Abstract
Impaired autophagic machinery is implicated in a number of diseases such as heart disease, neurodegeneration and cancer. A common denominator in these pathologies is a dysregulation of autophagy that has been linked to a change in susceptibility to cell death. Although we have progressed in understanding the molecular machinery and regulation of the autophagic pathway, many unanswered questions remain. How does the metabolic contribution of autophagy connect with the cell’s history and how does its current autophagic flux affect metabolic status and susceptibility to undergo cell death? How does autophagic flux operate to switch metabolic direction and what are the underlying mechanisms in metabolite and energetic sensing, metabolite substrate provision and metabolic integration during the cellular stress response? In this article we focus on unresolved questions that address issues around the role of autophagy in sensing the energetic environment and its role in actively generating metabolite substrates. We attempt to provide answers by explaining how and when a change in autophagic pathway activity such as primary stress response is able to affect cell viability and when not. By addressing the dynamic metabolic relationship between autophagy, apoptosis and necrosis we provide a new perspective on the parameters that connect autophagic activity, severity of injury and cellular history in a logical manner. Last, by evaluating the cell’s condition and autophagic activity in a clear context of regulatory parameters in the intra- and extracellular environment, this review provides new concepts that set autophagy into an energetic feedback loop, that may assist in our understanding of autophagy in maintaining healthy cells or when it controls the threshold between cell death and cell survival.
Keywords: ATP, autophagy, energy, energetics, metabolite, mitochondria, threshold
Introduction
Autophagy, from Greek “self-eating,” is a major protein degradation system that targets primarily long-lived cytoplasmic proteins. This catabolic process serves ubiquitously as a stress response and is conserved in all eukaryotic cells.1 Three main forms of autophagy exist: chaperone-mediated autophagy, microautophagy and macroautophagy. In chaperone-mediated autophagy, single cytosolic proteins are recognized by a chaperone complex that facilitates target-motif governed delivery, lysosomal membrane binding, translocation and subsequent internalization.2 Microautophagy delivers cytoplasmic materials by a process involving lysosomal limiting membrane invagination, protrusion or septation. Macroautophagy involves a dynamic subcellular membrane rearrangement whereby cytoplasmic proteins and organelles are sequestered in double-membrane vesicular structures (autophagosomes) that fuse with the lysosome or vacuole, where the material is catabolically degraded, recycled and utilized as an energy source. Variants in type of material and in the selectivity of cargo recognition exist, leading to the distinction between “bulk” or nonselective macroautophagy involving the sequestration of random cytosolic material, and selective types of macroautophagy including lipophagy, mitophagy, ribophagy, pexophagy, etc., where the autophagosome exclusively contains lipid droplets, mitochondria, ribosomes or peroxisomes, respectively. Although all three forms of autophagy differ in the pathway by which cargo material is delivered to the lysosome, they are similar with regard to the final and fundamental steps of lysosomal protein degradation by hydrolase exposure.
Macroautophagy (hereafter referred to as autophagy) is not only the most widely studied and best-understood process, but also the autophagic pathway where the most powerful connections have been identified between its course of action, cellular metabolism and cell death. However, the relationship between autophagy, catabolism and balancing cellular energetics on the one hand, and the integration of a cellular stress response on the other hand, remains largely unclear. Although substantial progress has been made in understanding the molecular control of autophagy and its regulation with regard to autophagosome initiation, formation, elongation and maturation, we are less able to answer questions that deal with the prediction of how and when the molecular machinery of autophagy affects cell death or cell survival. Central to this outcome is the cell’s capacity to generate sufficient ATP and its ability to meet metabolic demands. Therefore, in this review, we highlight the network that connects autophagy and energetic sensing with substrate provision, cellular energy demands and cell death. We assess the conditions that determine whether a change in autophagic activity is compatible with cell function and metabolic demands, or whether it leads to the induction of apoptosis or necrosis. We address how the governance of autophagy through metabolic sensing translates into a metabolic robustness that may act upon a framework of intracellular as well as extracellular parameters that together affect cellular susceptibility. Finally, we introduce autophagy as an energetic feedback loop that is anchored between energetic sensing and ATP consumption, thereby integrating autophagic flux and susceptibility to cell death in a coherent manner.
Autophagy: Anchored within an Energetic Feedback Loop
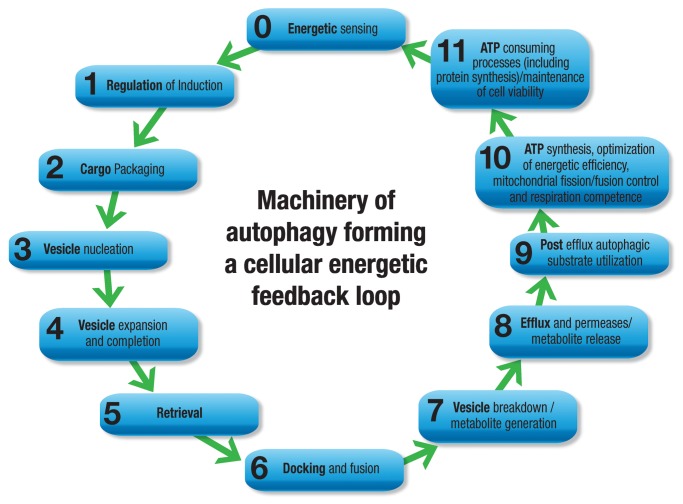
The complete autophagy process can be dissected into the eight functional aspects of (1) induction; (2) cargo selection and packaging; (3) vesicle nucleation; (4) vesicle expansion and completion; (5) retrieval; (6) vesicle targeting to, and docking and fusion with, the lysosome or vacuole; (7) intralysosomal vesicle breakdown; and last, (8) permease efflux. More than 30 genes and respective autophagy-related (Atg) proteins participate in the organization of functional complexes that control and regulate this process from the induction of autophagy to vesicle breakdown and efflux from permeases, and readers are referred to comprehensive reviews.1,3 Formation of the autophagosome is facilitated by the formation of a limiting membrane, characterized by the organization of modulatory and kinase complexes around membrane structures that are derived from organelle membranes such as that of the endoplasmic reticulum, Golgi apparatus and mitochondria.2 BECN1 and PIK3C3/VPS34, for example, are pivotal in the formation of a lipid kinase complex, which in turn allows the recruitment and shuttling of ATG proteins that contribute to the generation of the phagophore, the initial sequestering compartment. Next, the ubiquitin-like conjugation systems govern the formation of the ATG12–ATG5 complex as well as LC3–phosphatidylethanolamine, processes that drive membrane elongation. Finally, phagophore formation completes by sealing to form the autophagosome. This process is thought to occur randomly during nonselective (bulk cytoplasm) autophagy; however, specific organelles can be surrounded with the help of recognition proteins such as SQSTM1/p62 during selective autophagy.4 Subsequent docking and fusion with lysosomes occurs with the formation of an autolysosome, a process first proposed by de Duve and Wattiaux.5 Exposed to lysosomal hydrolases, the autophagosome and its cargo are completely degraded and their basic components are released through permeases into the cytosol, providing an additional source of metabolites.
However, to fully incorporate the primary role of autophagy, i.e., to protect cells under stress conditions, and to comprehend its variability in favoring or delaying cell death, we need to expand the currently known schematic model of the autophagy process and its machinery. Therefore, here we argue that it is essential to anchor the autophagic machinery within an energetic feedback loop that includes the regulatory components of energetic sensing, metabolite generation, substrate utilization and the demand for ATP consumption, thereby controlling the cellular energetic balance as a homeostatic feedback mechanism. We postulate that the molecular machinery of autophagy actively operates already prior to its “induction” through multifaceted energetic sensing pathways, and expands beyond the “final” step of “permease efflux”: Relative to the eight points above, a pre-induction point 0 summarizes a network of metabolic sensing mechanisms that governs the onset and magnitude of autophagic activity, which in turn tightly connects to the post-efflux processes (Fig. 1). Moreover, an additional point, 9, addresses the functional aspects of post-efflux autophagic utilization of substrate and synthesis of ATP. Point 10 attends to the optimization of energy efficiency, mitochondrial fission/fusion control and respiration competence; and last, point 11 concentrates on the final step that closes the functional feedback loop, the state of ATP consuming processes and the synthesis of survival proteins (Fig. 1). In this way the feedback loop is complete as ATP consumption, which is required to maintain cell viability and function, connects back to the metabolic sensor, enabling homeostatic control of the cellular stress response. This expanded scheme assists with the understanding of autophagic flux control, as it draws attention to the molecular machineries of distinct energy sensing pathways, their impact on ATP generating systems and the subsequent change in capacity to meet the cell’s energy requirements. A central theme that determines the likelihood of cell death during a stress response lies in the availability of ATP. When key molecular markers that connect the autophagic machinery to its function as a stress response mechanism are included, a molecular network emerges that spans densely around the cellular energetic balance system. It becomes clear that multiple energetic feedback loops exist that connect the extracellular environment with autophagic flux and intracellular metabolite conditions, controlling energetic efficiency and, concomitantly, susceptibility to undergo cell death.
Figure 1. When assessing autophagy in the context of its functional role in metabolism, the sequence of the machinery may be expanded in both directions in order to complete the energetic feedback loop. Prior to its induction we highlight its role of energetic sensing, whereas post efflux and permease release we suggest substrate utilization, ATP synthesis and energetic efficiency as well as protein synthesis/ATP consumption.
Autophagy: The Energetic Sensor that Creates Metabolic Robustness
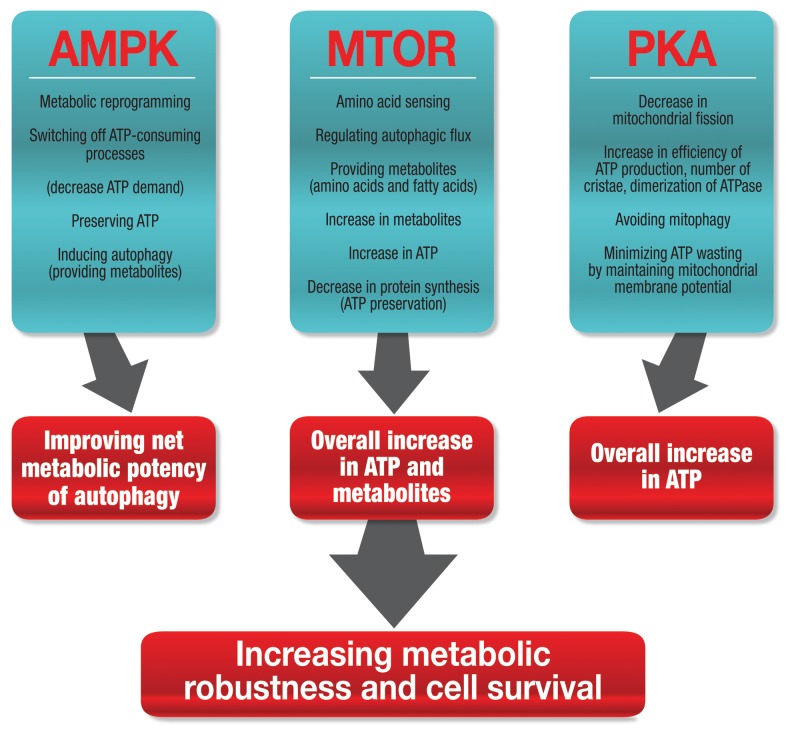
The autophagic machinery is more densely connected to the major energy sensors of the cell than previously thought. The recently discovered interplay between autophagy and metabolism reveals a profound dynamic feedback loop that controls the cellular energetic state and thereby closely interacts with the organization of the onset of cell death.6 The autophagic machinery and its activity is controlled by numerous extracellular and intracellular parameters that assess the metabolic or energetic state in a direct or indirect fashion. Here, we argue that three major roles can be assigned to the autophagic machinery that networks cellular metabolism in response to perturbations: first, multifaceted, convergent sensing of the energetic state; second, response with significant metabolite generation through the intricate tuning of autophagic flux; and third, complex integration of autophagic activity with ATP-consuming processes and efficiency of mitochondrial respiration (Fig. 2). Due to the central role of ATP in the dynamics of cell death onset and control of cell death subroutines, we propose that this network fundamentally governs the susceptibility to cell death.7,8
Figure 2. Three major energy-sensing pathways connect the cellular energy levels with the autophagic machinery that results in increased ATP availability, improved energy efficiency and respiration, culminating in the control of cellular metabolic robustness, which maximizes cell viability.
Three major energy-sensing pathways connect the cellular energy levels with the autophagic machinery, resulting in increased ATP availability, improved energy efficiency and cellular respiration (Fig. 2). Hence, the downstream elicited effects culminate in the control of cellular metabolic robustness, which in turn defines the threshold for cell viability. The primary cellular energy sensing kinase cascades involve PKA (protein kinase, cAMP-dependent), AMPK (protein kinase, AMPK-activated) and MTOR (mechanistic target of rapamycin). All three signaling systems are tightly networked and intricately linked to the regulation of the molecular machinery of autophagy.9,10 Protein synthesis and protein degradation are reciprocally controlled by the MTOR pathway, and it was initially thought that MTOR itself senses intracellular ATP concentration.11,12 In yeast, interaction between Atg1 and Atg13 may be controlled by TORC1-regulated Atg13 hyperphosphorylation, which initiates autophagosome formation.13 This interaction of Atg1-Atg13 appears to occur constitutively, as it is not altered by nutrient availability or MTOR inhibition.14 In the mammalian system, MTORC1 phosphorylates ULK1, an Atg1 homolog, which, in a complex with ATG13 and RB1CC1/FIP200 suppresses the initiation of autophagosome synthesis.15 TOR/MTOR is a central regulatory component that connects the cellular energetic environment with autophagic activity, thereby increasing metabolites for ATP generation. For example, as mentioned above, TOR hyperphosphorylates Atg13, resulting in an inhibition of autophagy.13 TOR activity is highly sensitive to the availability of nitrogen sources, such as L-glutamine or L-asparagine.16 MTOR sensing of amino acid availability is primarily based on the efflux of L-glutamine and influx of L-leucine as well as other essential amino acids through a bidirectional transport system.16 Leucine is one of the most potent amino acids in stimulating MTOR signaling.11 MTOR is therefore involved in a negative feedback loop that controls metabolite (amino acid) availability. The exact sensing mechanism, however, as well as the mechanism of MTOR dephosphorylation, is not fully understood.
AMPK is a sensor for cellular energy levels. It is required for autophagy, as AMPK mutants as well as AMPK inhibition suppress autophagy.17 AMPK is activated by ATP depletion, indicated by an increase in ADP and AMP, or glucose starvation.18 AMPK is able to modulate the autophagic machinery by affecting two crucial signaling complexes: by inhibiting MTOR and, as recently revealed, by directly phosphorylating ULK1.19-22 AMPK operates therefore in a “fail-safe” mechanism, regulating autophagic activity through two distinct molecular avenues, both controlling induction of autophagy and therefore metabolite provision for ATP generation. Importantly, at the same time, AMPK has a critical role in reprogramming overall cellular metabolism, thereby increasing the metabolic potency of induced autophagy.23 For example, by switching off ATP-dependent processes, AMPK contributes to the conservation of ATP (Fig. 2).24 This may be especially important if oxygen tension becomes low and ATP generation per mole oxygen needs to be maximized. AMPK plays therefore a dual role, first by providing metabolites that can serve as substrates for ATP synthesis, thereby increasing ATP availability, and second by decreasing ATP consumption and minimizing ATP “wastage” through switching off defined ATP-consuming processes (Fig. 2).
Upon starvation of cells from amino acids and serum, PKA mediates phosphorylation of DNM1L/DRP1, leading to an inhibition of fission, thereby disturbing the equilibrium between fission/fusion and shifting it toward the latter.25 This results in a greater mitochondrial network with more elongated mitochondria that exhibit increased efficiency in ATP synthesis. The higher energy production rate is brought about by increased dimerization of mitochondrial ATP synthase as well as a higher density and number of mitochondrial cristae (Fig. 2).26 If residual oxygen as well as metabolites are available, a cellular energetic state can be maintained that is more likely compatible with the current increased energetic demands. In that way, ATP is preserved, as chances for mitophagy of depolarized and fragmented mitochondria are minimized.10,27 Moreover, the cytosolic ATP that is available is also effectively used, as nonelongated mitochondria would use more cytosolic ATP to maintain their membrane potential and thereby deplete ATP stores, accelerating the onset of cell death.26 In addition, autophagy regulates cell survival during energy stress through controlling the clearance of damaged organelles and misfolded proteins, thereby affecting the load of functional mitochondria and decreasing the risk of toxic protein aggregate formation.21,28 Taken together these data indicate that the net metabolic efficiency can be powerfully modulated as a change in autophagic flux is induced.
Autophagy: Metabolite Provision that can be Effective
All eukaryotic systems with a lysosomal compartment are able to undergo the process of autophagy. A basal level of autophagic pathway activity is therefore crucial to maintain cellular housekeeping functions. Autophagy produces amino acids for the survival of the cell or organism, when nutrients become limited.29,30 However, direct evidence for a change in metabolite profile and its connection to cell viability remains limited. It is becoming increasingly clear that autophagy indeed plays a role of a powerful (and likely underestimated) provider of metabolite substrates. Protein catabolism was the first clearly defined metabolic function brought about by autophagy and its dynamic control by nutrient supply and amino acid availability has been stressed.31 The autophagic pathway is generally considered to degrade long-lived proteins, as opposed to the ubiquitin-proteasome system for short-lived proteins.32 As more than 99% of intracellular proteins are long lived, the pool size of the available protein for bulk degradation is substantial. After 24 h fasting we find a 25% net loss of protein in liver cells, and an average clearance rate of 2% per hour in hepatocytes and embryonic stem cells.32,33 This rate can be increased to up to 5% protein loss per hour, indicating the huge metabolic buffer capacity available through autophagy.33 However, it is largely unclear whether substrates generated by autophagy necessarily contribute with the same metabolic value to cellular metabolism. Because amino acid metabolism is cell-type dependent, it is likely that the regulation of autophagy by amino acids also varies among cell types.11
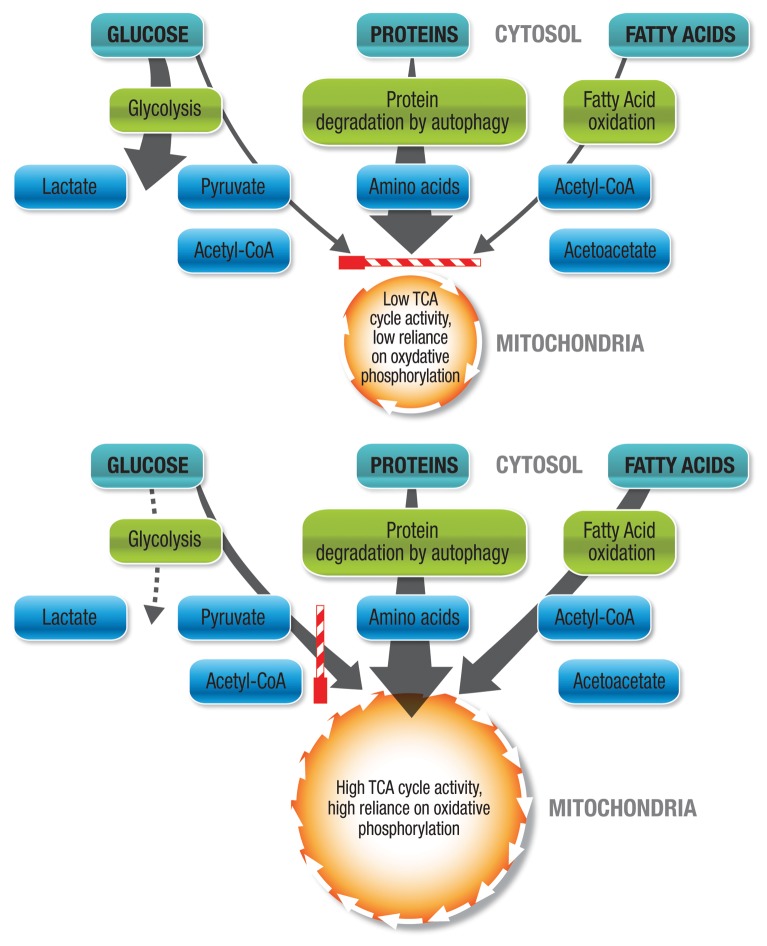
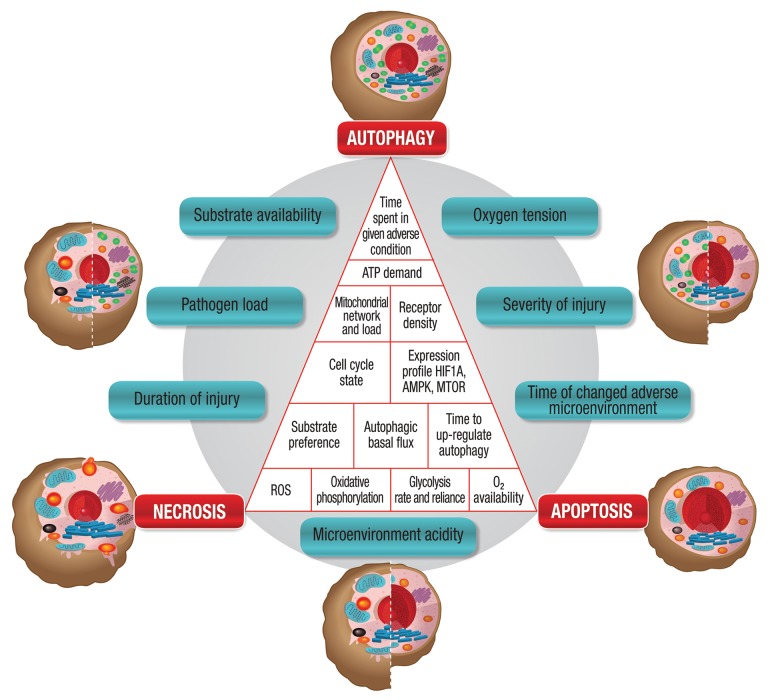
Although the regulatory role of autophagy in maintaining physiological amino acid levels has been demonstrated in both yeast and mammalian cells, studies that assess the alteration of metabolite profile during changes in autophagic flux remain limited.34,35 Indirectly, abundant evidence demonstrates that the metabolic profile affects the energetic state of the cell. The branched-chain α-ketoacid dehydrogenase complex, which is the regulatory enzyme to channel branched-chain amino acids such as leucine, isoleucine and valine into the tricarboxylic acid (TCA) cycle, is upregulated in starvation.35 Theoretically, both glucogenic and ketogenic amino acids generated through increased autophagic flux may significantly fuel the TCA cycle at multiple entry points. However, conditions for autophagy to contribute metabolically are not always favorable. Autophagy is able to contribute through substrate provision only if the cell’s substrate preference and basal metabolism are favorable for oxidative phosphorylation (Fig. 3). The extent of tissue reliance on anaerobic vs. aerobic metabolism, its glycolysis rate vs. mitochondrial respiration rate, as well as the substrate distribution and substrate preference are all key factors that determine the possible net contribution of autophagy to metabolism (Fig. 3). On the one hand, it may therefore be said that unfavorable conditions for increased autophagic flux to contribute to the metabolic balance sheet are low basal TCA activity, high basal anaerobic metabolism, low oxygen tension, a high degree of fissioned mitochondria, a large proportion of dysfunctional mitochondria, or low mitochondrial load (Fig. 3). On the other hand, favorable conditions for increased autophagic flux to metabolically contribute are a high basal TCA cycle activity, a high basal reliance on oxidative phosphorylation, sufficient residual oxygen availability, a sufficient number of functional mitochondria, a high degree of fused mitochondria, or a high mitochondrial load (Fig. 3).
Figure 3. Glucogenic and ketogenic amino acids generated through increased autophagic flux fuel the TCA cycle at multiple entry points. However, conditions for autophagy to contribute metabolically are not always favorable. Autophagy is only able to contribute through substrate provision if the substrate preference and basal metabolism are favorable for oxidative phosphorylation. Unfavorable conditions for increased autophagic flux to contribute metabolically are low basal TCA activity, high basal anaerobic metabolism, low oxygen tension, a high degree of fissioned mitochondria, dysfunctional mitochondria, or low mitochondrial load. Access of autophagically generated metabolites to the TCA cycle is not favored (striped bar in horizontal position). Favorable conditions for increased autophagic flux to contribute metabolically are high basal TCA cycle activity, minimal basal anaerobic metabolism, high basal reliance on oxidative phosphorylation, sufficient residual oxygen availability, sufficient functional mitochondria, high degree of fused mitochondria, or high mitochondrial load. Access of autophagically generated metabolites to the TCA cycle is favored (striped bar in vertical position).
It becomes clear that the cargo-specific forms of autophagy, in particular lipophagy and mitophagy, may both powerfully affect this regulation during energy stress by first providing particularly fatty acids to fuel the TCA cycle, and second by assuring the availability of actively respiring mitochondria with functional TCA cycle flux. Intracellular lipid droplets can operate as convergence sites for the autophagic pathway, but only recently was it directly demonstrated that macrolipophagy connects autophagy with lypolysis during energy stress by affecting the rates of β-oxidation, triglyceride stores and cellular cholesterol content.36-38 Mitophagy effectively regulates mitochondrial number and their quality control by sensing a decrease in the mitochondrial inner membrane electrochemical gradient, which indicates an inability to produce ATP. Here, depolarized mitochondria are recognized through an interplay between signaling pathways mediated by PINK1 and PARK2, a function that is impaired in Parkinson disease.39 Numerous examples indicate an important role of autophagy in the onset or progression of pathological conditions that are in particular characterized by high energy demands and energy stress. For example, altered autophagy has been observed in the ischemic, hypertrophied and failing heart, and has been shown to not only be required to maintain cardiac contractile function but also to limit infarct size and attenuate remodeling after a myocardial infarction.40-42 Similar to cardiac myocytes, neurons are characterized by a high energy demand. Here, the high autophagic protein clearance rate is required to maintain neural function and to prevent accumulation of aggregate-prone proteins associated with neurodegeneration such as is observed in Alzheimer or Huntington disease.28,43,44 Tumors are characterized by high energy demands to maintain proliferation even in nutrient-poor conditions. Metabolic stress is a highly common feature especially in solid tumors, where autophagy localizes to hypoxic regions and restricts necrotic cell death and inflammation.45,46
In context with the role of autophagy in the human pathologies described above, it is crucial to recognize that different tissue types show highly defined and distinct metabolic properties, such as the myocardium, which preferentially utilizes fatty acids, nervous tissue, which almost exclusively uses glucose, and many cancer cells, which upregulate glucose metabolism (Warburg effect). Importantly, this specific metabolic reliance is not static, but can change developmentally or be pathologically induced. Such changes are observed in the transition of the neonatal to the adult myocardium, indicating an additional level of complexity for autophagy to contribute metabolically. In fact, the magnitude of metabolic reliance on autophagy to provide amino acids is revealed in the fetal to neonatal transition, which provides a powerful experimental model to assess metabolism.47 Conditional knockout mice lacking Atg7 indicate the magnitude of the contribution to energy supply, as these mice do not survive longer than 12 h after birth due to severe nutrient deficiency.48,49 The targeted inactivation of specific Atg genes leads to a profound decrease in tissue and plasma amino acids, as well as hypoglycemia and hypolipidemia.49,50 The myocardium and the diaphragm show substantially increased autophagy, which supports the notion for high metabolic activity and reliance on fuels generated by autophagy.50 Likewise, fasted atg5−/− neonatal hearts display a severe ST segment elevation in the electrocardiogram pattern, suggesting a shortage in respiratory substrates.50 Also, myocardial tissue amino acids significantly decrease, indicating myocardial energy depletion due to a lack of substrates generated by autophagy. Moreover, AMPK is activated 10 h after birth in atg5−/− neonates, but not so in wild-type mice, indicating the increased metabolic demand of neonatal tissue due to the lack of autophagy.50 These data indicate strongly that autophagy can contribute significantly to the metabolic balance sheet of mammalian cells. Further data suggesting that autophagy may be required for maintaining cellular energetics have been derived from studies investigating myocardial ATP levels; both starvation and bafilomycin A1 treatment reduce myocardial ATP levels.51
Although glycogen is also delivered to lysosomes, its storage and consumption appear normal in Atg knockout models, indicating a minor role toward substrate generation.37 These findings confirm, in vitro and in vivo, that an additional metabolite fuel source is provided through autophagy. Adipocyte-specific Atg7 knockout produces mice with decreased white adipose tissue mass, but, importantly, the white adipocytes that are present show features of brown adipocytes. These data suggest a potential metabolic compensatory mechanism, due to the lack of autophagically-generated substrates.37 However, neonatal atg5−/− mice do not show differences in their lipolytic rates, suggesting that different nutritional conditions and metabolic demands control the contribution of autophagic degradation.50 The regulation of lipid droplet metabolism through autophagy broadens the cellular response repertoire to meet its energy demands in adverse conditions.52 Further studies will be necessary to delineate exact conditions that favor autophagy to contribute metabolically and to clarify the role of autophagy in amino acid and lipid metabolism in different cell types under defined metabolic perturbations.
Autophagy: The Integration of Metabolic Feedback that Controls Cell Death
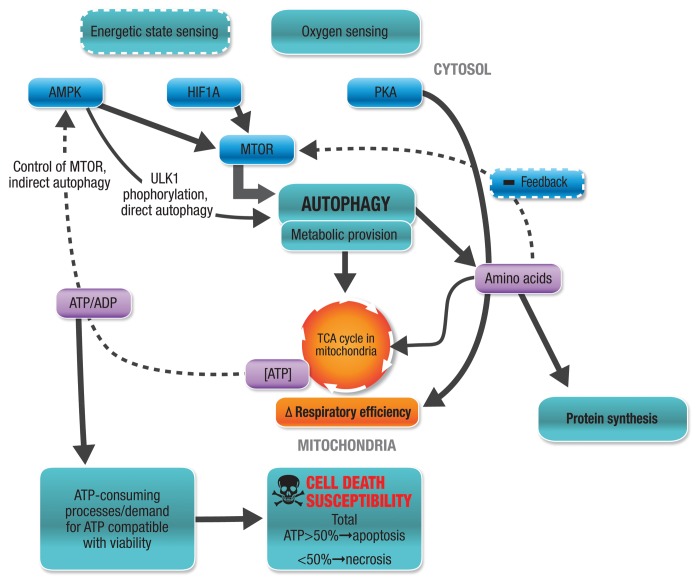
How does the cell’s energetic state and ATP availability connect with autophagic flux and onset of cell death? The likelihood of cell death during a stress response lies, among other things, in the cell’s ability to maintain intracellular ATP levels sufficient to maintain function. It is hence no surprise that ATP-generating systems as well as ATP-consuming processes affect susceptibility to cell death. The autophagic pathway is integrated in a complex feedback network on multiple levels to assure substrate availability and to assure maintenance of energetic state so that metabolic stress may be overcome and viability maintained (Fig. 4). Autophagic activity therefore contributes to the control of metabolite levels to allow sufficient ATP generation to take place to meet the cellular metabolic demands compatible with viability and function. In this section, the role of ATP as well as the balance of ATP generation and ATP consumption on the induction and execution of cell death will be discussed.
Figure 4. Relationship between energetic sensing, autophagic activity, ATP and cell death. Autophagic activity forms an integral part of a homeostatic metabolic feedback system that ensures the control of metabolite levels in order to allow sufficient ATP generation to take place that meets the current cellular metabolic demands to comply with cell viability.
The significance of the metabolic role of increased autophagic flux on susceptibility to cell death can only fully be understood in the context of the cellular energetic balance and cell death, the direct relationship between autophagy and onset of apoptosis or necrosis, as well as the more indirect relationship with necrosis via the ATP-dependent shift between apoptosis and necrosis. With the improvement in methodology, it became clear that there are many examples where biochemical or morphological hallmarks of more than one cell death subroutine can be shared within the same cell. Cell death is a highly dynamic response that may manifest in a spectrum of morphological overlap.7
Historically, cell death has primarily been classified as apoptotic, autophagic and necrotic. Often, intermediate forms are seen, indicating the dynamic nature of cell death. Apoptosis is morphologically characterized by chromatin condensation (pyknosis) and fragmentation (karyorrhexis), rounding up of the cell, and the retraction of pseudopods. The cell membrane remains intact, although the change in membrane polarity leads to phosphatidylserine exposure on the outer membrane. Necrosis was originally considered as an accidental cell death that operates rather uncontrolled and passively as a consequence of physical or chemical stress. However, recent data indicate that necrosis execution may be finely controlled by a network of signaling pathways.53 The term necroptosis has since been proposed to indicate this regulated form of necrotic cell death. RIPK1 [receptor (TNFRSF)-interacting serine-threonine kinase 1] has been implicated in many conditions leading to necrotic cell death in mammalian systems.54 Morphologically, necrosis is characterized by a loss in membrane integrity with subsequent rupture as well as cytoplasmic and organellar swelling. The cellular swelling prior to necrosis onset has been termed oncosis, derived from “oncos,” meaning swelling.55 Importantly, a rapid drop in ATP below 50% has been implicated in necrotic cell death.56 Most often, mitochondrial injury leads to necrosis, which can be rescued by restoring ATP synthesis through inducing glycolysis.56 Uncoupling of mitochondria leads to their depolarization, loss of ATP and subsequent necrosis of the cell, which can be rescued by preventing the uncoupling and thereby increasing ATP. Hence ATP availability is central in the induction and execution of necrosis. The cell membrane pumps, as well as lysosomal ATP-dependent transporters are constituents of the major cellular ATP-consuming processes, indicating a relationship between ATP availability, ATPases and cytoplasmic/organellar swelling.57 It has therefore been suggested that the signaling for necrosis takes place long before its manifestation in a morphologically visible necrotic cell death subroutine, involving a so called point-of-no-return (PONR).58 In addition, an ATP-dependent relationship between apoptosis and necrosis has been described. In fact, a progressive replacement of necrosis with apoptosis occurs when intracellular ATP becomes available again, in particular when ATP exceeds 50% of normal intracellular ATP stores.59 Mutations in autophagy genes partially inhibit necrosis by converging within the calpain-cathepsin pathway, which affects acidification of the cytosol.60 Spermidine, which induces autophagy, inhibits loss of membrane integrity and HMGB1 release.61 Similarly, induction of autophagy via acetic acid inhibits necrotic cell death in yeast.62
These data demonstrate very clearly that cell death is not static, and that ATP generated through increased autophagic flux may affect the onset of both apoptotic and necrotic cell death. In order to better understand the fundamental role of autophagy in susceptibility to cell death, it is important to remind ourselves that the term “programmed,” does not mean that the cellular fate per se is “pre-written,” but that merely the capacity exists to induce programmed cell death if present and past conditions as well as intra- and extracellular conditions are favorable for induction of cell death. That these conditions can change and rapidly influence the execution of cell death is made clear by the PONR in cellular injury, a concept that was introduced already more than 50 y ago.58 The PONR clearly indicates the dynamic behavior of cell death and reveals the identity of a restriction point for cell death as a time point when a cell is still salvageable. The PONR enables therefore the distinction of induction and execution of cell death, of the cell death process from the cell death endpoint, and therefore the difference between a living and an already dead cell. A number of parameters, such as mitochondrial membrane permeabilization, caspase activation, the dissipation of mitochondrial transmembrane potential ΔΨ or cytochrome c interactions have all been considered as the PONR in models of programmed cell death.63-67 In models of necrosis, lysosome permeabilization and subsequent plasma membrane rupture have been described as the PONR.68 The magnitude of autophagic activity may influence the position of the PONR due to its role in metabolite sensing, substrate provision and response integration (Fig. 4), thereby controlling the cellular energetic state and death onset.8
Autophagy: The Matching of Efflux Rate with ATP Demand that Controls Cell Death
In light of the specific role of autophagy in responding to shortage of metabolite substrate, it becomes clear that the magnitude of autophagic flux determines the extent of metabolite contribution. Especially in the metabolic context it is crucial to fully anticipate the fundamental role of autophagic flux. Both how many metabolites can be generated and how energetically efficient ATP consumption can be integrated, are dependent on autophagic flux. Based on the definition of autophagic flux that describes the rate of turnover of the autophagosome pool in steady-state where the size of the autophagosome pool remains constant, it becomes evident that a rate of synthesis or degradation can be assessed only by measuring multiple time points and by completely inhibiting autophagosomal-lysosomal fusion.69 Although data on baseline autophagic flux of various tissues and their metabolic capacity through autophagy are limited, from hepatocytes we know that the autophagosomal half-life is approximately 8 min, with a 25% net loss of protein following a 24 h fasting period.29 Importantly, especially when oxygen availability is a limiting factor, autophagy may become a very suitable provider of metabolites. Hepatic oxidation of 900 mmol amino acids would generate 5.8 mol ATP per O2 molecule utilized.70 When comparing this substrate utilization and oxygen efficiency with that of glucose or palmitic acid, we find to our surprise that amino acid oxidation has stoichiometrically an almost identical oxygen sparing effect to glucose oxidation. Moreover, when assessing the ATP yield per mol O2, an 11% increase of energy production is achieved through amino acid oxidation when compared with fatty acid oxidation.71 This may be of particular advantage in conditions that are metabolically perturbed through compromised oxygen availability. These findings also support the argument that autophagy indeed makes energetic sense in providing metabolites, by improving energy efficiency. During autophagy, the cell conserves ATP by decreasing general protein synthesis in a linear fashion, proportional to the degree of flux change that is increased by autophagy, while ensuring the expression of proteins required for execution of autophagy as well as proteins that are critical in adaptation to starvation.72-74
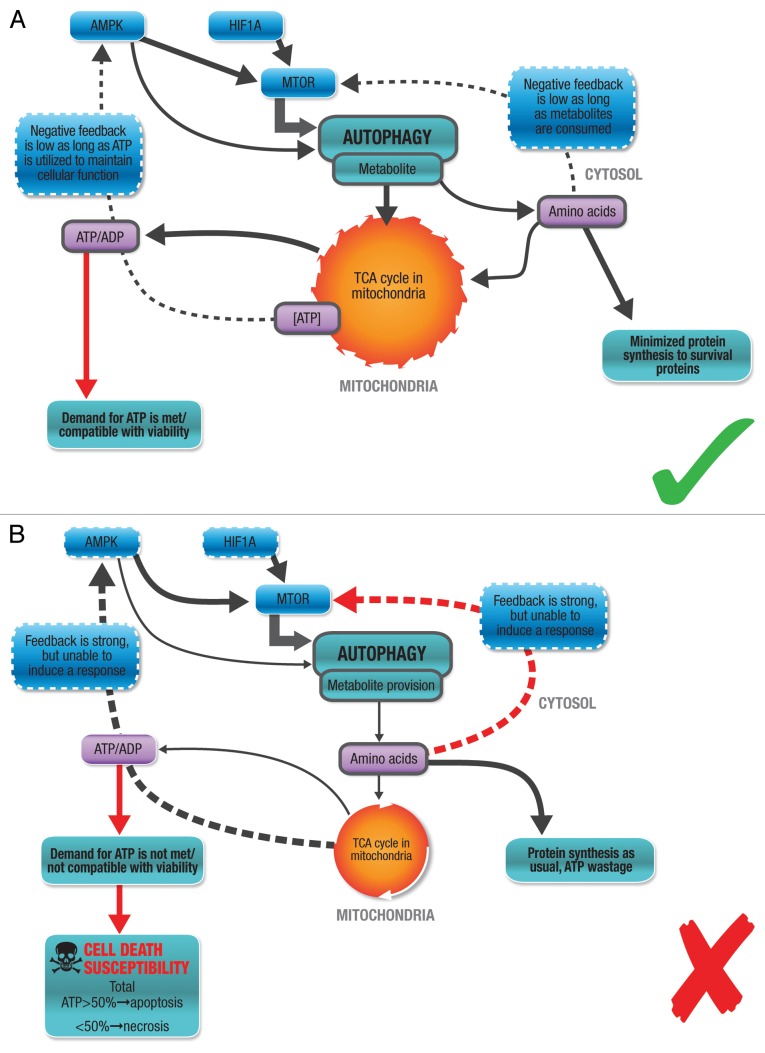
The autophagic machinery itself requires ATP levels to be sufficient, especially for successful initiation, the first step in the autophagic pathway.29,75 An ATP reduction by 30–50% decreases autophagic volume by 70%, suggesting that a minimal energetic threshold must be maintained for the cell to respond adequately through autophagy. The maintenance of the proton electrochemical gradient across the lysosomal membrane is another major metabolic expense that is required for functional autophagy.76 Hence, the higher the autophagic activity, the higher is the ATP cost to maintain elevated levels of autophagic flux. This might be one explanation why highly proliferative cells become metabolically more volatile, as autophagic activity is high as well as the cell’s metabolic demand. We know that the efflux of the amino acids that result from autophagic degradation, such as leucine and isoleucine, is mediated by vacuolar effluxers such as Atg22, indicating a high degree of specificity of the release of metabolites.77 However, although this stage is often seen as the final step of autophagy, the metabolic feedback loop is only completed when these metabolites are oxidized and contribute to the cellular ATP pool (Fig. 1). In the context of autophagy as part of an ATP supply chain and cellular function as part of an ATP demand system, the control of cellular energetics through tuning autophagic flux becomes clear: Increased autophagic activity is compatible with cellular metabolism and function when metabolic supply (efflux rate) matches cellular ATP demand at a given time of insult or stress, preserving viability (Fig. 5A). If the insult is mild, if residual oxygen tension is available and the increase in ATP demand is mild to moderate, no cell death is induced. In contrast, decreased autophagic flux or an impaired autophagic machinery is not compatible with cellular metabolism and function, as efflux rate does not meet the metabolic demands, leading to net loss in ATP (Fig. 5B). The negative feedback is primarily brought about through low metabolite (leucine) levels, low ATP/ADP ratio and low oxygen tension. If the stress or insult is severe, if oxygen availability is limited and the increased ATP demand is severe, the decrease in ATP induces apoptotic or necrotic cell death, depending on specific intracellular ATP levels (Fig. 5B).
Figure 5. Autophagy correlates with cellular metabolism. (A) Increased autophagic activity is compatible with cellular metabolism and function. Metabolic supply (efflux rate) matches cellular ATP demand at a given time of insult or stress, maintaining viability. The insult is mild, residual oxygen tension is available and the increase in ATP demand is mild to moderate. No cell death is induced. (B) Impaired autophagic machinery is not compatible with cellular metabolism and function. A delayed response, a decrease or inhibition of autophagic flux at a given time of insult or stress, leads to cell death. The negative feedback is primarily brought about by low metabolite (leucine) levels, low ATP/ADP ratio and low oxygen tension. Metabolic supply (efflux rate) does not match cellular ATP demand. The stress or insult is severe, oxygen availability is limited and the increased ATP demand is severe. The decrease in ATP induces apoptotic or necrotic cell death.
Autophagy and the Cellular Environment: Looking Back to Look Forward
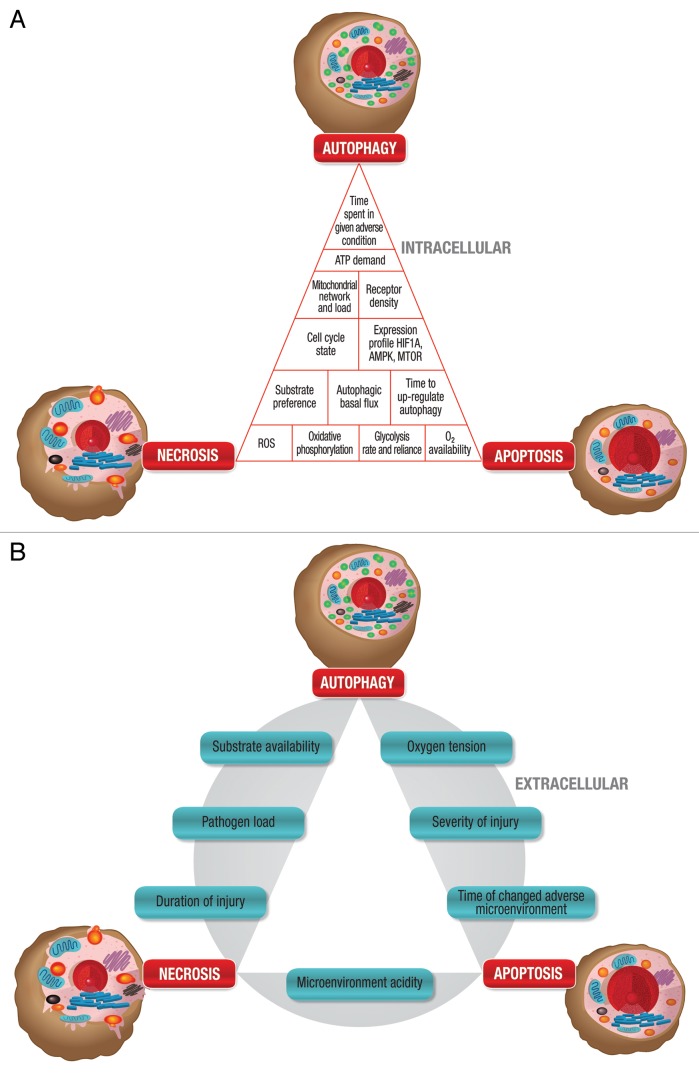
In the past 20 years, we have seen remarkable changes in the field of cell death. Although we know the machinery of cell death and the morphology it presents, we are only beginning to understand the complexity of the molecular origin and crosstalk that has led to the manifestation of a specific cell death subroutine. Very often we do not consider the context of a cell death subroutine and the contribution of both intracellular and extracellular parameters (Fig. 6A and B) that form the backdrop for autophagy. How many years has the cell spent in stress, brought about by metabolic perturbation (Fig. 6A)? How long has it been utilizing substrates that are not preferred metabolites? How long has its autophagic flux been slightly higher due to a mild hypoxic environment, or slightly lower? How much toxic protein aggregate has accumulated and how is its mitochondrial fission or fusion rate disturbed as a result of that? It becomes clear that in order to understand and predict cell death and cell survival, and in order to unravel the molecular mechanisms that result in morphological overlap of cell death, we will have to take into account the metabolic status and history of the cells, as well as the matrices in which they are found (Fig. 7). The impact of these parameters on autophagic activity and its relevance to cell death become particularly important when assessing the role of autophagy in conditions characterized by overnutrition or aging. For example, autophagy plays a role in adipogenesis, is upregulated in adipose tissue in human obesity and stimulated in β cells, deriving from a high energy status induced by a high-fat diet with a decrease in islet ATP production.78-80 Transcriptional downregulation of autophagy during aging enhances proteotoxic stress and increases apoptotic cell death.81-83 The fine balance between amino acid availability and autophagic activity affects not only cell death susceptibility but also influences longevity.84,85
Figure 6. Connections between autophagy, apoptosis and necrosis. (A) The intracellular condition (inner triangle) dynamically drives cell death. Each block contributes to cellular robustness and death susceptibility. (B) The extracellular microenvironment (outer circle) dynamically drives cell death. Each block contributes to robustness and, hence, susceptibility.
Figure 7. The intracellular state and the extracellular microenvironment dynamically drive cell death and together contribute to cellular robustness and cell death susceptibility. Due to the molecular crosstalk, a mixed morphology between autophagy, apoptosis and necrosis can be observed, depending on the individual contribution and magnitude of the intracellular and extracellular parameters.
Cells die in the context of their environment.86 Starting in the 1950s, experimentally derived data started to shape the idea of cells being able to self-destruct in a programmed and regulated fashion, if the microenvironment provided the signals to do so.87-89 Glücksmann already noted at that time that the cause of cell death is related to a combination of “imprecise induction and proliferation stimuli with a limited adaptability,” indicating the importance of a timely cellular response to a changing microenvironment.90 We know that a timely autophagic response to metabolic stress is crucial for a successful stress response; however, we do not consequently follow this line of evidence to the cellular susceptibility to undergo cell death. Glücksmann pointed out that the location of cells, a free surface vs. the restriction of movement by close packaging, contributes to morphogenetic sculpturing.90 A center to margin differentiation of cell death subroutines seems a common feature in the manifestation of cell death in tissues such as central pyknotic cells in retina development, the interdigital cell death during limb formation and the central occurrence of cell death in hypoxic tumor or infarcted tissue.90 The fact that cells die while attempting division indicates that the microenvironment plays a major role in cell death control. Back then it was hypothesized that so-called “maintenance factors” must exist that allow normally degenerating cells to survive under different conditions. We now propose that autophagic activity plays a fundamental role in the homeostasis of these “maintenance factors” and that autophagic flux and response time determine the robustness of a tissue in providing adequate “maintenance factors” during changing conditions or metabolic demands.
Abundant cell death, be it phylogenetic, histogenetic or morphogenetic in nature, is a fundamental part of early development.91 It becomes increasingly evident that autophagic activity is controlled similarly in a developmentally regulated manner and that it plays an essential role during development.92,93 Especially during embryogenesis, a rapid change from quiescence to enhanced metabolism facilitates proliferation, differentiation, migration and death. These cellular responses are highly dynamic and governed by both the microenvironment and the cells’ inherent genetic program. In the developing limb, timing and signaling gradients and resulting pattern formation is most evident. Gradients of signaling by SHH (sonic hedgehog) and WNT5A govern predominantly cellular behavior.94 Autophagy may negatively regulate WNT signaling, providing a potential molecular connection between metabolic sensing, morphogenesis and tissue sculpting.95 A relationship also exists between AMBRA1, a positive regulator of BECN1-dependent autophagy, and neuronal cell survival during the development of the nervous system, indicating a clear role of autophagy in cell fate during development.96 Moreover, it has recently been demonstrated that ULK3 expression levels correlate with SHH tissue activity in the fetal brain.97 These data suggest a role for autophagy in sensitizing cell death, where cell numbers need to be adjusted or structures deleted.
A common denominator in developmental cell death is an imbalance between ATP supply and demand. The demand may be driven by increased proliferation or a decrease in supply through, for instance, ischemic injury. As demand for ATP can be regulated through autophagy, activity of autophagy can determine whether supply and demand are in equilibrium. A close relationship exists between availability of growth factors, autophagic activity and the maintenance of ATP production from catabolism of autophagically generated substrates.98 Can a role of autophagic flux governed by the microenvironment be attributed to, for example, the waves of necrosis that sweep along the mesoderm during the sculpturing of the chick embryo limb? Can autophagic activity be part of the “epigenetic control of morphogenetic death”?91 We know that loss of interdigital cells during murine limb development can occur by cell death resembling necrosis, when caspases are inhibited.99 These findings suggest that another location of control for the regulation of onset of cell death exists, and alternative mechanisms have been proposed.100 In 1988 the experiment performed by Saunders, identifying an intrinsic “death clock” in the posterior necrotic zone, was taken one step further in terms of control of death, as it was found that cell death was reduced when explants were taken from younger donors. It was then formulated that control of cell death resides “in both the cellular environment and the developmental history of the cells.”101,102
We most often achieve cell death when tissues fold or invaginate, hence, when metabolic gradients are profoundly enhanced, leading to regions of hypoxia. When oxygen becomes limited to 0.1–3%, a fine balance exists between maintaining necessary cellular respiration and simultaneously limiting the accumulation of reactive oxygen species. Autophagic activity controls both metabolite substrate availability and mitochondrial autophagy. HIF1A [hypoxia inducible factor 1, α subunit (basic helix-loop-helix transcription factor)] upregulates autophagy of mitochondria, requiring HIF1A-dependent expression of BNIP3, ATG5 and BECN1.103 Hypoxia-induced autophagy is thought of as a survival response that stabilizes cell viability under adverse conditions.104 However, the severity of hypoxia and the concomitant metabolite availability play a crucial role in determining whether the cellular response will be successful.105 Insulin, for example, is important for glucose availability, but also for autophagy regulation due to the PI 3-kinase and AKT-mediated activation of MTOR. Insulin negatively regulates autophagy and suppresses its activity also in insulin resistance.106 We therefore need to consider to what extent the cell has become resistant to insulin, for how long it has been exposed to a hyperglycemic environment, and to what extent that has affected basal autophagic activity and autophagic responsiveness. Moreover, if autophagic metabolite generation provides ATP, cells compete for a necessary ATP availability that is compatible with viability. Control of autophagic flux is used to adjust the numbers of developing cells. Similarly, fetal neurons compete for limited amounts of nerve growth factor, with only some being able to escape induction of apoptosis by growth factor withdrawal.107 Also, the acidity of the microenvironment that may be brought about by an increase in anaerobic glycolysis has a profound effect on autophagic activity. Lower pH not only decreases basal autophagy, but also suppresses starvation-induced autophagy, which in turn sensitizes to cell death.108 Continued progress on the methodology of the assessment of intra- and extracellular parameters (Fig. 6A and B; Fig. 7) and quantification of autophagic flux will undoubtedly provide insights that delineate the exact conditions that favor autophagy to contribute metabolically and to affect cell death.
Summary and Future Outlook
Taken together, it becomes clear that the autophagic pathway is densely networked with the cell’s metabolism. Upstream, it plays a crucial role in metabolic sensing by signaling through the AMPK, MTOR and PKA pathways, thereby tuning cellular energy demands with autophagic flux, resulting in improved energy efficiency and respiration. Cell viability is likely to be maintained not only when the efflux rate matches cellular ATP demand at a time of insult or stress, but also if autophagically-generated glucogenic and ketogenic amino acids are enabled to fuel the TCA cycle at multiple entry points. It is therefore most crucial to assess equally the cell’s history, the type and severity of stress and how these may affect metabolic sensing and ATP-generating pathways, and ultimately determine the energetic state and function of the cell. This will require us to enlarge the footprint of the autophagic pathway and to integrate it into a cellular energetic feedback loop. Continued progress on methodology and a combined assessment and quantification of autophagic flux with the measurement of metabolite pool sizes, the cell’s substrate preference and its reliance on oxidative phosphorylation will be required. An increased reporting on oxygen availability and mitochondrial fission-fusion rate in conjunction with ATP availability and ATP-consuming processes in the context of the cell’s intra- and extracellular environment will undoubtedly provide insights that deepen our understanding on the variability of autophagy and cell death susceptibility. In doing so, this approach may result in the elucidation of the threshold that controls cell death and cell survival, providing a novel means to exploit the autophagic machinery for therapeutic purposes.
Glossary
Abbreviations:
- ATG
autophagy-related
- PONR
point-of-no-return
- TCA
tricarboxylic acid
Disclosure of Potential Conflicts of Interest
I confirm that this manuscript is not under consideration elsewhere and has not previously been published. I also confirm that all authors have read and approved this manuscript. There is no potential conflict of interest whatsoever regarding the content of this manuscript.
Acknowledgments
This work was supported by the National Research Foundation (NRF) and Medical Research Council (MRC), South Africa (to B.L.) and in part by funding from the NIH (NIAID) R15 AI094351-01 (to Z.Z.) and (NIGMS) GM053396 (to D.J.K).
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/25560
References
- 1.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez-Vicente M, Tallóczy Z, Wong E, Tang G, Koga H, Kaushik S, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–76. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–70. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–90. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 5.Deter RL, Baudhuin P, De Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol. 1967;35:C11–6. doi: 10.1083/jcb.35.2.C11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loos B, Engelbrecht AM. Cell death: a dynamic response concept. Autophagy. 2009;5:590–603. doi: 10.4161/auto.5.5.8479. [DOI] [PubMed] [Google Scholar]
- 8.Loos B, Genade S, Ellis B, Lochner A, Engelbrecht AM. At the core of survival: autophagy delays the onset of both apoptotic and necrotic cell death in a model of ischemic cell injury. Exp Cell Res. 2011;317:1437–53. doi: 10.1016/j.yexcr.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 9.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 10.Blackstone C, Chang C-R. Mitochondria unite to survive. Nat Cell Biol. 2011;13:521–2. doi: 10.1038/ncb0511-521. [DOI] [PubMed] [Google Scholar]
- 11.Meijer AJ. Amino acid regulation of autophagosome formation. Methods Mol Biol. 2008;445:89–109. doi: 10.1007/978-1-59745-157-4_5. [DOI] [PubMed] [Google Scholar]
- 12.Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G. Mammalian TOR: a homeostatic ATP sensor. Science. 2001;294:1102–5. doi: 10.1126/science.1063518. [DOI] [PubMed] [Google Scholar]
- 13.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–13. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kraft C, Kijanska M, Kalie E, Siergiejuk E, Lee SS, Semplicio G, et al. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 2012;31:3691–703. doi: 10.1038/emboj.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan EY. mTORC1 phosphorylates the ULK1-mAtg13-FIP200 autophagy regulatory complex. Sci Signal. 2009;2:pe51. doi: 10.1126/scisignal.284pe51. [DOI] [PubMed] [Google Scholar]
- 16.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–34. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meley D, Bauvy C, Houben-Weerts JHPM, Dubbelhuis PF, Helmond MTJ, Codogno P, et al. AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem. 2006;281:34870–9. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- 18.Matsui Y, Kyoi S, Takagi H, Hsu CP, Hariharan N, Ago T, et al. Molecular mechanisms and physiological significance of autophagy during myocardial ischemia and reperfusion. Autophagy. 2008;4:409–15. doi: 10.4161/auto.5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoki K, Zhu T, Guan K-L. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–90. doi: 10.1016/S0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 20.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–26. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–61. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–23. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam A, Lopaschuk GD. Anti-anginal effects of partial fatty acid oxidation inhibitors. Curr Opin Pharmacol. 2007;7:179–85. doi: 10.1016/j.coph.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Chang C-R, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem. 2007;282:21583–7. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- 26.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–98. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci U S A. 2011;108:10190–5. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 29.Blommaart EFC, Luiken JJFP, Meijer AJ. Autophagic proteolysis: control and specificity. Histochem J. 1997;29:365–85. doi: 10.1023/A:1026486801018. [DOI] [PubMed] [Google Scholar]
- 30.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–8. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortimore GE, Pösö AR, Kadowaki M, Wert JJ., Jr. Multiphasic control of hepatic protein degradation by regulatory amino acids. General features and hormonal modulation. J Biol Chem. 1987;262:16322–7. [PubMed] [Google Scholar]
- 32.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 33.Mortimore GE, Pösö AR, Lardeux BR. Mechanism and regulation of protein degradation in liver. Diabetes Metab Rev. 1989;5:49–70. doi: 10.1002/dmr.5610050105. [DOI] [PubMed] [Google Scholar]
- 34.Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–6. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- 35.Harris RA, Goodwin GW, Paxton R, Dexter P, Powell SM, Zhang B, et al. Nutritional and hormonal regulation of the activity state of hepatic branched-chain α-keto acid dehydrogenase complex. Ann N Y Acad Sci. 1989;573:306–13. doi: 10.1111/j.1749-6632.1989.tb15007.x. [DOI] [PubMed] [Google Scholar]
- 36.Ohsaki Y, Cheng J, Fujita A, Tokumoto T, Fujimoto T. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol Biol Cell. 2006;17:2674–83. doi: 10.1091/mbc.E05-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–5. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol. 2012;2012:282041. doi: 10.1155/2012/282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–24. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 41.Gottlieb RA, Mentzer RM., Jr. Autophagy during cardiac stress: joys and frustrations of autophagy. Annu Rev Physiol. 2010;72:45–59. doi: 10.1146/annurev-physiol-021909-135757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buss SJ, Muenz S, Riffel JH, Malekar P, Hagenmueller M, Weiss CS, et al. Beneficial effects of Mammalian target of rapamycin inhibition on left ventricular remodeling after myocardial infarction. J Am Coll Cardiol. 2009;54:2435–46. doi: 10.1016/j.jacc.2009.08.031. [DOI] [PubMed] [Google Scholar]
- 43.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 44.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, et al. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin S, White E. Role of autophagy in cancer: management of metabolic stress. Autophagy. 2007;3:28–31. doi: 10.4161/auto.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schiaffino S, Mammucari C, Sandri M. The role of autophagy in neonatal tissues: just a response to amino acid starvation? Autophagy. 2008;4:727–30. doi: 10.4161/auto.6143. [DOI] [PubMed] [Google Scholar]
- 48.Uchiyama Y, Shibata M, Koike M, Yoshimura K, Sasaki M. Autophagy-physiology and pathophysiology. Histochem Cell Biol. 2008;129:407–20. doi: 10.1007/s00418-008-0406-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 51.Kanamori H, Takemura G, Maruyama R, Goto K, Tsujimoto A, Ogino A, et al. Functional significance and morphological characterization of starvation-induced autophagy in the adult heart. Am J Pathol. 2009;174:1705–14. doi: 10.2353/ajpath.2009.080875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong H, Czaja MJ. Regulation of lipid droplets by autophagy. Trends Endocrinol Metab. 2011;22:234–40. doi: 10.1016/j.tem.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kepp O, Galluzzi L, Lipinski M, Yuan J, Kroemer G. Cell death assays for drug discovery. Nat Rev Drug Discov. 2011;10:221–37. doi: 10.1038/nrd3373. [DOI] [PubMed] [Google Scholar]
- 54.Festjens N, Vanden Berghe T, Vandenabeele P. Necrosis, a well-orchestrated form of cell demise: signalling cascades, important mediators and concomitant immune response. Biochim Biophys Acta. 2006;1757:1371–87. doi: 10.1016/j.bbabio.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 55.Majno G, Joris I. Apoptosis, oncosis, and necrosis. An overview of cell death. Am J Pathol. 1995;146:3–15. [PMC free article] [PubMed] [Google Scholar]
- 56.Lemasters JJ, Nieminen AL, Qian T, Trost LC, Elmore SP, Nishimura Y, et al. The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim Biophys Acta. 1998;1366:177–96. doi: 10.1016/S0005-2728(98)00112-1. [DOI] [PubMed] [Google Scholar]
- 57.Buttgereit F, Brand MD. A hierarchy of ATP-consuming processes in mammalian cells. Biochem J. 1995;312:163–7. doi: 10.1042/bj3120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Majno G, La Gattuta M, Thompson TE. Cellular death and necrosis: chemical, physical and morphologic changes in rat liver. Virchows Arch Pathol Anat Physiol Klin Med. 1960;333:421–65. doi: 10.1007/BF00955327. [DOI] [PubMed] [Google Scholar]
- 59.Leist M, Single B, Castoldi AF, Kühnle S, Nicotera P. Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis. J Exp Med. 1997;185:1481–6. doi: 10.1084/jem.185.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCall K. Genetic control of necrosis - another type of programmed cell death. Curr Opin Cell Biol. 2010;22:882–8. doi: 10.1016/j.ceb.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–14. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 62.Schauer A, Knauer H, Ruckenstuhl C, Fussi H, Durchschlag M, Potocnik U, et al. Vacuolar functions determine the mode of cell death. Biochim Biophys Acta. 2009;1793:540–5. doi: 10.1016/j.bbamcr.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 63.Galluzzi L, Morselli E, Kepp O, Kroemer G. Targeting post-mitochondrial effectors of apoptosis for neuroprotection. Biochim Biophys Acta. 2009;1787:402–13. doi: 10.1016/j.bbabio.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 64.Leber B, Lin J, Andrews DW. Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis. 2007;12:897–911. doi: 10.1007/s10495-007-0746-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galluzzi L, Zamzami N, de La Motte Rouge T, Lemaire C, Brenner C, Kroemer G. Methods for the assessment of mitochondrial membrane permeabilization in apoptosis. Apoptosis. 2007;12:803–13. doi: 10.1007/s10495-007-0720-1. [DOI] [PubMed] [Google Scholar]
- 66.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 67.Golbs A, Heck N, Luhmann HJ. A new technique for real-time analysis of caspase-3 dependent neuronal cell death. J Neurosci Methods. 2007;161:234–43. doi: 10.1016/j.jneumeth.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 68.Giusti C, Luciani MF, Klein G, Aubry L, Tresse E, Kosta A, et al. Necrotic cell death: From reversible mitochondrial uncoupling to irreversible lysosomal permeabilization. Exp Cell Res. 2009;315:26–38. doi: 10.1016/j.yexcr.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 69.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brosnan JT. Glutamate, at the interface between amino acid and carbohydrate metabolism. J Nutr. 2000;130(Suppl):988S–90S. doi: 10.1093/jn/130.4.988S. [DOI] [PubMed] [Google Scholar]
- 71.Loos B, Lochner A, Engelbrecht AM. Autophagy in heart disease: a strong hypothesis for an untouched metabolic reserve. Med Hypotheses. 2011;77:52–7. doi: 10.1016/j.mehy.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 72.Blommaart EFC, Luiken JJFP, Blommaart PJE, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem. 1995;270:2320–6. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 73.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–62. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 74.Kuma A, Mizushima N. Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Semin Cell Dev Biol. 2010;21:683–90. doi: 10.1016/j.semcdb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 75.Schellens JPM, Vreeling-Sindelárová H, Plomp PJAM, Meijer AJ. Hepatic autophagy and intracellular ATP. A morphometric study. Exp Cell Res. 1988;177:103–8. doi: 10.1016/0014-4827(88)90028-6. [DOI] [PubMed] [Google Scholar]
- 76.Van Dyke RW. Proton pump-generated electrochemical gradients in rat liver multivesicular bodies. Quantitation and effects of chloride. J Biol Chem. 1988;263:2603–11. [PubMed] [Google Scholar]
- 77.Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell. 2006;17:5094–104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–32. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106:19860–5. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kovsan J, Blüher M, Tarnovscki T, Klöting N, Kirshtein B, Madar L, et al. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab. 2011;96:E268–77. doi: 10.1210/jc.2010-1681. [DOI] [PubMed] [Google Scholar]
- 81.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–38. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2010;107:14164–9. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Caramés B, Taniguchi N, Otsuki S, Blanco FJ, Lotz M. Autophagy is a protective mechanism in normal cartilage, and its aging-related loss is linked with cell death and osteoarthritis. Arthritis Rheum. 2010;62:791–801. doi: 10.1002/art.27305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alvers AL, Fishwick LK, Wood MS, Hu D, Chung HS, Dunn WA, Jr., et al. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell. 2009;8:353–69. doi: 10.1111/j.1474-9726.2009.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grandison RC, Piper MDW, Partridge L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature. 2009;462:1061–4. doi: 10.1038/nature08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zakeri Z, Lockshin RA. Cell death during development. J Immunol Methods. 2002;265:3–20. doi: 10.1016/S0022-1759(02)00067-4. [DOI] [PubMed] [Google Scholar]
- 87.Lockshin RA, Williams CM. Programmed cell death. II. Endocrine potentiation of the breakdown of the intersegmental muscles of silkmoths. J Insect Physiol. 1964;10:643–9. doi: 10.1016/0022-1910(64)90034-4. [DOI] [Google Scholar]
- 88.Saunders JW, Jr., Gasseling MT, Cairns JM. Effect of implantation site on the development of an implant in the chick embryo. Nature. 1955;175:673–4. doi: 10.1038/175673a0. [DOI] [PubMed] [Google Scholar]
- 89.Kerr JFR, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–57. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Glücksmann A. Cell death in normal development. Arch Biol (Liege) 1965;76:419–37. [PubMed] [Google Scholar]
- 91.Saunders JW., Jr. Death in embryonic systems. Science. 1966;154:604–12. doi: 10.1126/science.154.3749.604. [DOI] [PubMed] [Google Scholar]
- 92.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15:344–57. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Juhász G, Csikós G, Sinka R, Erdélyi M, Sass M. The Drosophila homolog of Aut1 is essential for autophagy and development. FEBS Lett. 2003;543:154–8. doi: 10.1016/S0014-5793(03)00431-9. [DOI] [PubMed] [Google Scholar]
- 94.Towers M, Wolpert L, Tickle C. Gradients of signalling in the developing limb. Curr Opin Cell Biol. 2012;24:181–7. doi: 10.1016/j.ceb.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 95.Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781–90. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- 96.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–5. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 97.Maloverjan A, Piirsoo M, Michelson P, Kogerman P, Østerlund T. Identification of a novel serine/threonine kinase ULK3 as a positive regulator of Hedgehog pathway. Exp Cell Res. 2010;316:627–37. doi: 10.1016/j.yexcr.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 98.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–48. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 99.Chautan M, Chazal G, Cecconi F, Gruss P, Golstein P. Interdigital cell death can occur through a necrotic and caspase-independent pathway. Curr Biol. 1999;9:967–70. doi: 10.1016/S0960-9822(99)80425-4. [DOI] [PubMed] [Google Scholar]
- 100.Yuan J, Kroemer G. Alternative cell death mechanisms in development and beyond. Genes Dev. 2010;24:2592–602. doi: 10.1101/gad.1984410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Saunders JW, Jr., Gasseling MT. Cellular death in morphogenesis of the avian wing. Dev Biol. 1962;5:147–78. doi: 10.1016/0012-1606(62)90008-8. [DOI] [PubMed] [Google Scholar]
- 102.Brewton RG, MacCabe JA. Ectodermal influence on physiological cell death in the posterior necrotic zone of the chick wing bud. Dev Biol. 1988;126:327–30. doi: 10.1016/0012-1606(88)90142-X. [DOI] [PubMed] [Google Scholar]
- 103.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 104.Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouysségur J, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–81. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mazur M, Miller RH, Robinson S. Postnatal erythropoietin treatment mitigates neural cell loss after systemic prenatal hypoxic-ischemic injury. J Neurosurg Pediatr. 2010;6:206–21. doi: 10.3171/2010.5.PEDS1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu HY, Han J, Cao SY, Hong T, Zhuo D, Shi J, et al. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484–92. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–54. doi: 10.1016/S0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 108.Xu T, Su H, Ganapathy S, Yuan ZM. Modulation of autophagic activity by extracellular pH. Autophagy. 2011;7:1316–22. doi: 10.4161/auto.7.11.17785. [DOI] [PMC free article] [PubMed] [Google Scholar]