Abstract
Infiltrating T-lymphocytes are frequently found in malignant tumors and are suggestive of a host cancer immune response. Multiple independent studies have documented that the presence and quantity of tumor-infiltrating T-lymphocytes (TILs) are strongly correlated with increased survival. However, due to methodological factors, the exact effect of TILs on prognosis has remained enigmatic, and inclusion of TILs in standard prognostic panels has been limited. For example, some reports enumerate all CD3+ cells, some count only cytotoxic CD8+ T cells, and the criteria used to score tumors as TIL positive or negative are inconsistent among studies. To address this limitation, we introduce a robust digital DNA-based assay, termed QuanTILfy, to reliably and inexpensively count TILs and assess T cell clonality in tissue samples, including tumors. We demonstrate the clonal specificity of this approach by the diagnosis of T-lineage acute lymphoblastic leukemia (T-ALL) and the accurate, sensitive, and highly reproducible measurement of TILs in primary and metastatic ovarian cancer. Our experiments demonstrate an association between higher TIL counts and improved survival among women with ovarian cancer, and are consistent with prior observations that the immune response against ovarian cancer is a meaningful and independent prognostic factor. Surprisingly, the TIL repertoire is diverse for all tumors in the study with no notable oligoclonal expansions. Furthermore, as variability in the measurement and characterization of TILs has limited their clinical utility as biomarkers, these results highlight the significant translational potential of a robust, standardizable DNA-based assay to assess TILs in a variety of cancer types.
Introduction
The human cellular adaptive immune system identifies and destroys cells expressing aberrant proteins or protein fragments. The source of the abnormal protein fragments can include intracellular pathogenic infection, genomic mutations, or deregulation of gene expression. Cancerous cells often express such aberrant peptides, prompting a cellular adaptive immune response. These peptides are presented on the surface of cells by HLA molecules for binding by T cell receptors on the surface of T-lymphocytes, the primary mediators of the cellular adaptive immune response.
Tumor infiltrating lymphocytes (TILs) have been shown to directly attack tumor cells in a variety of types of cancer (1), and multiple independent studies have demonstrated that the presence of TILs are strongly correlated with increased survival (2-6). For both colorectal and ovarian carcinoma patients, the presence or absence of TILs provides a strong prognostic marker for survival independent of current staging methods (6, 7). However, existing assays and pathology tests to measure TILs are cumbersome, have inherent variability, are mostly restricted to research studies, and are not utilized for clinical decision-making. Currently, the anti-CD3 antibody immunohistochemistry (IHC) test is the most common method used to measure TILs. However, data interpretation is operator- and laboratory-dependent and only represents a small, two-dimensional cross section of the tumor. Consequently, IHC tests for TILs are poorly standardized and qualitative in nature. To achieve a quantitative measurement of TILs, some research labs use flow sorting. However, flow cytometric quantification using T cell-specific surface markers is technically challenging within a solid tumor. Methods used to disaggregate solid tumors into a single cell suspension utilize mechanical disruption followed by treatment with nucleases and proteases, but this can result in a low cellular yield and the potential digestion of surface markers, compromising the fundamental endpoint used for quantification (8). Furthermore, tumor surface marker expression is inherently heterogeneous between patients, preventing standardization of CD3 expression-based TIL measurement (8).
As the importance of TILs gains appreciation, particularly given their potential utility for cancer prognostication and their role in immunotherapeutic response, new technologies to quantitatively measure TILs are needed. Fortunately, adaptive immune cells have a molecular signature that can be exploited for direct measurement. T cells have gene rearrangements in their T cell receptor (TCR) loci. The nucleotide sequences that encode the TCR regions are generated by somatic rearrangement of noncontiguous variable (V), diversity (D), and joining (J) region gene segments for the β chain, and V and J segments for the α chain. The existence of multiple V, D, and J gene segments in germline DNA permits substantial combinatorial diversity in receptor composition, and receptor diversity is further increased by the deletion of nucleotides adjacent to the recombination signal sequences (RSS) of the V, D, and J segments, and template-independent insertion of nucleotides at the Vβ-Dβ, Dβ-Jβ, and Vα−Jα junctions. We have developed QuanTILfy to measure the number of T-lymphocytes and assess clonality in a tissue using droplet digital PCR (ddPCR) technology. The massive sample partitioning is a key aspect of the ddPCR technique, and a vital component of the QuanTILfy assay. DdPCR surpasses the performance of earlier techniques by introducing a scalable implementation of digital PCR, where the creation of tens of thousands of droplets allows for the generation of tens of thousands of data points, bringing the power of statistical analysis inherent to digital PCR into practical application.
Results
Digital quantification of T-lymphocytes
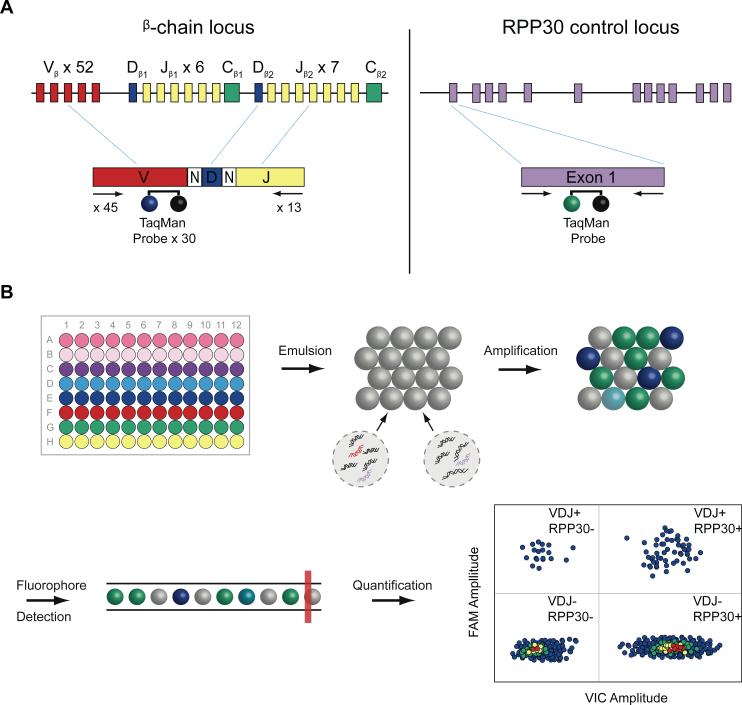
We applied this technology to ovarian cancer, where we addressed heterogeneity of TILs in different locations within a tumor. Additionally, we applied the technology to a set of primary and metastatic tumors from 30 stage III and IV ovarian patients to assess the relationship between TIL count and survival among stage-matched patients. We have quantified T-lymphocytes using a multiplex ddPCR system (Fig. 1), which amplified rearranged TCRβ loci from genomic DNA using 45 forward primers (Supplemental Table 1), each specific to one or multiple functional TCR Vβ segments, and 13 reverse primers (Supplemental Table 2), each specific to a TCR Jβ segment (9). The multiplex reaction also included one of a series of 35 minor groove binder (MGB) 6-carboxyfluorescein (FAM) TaqMan® probes complementary to 52 different Vβ gene segments (Supplemental Table 3), and a VIC® probe complementary to ribonuclease P protein subunit p30 (RPP30), which served as a reference gene to permit normalized quantification (Supplemental Table 4). To enhance throughput, yet retain the ability to identify clonal expansion of TCR genes, we designed 8 Vβ gene segment subgroups, each of which contains the forward primers and TaqMan® probes specific to a non-overlapping subset of Vβ gene segments (Supplemental Table 5). Each Vβ gene subgroup was combined with all 13 Jβ gene segment primers, as well as the RPP30 primers and probe, effectively creating an 8-well, multiplex ddPCR assay for TCRβ rearrangement detection. Each TCRβ gene was measured once in exactly one of the eight wells. So, the sum of counts from all eight wells gave a precise digital count of the total number of rearranged TCRβs in the sample. The subdivision into eight wells allowed detection of one or more large clonal expansions, as measured by a disproportionate fraction of counts in one or a few of the wells.
Figure 1.
Digital quantification and profiling of tumor-infiltrating lymphocytes. (A) Strategy for droplet digital PCR (ddPCR) amplification and probe hybridization of rearranged TCRβ CDR3 regions and RPP30. A generic rearranged TCR β CDR3 region PCR product is shown, indicating the constituent Variable (Vβ) segment, Diverse (Dβ) segment, Joining (Jβ) segment, and the nontemplated nucleotides inserted at the Vβ-Dβ and Dβ-Jβ junctions. Primers and Taqman® probes were designed for T-cell receptor β-chain loci and RPP30 control locus detection by ddPCR. To identify and quantify all possible TCR β chain loci resulting from somatic recombination, 45 forward primers were designed, each specific to a single functional Vβ segment or a small family of Vβ segments. The 3′ end of each Vβ forward primer is anchored at position −43 in the Vβ segment, relative to the recombination signal sequence, thereby providing a unique Vβ tag sequence within the amplified region. Thirty TaqMan® probes, each with a FAM fluorophore, were designed to bind to the 52 possible Vβ gene segments, while 13 reverse primers specific to each Jβ segment are anchored in the 3′ intron To measure the frequency of TCR β-chain loci in a tissue, we normalize to a reference gene, RPP30, a subunit of Ribonuclease P. Primers and a TaqMan® probe, with a VIC® fluorophore, were designed to specifically amplify and detect exon 1 of RPP30, allowing genome-normalized quantification. (B) ddPCR-based quantification of TCR β-chain and RPP30 loci. Primers and probes for Vβ gene segments are divided into 8 subgroups. In a 96 well format, these Vβ subgroup primers and probes are multiplexed with the 13 reverse Jβ primers, along with the RPP30-VIC® reference gene assay. These are combined ZLWK 3&5 PDVWHUPL[ DQG VDPSOH '1$ DQG WKHQ HPXOVLILHG LQWR D PL[WXUH RI ZDWHU LQ RLO droplets (~20,000 droplets per well), each droplet serving as an individual reaction chamber for PCR. These droplets are thermally cycled before being passed through a modified flow cytometer, where FAM and VIC® fluorescence is measured for each droplet. A FAM vs. VIC® plot allows gating of droplets with and without TCR β chain and RPP30 loci, and then Poisson statistics are applied to these populations for accurate quantification.
The number of target molecules present was calculated via droplet digital PCR (ddPCR) by sequestering amplicons into homogenous, one-nanoliter, water-in-oil emulsion droplets and subjecting the droplets to normal PCR amplification. With the inclusion of TaqMan® reporter chemistry, droplets bearing amplified templates were readily distinguished by their fluorescence amplitude using a flow cytometry system. Because the droplet volumes were highly uniform, Poisson statistics could be applied to calculate the average number of target molecules per droplet, and the absolute concentration of TILs could be determined with high precision and accuracy(10). This allowed enumeration of the absolute number of T cell-receptor rearrangements per genome in any tissue sample. By utilizing ddPCR, differing primer amplification efficiencies were overcome via single molecule end-point amplifications.
To assess the accuracy and sensitivity of the QuanTILfy assay, human T cells were purified from blood and mixed with MRC5 normal human lung fibroblasts at different ratios across several orders of magnitude (Fig. 2a). Cells were quantified with a TC10 Automated Cell Counter (Bio-rad), and then the T cells were serially diluted and mixed with MRC5 cells at the appropriate ratios. DNA was extracted and ddPCR-based TCRβ detection was performed in duplicate for each cell mixture. The RPP30-normalized ratio of TCRs per diploid genome had a strong linear correlation with the expected fraction of T cells in the mixture (Linear regression, y = 1.0998x – 0.0007, R2 = 0.9926, n=6), demonstrating scalable detection across 4 orders of magnitude. The slope of this line indicates that the detected TCRs exceeded the expected number of T-cells by 9.98%, but this is consistent with previous findings that show that in normal blood, up to 15% of T-cells have both TCR alleles rearranged (9). In the absence of T cells, a false positive frequency of 5 TCRs was detected in a background of approximately 26,000 human fibroblast cells, across the 8-well assay (Fig. 2a).
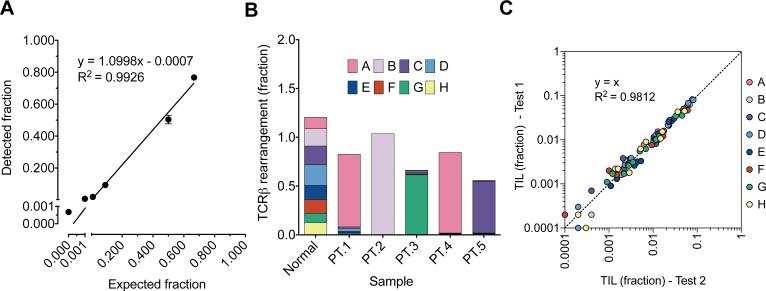
Figure 2.
Spike-in validation of digital TCR quantification and profiling. (a) QuanTILfy was performed on mixtures of T-cells and lung fibroblasts with known ratios. A linear relationship was observed between the expected ratio of T-cells to total cells and the actual ratio of TCR to total cells (Linear regression, y = 1.0998x – 0.0007, R2 = 0.9926, n=6), illustrating the accurate quantification of TCR across several orders of magnitude. (b) To validate the profiling capabilities of QuanTILfy, TCRs were measured in DNA from T-ALL patient bone marrow. In each patient screened, a single TCRβ subgroup was found to dominate the T-cell population (>90%), corresponding perfectly with direct sequencing results. DNA from normal T-cells, however, displayed a relatively even distribution of TCRβ subgroups. (c) To illustrate the reproducibility of the assay, QuanTILfy was performed on 11 ovarian cancer tissues, which exhibit a wide range of TIL counts, in 2 independent experiments. The TIL detected per cell for experiment 1 and experiment 2 were plotted per TCRβ subgroup for each sample, and the data was assessed for conformation to a linear model of ideal reproducibility (R-squared/coefficient of determination to y=x, R2=0.9812, n=88).
To test the specificity of the eight assay subgroups on clinical specimens, QuanTILfy was performed on bone marrow blast samples from five patients with T-cell acute lymphoblastic leukemia (T-ALL) (Fig. 2b). In each case, a single QuanTILfy assay subgroup was observed to represent the vast majority (90.2 – 99.8%) of detectable TCRβ rearrangements, indicative of clonal T-cell expansion. Contrastingly, T-cells from a healthy donor exhibited an even distribution of all QuanTILfy subgroups, with each subgroup representing between 8.0 and 17.5% of total TCR detected. T-ALL patient TCRβ rearrangements were then interrogated by deep sequencing, which verified that, for each patient, the clonal subgroup identified by QuanTILfy matched the clonal TCRβ identified by sequencing, demonstrating that the QuanTILfy assay can specifically and accurately characterize clonal T-cell expansion in patient samples (11) (Supplemental Table 6). To test the reproducibility of the QuanTILfy assay, we interrogated TIL in ovarian tumors, which harbored a range of varying infiltrate fractions, in two independently performed tests (Fig. 2c). The results of the initial and repeat analyses illustrate the high reproducibility (y=x, R2=0.9812, n=88) of the QuanTILfy assay to precisely enumerate TCRβ subclones across several orders of magnitude (Fig. 2c).
Assessing heterogeneity of TILs
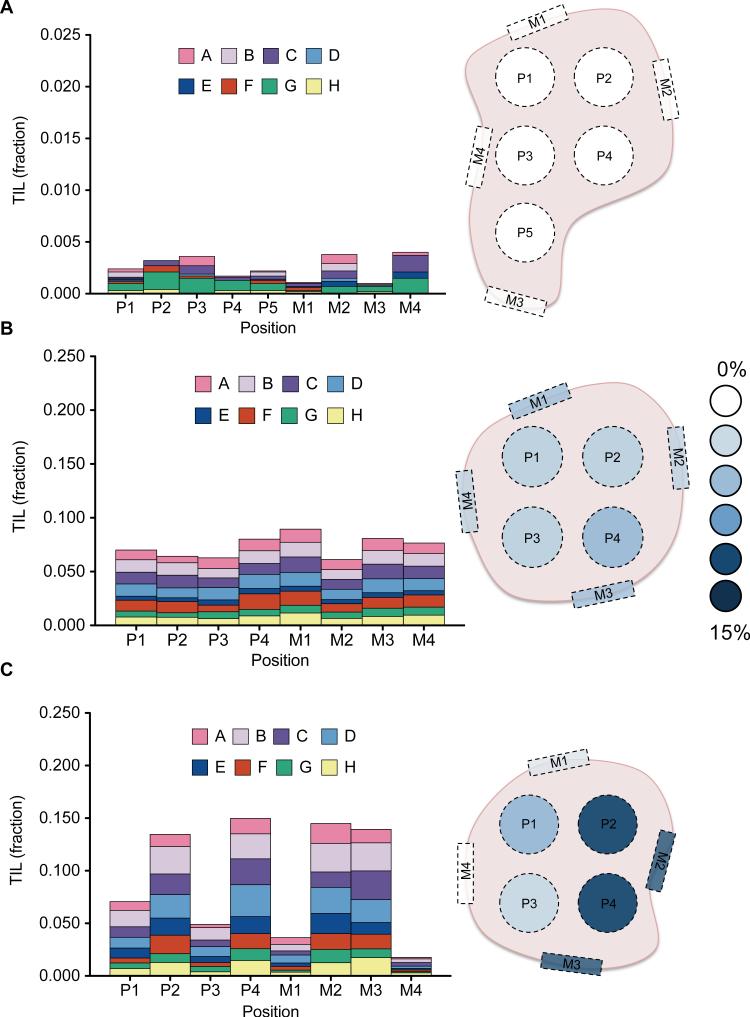
Primary ovarian carcinomas can be large, and the spatial distribution of TILs within these tumors is unknown. Understanding the heterogeneity of TILs within these tumors is vital to design a sampling strategy for clinical applications. To assess the spatial distribution of TILs in primary ovarian tumors, we employed a comprehensive sampling scheme that allowed us to systematically biopsy throughout the tumors of three patients, each diagnosed with late-stage serous ovarian carcinoma. Longitudinal slices, approximately one centimeter in width, were taken from each primary tumor. We then created a two dimensional lattice with two centimeter spacing on top of each slice. At each lattice point, we took a 1.5 cm punch biopsy. In addition, a punch biopsy was taken at four points around the periphery of the tissue slice to assess heterogeneity of the tumor surface compared to the interior. We snap froze a portion of each punch, isolated DNA and performed QuanTILfy on each sample. Our ddPCR results revealed that the fraction of TILs, as a percent of total cells, ranged from 0-15% among the tumor punches (Fig. 3). None of the samples appeared to exhibit clonal T cell expansion, with each assay subgroup displaying consistent fractions of the total infiltrate both within each tumor and between different tumors (Fig. 3). Patient 1's tumor had less than 0.4% TILs for all nine punches, including those from the tumor margins and interior (Fig 3a). For patient 2's tumor, TILs ranged between 5 and 10% for each of eight locations (Fig 3b). The tumor of patient 3 appeared to have differing immune response in its two halves (Fig. 3c). One half had four punches with low TIL percent of 2.5-7.5%, while the other half had a consistently high TIL percent of 12.5-15% for four punches. Thus, using a single small biopsy may potentially be misleading in some patients, where larger or multiple biopsies will more accurately capture a tumor's mean TIL value. The TIL percent at the tumor margins, however, was consistent with the TIL percent within the tumor interior for every case studied (Fig. 3).
Figure 3.
Spatial characterization of TILs in primary ovarian carcinoma. To assess the heterogeneity of TIL frequency and profile throughout a tumor, a two-dimensional lattice was made for slices of primary tumors from 3 patients (a,b,c), with punch biopsies taken at one cm intervals from the interior (P) and margins (M), as indicated in the diagrams. DNA was isolated, and digital TIL quantification was performed for each biopsy. (a) In patient 1, a consistently low TIL fraction (< 0.5%) was observed across all biopsies. (b) Patient 2 displayed higher, but similarly homogenous TIL frequencies (6 – 8%). (c) Patient 3, however, showed a high degree of variability between biopsies, ranging from 2 to 15% TILs. Plotting these TIL fractions on the tumor diagram reveals that the biopsies with higher TIL fractions were all from one side, and the biopsies with lower TIL fractions were from the other.
T-cell immune response in primary and metastatic tumors
From the Pacific Ovarian Cancer Research Consortium Ovarian SPORE repository, we obtained matched primary and metastatic omentum tumor tissue from 18 women with late-stage serous ovarian carcinoma for TIL analysis. Using our QuanTILfy assay, we directly compared the TIL percent in each primary tumor versus that of the paired metastasis. The average TIL fraction of the metastases was more than two-fold higher than that of the paired primary, 0.107±0.015 and 0.052± 0.012, respectively (Supplemental Fig. 1). These results demonstrate that on average, cellular immune response is statistically different and greater in metastatic tumors within the same individual when compared to the primary tumor (n= 18, two-tailed paired t test, p = 0.0007), where 78% (14/18) of patients exhibited a higher TIL count within their metastatic tumor.
Association between TILs counts and improved survival in ovarian cancer patients
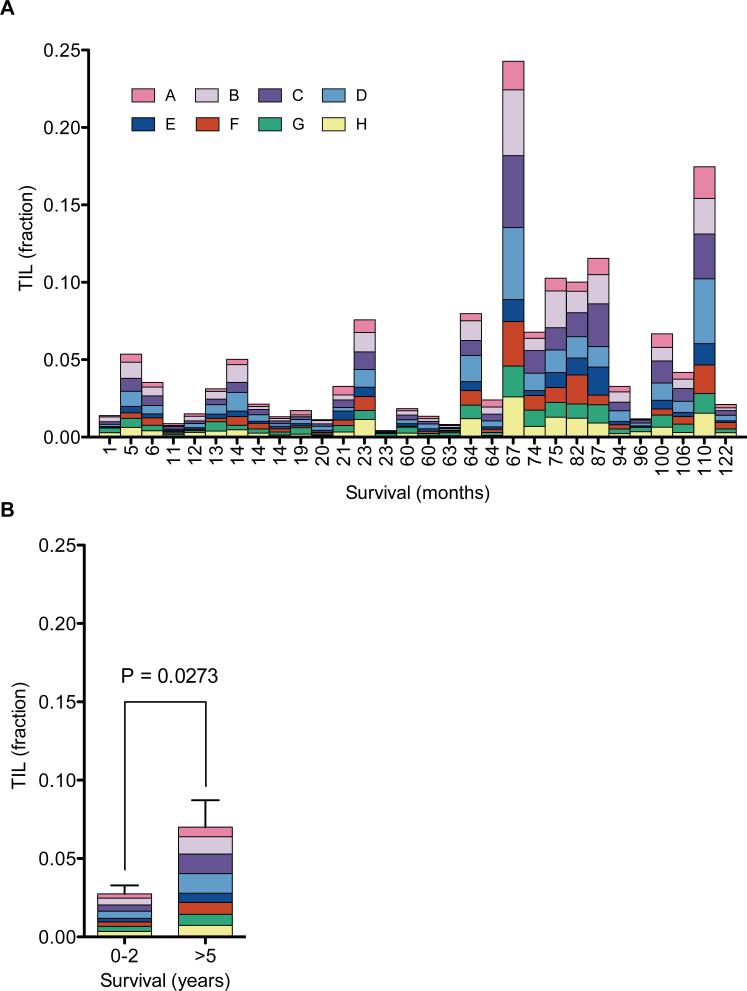
Focusing on primary tumors, we assessed whether mean TIL percentage correlated with survival for ovarian cancer patients with stage III or IV disease, as multiple studies suggest based on IHC data (6). From the Ovarian SPORE repository, we selected patients diagnosed with stage III or IV ovarian carcinoma that were treated with primary debulking surgery followed by platinum-taxane combination chemotherapy, with survival of less than two years (14 patients) or more than five years (16 patients) (Table 1). The frequency of TILs was measured in DNA extracted from frozen samples of each resected tumor (Fig 4a). The clonality of each sample was assessed as the coefficient of variation of the eight V-segment subgroups. Although there was a slight trend of lower clonality (or variation) among the five-year survivors (Fig. 4b), the effect was not statistically significant. However, the total TIL percent was approximately three-fold higher on average for the patients with > 5 year survival (P = 0.0273, two-tailed T test) (Fig. 4b). These results show that higher TIL levels correlate positively with outcome, consistent with the hypothesis that T cells play an active role in suppressing tumorigenesis (12). However, there was some variation between patients, notably a few > 5 year survivors who have a low TIL percent.
Table 1.
Patient data.
| Variable | Short-term | Long-term | Overall |
|---|---|---|---|
| survival (n = 14) | survival (n = 16) | (N = 30) | |
| Age (years) | |||
| Mean | 61.07 | 60.38 | 60.7 |
| SD | 11.28 | 12.78 | 11.9 |
| Range | 45–79 | 40–88 | 40–88 |
| Progression-free interval (months) | |||
| Mean | 5.78 | 39.38 | 23.7 |
| SD | 5.67 | 32.73 | 29.31 |
| Range | 0–15 | 0–94 | 0–94 |
| Survival (months) | |||
| Mean | 14 | 85.44 | 52.1 |
| SD | 6.79 | 19.3 | 39.08 |
| Range | 1–23 | 60–126 | 1–126 |
| Stage | |||
| IIIA | 0 | 1 | 1 |
| IIIB | 1 | 0 | 1 |
| IIIC | 10 | 13 | 23 |
| IV | 3 | 2 | 5 |
| Residual disease (cm) | |||
| 0 | 4 | 6 | 10 |
| <1 | 6 | 10 | 16 |
| 1–<2 | 2 | 0 | 2 |
| ≥2 | 2 | 0 | 2 |
Figure 4.
TIL fraction in ovarian carcinoma and survival. (a) QuanTILfy was performed on primary tumors from 30 ovarian carcinoma patients with known survival outcomes, ranging from 1 to 122 months. A high degree of variability in TIL percent was observed among patients, ranging from 1 to 25%. Notably, no clonal expansion of specific T-cell populations was observed in the tumors, as each sample contained a normal distribution of TCRβ subgroups. (b) TIL fractions were averaged for tumors from patients with short-term (< 2 yrs., n = 14) and long-term survival (> 5 yrs. n = 16). A 2.5-fold higher average TIL fraction was observed for > 5 yr. survivors (two-tail unpaired t test, n=30, p = 0.0273).
Discussion
We have presented a robust, precise technology to measure tumor-infiltrating T-lymphocytes. As TIL count is increasingly recognized as an independent informative factor for prognosis for a wide variety of tumors and as a potential predictive biomarker for response to immunotherapy, there is a growing need for new technologies. Recent evidence suggests that scoring both total T cell and CD8+ T cell counts is a better prognostic for colorectal cancer (CRC) patients than the accepted TNM staging (7). An international effort is underway to modify the CRC staging guidelines to include TIL measurements. The importance of TILs in prognosis of melanoma patients has also been known for many years (13). More recently, with the success of CTLA-4 inhibitors in the treatment of metastatic melanoma, multiple groups have postulated that TIL counts can act as a predictor of efficacy of these immunotherapeutic treatments.
Presently, TIL counts are primarily measured by immunohistochemistry (IHC). A major limitation of IHC is the two-dimensional nature of stained slides. Assaying a two-dimensional cross section of a three-dimensional tumor severely limits the information sampled and increases variation. A second key shortcoming of IHC is the difficulty in standardization between laboratories. Staining intensity and specificity varies significantly between labs even when the same antibodies are used with a shared operating procedure. Additionally, IHC is not capable of addressing clonality. Each clone is defined by the sequence of its rearranged TCR gene, which is not captured by IHC. While our data did not show significant correlation between T cell clonality and outcomes in this small study, we believe this measurement has potential utility. In particular, oligoclonal expansion might be useful for predicting immunotherapeutic response.
Although our ddPCR assay has advantages over IHC in standardizability and quantitative accuracy for measuring total TIL count, the two assays each provide complementary information that is not attainable in the other. IHC can be used to stain for multiple markers, which provide information on T cell subsets. There is presently a debate about whether total TIL count or the ratio of CD4+ /CD8+ T cell counts are more predictive of outcome in ovarian cancer patients (6). Also, the ddPCR technology presented cannot replace IHC, which provides potentially useful prognostic information from T regulatory counts, the identification of tertiary lymphoid structures (TLS), and spatial relationships that are not possible to enumerate with the QuanTILfy technology. On the other hand, the ddPCR assay is able to assess clonality of the TILs, which is not possible by IHC. As prior technologies were unable to accurately measure this quantity, its correlation with clinical outcomes has not been addressed yet in larger studies. We hypothesize that clonality will predict response to immunotherapy, as clonal expansion is the hallmark of an adaptive immune response.
An additional alternative is the use of high-throughput sequencing (HTS) to assess TIL (9, 14). Although HTS has substantially higher clonal resolution, the level of quantitative accuracy of the QuanTILfy method has not been demonstrated for any sequencing-based assay. Moreover, as the average difference in TIL count between patients with long and short-term survival is less than a factor of three (Fig. 4b), accurate quantitation is important for prognosis. Moreover, the ddPCR assay is faster and less expensive than HTS, although technology can improve rapidly.
As a third alternative, quantitative PCR (qPCR) can be used to measure V diversity. However, there are considerable drawbacks to qPCR, including amplification biases due to template size and GC-content, as well as the need for a standard curve to estimate the absolute quantity of DNA (15). Creating a standard curve for each primer-probe pair (45 × 13 = 585) is impractical and in itself an indeterminate process that leads to inaccuracies in measurements of absolute target quantity (16, 17). Thus, a qPCR-based T cell diversity assay would be challenging to standardize across laboratories.
As the ovarian cancer data show, the difference in TIL counts between the bottom quartile and top quartile of TIL counts can be less than a factor of three. Thus, utilization of TIL count for prognosis and prediction requires precise quantitation. However, we have shown that in some cases, using a single small biopsy to assess TIL may potentially be misleading, whereas larger or multiple biopsies will more accurately capture a tumors mean TIL value. Such an approach is clinically feasible especially for cancers like ovarian, where complete surgical excision is part of standard care. For other cancers, multiple core biopsies can be obtained via image guided percutaneous approaches. Future studies, however, are needed to determine the degree of spatial diversity of TIL frequency within other cancers, and to establish the size and number of biopsies required for accurate patient prognosis. In any case, the quantitative precision, ability to track clonality, and assay robustness offered by the QuanTILfy technology are vital improvements over IHC and other detection alternatives, and should facilitate the inclusion of TIL characterization in clinical cancer prognostic panels.
Materials and Methods
Tissue samples
Patient Samples
Tissue specimens were collected from women diagnosed with late-stage serous ovarian cancer at the time of primary debulking surgery. None of these patients had received prior chemotherapy. The debulking surgeries and tissue collections were performed at the Swedish Medical Center – First Hill Campus (Seattle, WA). All patient recruitment and enrollment, specimen collection and processing were conducted through the Pacific Ovarian Cancer Research Consortium (POCRC) as approved by the Fred Hutchinson Cancer Research Center's (FHCRC) Institutional Review Board (FHCRC IR file numbers #4771 and #4563). The initial diagnosis was made intrasurgically by the on-call pathologist, followed by confirmation by the POCRC's research pathologist, who corroborated the diagnosis of late-stage, high-grade serous ovarian cancer and that the study samples had at least 50% malignant epithelium present. Tissues, both primary and metastatic (omentum), were typically processed and frozen within 30 minutes of excision from the patient and stored in liquid nitrogen until DNA extraction. DNA was isolated from approximately 30 mg of frozen tissue using the Qiagen TissueLyser and DNeasy Blood and Tissue kit (Qiagen, Valencia, CA), and quantified using the Quant-IT dsDNA kit (Invitrogen, Life Technologies).
Collection scheme for assessment of tumor heterogeneity
Tissue was collected from the primary ovarian tumor with the guidance of the on-call pathologist. When possible, the tumor surface was inked to allow for identification of the surface in later steps. Longitudinal slices, approximately one centimeter in width, were taken from the resected tumor tissue. Each tumor slice was laid onto a sterile surface and 1.5 cm tumor punches were taken from throughout the slice in a gridded pattern where the punches were spaced 2 cm apart. Four punches were also taken from around the periphery of the tumor slice (i.e., surface punches) at 12, 3, 6 and 9 o'clock. Each punch was then divided into thirds; the middle third was Formalin Fixed and Paraffin Embedded (FFPE) and reserved for other studies while the other two thirds were snap-frozen in foil on dry ice and reserved for DNA analysis. DNA was isolated from frozen tissue samples using the Qiagen TissueLyser and DNeasy Blood and Tissue kit (Qiagen, Valencia, CA), and quantified using the Quant-IT dsDNA kit (Invitrogen, Life Technologies).
Bone marrow aspirate samples were obtained from patients diagnosed with T-ALL. Samples were submitted for flow cytometry analysis at the University of Washington as part of the AALL0434 COG trial protocol. For a normal T-cell control sample, whole blood was drawn from a healthy volunteer donor. Peripheral blood mononuclear cells (PBMCs) were isolated using ficoll, and stained on ice for 20 minutes with anti-CD3 FITC (BD Biosciences, San Diego, CA). Live CD3+ singlet cells were sorted using a BD FACS Aria II cell sorter. All patients provided informed consent as part of the COG trial.
Detection and quantification of T-cell rearrangements (TCRs) by droplet digital PCR
TCRβ loci were measured by using multiplexed Taqman® chemistry in a droplet digital PCR (ddPCR) platform. Primer sequences were adapted from those described previously (9), with 45 forward primers, each specific to one or more TCRβ Variable (V) genes (Supplemental Table 1), and 13 reverse primers, each specific to a TCRβ Joining (J) gene (Supplemental Table 2). Additionally, 35 Taqman® probes were designed, each with a FAM fluorophore, and with specificity to one or more TCRβ V-genes (Supplemental Table 3). TCRβ V-gene primers and probes were assigned to 8 assay subgroups (Supplemental Table 5), grouped together based on similar individual background fluorescence amplitude. To obtain a snapshot of TCRβ quantity and diversity in a sample, DNA samples are run with all eight of the assay subgroups, each in a separate well. To concurrently measure total number of cells screened for normalization, primers and a Taqman® probe with a VIC® fluorophore were designed specific to the reference gene RPP30, which is present at two copies per diploid genome (Supplemental Table 4).
Droplet digital PCR
Reaction mixtures were prepared with ddPCR SuperMix (Bio-rad), the appropriate TCRβ V-region primers (forward) at 900 nM each, all TCRβ J-region primers (reverse) at 900 nM each, appropriate TCRβ V-region Taqman probes at 250 nM each, RNaseP primers at 900 nM each, RNaseP Taqman® probe at 250 nM, and 20-100 ng of DNA per 20 μL reaction volume. We note, however, that in the present reiteration 100 ng of DNA is suggested per subgroup, or 800 ng total from 5 mg of tumor tissue to ensure optimum statistical power. Bulk reaction volumes were partitioned into 1 nL aqueous-in-oil immersions with the QX100 ddPCR System Droplet Generator (Bio-rad). Droplets were transferred into a 96-well PCR plate, and then cycled with the following conditions: 95°C for 10 min, followed by 50 cycles of 94°C for 30 sec and 61°C for 1 min.
Droplets were individually analyzed for fluorescence by a QX100 ddPCR System Droplet Reader (Bio-rad). Droplets with fluorescence in the FAM or VIC® channel were counted, and concentrations of target molecules were calculated by QuantaSoft software (Biorad) via Poisson statistics to quantify TCRs (FAM) and RPP30 (VIC®) in copies per μL of reaction volume. Thus, TCRβ per diploid genome values were calculated individually for each assay subgroup as follows: TCRs/diploid genome (subgroup X) = [TCR countsubgroup X] ÷ (2 × [RPP30 count]).
Validation of digital TCRβ quantification in cell mixtures
To obtain pure T-cells, whole blood was drawn from a healthy volunteer donor. Peripheral blood mononuclear cells (PBMCs) were isolated using ficoll, and stained on ice for 20 minutes with anti-CD3 FITC (BD Biosciences, San Diego, CA). Live CD3+ singlet cells were sorted using a BD FACS Aria II cell sorter. MRC5 human lung fibroblasts were grown at 37°C in Minimum Essential Medium with Earle's Balanced Salts (MEM/EBSS) supplemented with 10% Fetal Bovine Serum (FBS) and 1X penicillin/streptomycin. T-cells and MRC5 cells were resuspended with 1X PBS and counted with a TC10 Automated Cell Counter (Bio-rad). T-cells were serially diluted and mixed with MRC5 cells at specific ratios across four orders of magnitude. An MRC5-only sample was also prepared as a negative control. DNA was extracted from the cell mixtures with the PicoPure® DNA Extraction Kit (Arcturus) and then subjected to digital TCRβ quantification, as described above. TCRβ concentrations were summed from the 8 QuanTILfy subgroup wells to assess the total fraction of TCRβ per diploid genome, and this value was compared directly with the expected fraction of T-cells to total cells for each cell mixture.
Detection of clonal T-cell expansion in clinical samples
Bone aspirate samples were obtained from T-cell acute lymphoblastic leukemia patients, and T-cell DNA was isolated as described above, as well as T-cell DNA from a healthy patient as a normal control. The QuanTILfy assay was performed on each patient sample, and TCRβ counts were determined for each assay subgroup. ddPCR-based TCRβ profile data for each patient was compared to previous high-throughput sequencing results to validate the accuracy and specificity of QuanTILfy's profiling capabilities.
High-throughput CDR3 sequencing
TCRβ CDR3 regions were amplified and sequenced from 2 μg of DNA. Amplification and sequencing of TCRB CDR3 regions was carried out using the ImmunoSEQ platform at Adaptive Biotechnologies (9). The sequences for both the TCRB CDR3 regions were delineated according to the definition established by the International ImMunoGeneTics collaboration (18). Sequences that did not match CDR3 sequences were removed from the analysis. A standard algorithm was used to identify which V, D, and J segments contributed to each TCRB CDR3 sequence (18).
Supplementary Material
One Sentence Summary.
We introduce a robust digital DNA-based assay to enumerate TILs and assess T cell clonality in tissue samples; and demonstrate an association between higher TIL counts and improved survival among women with ovarian cancer.
Acknowledgments
Funding: The authors acknowledge support from the Listwin Family Foundation (to J.H.B.), an Ellison Medical Foundation New Scholar award (AG-NS-0577-09, to J.H.B), an Outstanding New Environmental Scientist Award (ONES) (R01) from the National Institute of Environmental Health Sciences (R01ES019319, to J.H.B.), a grant from the Congressionally Directed Medical Research Programs/U.S. Department of Defense (W81XWH-10-1-0563, to J.H.B.), the Pacific Ovarian Cancer Research Consortium Ovarian Cancer SPORE Award (P50 CA083636), a Department of Defense Ovarian Cancer Idea Award (OC093221, to M.T.), a Susan G. Komen postdoctoral fellowship (to J.G.), and from the Canary Foundation (to M.T.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute, the National Institute of Health, nor of any of the other granting agencies.
Footnotes
Author contributions: H.S.R. helped designed the experiments and co-wrote the manuscript. N.G.E. performed the majority of the digital genomic quantification of T-lymphocytes and data analysis. M.T. designed the collection of, and J.G. processed the biospecimens used to assess T-cell heterogeneity in ovarian cancer. C.W.D. surgically removed, and with K.C.O. and M.T., oversaw the collection of biospecimens, their processing, storage, annotation, and distribution. J.H.B. designed the experiments, developed the QuanTILfy assay, performed digital genomic quantification of T-lymphocytes during assay development, analyzed the data, and co-wrote the manuscript. All authors reviewed and edited the manuscript.
Competing interests: J.H.B. has equity in Adaptive Biotechnologies. H.S.R. has consultancy, equity ownership, patents, and royalties with Adaptive Biotechnologies. The Fred Hutchinson Cancer Research Center and Adaptive Biotechnologies have jointly filed for a patent application on this technology, entitled “Quantification of Adaptive Immune Cell Genomes in a Complex Mixture of Cells” (US 13/656,265).
References and Notes
- 1.Igney FH, Krammer PH. Immune escape of tumors: apoptosis resistance and tumor counterattack. Journal of leukocyte biology. 2002 Jun;71:907. [PubMed] [Google Scholar]
- 2.Galon J, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006 Sep 29;313:1960. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 3.Leffers N, et al. Prognostic significance of tumor-infiltrating T-lymphocytes in primary and metastatic lesions of advanced stage ovarian cancer. Cancer immunology, immunotherapy : CII. 2009 Mar;58:449. doi: 10.1007/s00262-008-0583-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato E, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proceedings of the National Academy of Sciences of the United States of America. 2005 Dec 20;102:18538. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. The New England journal of medicine. 2003 Jan 16;348:203. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 6.Hwang WT, Adams SF, Tahirovic E, Hagemann IS, Coukos G. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: a meta-analysis. Gynecologic oncology. 2012 Feb;124:192. doi: 10.1016/j.ygyno.2011.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galon J, et al. The immune score as a new possible approach for the classification of cancer. Journal of translational medicine. 2012;10:1. doi: 10.1186/1479-5876-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang Q, Hedley D. Emerging applications of flow cytometry in solid tumor biology. Methods. 2012 Jul;57:359. doi: 10.1016/j.ymeth.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Robins HS, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009 Nov 5;114:4099. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinheiro LB, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012 Jan 17;84:1003. doi: 10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu D, et al. High-throughput sequencing detects minimal residual disease in acute T lymphoblastic leukemia. Sci Transl Med. 2012 May;416:134ra63. doi: 10.1126/scitranslmed.3003656. [DOI] [PubMed] [Google Scholar]
- 12.Egen JG, Kuhns MS, Allison JP. CTLA-4: new insights into its biological function and use in tumor immunotherapy. Nature immunology. 2002 Jul;3:611. doi: 10.1038/ni0702-611. [DOI] [PubMed] [Google Scholar]
- 13.Azimi F, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012 Jul 20;30:2678. doi: 10.1200/JCO.2011.37.8539. [DOI] [PubMed] [Google Scholar]
- 14.Sherwood AM, et al. Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T cell receptor sequences that differ from the T cells in adjacent mucosal tissue. Cancer immunology, immunotherapy : CII. 2013 Jun 16; doi: 10.1007/s00262-013-1446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valasek MA, Repa JJ. The power of real-time PCR. Advances in physiology education. 2005 Sep;29:151. doi: 10.1152/advan.00019.2005. [DOI] [PubMed] [Google Scholar]
- 16.White RA, 3rd, Blainey PC, Fan HC, Quake SR. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC genomics. 2009;10:116. doi: 10.1186/1471-2164-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yun JJ, et al. Genomic DNA functions as a universal external standard in quantitative real-time PCR. Nucleic acids research. 2006;34:e85. doi: 10.1093/nar/gkl400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yousfi Monod M, Giudicelli V, Chaume D, Lefranc MP. IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics (Oxford, England) 2004 Aug 4;20(Suppl 1):i379. doi: 10.1093/bioinformatics/bth945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.