational link of these formulations to the clinic.
Acknowledgements
We are grateful to JA Taylor and JR Talaska (Confocal Laser Scanning Microscope Core Facility, the University of Nebraska Medical Center, USA) for providing assistance with confocal microscopy, the Nebraska Research Initiative and the Eppley Cancer Center (USA) for their support of the Core Facility, and L Shlyakhtenko for assistance with atomic force microscopy studies. We also thank D Filonova for help in schematic representation of the nanozyme structure.
This study was supported by the US NIH grants 1RO1 NS057748 (to EV Batrakova), RR021937 (to AV Kabanov), 2R01 NS070190 (to RL Mosley), 2R01 NS034239, 2R37 NS36126, P01 NS31492, P20RR 15635, P01 MH64570 and P01 NS43985 (to HE Gendelman), and Ministry of Education and Science of Russian Federation grants 11.G34.31.0004 and 02.740.11.5232 (both to AV Kabanov).
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Contributor Information
Natalia L Klyachko, Laboratory for Chemical Design of Bionanomaterials & the Department of Chemical Enzymology, Faculty of Chemistry, MV Lomonosov Moscow State University, Moscow, Russia and Department of Pharmaceutical Sciences, Center for Drug Delivery & Nanomedicine, University of Nebraska Medical Center, Omaha, NE, USA.
Matthew J Haney, Department of Pharmaceutical Sciences, Center for Drug Delivery & Nanomedicine, University of Nebraska Medical Center, Omaha, NE, USA.
Yuling Zhao, Department of Pharmaceutical Sciences, Center for Drug Delivery & Nanomedicine, University of Nebraska Medical Center, Omaha, NE, USA.
Devika S Manickam, Department of Pharmaceutical Sciences, Center for Drug Delivery & Nanomedicine, University of Nebraska Medical Center, Omaha, NE, USA.
Vivek Mahajan, Department of Pharmaceutical Sciences, Center for Drug Delivery & Nanomedicine, University of Nebraska Medical Center, Omaha, NE, USA and Department of Pathology & Microbiology, University of Nebraska Medical Center, Omaha, NE, USA and Department of Pharmacology & Experimental Neuroscience, Center for Neurodegenerative Disorders, University of Nebraska Medical Center, Omaha, NE, USA.
Poornima Suresh, Department of Pharmaceutical Sciences, Center for Drug Delivery & Nanomedicine, University of Nebraska Medical Center, Omaha, NE, USA and Department of Pharmacology & Experimental Neuroscience, Center for Neurodegenerative Disorders, University of Nebraska Medical Center, Omaha, NE, USA.
Shawn D Hingtgen, Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
R Lee Mosley, Department of Pharmacology & Experimental Neuroscience, Center for Neurodegenerative Disorders, University of Nebraska Medical Center, Omaha, NE, USA.
Howard E Gendelman, Department of Pharmacology & Experimental Neuroscience, Center for Neurodegenerative Disorders, University of Nebraska Medical Center, Omaha, NE, USA.
Alexander V Kabanov, Laboratory for Chemical Design of Bionanomaterials & the Department of Chemical Enzymology, Faculty of Chemistry, MV Lomonosov Moscow State University, Moscow, Russia and Department of Pharmaceutical Sciences, Center for Drug Delivery & Nanomedicine, University of Nebraska Medical Center, Omaha, NE, USA.
Elena V Batrakova, Department of Pharmaceutical Sciences, Center for Drug Delivery & Nanomedicine, University of Nebraska Medical Center, Omaha, NE, USA and Center for Nanotechnology in Drug Delivery, Division of Molecular Pharmaceutics, Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Genetic Medicine Building, 120 Mason Farm Road, Chapel Hill, NC 27599–7362, USA.
References
- 1.Bachis A, Mocchetti I. Brain-derived neurotrophic factor is neuroprotective against human immunodeficiency virus-1 envelope proteins. Ann. NY Acad. Sci. 2005;1053:247–257. doi: 10.1196/annals.1344.022. [DOI] [PubMed] [Google Scholar]
- 2.Ying Wang J, Peruzzi F, Lassak A, et al. Neuroprotective effects of IGF-I against TNFalpha-induced neuronal damage in HIV-associated dementia. Virology. 2003;305(1):66–76. doi: 10.1006/viro.2002.1690. [DOI] [PubMed] [Google Scholar]
- 3.Koliatsos VE, Clatterbuck RE, Nauta HJ, et al. Human nerve growth factor prevents degeneration of basal forebrain cholinergic neurons in primates. Ann. Neurol. 1991;30(6):831–840. doi: 10.1002/ana.410300613. [DOI] [PubMed] [Google Scholar]
- 4.Dogrukol-Ak D, Banks WA, Tuncel N, Tuncel M. Passage of vasoactive intestinal peptide across the blood-brain barrier. Peptides. 2003;24(3):437–444. doi: 10.1016/s0196-9781(03)00059-7. [DOI] [PubMed] [Google Scholar]
- 5.Perry VH, Bell MD, Brown HC, Matyszak MK. Inflammation in the nervous system. Curr. Opin. Neurobiol. 1995;5(5):636–641. doi: 10.1016/0959-4388(95)80069-7. [DOI] [PubMed] [Google Scholar]
- 6.McGeer PL, Itagaki S, Boyes BE, McGeer EG. Reactive microglia are positive for HLA-DR in the substantia nigra of Parkinson’s and Alzheimer’s disease brains. Neurology. 1988;38(8):1285–1291. doi: 10.1212/wnl.38.8.1285. [DOI] [PubMed] [Google Scholar]
- 7.Busciglio J, Yankner BA. Apoptosis and increased generation of reactive oxygen species in Down’s syndrome neurons in vitro . Nature. 1995;378(6559):776–779. doi: 10.1038/378776a0. [DOI] [PubMed] [Google Scholar]
- 8.Ebadi M, Srinivasan SK, Baxi MD. Oxidative stress and antioxidant therapy in Parkinson’s disease. Prog. Neurobiol. 1996;48(1):1–19. doi: 10.1016/0301-0082(95)00029-1. [DOI] [PubMed] [Google Scholar]
- 9.Wu DC, Teismann P, Tieu K, et al. NADPH oxidase mediates oxidative stress in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine model of Parkinson’s disease. Proc. Natl Acad. Sci. USA. 2003;100(10):6145–6150. doi: 10.1073/pnas.0937239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maruyama W, Akao Y, Carrillo MC, Kitani K, Youdium MB, Naoi M. Neuroprotection by propargylamines in Parkinson’s disease: suppression of apoptosis and induction of prosurvival genes. Neurotoxicol. Teratol. 2002;24(5):675–682. doi: 10.1016/s0892-0362(02)00221-0. [DOI] [PubMed] [Google Scholar]
- 11.Batrakova EV, Li S, Reynolds AD, et al. A macrophage-nanozyme delivery system for Parkinson’s disease. Bioconjug. Chem. 2007;18(5):1498–1506. doi: 10.1021/bc700184b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brynskikh AM, Zhao Y, Mosley RL, et al. Macrophage delivery of therapeutic nanozymes in a murine model of Parkinson’s disease. Nanomedicine (Lond.) 2010;5(3):379–396. doi: 10.2217/nnm.10.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Haney MJ, Klyachko NL, et al. Polyelectrolyte complex optimization for macrophage delivery of redox enzyme nanoparticles. Nanomedicine (Lond.) 2011;6(1):25–42. doi: 10.2217/nnm.10.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Haney MJ, Mahajan V, et al. Active targeted macrophage-mediated delivery of catalase to affected brain regions in models of Parkinson’s disease. J. Nanomed. Nanotechnol. 2011;S4 doi: 10.4172/2157-7439.S4-003. pii:003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batrakova EV, Gendelman HE, Kabanov AV. Cell-mediated drug delivery. Expert Opin. Drug Deliv. 2011;8(4):415–433. doi: 10.1517/17425247.2011.559457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas SR, Witting PK, Drummond GR. Redox control of endothelial function and dysfunction: molecular mechanisms and therapeutic opportunities. Antioxid. Redox Signal. 2008;10(10):1713–1765. doi: 10.1089/ars.2008.2027. [DOI] [PubMed] [Google Scholar]
- 17.Muzykantov VR. Targeting of superoxide dismutase and catalase to vascular endothelium. J. Control. Release. 2001;71(1):1–21. doi: 10.1016/s0168-3659(01)00215-2. [DOI] [PubMed] [Google Scholar]
- 18.Supinski GS, Callahan LA. Polyethylene glycol-superoxide dismutase prevents endotoxin-induced cardiac dysfunction. Am. J. Respir. Crit. Care Med. 2006;173(11):1240–1247. doi: 10.1164/rccm.200410-1346OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hood E, Simone E, Wattamwar P, Dziubla T, Muzykantov V. Nanocarriers for vascular delivery of antioxidants. Nanomedicine (Lond.) 2011;6(7):1257–1272. doi: 10.2217/nnm.11.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thiele L, Merkle HP, Walter E. Phagocytosis and phagosomal fate of surface-modifed microparticles in dendritic cells and macrophages. Pharm. Res. 2003;20(2):221–228. doi: 10.1023/a:1022271020390. [DOI] [PubMed] [Google Scholar]
- 21.Klyachko NL, Manickam DS, Brynskikh AM, et al. Cross-linked antioxidant nanozymes for improved delivery to CNS. Nanomedicine. 2012;8(1):119–129. doi: 10.1016/j.nano.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinogradov S, Bronich T, Kabanov A. Self-assembly of polyamine-poly(ethylene glycol) copolymers with phosphorothioate oligonucleotides. Bioconjug. Chem. 1998;9(6):805–812. doi: 10.1021/bc980048q. [DOI] [PubMed] [Google Scholar]
- 23.Dou H, Destache CJ, Morehead JR, et al. Development of a macrophage-based nanoparticle platform for antiretroviral drug delivery. Blood. 2006;108(8):2827–2835. doi: 10.1182/blood-2006-03-012534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevenson HC, Katz P, Fauci AS. Purification of human monocytes by counter-current centrifugation elutriation. Cell Immunol. 1980;53(1):94–103. doi: 10.1016/0008-8749(80)90429-3. [DOI] [PubMed] [Google Scholar]
- 25.Ferrari M, Fornasiero M, Isetta A. MTT colorimetric assay for testing macrophage cytotoxic activity in vitro . J. Immunol. Methods. 1990;131(2):165–172. doi: 10.1016/0022-1759(90)90187-z. [DOI] [PubMed] [Google Scholar]
- 26.Batrakova E, Han H, Miller D, Kabanov A. Effects of pluronic p85 unimers and micelles on drug permeability in polarized BBMEC and Caco-2 cells. Pharm. Res. 1998;15(10):1525–1532. doi: 10.1023/a:1011942814300. [DOI] [PubMed] [Google Scholar]
- 27.Batrakova EV, Vinogradov SV, Robinson SM, Niehoff ML, Banks WA, Kabanov AV. Polypeptide point modifications with fatty acid and amphiphilic block copolymers for enhanced brain delivery. 2005;16(4):793–802. doi: 10.1021/bc049730c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haney MJ, Zhao Y, Li S, et al. Cell-mediated transfer of catalase nanoparticles from macrophages to brain endothelial, glial and neuronal cells. Nanomedicine (Lond.) 2011;6(7):1215–1230. doi: 10.2217/nnm.11.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haney MJ, Zhao Y, Harrison EB, et al. Specific transfection of inflamed brain by macrophages: a new therapeutic strategy for neurodegenerative diseases. PLoS ONE. 2013;8(4):e61852. doi: 10.1371/journal.pone.0061852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papadopoulos S, Jurgens KD, Gros G. Protein diffusion in living skeletal muscle fibers: dependence on protein size, fiber type, and contraction. Biophys. J. 2000;79(4):2084–2094. doi: 10.1016/S0006-3495(00)76456-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solomatin SV, Bronich TK, Eisenberg A, Kabanov VA, Kabanov AV. Nanomaterials from ionic block copolymers and single-, double-, and triple-tail surfactants. Langmuir. 2007;23(5):2838–2842. doi: 10.1021/la062693o. [DOI] [PubMed] [Google Scholar]
- 32.Kigerl KA, Gensel JC, Ankeny DP, Alexander JK, Donnelly DJ, Popovich PG. Identification of two distinct macrophage subsets with divergent effects causing either neurotoxicity or regeneration in the injured mouse spinal cord. J. Neurosci. 2009;29(43):13435–13444. doi: 10.1523/JNEUROSCI.3257-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freundt EC, Czapiga M, Lenardo MJ. Photoconversion of LysoTracker Red to a green fluorescent molecule. Cell Res. 2007;17(11):956–958. doi: 10.1038/cr.2007.80. [DOI] [PubMed] [Google Scholar]
- 34.Haney MJ, Suresh P, Zhao Y, et al. Blood-borne macrophage-neural cell interactions hitchhike endosome networks for cell-based nanozyme brain delivery. Nanomedicine (Lond.) 2012;7(6):815–833. doi: 10.2217/nnm.11.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stone DK, Reynolds AD, Mosley RL, Gendelman HE. Innate and adaptive immunity for the pathobiology of Parkinson’s disease. Antioxid. Redox Signal. 2009;11(9):2151–2166. doi: 10.1089/ars.2009.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nowacek AS, Miller RL, Mcmillan J, et al. NanoART synthesis, characterization, uptake, release and toxicology for human monocyte–macrophage drug delivery. Nanomedicine (Lond.) 2009;4(8):903–917. doi: 10.2217/nnm.09.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gbadamosi JK, Hunter AC, Moghimi SM. PEGylation of microspheres generates a heterogeneous population of particles with differential surface characteristics and biological performance. FEBS Lett. 2002;532(3):338–344. doi: 10.1016/s0014-5793(02)03710-9. [DOI] [PubMed] [Google Scholar]
- 38.Mizushima Y, Hamano T, Yokoyama K. Tissue distribution and anti-inflammatory activity of corticosteroids incorporated in lipid emulsion. Ann. Rheum. Dis. 1982;41(3):263–267. doi: 10.1136/ard.41.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hillaireau H, Couvreur P. Nanocarriers’ entry into the cell: relevance to drug delivery. Cell. Mol. Life Sci. 2009;66(17):2873–2896. doi: 10.1007/s00018-009-0053-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hendriks JJ, Teunissen CE, De Vries HE, Dijkstra CD. Macrophages and neurodegeneration. Brain Res. Rev. 2005;48(2):185–195. doi: 10.1016/j.brainresrev.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Shechter R, Schwartz M. Harnessing monocyte-derived macrophages to control central nervous system pathologies: no longer ‘if’ but ‘how’. J. Pathol. 2013;229(2):332–346. doi: 10.1002/path.4106. [DOI] [PubMed] [Google Scholar]
- 42.Lassmann H, Schmied M, Vass K, Hickey WF. Bone marrow derived elements and resident microglia in brain inflammation. Glia. 1993;7(1):19–24. doi: 10.1002/glia.440070106. [DOI] [PubMed] [Google Scholar]
- 43.Vass K, Hickey WF, Schmidt RE, Lassmann H. Bone marrow-derived elements in the peripheral nervous system. An immunohistochemical and ultrastructural investigation in chimeric rats. Lab. Invest. 1993;69(3):275–282. [PubMed] [Google Scholar]
- 44.Pawlowski NA, Kaplan G, Abraham E, Cohn ZA. The selective binding and transmigration of monocytes through the junctional complexes of human endothelium. J. Exp. Med. 1988;168(5):1865–1882. doi: 10.1084/jem.168.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lossinsky AS, Shivers RR. Structural pathways for macromolecular and cellular transport across the blood–brain barrier during inflammatory conditions. Review. Histol. Histopathol. 2004;19(2):535–564. doi: 10.14670/HH-19.535. [DOI] [PubMed] [Google Scholar]
- 46.Muller WA. Getting leukocytes to the site of inflammation. Vet. Pathol. 2013;50(1):7–22. doi: 10.1177/0300985812469883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Persidsky Y, Ghorpade A, Rasmussen J, et al. Microglial and astrocyte chemokines regulate monocyte migration through the blood–brain barrier in human immunodeficiency virus-1 encephalitis. Am. J. Pathol. 1999;155(5):1599–1611. doi: 10.1016/S0002-9440(10)65476-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blamire AM, Anthony DC, Rajagopalan B, Sibson NR, Perry VH, Styles P. Interleukin-1beta -induced changes in blood–brain barrier permeability, apparent diffusion coefficient, and cerebral blood volume in the rat brain: a magnetic resonance study. J. Neurosci. 2000;20(21):8153–8159. doi: 10.1523/JNEUROSCI.20-21-08153.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anthony DC, Blond D, Dempster R, Perry VH. Chemokine targets in acute brain injury and disease. Prog. Brain Res. 2001;132:507–524. doi: 10.1016/s0079-6123(01)32099-x. [DOI] [PubMed] [Google Scholar]
- 50.Mahmood A, Lu D, Lu M, Chopp M. Treatment of traumatic brain injury in adult rats with intravenous administration of human bone marrow stromal cells. Neurosurgery. 2003;53(3):697–702. doi: 10.1227/01.neu.0000079333.61863.aa. [DOI] [PubMed] [Google Scholar]
- 51.Zlokovic BV. The blood–brain barrier in health and chronic neurodegenerative disorders. Neuron. 2008;57(2):178–201. doi: 10.1016/j.neuron.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 52.Alvarez JI, Cayrol R, Prat A. Disruption of central nervous system barriers in multiple sclerosis. Biochim. Biophys. Acta. 2011;1812(2):252–264. doi: 10.1016/j.bbadis.2010.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.