Abstract
Autophagy is induced by many cytotoxic stimuli but it is often unclear whether, under specific conditions, autophagy plays a prosurvival or a prodeath role. To answer this critical question we developed a novel methodology that employs automated live microscopy and image analysis to measure autophagy and apoptosis simultaneously in single cells. We used this approach to perform a systems-level analysis of pathway dynamics for both autophagy and apoptosis. We found that induction of autophagy in response to different stimuli is uniformly unimodal; in contrast, cells induce apoptosis in an all-or-none bimodal fashion. By tracking the fate of single cells we found that autophagy precedes apoptosis, and that within the same population apoptosis is delayed in cells that mount a stronger autophagy response. Inhibition of autophagy by knocking down ATG5 promoted apoptosis, thus confirming that autophagy plays a protective role. We anticipate that our single-cell approach will be a powerful tool for gaining a quantitative understanding of the complex regulation of autophagy, its influence on cell fate decisions and its relationship with other cellular pathways.
Keywords: autophagy, single cell, live cell, apoptosis, crosstalk, image analysis, cell tracking
Introduction
Autophagy, a lysosome-dependent intracellular degradative process, is an essential part of cellular homeostasis and a protective mechanism against a variety of diseases, including neurodegeneration, cancer and aging.1 Autophagy occurs constitutively at the basal level but can be readily induced in response to both physiological2 and pathological stimuli.1 Thus far, induction of autophagy has been studied primarily at the population level. However, even cultured isogenic cells often exhibit large cell-to-cell variability in their response to extracellular cues, such as changes in nutrient and growth factor availability or pharmacological treatments. Such large variability makes the average of the population insufficient or even misleading to represent individual cell responses. On the other hand, at a given time point and with a given treatment, the cell-to-cell variability in a population of cells results in a distribution of responses, and the shape of such distribution can be informative for understanding the nature of the underlying cellular pathways. For example, signaling networks that make binary cell fate decisions (such as life vs. death) lead to bimodal distribution of cellular responses,3,4 whereas networks designed for broad dynamic ranges and proportionate response give rise to unimodal distributions.5
More importantly, the single-cell approach can be used to directly investigate the role of autophagy in decision making by individual cells. Autophagy has a complex crosstalk with cell death.6 Although under many conditions autophagy is considered a survival mechanism, under some circumstances, especially when apoptosis is blocked, it may function as an alternative form of programmed cell death.7 Since many cytotoxic reagents trigger both autophagy and apoptosis, it often remains a matter of intense debate whether under particular conditions autophagy contributes to cell death or survival. Single-cell approach can provide a more accurate way to monitor the interplay of these two pathways in real time and better clarify how they jointly contribute to cell fate decisions.
In this study, we present a quantitative single-cell analysis approach comprised of high-throughput imaging of fluorescent reporters over time and subsequent automated single-cell analysis of autophagy levels and cell death. We validated this method in a diverse set of cell lines (H4 human neuroglioma, L929 mouse connective tissue cells and HeLa human adenocarcinoma) and in response to different proautophagy and prodeath stimuli. Finally, we used this approach to dissect the basic characteristics of autophagy dynamics in single cells in response to starvation, inactivation of the MTOR pathway by small molecule inhibitors, or induction of autophagy and cell death by proapoptotic stimuli. Our data demonstrated that deprivation of glucose or serum both induce autophagy response that forms unimodal distributions, but only the former is accompanied by apoptosis. Inhibition of MTOR with rapamycin or Torin 1 also leads to unimodal distribution of autophagic responses, indicating a graded pathway with a broad dynamic range. When apoptosis is induced with staurosporine (STS), autophagy is induced with faster kinetics and reaches higher levels, but the distributions remains unimodal. We further used live-cell analysis to address how STS-induced autophagy interplays with apoptosis in individual cells over time. Our data indicated that autophagy is a prosurvival mechanism in STS-treated cells. Autophagy and apoptosis are consecutively induced in the same cells; however, levels of autophagy decline prior to cell death. Our data illustrated utility of image based single-cell analysis for deciphering both autophagy dynamics as well as its influence on cell fate decisions such as cell death.
Results
Quantitative image-based single-cell analysis of autophagy and cell death
In order to simultaneously evaluate levels of autophagy and apoptosis in single cells, we created stable H4 human neuroglioma,2 HeLa human adenocarcinoma and L929 mouse connective tissue cells expressing both the autophagy marker GFP-LC3 and the histone cluster 2 (HIST2H2BE)-RFP (H2B-RFP) chromatin marker (LC3-H2B cells). Single cell clones were selected to exclude heterogeneity in expression levels due to differential incorporation of exogenous genes. All experiments were repeated in multiple clones of LC3-H2B cells.
The H2B-RFP reporter was used as both a nuclear marker necessary for automated cell identification and tracking8 and as an apoptosis marker. To identify individual cells with high fidelity, we took advantage of the well-defined boundaries of nuclei marked with H2B-RFP. A hallmark of apoptosis is the condensation and cleavage of DNA. When DNA is fluorescently labeled either directly by an intercalating dye or indirectly by histone-fluorescent protein fusion, this process can be easily observed as shrinkage and increased intensity of the nuclei, followed by their fragmentation. Therefore, we were able to use the H2B-RFP reporter as both a nuclear and an apoptosis marker. Compared with the use of a separate specific marker for apoptosis, such as loss of plasma membrane integrity, our approach requires one less fluorescent channel. This simplifies derivation of stable clones, reduces necessary perturbations to the cells, and simplifies the imaging process and image analysis. The GFP-LC3 reporter was used both as a marker of autophagy as well as to delineate cell boundaries. The procedure for automated image analysis for autophagy and apoptosis is illustrated in Figure 1.
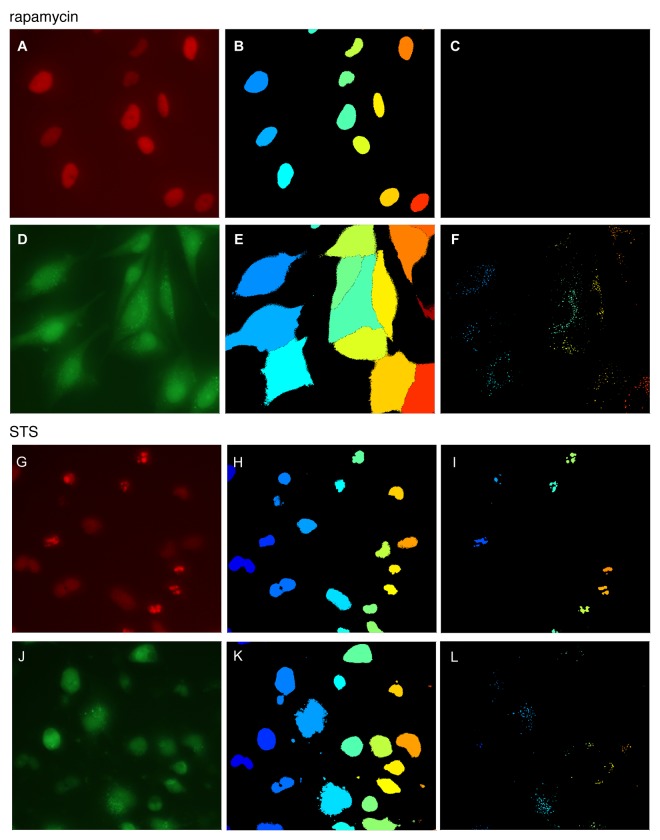
Figure 1. Image analysis of autophagy and apoptosis. H4 cells stably expressing GFP-LC3 autophagy reporter and H2B-RFP chromatin marker were treated with 20 nM rapamycin for 6 h (A–F) or 1 μM STS for 2 h (G–L). Live cells were imaged at 20× magnification and images were processed and quantified as described in text. (A and G) H2B-RFP image, (B and H) nuclear identification, (C and I) detection of apoptotic puncta; (D and J) GFP-LC3 image, (E and K) results of cell morphology segmentation, (F and L) detection of autophagic puncta.
1. Nuclei were identified by mean-filter local adaptive thresholding of nuclear marker images (Fig. 1A and B, and G and H).
2. Mean-filter local adaptive thresholding was applied to the GFP-LC3 channel (Fig. 1D and E, J and K) to separate cells from background. Nuclei identified in step 1 were superimposed on the GFP-LC3 image and watershed algorithm was applied to generate cell boundaries. This separated cells in contact and determined the morphology of individual cells. (Fig. 1E and K)
3. Punctate structures were detected in the H2B-RFP and GFP-LC3 channels by the Tophat algorithm. These correspond to DNA condensation (Fig. 1C and I) and autophagosomes or autolysosomes (Fig. 1F and L), respectively.
4. Quantitative single-cell information was extracted from segmented cells, including cell area, total expression level of both reporters, and number and intensity of puncta formed by GFP-LC3 and H2B-RFP. Puncta score for a given cell is defined as the ratio between total intensity in the puncta and the total intensity in the cell. The use of ratios minimizes influence of differences in reporter expression levels between individual cells and cell populations. Such scores for GFP-LC3 and H2B-RFP were used to report autophagic responses (autophagosome score) and apoptotic responses (cell death score), respectively. Image analysis for autophagy and cell death was validated following knockdown of MTOR and ATG5 or treatment of cells with STS, respectively (Fig. S1).
5. Consistent with high cell viability, very few H2B-RFP puncta were detected in rapamycin-treated cells (Fig. 1C), whereas many appeared after STS treatment (Fig. 1I) indicating extensive cell death. Nuclei of mitotic cells are also condensed and can occasionally be interpreted as “puncta” by this algorithm. One way to distinguish these two processes is by the number, size and morphology of the detected particles in the nuclear zone. The chromatin of mitotic cells prior to division is condensed but not fragmented, whereas cells entering death program are typically characterized by multiple small puncta. We demonstrate this in the supplementary materials (Fig. S2). Alternatively, apoptotic and mitotic cells can also be readily distinguished by the time scale of puncta formation in live-cell imaging, as illustrated in step 7.
6. In the case of live-cell imaging, multiple frames of GFP-LC3 and H2B-RFP images were segmented and analyzed. We then tracked the motion of cells and generated trajectories using an automated tracking program adapted from published programs (Fig. S3).9 Broken trajectories (for example due to a cell migrating out of the field of view) were automatically removed.
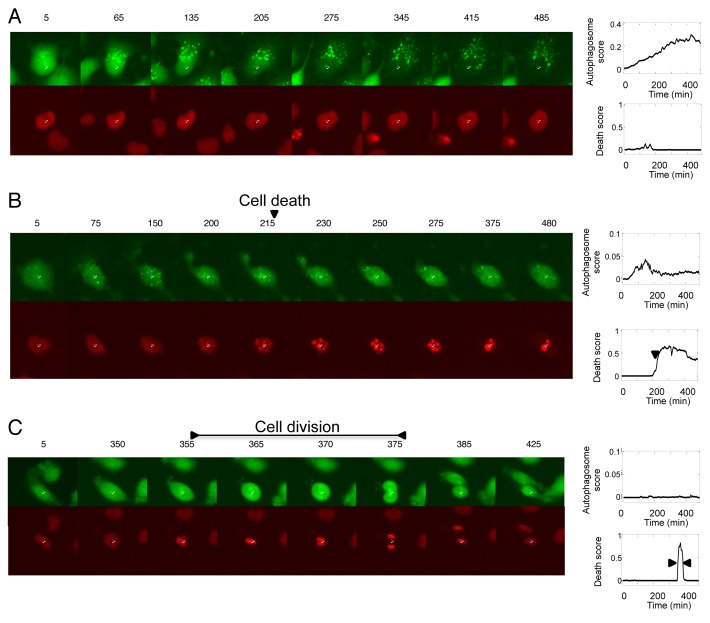
7. The expression levels and puncta scores were then plotted as a function of time for each individual cell (Fig. 2). The dynamics of H2B fragmentation score were used to distinguish between cell death (Fig. 2B) and cell division (Fig. 2C). Cell death and cell division are both characterized by formation of puncta, but as discussed in step 5 and Figure S2, the number and size of puncta are different in these two cases. Timelapse data also reveal that nuclear condensation during normal cell division is transient (10 to 30 min) as compared with the nuclear fragmentation during cell death (lasts hours after the initiation at 215 min). Therefore, in live-cell experiments in this study, cell death and cell division were distinguished by the duration of the high H2B fragmentation score.
Figure 2. Analysis of single-cell dynamics of autophagy, apoptosis and cell division in time-lapse data. H4 cells stably expressing GFP-LC3 and H2B-RFP markers were treated with (A and B) 0.5 μM STS or (C) left untreated. Images were acquired. Time after treatment (min) is labeled above each image. The white cross in each image indicates center of the nucleus of the tracked cell. Following image analysis and quantification, autophagy and cell death scores were plotted against time for individual cells. Representative examples of cells undergoing autophagy, apoptosis and cell division are presented in (A–C), respectively.
Basic characteristics of autophagy dynamics in single cells
In order to better understand the basic dynamics of autophagy as well as its relationship to cell death, we compared the distributions of autophagy and apoptosis levels induced by several conditions. We first tested starvation-induced autophagy by depriving H4 cells of serum (Fig. 3A–D; Fig. S4A) or glucose (Fig. 3E–H; Fig. S4B). We calculated the autophagosome score and the death score in single cells by dividing, respectively, the intensity of GFP-LC3 or H2B-RFP puncta in a cell by the corresponding total intensity of that cell. We then calculated the probability density functions of both scores from their single-cell measurements (Fig. 3B, D, F and H).
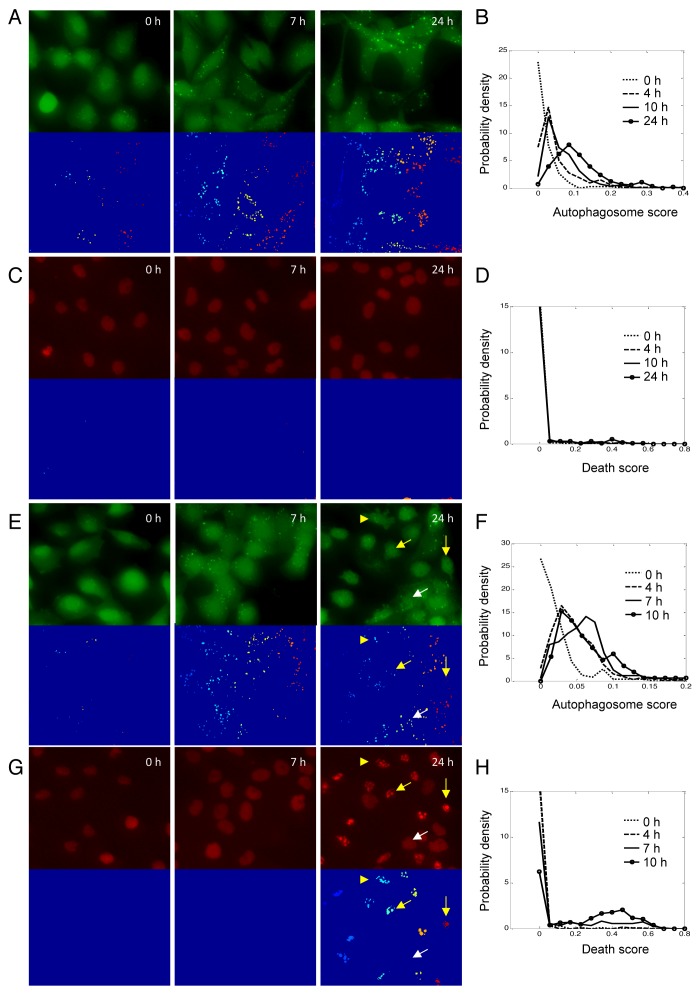
Figure 3. Autophagy and apoptotic responses in H4 cells during starvation. (A) Images of GFP-LC3 and the corresponding segmentation results after serum deprivation for indicated periods of time. (B) Distributions of autophagosome scores after serum deprivation. (C) Images of H2B-RFP and the corresponding segmentation results after serum deprivation for indicated periods of time. (D) Distributions of death scores after serum starvation. (E) Images of GFP-LC3 and the corresponding segmentation results after glucose deprivation for indicated periods of time. (F) Distributions of autophagosome scores after glucose deprivation. (G) Images of H2B-RFP and the corresponding segmentation results after glucose deprivation for indicated periods of time. (H) Distributions of death scores after glucose starvation. To make the height of the curve insensitive to the size of bins used, the total area under each density function curve was normalized to 1.
In untreated H4 cells (0 h), GFP-LC3 puncta were observed only in a small subpopulation of cells (Fig. 3A and E, left panels). The resulting autophagosome score distribution had a long tail to the right, but the population median was nearly zero (Fig. 3B and F, dotted lines). When serum was removed, autophagy was increased within a few hours. We followed induction of autophagy over time in individual cells under those conditions (Fig. S4A). Cells with no or few autophagosomes developed more puncta within 4 h (yellow arrows), whereas in cells that already had a number of autophagosomes, the intensity of the puncta increased (white arrows). The resulting autophagosome score distribution at 4 h (Fig. 3B, dash line) showed a single peak at 0.02, indicating that on average about 2% of total GFP-LC3 intensity resided in puncta, but there was a substantial cell-to-cell variability. The peak of the autophagosome score distribution continued shifting rightward with time. At 24 h, autophagosomes could be detected in essentially all cells (Fig. 3A, right panel), and a broad unimodal distribution was observed (Fig. 3B, solid line with circles). We also noticed that due to autophagic degradation, the total intensity of GFP-LC3 signal in cells was substantially reduced as compared with untreated cells. No nuclear fragmentation and cell death was observed up to 24 h. of serum starvation. (Fig. 3C and D)
When the cells were deprived of glucose, we observed induction of both autophagy and apoptosis. At the early time points, the autophagy response was similar to the response observed following serum starvation, with both the number and intensity of puncta increased (Fig. 3E; Fig. S4B). The probability density functions of the autophagosome score also remained unimodal, with peaks shifting rightward from 0 to 7 h (Fig. 3F).
From 7 to 10 h, many cells began initiating apoptotic program and cell blebbing was frequently observed (Fig. 3E, 10 h, yellow arrow-head). We also detected many cells with condensed and fragmented chromatin, as indicated by the presence of multiple H2B-RFP puncta. Presence of H2B puncta in a given cell indicates that this cell has initiated the irreversible apoptotic death program (Fig. 3G); therefore, we considered any cell with a non-zero H2B puncta score as dying or dead. As expected, the apoptotic program made binary decisions for cell survival or death, so the observed probability function of death was bimodal, with live cells forming a sharp peak at the death score equal to 0, and apoptotic cells in a second, broader peak around 0.5 (Fig. 3H, solid line with circles).
At 10 h we also observed an interesting trend in the autophagy response. The cells that entered apoptotic death program (Fig. 3E and G, yellow arrows) showed lower levels of autophagy as compared with the surviving cells (white arrows). This could not be accounted for by cell rounding as additional puncta could not be detected by adjusting the focal plane (data not shown). Even more strikingly, the autophagosome score of surviving cells was also reduced as compared with the 7 h time point (Fig. 3F, solid line with circles). These observations of reduced autophagy in dying cells suggest that autophagy and apoptosis may be mutually exclusive in individual cells during glucose starvation and that cells may downregulate levels of autophagy prior to or at the time of initiation of apoptosis under those conditions.
Our data indicated that induction of autophagy in response to either serum or glucose deprivation is unimodal, indicating a signaling network designed for broad dynamic ranges and proportionate response. Induction of autophagy in response to starvation is known to incorporate signals from many pathways. Next, we wanted to determine whether the unimodal response could be generated by an individual proautophagy pathway. Since the MTOR pathway is known to play an essential role in regulation of autophagy in response to changes in extracellular nutrient and/or growth factor levels, we focused on this pathway by using MTOR inhibitors rapamycin and/or Torin 1 in H4, L929 and HeLa cells.
Consistent with previously published data,2 robust autophagy but no increase in apoptosis was observed following rapamycin treatment of H4 cells (Fig. S5). The autophagosome score changed with both time of treatment and rapamycin concentration (Fig. 4A–D). Rapamycin raised the autophagosome score in virtually all cells but the kinetics was slow even at the highest dose (Fig. 4B, representative kinetics of population median). We observed a similar trend when H4, L929 or HeLa cells were treated with another MTOR inhibitor, Torin 1 (Figs. S6–S8). The autophagy levels in response to Torin 1 in all cell types tested continued to increase up to 30 h after treatment. The HeLa cells showed much higher basal autophagy levels as compared with H4 and L929 cells, but treatment with Torin 1 still led to a robust increase in the number and intensity of GFP-LC3 puncta (Fig. S8).
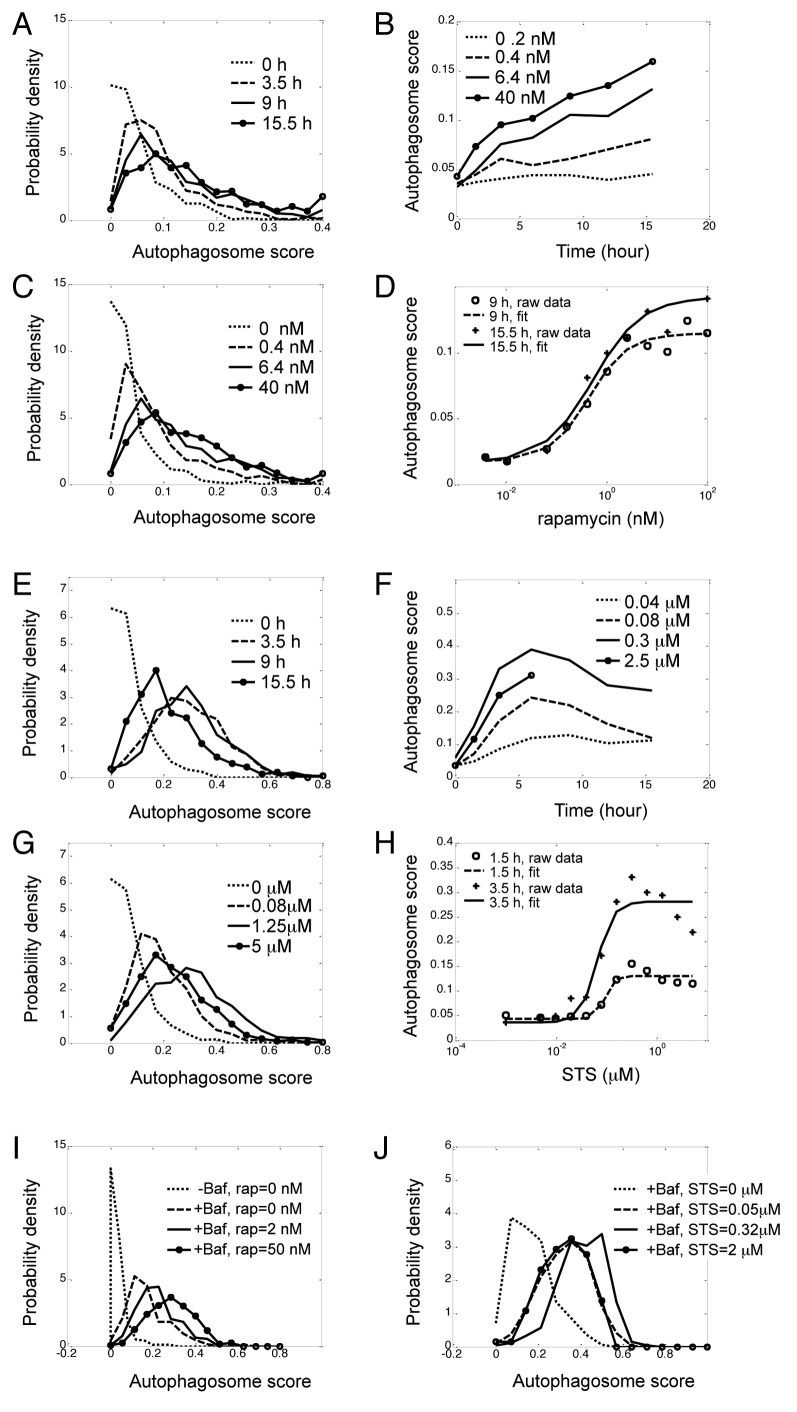
Figure 4. Unimodal dose response and kinetics of autophagy in response to rapamycin and STS. H4 cells expressing GFP-LC3 and H2B-RFP were treated with indicated doses or rapamycin (A–D) or STS (E–H) and imaged over time. (A) Kinetics of distributions of rapamycin-induced autophagy. Concentration of rapamycin was 6.4 nM. (B) Kinetics of population average (median) of rapamycin-induced autophagy. Rapamycin concentrations are indicated in legends. (C) Dose-response of distributions of rapamycin-induced autophagy at 9 h, (D) Dose-response of population median of rapamycin-induced autophagy, which was fitted with the Hill equation (see ref. 10). (E) Kinetics of distributions of STS (0.16 μM)-induced autophagy. (F) Kinetics of population median of STS-induced autophagy at indicated doses. (G) Dose-response of distributions of STS-induced autophagy at 3.5 h. (H) Dose-response of population median of STS-induced autophagy, which was fitted with the Hill equation. (I and J) Dose-response of induction of autophagy by rapamycin (I) and STS (J) in presence of Baf (100 nM). Again, the total area under each density function curve was normalized to 1.
At a given time point, as rapamycin or Torin 1 concentration increased, the peak of the distribution shifted to the right but the autophagic response remained unimodal and long tailed (Fig. 4C and D; Fig. S6C). This is in contrast to the situation of switch-like response where cells are redistributed between two peaks when the dose of stimuli is changed (Fig. S9).4,5 To determine if the graded induction of autophagy was due to the nature of MTOR signaling following inhibition with rapamycin, we evaluated phosphorylation of the downstream target of MTOR, the RPS6, ribosomal protein S6. In response to rapamycin RPS6 phosphorylation (pRPS6) was reduced rapidly within 2 to 3 h (Fig. S10A). In contrast to the unimodal distribution observed for autophagy, we observed a steep dose-response curve (Fig. S10B) and bimodal distribution of pRPS6 (Fig. S10C). Therefore, inhibition of MTOR by rapamycin in H4 cells can trigger graded response of autophagy but the concurrent suppression of RPS6-protein phosphorylation is switch-like.
Next we treated H4, L929 and HeLa cells with the pan-kinase inhibitor Staurosporine (STS), a potent inducer of apoptotic cell death.11,12 The autophagic response following STS treatment in all cell types was faster and reached higher maximum levels than what was observed after rapamycin or Torin 1 treatment (Fig. 4E and F; Figs. S11–13). For example, it took over 5 h for autophagosome score to reach 0.1 even after the highest rapamycin dose (Fig. 4B) but only approximately 1 h after intermediate (0.3 μM) STS treatment (Fig. 4F). Additionally, unlike the continuous increase of autophagy response when cells were treated with rapamycin or Torin 1, the kinetics of STS-induced autophagy in H4 cells reached a maximum around 5 h. Similarly to the situation observed following glucose deprivation (Fig. 3E and F) once the maximal autophagy response was reached, it rapidly decreased again (Fig. 4E and F). Such biphasic response was also observed in dose-response. The autophagosome score reached its maximum in the sub-micromolar range, but higher concentrations of STS reduced autophagy (Fig. 4G and H, note the apparent discrepancy between the data and the monotonic Hill equation fit10). As the majority of cells were still alive and the integrity of their nuclei was not compromised at 5 h, the biphasic response was not due to loss of dead cells. We hypothesize that this late decrease in autophagy may coincide with the commitment to apoptosis, including shut down of normal cellular function and initiation of the apoptotic program preceding overt cell death.13 A similar trend was observed in STS treated HeLa cells (Fig. S13).
Despite much higher and more prompt response, distribution of STS-induced autophagy remained unimodal, with a single discernible peak (Fig. 4E and G; Figs. S12 and S13). STS also induced apoptotic cell death (Figs. S11–S14). Similar to the case of glucose starvation, apoptosis scores formed bimodal distributions (Fig. S14A), and the fraction of dead cells changed rapidly with STS concentration (Fig. S14B) and treatment time (Fig. S14C). Unlike autophagy, which was induced almost immediately after STS treatment, little cell death was observed until after 4 h (Fig. S14C). At that time most cells had already downregulated autophagy from its maximum. This confirmed that the measurement of autophagic dynamics was not directly affected by loss of dead cells.
The number of GFP-LC3-positive autophagosomes present in a cell at any given time is a function of both autophagosome formation and the rate of lysosomal degradation. To test whether the autophagosome formation per se was also unimodal, we blocked lysosomal degradation by treating H4 cells with bafilomycin A1 (Baf). The resulting distribution, reflecting the cell-to-cell variation in basal rate of autophagosome formation, was also unimodal (Fig. S15). Adding rapamycin or STS together with Baf to the cells further increased the number of autophagosomes but the distributions remained unimodal (Fig. 4I-J; Fig. S16). Therefore, autophagosome formation itself is also a graded response. Furthermore, treating cells with Baf did not reverse inhibition of autophagy by high concentration of STS (Fig. 4J; Fig. S16C and S16D), confirming it was not due to increased rate of lysosomal degradation.
We also treated H4 cells with another proapoptotic drug, the ER stress inducer tunicamycin. In this case, relatively low levels of autophagy were induced, and even at a saturating dose of tunicamycin (> 2 μg/ml), cell death was not observed until at least 10 h after treatment (Fig. S17). At about 36 h, the entire cell population entered apoptosis. Therefore, although both induced cell death, the effect of tunicamycin was much slower and had a smaller effect on autophagy in this cell type as compared with STS.
Finally, to confirm our results were not an artifact due to how the autophagosome score was defined (ratio between puncta intensity and the total cellular GFP-LC3 intensity), we also tested the distributions of other ways of scoring for autophagy, including number of puncta, total area of puncta and average brightness of puncta. They all reflect the autophagic response, and none of them showed any signs of bimodality for any of the stimuli or cell types tested.
Interplay between autophagy and cell death in single cells after STS treatment
Our next goal was to further examine the interaction between autophagy and apoptosis. The kinetics of distributions demonstrated that autophagosome formation and DNA condensation have two distinct time scales, so these two biological events cannot be simultaneously measured in the same cell at a single time point. Consequently, fixed time point data can be difficult to interpret. For instance, if autophagy response and apoptosis are mutually exclusive in a population of cells at a given time point, it is hard to know whether a dead cell upregulated autophagy earlier, and whether a cell with high autophagy will die later. We also would not know whether cells that died sooner had higher or lower autophagy responses compared with the surviving cells.
To circumvent the above difficulties we performed live-cell microscopy and tracked individual cell trajectories, which enabled us to measure autophagy dynamics and apoptosis in tandem in the same cells over time (Fig. 2A and B; Fig. 5).
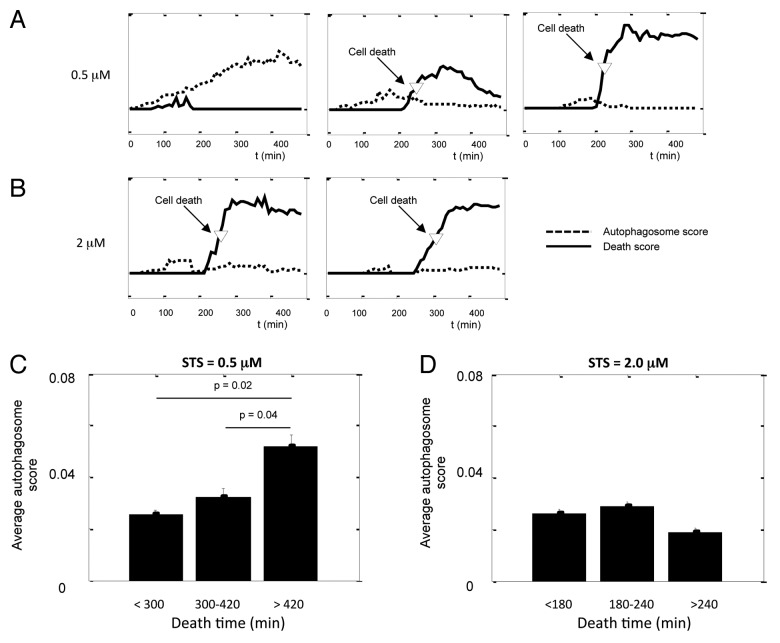
Figure 5. Single-cell analysis indicates a protective function of autophagy against STS-induced apoptosis. (A and B) Kinetics of autophagosome score and cell death score in cells treated with (A) 0.5 μM and (B) 2.0 μM STS. The movies were taken on the Nikon TE2000 system, one frame every 5 min. Autophagosome scores were rescaled so they can fit in the same graph with the death scores. (C and D) The average autophagosome score of cells binned by their death time for (C) 0.5 μM and (D) 2.0 μM STS. Error bars are standard error of mean; p value was calculated by unpaired Student’s t-test; n = 10 to 30.
At intermediate doses of STS (0.5 μM, Fig. 5A), autophagy alone or both autophagy and cell death were consecutively observed in individual H4 cells. Consistent with population measurements, autophagosomes rapidly formed following STS treatment, and many cells reached maximal autophagosome scores in approximately 3 h (180 min). Soon afterwards the autophagosome scores started decreasing. As autophagy decreased, the H2B-RFP pattern in the same cell often became brighter; the puncta started forming and quickly expanding, indicating that the cell was undergoing apoptosis. The cell death time, defined as the time when the nuclear fragmentation score reached half maximum, was automatically determined from dynamics of nuclear fragmentation and labeled as an arrow (Fig. 5A). By that time, autophagy was undetectable in many cells. Therefore, STS could stimulate autophagy and apoptosis in the same individual cells, but autophagy was suppressed before the cell death.
Furthermore, we witnessed an interesting trend: cells dying sooner had lower autophagy as compared with cells dying later in the course of experiment or those able to survive. Therefore, high levels of autophagy and cell death seemed to be inversely correlated or mutually exclusive, suggesting potential protective function for autophagy. To further quantify the relationship between autophagy and cell death, we calculated the time-average autophagosome score for each individual cell before its death. For example, if a cell died after 250 min of treatment, we took the time-average value of autophagy kinetics for that cell before 250 min. This largely reflects the history of autophagic response before the death of a given cell. We then binned the cells based on their time of death (Fig. 5C), Consistent with the protective function of autophagy, cells dying soonest had significantly lower autophagic response (p = 0.02) whereas cells that died latest had the highest autophagosome score.
At high concentration of STS (2 μM, Fig. 5B), almost all cells initiated apoptosis within 250 min and overall very low levels of autophagy were observed (Fig. 5B and D). Additionally, although STS at high dose failed to substantially induce autophagy, we did not observe loss of cells at the early stages of treatment. Since we recorded the complete history of autophagy before cell death, we were able to rule out that our data was biased by dying cells.
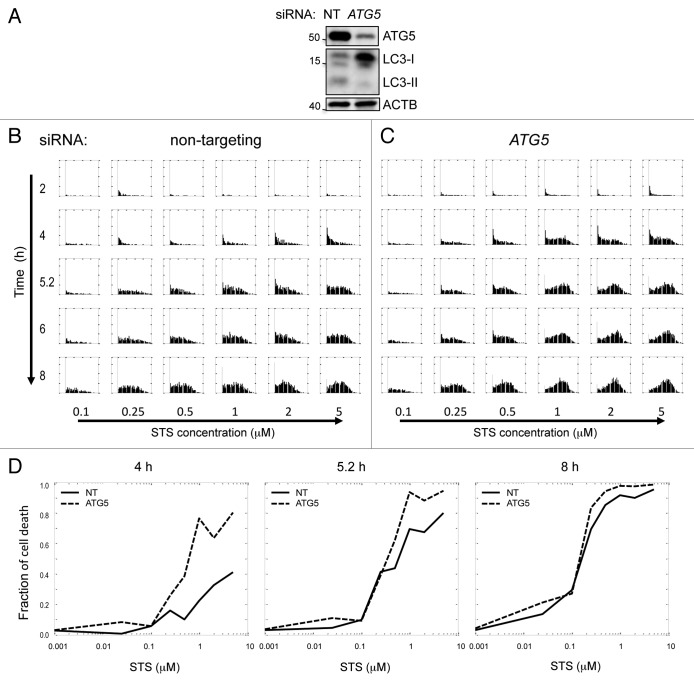
The time-lapse cell data confirmed the inverse-correlation between autophagy and apoptosis. To determine whether this was due to the suppression of apoptosis by autophagy (ie. causative), or merely coincidental, we knocked down the essential autophagy gene ATG5 by siRNA. To obtain sufficient data for statistical analysis, we examined the fraction of cell death in a large population of cells as a function of time rather than by cell tracking.
As expected, siRNA against ATG5 reduced basal levels of autophagy as well as the formation of autophagosomes after cells were treated with STS (Fig. 6A; Fig. S18). We compared the distributions of cell death scores in the nontargeting control (NT) and ATG5-knockdown cells (Fig. 6B and C). At multiple time points and concentrations of STS, more ATG5 knockdown cells initiated apoptosis as compared with controls (Fig. 6B and C). The effect of ATG5 knockdown on apoptosis was more significant at early time points (2–4 h) and medium STS concentrations (Fig. 6D). Between 5 to 6 h, there were still obvious differences in the fraction of cell death between NT and ATG5-knockdown cells treated with 2 μM STS, but at maximal STS concentration that fraction was similar for both. Therefore, ATG5-knockdown cells appeared to have faster kinetics and a lower STS threshold for death, but the maximal death fraction was similar to the NT cells. Finally, the two curves became less distinguishable after 6 to 8 h of treatment. This indicates that the suppression of autophagy sensitizes cells to STS-induced apoptosis; however, autophagy only slows down cell death rather than completely blocking the apoptotic program.
Figure 6. Suppression of autophagy potentiates STS-induced cell death. (A) siRNA mediated knockdown of ATG5 suppressed levels of basal autophagy in H4 cells. Cells were transfected with nontargeting siRNA (NT) or siRNA against human ATG5 for 48 h and treated with 10 μg/ml E64d for 4 h prior to lysis. (B and C) Kinetics and dose response of cell death score in NT and ATG5 knockdown cells. Cells were transfected with siRNA for 48 h, then treated with indicated doses of STS. Images were taken on the high throughput CellWoRx microscope. Each row is a different time point, and each column corresponds to different STS concentration. For each individual panel, the X-axis is cell death score, and the Y-axis is probability density of the distribution. (D) Dose-response of fraction of cell death at indicated time points after STS treatment. Fraction of cell death was calculated by counting cells with positive cell death score.
Discussion
In this article, we present a novel approach for studying autophagy by measuring and characterizing single-cell dynamics. We illustrate several applications of this single-cell approach.
First of all, we found no indication of bimodality in induction of autophagy after either serum or glucose deprivation, or in response to inhibition of the MTOR pathway in any of the cell types tested. On the other hand, inhibition of the RPS6 ribosomal protein phosphorylation by rapamycin was bimodal and switch-like. Therefore, graded induction of autophagy is not simply due to the nature of MTOR signaling. Instead, it may arise from the fact that autophagy is jointly regulated by multiple pathways.14 The cells constantly take into account a variety of extracellular inputs, including levels of nutrients, cytokines and growth factors and redox conditions, which all can affect levels of autophagy. We hypothesize that the benefit of graded autophagy response with a relatively broad dynamic range is that cells, either individually or as a population, can better adjust autophagy levels to such changes in environmental conditions to maintain cellular homeostasis.
Although STS-induced autophagy is also unimodal, it is more acute than starvation or MTOR inhibition-induced autophagy, suggesting that autophagy response can be upregulated more rapidly in an emergency situation such as presence of a cyctotoxic agent. We hypothesize that the other inputs that enable the slow graded response may be overridden under such circumstances. The different kinetics of the autophagic response may therefore reflect different roles of physiological and stress-induced autophagy.
The autophagy pathway and the apoptosis pathway have a complex crosstalk,6,15,16 and a single cytotoxic reagent can trigger both responses in a population of cells. Using single-cell analysis of live cells allowed us to look not only at the extent of cell death, but also to monitor the history of autophagy prior to cell death in individual cells. Our data demonstrate that in response to a cytotoxic agent such as STS the same cell can consecutively induce both autophagy and apoptosis. Therefore, autophagy and apoptotic cell death are mutually exclusive only at the time of occurrence, but not during the entire history of an individual cell. This is a highly dynamic view of the interplay between autophagy and the apoptotic cell death, indicating a dynamic decision-making process. This study clearly reveals the unique advantage of the live-cell microscopy method over other single-cell approaches such as fixed time point microscopy or flow cytometry, which can only measure population distributions instead of real time dynamics in individual cells.13,17
Furthermore, in the past it has often proven difficult to determine whether autophagy plays a protective or detrimental function under stress conditions. Our multiplexed live-cell data demonstrated that at intermediate doses of STS, increased levels of autophagy correlated with better survival, suggesting a protective function for autophagy. SiRNA knockdown of ATG5 further confirmed that induced autophagic response is indeed prosurvival in STS-treated H4 cells. However, autophagy only delays STS-induced apoptosis but cannot fully prevent it. This supports a “buffering” role for autophagy.
Interestingly, high doses and/or long exposure to STS or long-term glucose deprivation inhibited autophagy in H4 cells. This could be due to either a passive loss-of-function of autophagy in irreparably damaged cells or to a direct inhibition during cell death. The latter may occur if cells are already committed to apoptosis and protective autophagy becomes undesirable. This trend is also consistent with a protective function for autophagy following both STS treatment and glucose deprivation. Interestingly, we failed to observe similar phenomena from the distribution data of tunicamycin-treated H4 cells. Autophagy has been previously reported to be protective following ER stress.18 However, in response to tunicamycin, H4 cells induced autophagy much more slowly and to a lesser extent than what we observed after exposure to STS or glucose deprivation. A similar trend was observed in STS-treated L929 cells, where we also failed to detect a decrease in autophagy preceding cell death. Therefore, the cytoprotective properties of autophagy may be proportional to its kinetics and induction levels produced by a particular stimulus in a particular cell type. Our high-throughput single-cell image analysis method described in this paper will be particularly suitable to address these types of future questions.
Additionally, this method has the potential for addressing other novel biological questions. For example, in addition to apoptosis, it can be easily adjusted to investigate the crosstalk of autophagy with other types of cell death as well as cell proliferation and differentiation. Additionally, in combination with live-cell probes for lysosomes, mitochondria, redox or intracellular signaling intermediates such as PtdIns3P, the current approach can be extended to study the influence of other cellular processes and signaling pathways on autophagy. The single-cell dynamics of these reporters can be quantified by parameters such as the total response, maximal response, rate of increase, etc., and these parameters can be used for scoring the effects of different molecular perturbation and pharmacological interventions on autophagy. Ultimately, we hope this knowledge of single-cell dynamics will be useful for building a more comprehensive system-level model of autophagy.
Materials and Methods
Autophagy and apoptosis reporter cell lines
All cells were cultured in Dulbecco’s Modification of Eagle’s Medium (Cellgro, 10-017-CV) with 10% FBS (Gemini Bio, 100-106). H4 cells stably expressing GFP-LC3 have been previously described.2 To generate double GFP-LC3 H2B-RFP (LC3-H2B) cells, these cells were transfected with the GFP-LC3 and H2B-RFP vector and FACS sorted to obtain population with detectable levels of expression of both markers. Single cell clones were then selected to exclude the heterogeneity due to differential incorporation of exogenous genes. All experiments were repeated in multiple clones of LC3-H2B cells.
Drug treatments
For fixed time point experiments LC3-H2B cells were plated in either standard or glass-bottom 96 well plates with about 800 cells per well for H4, 600 for HeLa and 500 for L929 cells. For time-lapse experiments H4 cells were plated in 24-well plates at 3000 cells/well. Cells were allowed to grow at 37°C and in 10% CO2 for 36 h before treatment. 10% of CO2 significantly reduced the basal accumulation of GFP-LC3 positive autophagosomes and did not otherwise affect the proliferation of H4 cells. The cells were then treated with indicated dilutions of rapamycin (Sigma, R8781), staurosporine (Calbiochem, 569396), tunicamycin (Sigma, T7765) or Torin 1 (Tocris, 4247) to induce autophagy or/and apoptosis. Lysosomal degradation was blocked by 100 nM of bafilomycin A1 (Sigma, B1793). Glucose-free DMDM was obtained from Invitrogen (A14430-01).
RNAi knockdown
H4 cells were transfected with nontargeting siRNA #2 or siRNA against human ATG5 (Dharmacon, D-004374-01) or MTOR (Dharmacon, D-003008-23) using reverse transfection with HiPerfect reagent (Qiagen, 301704) as previously described.2 Cells were split 24 h after transfection and grew for additional 24 to 48 h before drug treatments as described above.
Image acquisition
For time-lapse acquisition a Nikon TE2000 motorized inverted microscope (Nikon Inc.), equipped with an environmental chamber was used. The images were taken at 20× magnification. Auto-focus was based on phase contrast imaging. Number of cells per imaging field was between 20 and 40. In each experiment two fields were imaged per condition. High-throughput images were acquired on CellWoRx high-throughput automated microscope (Applied Precision) with 10× objective. Two fields were imaged per well with 250 to 500 cells per field.
Image analysis
All image analysis codes were written in Matlab. On a PC with Intel Core i3 CPU (3 GHz) and 2 GB of RAM it took about 3 to 5 h to analyze a 96-well plate in a fixed cell study, and about 30 min to 1 h to analyze 100 frames of live-cell images.
Statistical analysis
We used probability density function to show the distribution of single-cell responses. In the plots, the total area under the density function curve is normalized to 1, so the height of the curve is not sensitive to the size of bins used. This makes the probability density function plot different from a common histogram, where the heights of the bars, either defined as number of cells or percentage of cells, depend on the size of the bin. We used unpaired t-test to compare the autophagosome scores of subgroups of cells with different death time. The standard error of the mean for each subgroup of cells was reported as error bars.
Western blot analysis
Protein samples were prepared as previously described2 and resolved on 4–15% gradient SDS-PAGE (BioRad, 4561086). For endogenous LC3 analysis cells were treated with 10 μg/ml lysosomal protease inhibitor E64d (Sigma, E8640), 5 μg/ml pepstatin A (Sigma, P5318) or Baf for 4 to 6 h prior to lysis. The following antibodies were used: LC3 (Novus Biologicals, NB100-2220), ATG5 (Sigma, A0731), cleaved CASP3 (Cell Signaling, 9664), SPTAN1 (Enzo Life Sciences, BML-FG6090), SQSTM1 (Chemicon, 610832), all at 1:1,000, ACTB (Sigma, A5316) at 1:10,000. LC3-II levels were compared with loading control (ACTB).19
Supplementary Material
Acknowledgments
We thank Dr. Daniel Needleman for stimulating discussion on live-cell tracking, Dr. Randy King for providing the HeLa-H2B-GFP cell line, and Dr. Chinmoy Sarkar for technical assistance. The high throughput imaging was performed at the ICCB-Longwood, HMS Boston MA. Time-lapse microscopy was performed at the Nikon Imaging Center, HMS Boston MA. This work was supported by P01 AG027916 to J.Y. and startup funds from the Department of Anesthesiology, UMSOM Baltimore MD to M.M.L.
Glossary
Abbreviations:
- ACTB
actin, beta
- ATG5
autophagy-related 5
- Baf
bafilomycin A1
- CASP3
caspase 3, apoptosis-related cysteine peptidase
- GFP
green fluorescent protein
- HIST2H2BE
histone cluster 2, H2be
- H2B
histone cluster 2, H2be
- LC3
microtubule-associated protein 1 light chain 3
- MTOR
mechanistic target of rapamycin
- PtdIns3P
phosphatidylinositol 3-phosphate
- RFP
red fluorescent protein
- RPS6
ribosomal protein S6
- SPTAN1
spectrin, alpha, non-erythrocytic 1
- SQSTM1
sequestosome 1
- STS
staurosporine
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/25080
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/25080
References
- 1.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipinski MM, Hoffman G, Ng A, Zhou W, Py BF, Hsu E, et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev Cell. 2010;18:1041–52. doi: 10.1016/j.devcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevense M, Muramoto T, Müller I, Chubb JR. Digital nature of the immediate-early transcriptional response. Development. 2010;137:579–84. doi: 10.1242/dev.043836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrell JE, Jr., Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–8. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 5.Biggar SR, Crabtree GR. Cell signaling can direct either binary or graded transcriptional responses. EMBO J. 2001;20:3167–76. doi: 10.1093/emboj/20.12.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 7.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Zhou X, Wong ST. Automated segmentation, classification, and tracking of cancer cell nuclei in time-lapse microscopy. IEEE Trans Biomed Eng. 2006;53:762–6. doi: 10.1109/TBME.2006.870201. [DOI] [PubMed] [Google Scholar]
- 9.Blair D, Dufresne E. http://physics.georgetown.edu/matlab/
- 10. http://en.wikipedia.org/wiki/Hill_equation_(biochemistry)
- 11.Rüegg UT, Burgess GM. Staurosporine, K-252 and UCN-01: potent but nonspecific inhibitors of protein kinases. Trends Pharmacol Sci. 1989;10:218–20. doi: 10.1016/0165-6147(89)90263-0. [DOI] [PubMed] [Google Scholar]
- 12.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–32. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 13.de la Calle C, Joubert PE, Law HK, Hasan M, Albert ML. Simultaneous assessment of autophagy and apoptosis using multispectral imaging cytometry. Autophagy. 2011;7:1045–51. doi: 10.4161/auto.7.9.16252. [DOI] [PubMed] [Google Scholar]
- 14.Lipinski MM. Towards the global understanding of the autophagy regulatory network. Autophagy. 2010;6:1218–20. doi: 10.4161/auto.6.8.13772. [DOI] [PubMed] [Google Scholar]
- 15.Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 16.Eisenberg-Lerner A, Bialik S, Simon H-U, Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16:966–75. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- 17.Larsen KB, Lamark T, Øvervatn A, Harneshaug I, Johansen T, Bjørkøy G. A reporter cell system to monitor autophagy based on p62/SQSTM1. Autophagy. 2010;6:784–93. doi: 10.4161/auto.6.6.12510. [DOI] [PubMed] [Google Scholar]
- 18.Høyer-Hansen M, Jäättelä M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14:1576–82. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- 19.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.