Abstract
Inflammatory bowel disease (IBD) is a chronic relapsing inflammatory disorder of the bowel. The etiology remains unknown, but IBD is immune-driven and multiple factors including genetic, environmental, and microbiological components play a role. Recombinase-activating gene-2-deficient (Rag2−/−) mice infected with Helicobacter hepaticus (H. hepaticus) have been developed as an animal model to imitate naturally occurring inflammatory events and associated key features of chronic inflammatory responses in humans. In this study, we have combined mass spectrometry-based metabolomics and peptidomics to analyze serum samples of Rag2−/− mice infected with H. hepaticus. Metabolomics profiling revealed that H. hepaticus infection dramatically changed numerous metabolite pathways, including tryptophan metabolism, glycerophospholipids, methionine-homocysteine cycle, citrate cycle, fatty acid metabolism and purine metabolism, with the majority of metabolites being down-regulated. In particular, there were notable effects of gut microflora on the blood metabolites in infected animals. In addition, the peptidomics approach identified a number of peptides, originating from proteins, including fibrinogen, complement C4 and alpha-2-macroglobulin, with diverse biological functions with potentially important implications for the progress of IBD. In summary, the strategy of integrating a relevant animal model and sensitive mass spectrometry-based profiling may offer a new perspective to explore biomarkers and provide mechanistic insights into IBD.
Keywords: metabolomics, inflammatory bowel disease, mouse model, Helicobacter hepaticus, microbiome
1. Introduction
Inflammatory bowel disease (IBD) is a chronic relapsing inflammatory disorder of the bowel, with more than a million patients in U.S. suffering from this debilitating disease1. IBD significantly increases the risk of colorectal cancer in humans 2. Many factors contribute to the occurrence and perpetuation of this disease and both genetic and environmental factors may play an important role3,4. The etiology remains unclear, although intestinal microbiota and host immunity are pivotal in the disease progression in the lower bowel 5–8. Commensal enteric bacteria are important factors that drive the progression of IBD 6; animal models have shown that intestinal bacterial flora are also important in the development of this disease 9–11 and altered gut microflora in IBD patients are a common component of the pathophysiology12–15.
Recombinase-activating gene-2-deficient (Rag2−/−) mice lack functional lymphocytes16 and have been used as an animal model to investigate IBD17–19. Rag2−/− mice infected with Helicobacter hepaticus (H. hepaticus) have been shown to emulate inflammatory events in humans, including characteristic key features such as accumulation of macrophages and neutrophils in the colon, generation of nitric oxide, up-regulation of inducible NO synthase and increased expression tumor necrosis factor-alpha (TNF-alpha) 17–19.
Early detection, mechanistic insights, and development of therapeutic targets largely depend on available biomarkers, and metabolomics is emerging as a powerful tool for characterizing and developing biomarkers associated with human diseases, including IBD. Nuclear magnetic resonance (NMR) and mass spectrometry (MS) are widely used to profile metabolites in diverse biological samples, and several studies have been conducted in animal models and IBD patients to analyze metabolites in urine, fecal extracts and biopsy samples, with NMR being a major tool20–25. Recently, however, MS-based metabolomics profiling is becoming attractive because of its high sensitivity, the ability to detect molecules with diverse structures, a wide dynamic range, quantitative capability and the ease of interfacing with other separation techniques such as gas and liquid chromatography.
In this study, we applied a liquid chromatography-mass spectrometry (LC-MS) metabolomics approach to analyze global metabolite changes in Rag2−/− mice infected with H. hepaticus for different time periods (10 week post-infection and 20 week post-infection). This non-targeted metabolomics profiling revealed a notable effect of H. hepaticus infection in Rag2−/− mice, with large numbers of metabolites being down-regulated. Our data also clearly illustrated large effects of gut flora on blood metabolites during infection. In addition, we have complemented our metabolomics analysis with peptidomics and identified a number of serum peptides that originate from up-regulated proteins. These precursor proteins, with diverse biological functions, may have important implications for the progress of IBD.
2. Experimental Section
Animals
Experiments were conducted in 129/SvEv Rag2−/− mice housed in the MIT Department of Comparative Medicine in static microisolator cages and were approved by the Institutional Animal Care and Use Committee. Each grouping was comprised of 10 mice unless otherwise stated. Mice manipulation and histopathologic evaluation were conducted as described previously26.
Helicobacter hepaticus
H. hepaticus (strain 3B1, ATCC 51449) was grown and the purity was confirmed as described previously17. At age 6–8 weeks, mice were dosed with H. hepaticus by gavage every other day for a total of 3 doses. Control mice were dosed with vehicle alone. H. hepaticus infection was confirmed by examining cecum and colon with PCR using H. hepaticus-specific primers. Serum and tissues were collected at 10 and 20 weeks post-infection.
Metabolite extraction
Metabolites were isolated from serum using methanol. Internal standard stock solution containing a synthetic peptide (10 FL) and cold methanol (80 FL) were added to 10 FL of serum. After vortexting for 1 minute, the samples were incubated at 4 °C for 20 minutes and then centrifuged for 10 minutes at 12,000 rpm. The supernatant was collected, dried in a SpeedVac, and then resuspended in 30 FL of 98:2 water:acetonitrile for MS analysis.
Metabolomics profiling by LC-MS
LC-MS analyses were performed on a LC/MSD time-of-flight (TOF) mass spectrometer from Agilent (Santa Clara, CA) with an electrospray ionization (ESI) source. The mass spectrometer was interfaced with a 1290 Ultra Performance Liquid Chromatography system from Agilent (Santa Clara, CA). The ESI-TOF was calibrated daily using the standard tuning solution from the Agilent Technologies. During analyses, the instrument was calibrated using two different reference masses with constant infusion. The typical mass accuracy of ESI-TOF was less than 5 ppm. Metabolites were analyzed in both positive and negative modes using a C18 T3 reverse phase column from Waters (Milford, MA). For positive mode experiments, the mobile phases were water with 0.1% formic acid (A) or acetonitrile with 0.1% formic acid (B). A linear gradient was run from 2% to 90% B over 60 minutes, at 200 FL/min. The ESI source parameters were: spray voltage, 3.5 kV; gas temperature, 325 °C; drying gas, 12 L/min; nebulizer, 35 psig; fragmentor, 160 V. In negative mode, the mobile phases were water with 1 mM ammonium fluoride (A) or acetonitrile (B). Similarly, a linear gradient was run from 2% to 90% B over 45 minutes, with a flow rate of 200 FL/min. The ESI source was set as follows: spray voltage, 3.0 kV; gas temperature, 325 °C; drying gas, 11 L/min; nebulizer, 30 psig; fragmentor, 165 V.
Data processing and analysis
Data acquired in Agilent. d format were converted to mzXML using MassHunter Workstation software from Agilent (Santa Clara, CA). Data were filtered by intensity; only signals with intensities larger than 1000 were considered. The converted data were processed by both MassProfiler Professional (MPP) software (Agilent, Santa Clara, CA) and XCMS (Scripps, La Jolla, CA) for peak picking, alignment, integration and extraction of the peak intensities. To profile individual metabolite differences between uninfected and infected groups, a 2-tailed Welch’s t-test was used (p < 0.05). The exact masses of molecular features with significant changes were searched against the Human Metabolome Database (HMDB) (http://www.hmdb.ca), METLIN (http://metlin.scripps.edu) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (http://www.genome.jp/kegg). The matched exact masses were stored and used for the generation of MS/MS data to further identify the metabolites.
Metabolite identification
MS/MS was generated on an Agilent Q-TOF 6510 mass spectrometer (Santa Clara, CA) to further confirm the identity of the metabolites. The Q-TOF was calibrated daily using the standard tuning solution from the Agilent Technologies. The instrument was calibrated using two different reference masses with constant infusion during the analyses. The typical mass accuracy was less than 5 ppm. Metabolites were extracted from 30 FL of pooled serum samples for MS/MS experiments. The column and gradients were the same as those used for metabolite profiling by ESI-TOF. The ESI source was set as follows: spray voltage, 3.5 kV; gas temperature, 325 °C; drying gas, 10 L/min; nebulizer, 32 psig; fragmentor, 150 V. A target list, which included previously determined exact masses according to the database search results, was generated for fragmentation. The retention-time differences between ESI-TOF and Q-TOF for each compound were corrected by manual alignment.
Statistical analysis
Data analysis was conducted with multivariate statistical methods. Principle component analysis (PCA) was performed to examine intrinsic clusters and obvious outliers within the observations. To identify potential outliers in samples, 95% confidence interval of all samples was used as the threshold. In addition, heat maps were generated using a hierarchical clustering algorithm to visualize the metabolite difference within the dataset. All data handling and statistical analysis were performed using the statistical R package.
Identification of serum peptides
The charge states of peptides with significant changes were determined from the isotope clusters. The exact masses, charge states and corresponding retention times were used to make a target list for Q-TOF MS/MS experiments. The collision energies were dependent on the size and charge of peptides, ranging from 13 to 25V. The resultant tandem mass spectra were extracted and searched against the NCBInr database with Agilent Spectrum Mill Software (Santa Clara, CA) to identify the sequences and potentially matched proteins. Spectrum Mill MS/MS search parameters were set as follows: database, NCBInr.fasta; species, Mus Musculus; protein pI, 3 to 10; modification, none; digestion, no enzyme; precursor mass tolerance, 20 ppm; product mass tolerance, 200 ppm; batch size, 25; search mode, identity. Peptides with Scores > 10, Scored Peak Intensity Percent (%SPI) > 65 and Fwd-Rev Score >2 were considered tentative identification27,28 and two peptide sequences were further validated using authentic synthetic peptides.
3. Results
3.1 Method development and LC-MS chromatograms
Figure 1 shows the experimental workflow for the initial metabolite profiling. Briefly, ten serum samples each from uninfected or infected Rag2−/− mice were extracted and analyzed by ESI-TOF, over a range of 80–1000m/z, in both positive and negative polarities, using a C18 reverse phase analytical column. Molecular features, i.e., all signals associated with a given analyte, with intensities larger than 1000 were further processed and statistically analyzed with MPP or XCMS software to profile metabolites with significant changes (1.5 fold change, p<0.05) between the uninfected and infected animals. The resultant peak list with exact masses was searched against metabolite databases including HMDB and METLIN, with a 10 ppm mass accuracy threshold. Next, the matched exact masses and associated retention times were used to generate MS/MS spectra to confirm the metabolite identities, followed by metabolic pathway or function analysis with the KEGG database.
Figure 1.
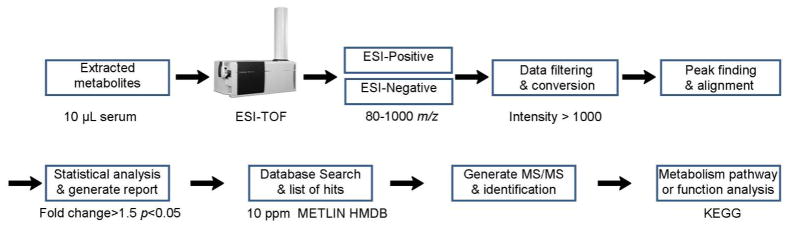
The experimental workflow for the metabolite profiling performed in this study. It starts with the metabolite extraction with methanol, followed by MS profiling under both positive and negative ionization modes, data processing, statistical analysis, database search, metabolite identification by tandem MS/MS and pathway analysis.
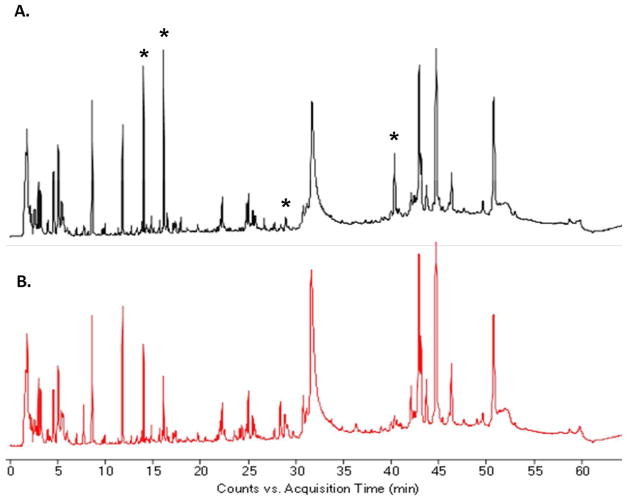
Figure 2 shows typical LC-ESI-TOF total-ion chromatograms of serum metabolites extracted from uninfected and infected animals, acquired under ESI positive mode. Several dominant compounds can be noted by inspection. For example, the intense peak eluting at 11.87 min is tryptophan. The strong peaks at 40–45 min are attributed to lysophosphatidyl cholines (Lyso-PC). The wide peaks between 30–35 min are largely residual serum proteins after methanol extraction.
Figure 2.
Typical LC-ESI-TOF total ion chromatograms of serum metabolites extracted from uninfected (A) and infected animals (20 week post-infection) (B). Data were acquired under positive ionization mode (80–1000 m/z).
Several peaks with different intensities, marked by asterisks in the TIC, could be readily observed between the uninfected and infected animal, while molecular feature extraction revealed an additional large number of metabolites with significantly-changed concentrations. The reproducibility of the LC-MS system, with less than 20-second retention-time shifts from run to run, facilitated good alignments within the datasets (Figure S1). From a typical positive LC-ESI-TOF chromatogram, more than 10,000 molecular features could be extracted (Table 1), although a given metabolite could be represented by several different molecular features. For example, it may have several isotope peaks and nonspecific or unknown adduct ions. Moreover, much fewer metabolites were captured by negative ESI-TOF (Table 1); some typical total ion chromatograms are shown in Figure S2.
Table 1.
Summary of metabolite profiling and comparison between the uninfected and infected group, acquired by LC-MS under both positive and negative modes.
| Ionization mode | Total features | Total significantly changed features* | Total decreased features# | Total increased Features# | |
|---|---|---|---|---|---|
| 10 week | LC-ESI+ | 10070 | 1055 | 857 | 198 |
| LC-ESI− | 3634 | 330 | 260 | 70 | |
| 20 week | LC-ESI+ | 10071 | 2026 | 1583 | 443 |
| LC-ESI− | 3058 | 618 | 514 | 104 |
Significantly changed features denote those with more than 1.5 fold changes (p<0.05).
Decreased or increased features are relative to the corresponding uninfected groups.
An initial assessment of the effect of H. hepaticus infection in Rag2−/− mice revealed a significant effect on the metabolite profiles (Table 1). Approximately 10% and 20% of the total features were changed for 10 week and 20 week infection groups, respectively, with the majority (~80%) being down-regulated.
3.2 Multivariate statistical analysis
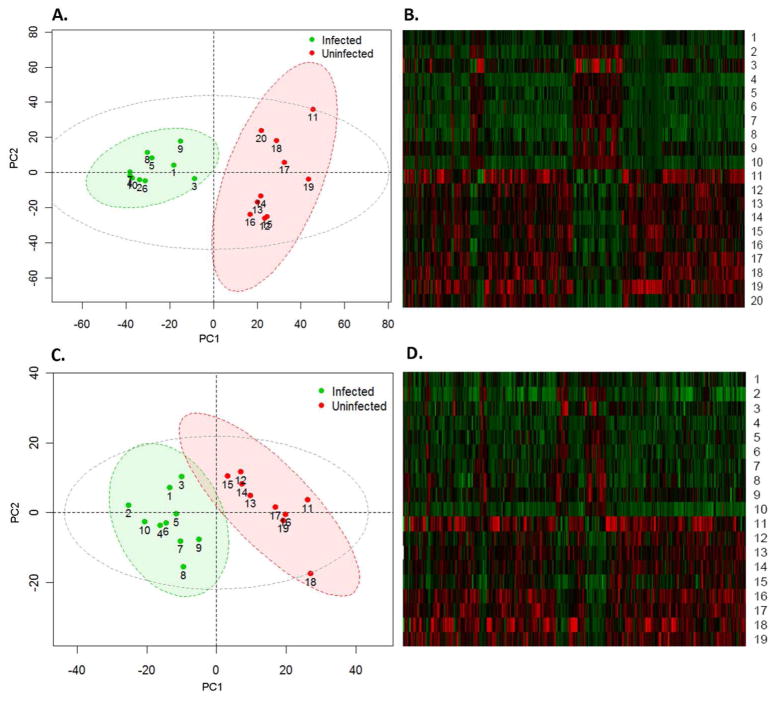
We next used multivariate statistics to examine the intrinsic clusters within each group and whether the uninfected and infected groups could be differentiated using metabolite profiles. PCA revealed excellent separations of the uninfected and infected animals under both positive and negative modes, as illustrated in Figure 3 A and 3C. The hierarchical clustering heat maps in Figure 3B and 3D show similar patterns within each group and confirm that the majority of metabolites were down-regulated in infected animals. A good separation was also observed for the 10-week samples, with one animal in the uninfected group identified as an outlier using the 95% confidence interval as the threshold (Figure S3A and S3C). The different pattern of that sample revealed by the hierarchical clustering heat map also supports this identification (Figure S3B and S3D). Interestingly, the metabolite features of infected animals are more homogeneous than those of uninfected animals, as illustrated by closer clustering in Figure 3A, suggesting that cellular responses to H. hepaticus infections are largely driving the biology in infected animals.
Figure 3.
Separation of uninfected and infected 20 week Rag2−/− mice by principle component analysis (A. positive-ESI; C. negative-ESI) and the hierarchical clustering heat maps constructed using molecular features with 1.5 fold changes (p<0.05) (B. positive-ESI; D. negative-ESI). The ellipses represent 95% confidence interval of the uninfected (Red), infected (Green) and all samples (Grey), respectively.
3.3 Identification of metabolites
Identifying the compounds that show significant differences between the uninfected and infected mice is the most laborious and time-consuming aspect of non-targeted metabolomics, involving database searches with exact masses followed by interpretation of MS/MS product-ion spectra. Figure S4 illustrates the process for the identification of metabolites with hippuric acid as an example, which decreased 3-fold in the 20-week infected samples. Database searching, using the exact mass at m/z 180.0645, gave 3–5 hits depending on the database. The product-ion spectrum arising from m/z 180.0645 at 15.1 min included characteristic hippuric acid fragments at m/z 77.0391 and m/z 105.0331 (Figure S4). Comparison of the retention time and fragmentation pattern with an authentic standard further confirmed the identity.
Table 2 summarizes the selected metabolites with greater than 1.5 fold changes (increase or decrease relative to uninfected) between the uninfected and infected mice. More-complete lists of identified metabolites in the 10 week and 20 week groups are shown in Table S1 and S2. We identified totally 27 and 50 significantly changed metabolites in the 10 week and 20 week post-infection groups, respectively. The variety of these structures, including lipids, amino acid derivatives, peptides, fatty acids, and nucleoside conjugates, highlight the capability and advantages of non-targeted mass spectrometry-based metabolomics profiling.
Table 2.
Selected significantly changed metabolites in Rag2−/− mice infected with H. hepaticus at 20 week post-infection.
| HMDB | Metabolites | Pathway | Fold | p-value | m/z | RT(min) | KEGG |
|---|---|---|---|---|---|---|---|
| 00671 | Indolelactic acid | Tryptophan metabolism | −7.06 | 0.00135 | 206.0796 | 21.8 | C02043 |
| 00763 | 5-Hydroxyindoleacetic acid | Tryptophan metabolism | −6.51 | 0.00029 | 192.0645 | 17.4 | C05635 |
| 02096 | 3-Indolebutyric acid | Tryptophan metabolism | −3.30 | 0.02198 | 204.1039 | 19.2 | C11284 |
| 00734 | Indoleacrylic acid | Tryptophan metabolism | −1.86 | 0.00246 | 188.0697 | 25.7 | C00331 |
| 02302 | 3-Indolepropionic acid | Tryptophan metabolism | −1.93 | 0.038 | 188.0720 | 13.3 | |
| 00898 | 1-Methylhistamine | Histidine metabolism | +3.03 | 0.00358 | 126.1020 | 1.7 | C05127 |
| 03045 | Ergothioneine | Histidine metabolism | −4.43 | 0.00106 | 230.0948 | 2.3 | C05570 |
| 00714 | Hippuric acid | Phenylalanine metabolism | −3.45 | 4.88E-07 | 180.0645 | 15.1 | C01586 |
| 00821 | Phenylacetylglycine | Phenylalanine metabolism | +1.79 | 0.00912 | 194.0801 | 16.9 | C05598 |
| 00939 | S-Adenosylhomocysteine | Cysteine and methionine | −1.90 | 0.01773 | 385.1272 | 5.9 | C00021 |
| 10381 | LysoPC(15:0) | Glycerophospholipid | −1.68 | 7.31E-05 | 482.3223 | 42.0 | C04230 |
| 10383 | LysoPC(16:1) | Glycerophospholipid | −1.86 | 6.10E-07 | 494.3225 | 41.0 | C04230 |
| 10389 | LysoPC(18:4) | Glycerophospholipid | −3.62 | 2.62E-05 | 516.3112 | 38.3 | C04230 |
| 10395 | LysoPC(20:4) | Glycerophospholipid | −1.80 | 0.0017 | 544.3388 | 42.4 | C04230 |
| 11475 | LysoPE(18:1) | Glycerophospholipid | −1.76 | 0.00798 | 478.2897 | 30.4 | |
| 11482 | LysoPE(20:1) | Glycerophospholipid | −1.63 | 0.00074 | 506.3284 | 34.4 | |
| 11494 | LysoPE(22:5) | Glycerophospholipid | −1.66 | 0.0205 | 526.2897 | 30.0 | |
| 00094 | Citric acid | Citrate cycle (TCA cycle) | −3.69 | 0.00747 | 191.0201 | 1.7 | C00158 |
| 01248 | FAD | Riboflavin/TCA cycle | +1.73 | 0.00052 | 784.1487 | 9.9 | C00016 |
| 00673 | Linoleic acid | Fatty acids | −2.25 | 0.02427 | 281.2456 | 60.6 | C01595 |
| 01388 | Alpha-Linolenic acid | Fatty acids | −1.67 | 0.01169 | 279.2299 | 57.1 | C06427 |
| 02183 | Docosahexaenoic acid | Fatty acids | −1.82 | 0.00907 | 329.2453 | 59.1 | |
| 01999 | Eicosapentaenoic acid | Fatty acids | −2.45 | 0.03266 | 303.2295 | 56.7 | C06428 |
| 01043 | Arachidonic acid | Fatty acids | −3.87 | 0.04086 | 305.2453 | 60.0 | C00219 |
| 00034 | Adenine | Purine metabolism | −1.74 | 0.04098 | 136.0609 | 8.9 | C00147 |
| 00071 | Deoxyinosine | Purine metabolism | −1.66 | 0.04146 | 253.0920 | 9.7 | C05512 |
| 03312 | Daidzein | Isoflavonoid biosynthesis | +2.08 | 0.00589 | 253.0522 | 19.8 | C10208 |
Fold change: + represents up-regulated metabolites in infected animals, while – denotes down-regulated metabolites in infected animals.
3.4 Effect of infection period on the metabolite profile
The infection period of H. hepaticus has a significant impact on the metabolite profile in Rag2−/− mice, which is reflected not only by a doubling of the number of changed metabolites in the 20 week groups (Table 1), but also by larger fold changes of specific metabolites in these mice. Figure 4 illustrates the influence of infection period on the fold changes of some metabolites. Typically, 20-week infected samples have equivalent or higher fold changes for the same metabolite compared to 10 week infected mice. These metabolite changes are accompanied by the progressive development of colonic inflammation, hyperplasia, dysplasia and colon adenocarcinoma caused by H. hepaticus infection in this animal model17,19,26. Specifically, inflammation, edema, epithelial defects, crypt atrophy, hyperplasia and dysplasia were common histopathologic changes for both 10-week and 20-week infected mice, with ileocecocolic junction and proximal colon being the most severely damaged locations. However, progression of dysplasia to intramural carcinoma was observed in selected 20-week H. hepaticus infected mice, which was characterized by invasion of neoplastic glands below the muscularis mucosae layer. Portal and lobular hepatitis were clearly identified in the liver tissues of both 10-week and 20-week H. hepaticus infected Rag2−/− mice26. In addition, a previous study also demonstrated that H. hepaticus infection induced progressive hepatitis in SCID/NCr mice including Kupffer, Ito, oval cell hyperplasia and multifocal to coalescing coagulative hepatocyte necrosis29.
Figure 4.
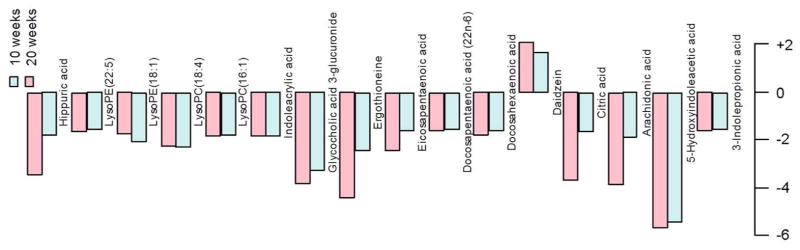
The effect of the length of H. hepaticus infection on fold changes of typical metabolites in Rag2−/− mice relative to the uninfected groups (Fold change: + represents up-regulated metabolites in infected animals, while – denotes down-regulated metabolites in infected animals). Mice infected for 20 weeks have equivalent or higher fold changes for the same metabolites compared to ten-week infected mice.
3.5 Effects of gut microflora on the metabolites in Rag2−/− mice infected with H. hepaticus
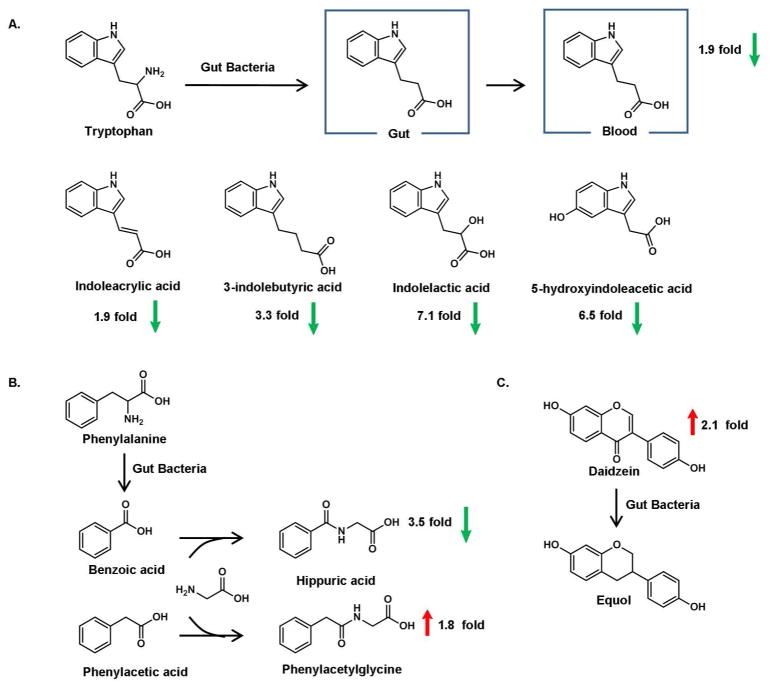
A number of indole derivatives were down-regulated in H. hepaticus infected Rag2−/− mice, as shown in Figure 5A. Among these indole-containing compounds, 3-indolepropionic acid (IPA) is of particular interest since this metabolite can only be generated by certain types of bacteria 30,31 The decreased amount of IPA suggests that the gut microflora were perturbed by the H. hepaticus infection. The effect of intestinal flora is further indicated by the changes of other gut microbe-regulated metabolites including hippuric acid and phenylacetylglycine, as illustrated in Figure 5B. Likewise, an increased amount of daidzein suggests down-regulated metabolic activity that also involves gut bacteria, as shown in Figure 5C. Taken together, these results clearly implicate the involvement of gut microflora on the metabolite profiles in H. hepaticus infected Rag2−/− mice.
Figure 5.
The effect of gut microflora on blood metabolites in H. hepaticus infected Rag2−/− mice. (A.) Indole-containing metabolites (B.) Glycine conjugates (C.) The metabolism of Diadzein to Equol.
3.6 Altered lipid metabolism and methionine-homocysteine cycle in Rag2−/− mice infected by H. hepaticus
N-methyl-histamine, which was synthesized by histamine N-methyltransferase using S-adenosylmethionine (SAM) as the methyl donor, increased approximately 3-fold in infected mice, as shown in Figure 6,. Meanwhile, we detected decreased amounts of S-adenosylhomocysteine (SAH), which is generated by SAM during methylation. Reduced levels of SAH, a central intermediate of the methionine-homocysteine cycle, suggests that other methylation pathways have been inhibited during infection. Previous work established that phosphatidylethanolamine methyltransferase (PEMT) converts phosphatidylethanolamine (PE) to phosphatidylcholine (PC) using SAM as a methyl donor with SAH as a by-product and accounts for 20~40% of normal hepatic PC synthesis 32. As shown in Table 2 and Table S1–2, a number of phospholipids were significantly down-regulated in our model. Therefore, a link may exist between altered phospholipid metabolism and the impaired methionine-homocysteine cycle in H. hepaticus infected Rag2−/− mice as illustrated in Figure 6.
Figure 6.

The potential link between altered phospholipid metabolism and disturbed methionine-homocysteine cycle in H. hepaticus infected Rag2−/− mice.
3.7 Altered energy metabolism and fatty acid pathways
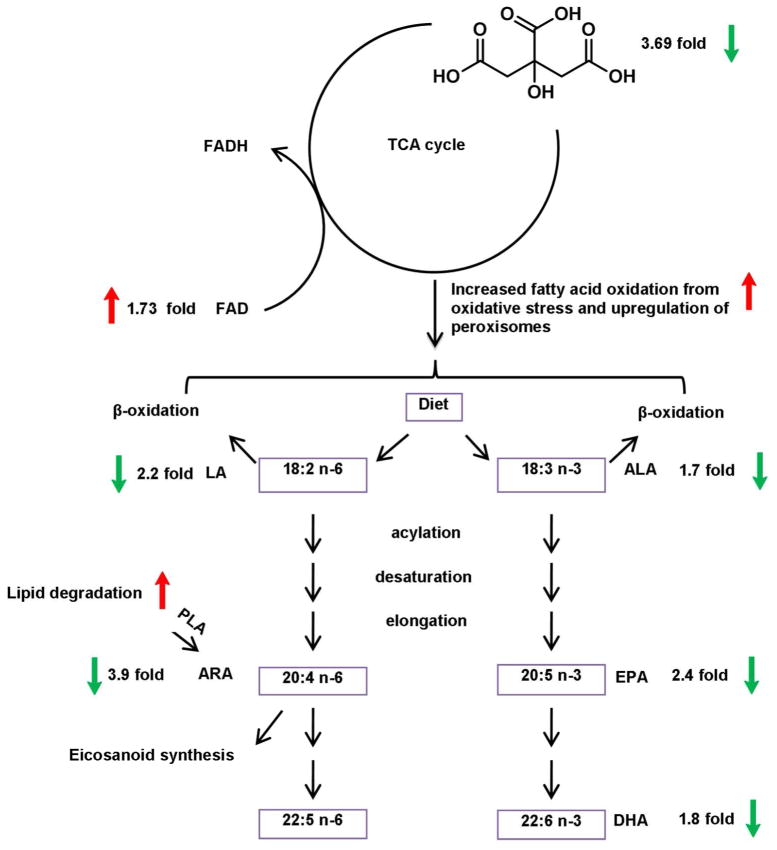
Our metabolomics profiling has shown that intermediates and cofactors of the TCA cycle change during inflammation. Decreased citric acid (−3.69 fold) and increased flavin adenine dinucleotide (FAD) (+1.73 fold) support the down-regulation of energy metabolism in our model. In accordance with this observation, a number of unsaturated fatty acids decreased upon infection, which may be the consequence of increased fatty acid oxidation induced by oxidative stress and upregulation of peroxisomes. The interplay between energy metabolism and fatty acid pathways is illustrated in Figure 7.
Figure 7.
Inhibited citric acid cycle and altered fatty acid pathways in H. hepaticus infected Rag2−/− mice. Linoleic acid: LA, alpha-linoleic acid: ALA, arachidonic acid: ARA, eicosapentaenoic acid: EPA, docosahexaenoic acid: DHA, phospholipase A2: PLA2, flavin adenine dinucleotide: FAD.
3.8 Increased endogenous peptides in Rag2−/− mice infected by H. hepaticus
Although H. hepaticus down-regulates the majority of metabolites in Rag2−/− mice, a small fraction of molecular features nonetheless is elevated during infection. A striking observation was that multiply-charged ions predominated among these species, suggesting that they were peptides, and we extended our standard metabolomics approach to peptidomics, i.e., targeted peptide sequencing to identify potential precursor proteins which share and, consequently, may generate, these sequences by protease processing. We again relied on the exact masses, tandem MS/MS spectra and database searching; an example is shown in Figure S5. Some selected peptides that were identified, as well as corresponding presumed precursor proteins, are listed in Table 3. The annotated MS/MS spectra with matched ions and fragments are listed in Figure S6.
Table 3.
Selected identified peptides that significantly elevated in H. hepaticus infected Rag2−/− mice 20 week post-infection and their corresponding protein sources.
| Peptide sequence | Score | Fold | Protein |
|---|---|---|---|
| (T)TDTEDKGEFLSEGGGV(R) | 98 | +3.3 | Fibrinogen, alpha polypeptide isoform 2 |
| (T)TDTEDKGEFLSEGGGVR(G) | 89 | +2.1 | Fibrinogen, alpha polypeptide isoform 2 |
| (T)DTEDKGEFLSEGGGV(R) | 80 | +2.6 | Fibrinogen, alpha polypeptide isoform 2 |
| (A)ADDDYDEPTDSLDA(R) | 96 | +2.7 | Fibrinogen, beta chain precursor |
| (A)DDPSVPLQPVTPLQLFEGRRS(R) | 78 | +4.3 | Complement C4 |
| (K)ALSFYQPRAPSAEVEMTAYVLLAYLTSESSRPT(R) | 84 | +2.1 | Alpha-2-macroglobulin |
| (V)PQINDALGADESLLNRLYGFLQSGDSLNPLLASF(F) | 73 | +2.1 | Serine/threonine-protein phosphatase 6 regulatory subunit 1 |
| (Q)QPPQPEGEEDASDGGRKRGQAGWEGGYPE(I) | 68 | +2.4 | Myc-induced SUN domain containing protein |
4. Discussion and conclusion
A large number of metabolites were significantly changed in Rag2−/− mice following infection with H. hepaticus, an IBD animal model, with the majority being down-regulated in infected animals. Database searching using exact masses coupled with tandem MS/MS analysis identified a number of significantly changed metabolites. Pathway analysis revealed that tryptophan metabolism was the major perturbed pathway, followed by phenylalanine and linoleic acid metabolism (Figure S7). Also, we unambiguously demonstrated the involvement of gut microflora on the blood metabolite profiles in these animals. Moreover, we identified many elevated serum peptides, which arise from proteins with diverse functions and important implications for the development and progression of IBD. These peptides are undoubtedly generated by unknown proteases in the serum.
We have found that a number of gut microbiome-regulated serum metabolites changed during infection, which could be the consequence of impaired gut microflora induced by enteric H. hepaticus. An elegant study using germ-free and conventional mice has revealed the large effects of gut microflora on mammalian blood metabolites31. Another study also found that the metabolic rate in germ-free mice is lower than in their conventional counterparts33, highlighting the important role of gut microflora on the metabolic activity of the host. Notably, previous studies have demonstrated that helicobacter infections perturb the gut microflora in mice34,35. Particularly, the colonization dynamics of Altered Schaedler Flora is influenced by H. hepaticus and H. trogontum infection in Swiss Webster mice and B6.129P2-IL10 tm1Cgn mice, respectively34,35.
Specifically, we identified several indole-containing metabolites that decreased severalfold in infected mice. IPA appears to be a specific indicator of imbalanced gut bacteria since a gut bacterial metabolic process is needed to synthesize this compound 30. A recent study identified Clostridium sporogenes as the only species, among 24 representative intestinal flora tested, to produce IPA31. Thus, lower amounts of IPA may indicate that Clostridium sporogenes or species with similar functions have been suppressed. Hippuric acid is another gut bacteria-regulated metabolite, which decreased approximately 3-fold in mice infected for 20 weeks. This observation is in good agreement with previous reports of reduced hippurate synthesis in IBD patients36. An alteration in the intestinal microbiota has been proven responsible for the reduced synthesis of hippuric acid, although the relevant gut flora have not been identified 36. Another example to support the role of gut bacteria on the blood metabolite profile is the metabolism of daidezin, i.e., its conversion to equol by intestinal bacteria 37. The increased levels of daidezin detected in this study suggest a reduced growth or decreased enzymatic activity of metabolizing bacteria.
The large effects of gut microflora may also be reflected by down-regulated phospholipid synthesis. We detected a number of decreased Lyso-PCs and Lyso-PEs in the serum of infected animals. Although synthesis and regulation of lipids are complicated, phospholipids in blood could be impacted by the changes of gut microflora. The gut microbiota modulates lipid metabolism, and germ-free mice have lower PC species and other phospholipids in blood than do conventionally raised animals 38. Thus, the decreased amounts of phospholipids in this study may mirror another aspect of the influence of gut microflora on biochemical reactions.
We also discovered that N-methyl-histamine increased 3-fold in our animal model, which is in accordance with the previous report that IBD patients have higher N-methyl-histamine 39. Histamine N-methyltransferase catalyzes the methylation of histamine, primarily released from mast cells, in the presence of SAM 39. SAM serves as a methyl donor and is involved in methionine-homocysteine cycle. SAH is generated after losing the methyl group from SAM and is tightly regulated by this cycle. Formation of higher amounts of N-methyl-histamine should induce higher levels of SAH. Contrary to our expectation, SAH dropped approximately 2 fold in infected animals. This evidence may point to other regulated methyl transfer pathways, which also use SAM as a methyl donor. The PC synthesis via PEMT is a major user of the methyl group of SAM and regulates plasma homocysteine 32. Inhibited synthesis of PC from PE via methylation may contribute to decreased SAH. Thus, reduced synthesis of PC from PE could correlate with the methionine-homocysteine cycle and account for the down-regulation of SAH due to lower availability of SAM as a methyl donor. There is increasing appreciation of the possible association between lipid synthesis and methylation in human nutrition and diseases40,41. Reduced synthesis of PC contributes to an altered methionine-homocysteine cycle, although this does not rule out the possibility that other methylation pathways are also regulated in infected mice. In addition, an induced expression of SAH hydrolase or inhibited SAM synthetase may also lead to a lower level of SAH 42.
We have shown that the TCA cycle is inhibited in infected mice, as supported by a decreased intermediate and increased cofactor. Accumulating evidence has highlighted the important roles of gut microbiota in energy harvest and metabolism in hosts 33,43,44, which are thought to be associated with a number of diseases such as obesity, hypertension, diabetes and cardiovascular diseases. We detected decreased amounts of citric acid or isocitric acid in infected animals, suggesting that the citric cycle was inhibited to some extent. The amount of FAD concurrently increased, which is consistent with reduced citric acid cycle, as FAD is an enzyme cofactor for the conversion of succinate to fumarate, two intermediates of the TCA cycle. A recent report noted decreased amounts of amino acids and metabolites of the citric acid cycle in IBD tissue samples 45, which is in a good agreement with our observation in this animal model. There is also evidence that IBD patients have mitochondria with reduced functions, reduced ATP generation, and reduced production of antioxidants46.
Significantly altered energy metabolism may also account for decreased amounts of polyunsaturated fatty acids, including linoleic acid (LA, −2.2 fold), alpha-linoleic acid (ALA, −1.7 fold), arachidonic acid (ARA, −3.8 fold), eicosapentaenoic acid (EPA, −2.4 fold) and docosahexaenoic acid (DHA, −1.8 fold). Germ-free mice have increased fatty acid oxidation compared to conventionally raised mice33,47, and this may generate more acetyl-CoA, the entry molecule for the TCA cycle, which compensates reduced energy production in malfunctioned mitochondria or increased energy uptake during inflammation. Therefore, lower amounts of LA and ALA, as the consequence of increased oxidation, result in decreased amounts of other downstream fatty acids such as ARA, EPA and DHA. On the other hand, many functions of these fatty acids are mediated through their roles as acyl moieties of membrane phospholipids and they are released to serve as substrates for further metabolism by several important enzymes, such as cyclooxygenases etc. Thus, a greater fold change of ARA may point to its higher involvement in eicosanoid synthesis, which is stimulated by chronic inflammatory responses.
Aside from the contribution of gut microflora, it should be noted that inflammation-associated hypoxia may also play a role in defining energy demand and affecting metabolism of related pathways as discussed above48. During inflammation, a large number of immune cells are recruited to inflammatory lesions. Cell migration, phagocytosis, bacterial killing and stimulated cell proliferation have high energy demands 49,50. Neutrophils, macrophages and dendritic cells primarily use glycolytic pathways to obtain energy, while B and T cells mainly use amino acids, glucose and lipids to generate energy during oxidative phosphorylation 48. Nutrition and oxygen can be rapidly limited or depleted in tissues with high inflammatory lesions and elicited immune activities. Dysregulation of gut microflora and inflammatory hypoxia may therefore have a synergistic effect on defining the overall outcome of energy production and uptake in H. hepaticus infected Rag2−/− mice.
Taken together, the effects of gut microbiota on blood metabolites in the Rag2 model have been revealed by a variety of altered metabolites and metabolic pathways, including indole-containing compounds, glycine conjugates, lipids, critic acid and fatty acids. These observations are in good accordance with the accumulating evidence for the role of intestinal bacteria during the progress of IBD. A number of studies have shown that the commensal bacteria community is disturbed in IBD, which may have significant biological consequences on the development of this disease. Here, we provide evidence for the direct impact of gut microflora on blood metabolites in an animal IBD model. Metabolomic analysis of human IBD serum samples are currently under investigation in our laboratory, which could determine whether gut microflora have the same or similar effects on blood metabolite profiles in IBD patients as it does in these mice.
We have examined both metabolomic and peptidomic profiling in our mouse model. Peptidomics explores the peptidome that is mechanistically linked to the proteome. We have discovered a number of elevated peptides, which could be enzymatically processed products generated by proteases (Figure S8), a complex family of more than 500 enzymes. Analysis of serum peptides offers a new perspective to explore chronic inflammatory responses and to develop potential blood biomarkers. Specifically, pathway analysis revealed several precursor proteins, including fibrinogen, complement C4 and alpha-macroglobulin, that were associated with acute phase and inflammatory responses (Figure S9 A). Increases of these acute-phase proteins is a well-documented inflammatory response of the liver to cytokines secreted by activated neutrophils, macrophages and lymphocytes51. Through independent analysis using multiplex sample processing, we demonstrated that fibrinogen and alpha-2-macroglobulin were also increased in serum at the protein level. In addition, several other serum acute phase proteins were up-regulated in our animal model (Figure S9 B). These elevated serum peptides are primarily from several common proteins, such as fibrinogen and complement C4, which is consistent with the findings by Tempst, colleagues and others52–54. Particularly, DTEDKGEFLSEGGGV, DTEDKGEFLSEGGGV and TDTEDKGEFLSEGGGVR all belong to the alpha chain of fibrinogen. The difference of a single amino acid suggests a diverse enzymatic processing by proteases. The elevated peptides in our animal model could arise from increased acute phase protein expression, or increased protease activities in serum or both. However, the activity of exoproteases could mask any tissue specific or disease specific protease activity. Nevertheless, Tempst, et al., found tumor-specific serum peptidome patterns by differential exoprotease activities53. Likewise, our results may also suggest a different pattern of regulated serum protease activities in infected animals. It is well documented that protease activities are up-regulated in tissues with ongoing inflammation in IBD. For example, a previous study, using a rat IBD model induced by dinitrobenzenesulfonic acid, has revealed 6–10-fold increased levels of serine protease activity in colon tissue compared to control55. Elevated levels of protease activity in tissue samples in UC patients were also reported 55. Increased protease activities may have a key role in eliciting increased intestinal permeability, an important physiopathology phenomenon commonly observed in IBD patients. Currently, it remains unclear whether serum protease activities are regulated in IBD and whether these elevated peptides are generated in vivo, or ex vivo or both. Further investigation will shed light on this intriguing question.
In summary, we have combined metabolomics and peptidomics to analyze serum samples of H. hepaticus infected Rag2−/−. Metabolomic profiling revealed that H. hepaticus infection dramatically changed many global metabolite features, with the majority of metabolites being down-regulated. We have demonstrated the involvement of gut microflora on the blood metabolites in infected animals. The peptidomics approach identified some elevated peptides that share sequences with proteins having diverse biological functions and important implications for the progress of IBD. Taken together, the strategy of integrating a relevant animal model with sensitive MS-based profiling should provide mechanistic insights into IBD, and may suggest potential biomarkers for the diagnosis and progression of these disorders.
Supplementary Material
Acknowledgments
This work was supported by NIH/NCI Grant No. CA26371. The authors thank the MIT Center for Environmental Health Sciences for financial support by NIEHS grant (Grant No. ES002109). Charles G Knutson was supported by the MIT-Merck Fellowship. We thank Agilent Technologies for graciously providing access to the tandem quadrupole mass spectrometer and the 1290 Infinity LC system.
Abbreviations
- IBD
Inflammatory bowel disease
- H. hepaticus
Helicobacter hepaticus
- MS
Mass spectrometry
- LC-MS
Liquid chromatography-mass spectrometry
- NMR
Nuclear magnetic resonance
- Rag2−/−
Recombinase-activating gene-2-deficient
- TNF-alpha
Tumor necrosis factor-alpha
- TOF
Time-of-flight
- ESI
Electrospray ionization
- HMDB
Human Metabolome Database
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PCA
Principle component analysis
- TIC
Total ion chromatogram
- Lyso-PC
Lysophosphatidyl cholines
- SAH
S-adenosylhomocysteine
- SAM
S-adenosylmethionine
- PE
Phosphatidylethanolamine
- PC
Phosphatidylcholine
- IPA
3-indolepropionic acid
- LA
Linoleic acid
- ALA
Alpha-linoleic acid
- ARA
Arachidonic acid
- EPA
Eicosapentaenoic acid
- DHA
Docosahexaenoic acid
- PLA2
Phospholipase A2
- FAD
Flavin adenine dinucleotide
- (PEMT)
Phosphatidylethanolamine methyltransferase
- PP6
Serine/threonine-protein phosphatase 6
Footnotes
The authors declare that no competing interests exist.
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Kun Lu, Email: kunlu@mit.edu.
Charles G. Knutson, Email: knutson@mit.edu.
John S. Wishnok, Email: wishnok@mit.edu.
James G. Fox, Email: jgfox@mit.edu.
References
- 1.Kappelman MD, Rifas-Shiman SL, Kleinman K, Ollendorf D, Bousvaros A, Grand RJ, Finkelstein JA. The prevalence and geographic distribution of Crohn’s disease and ulcerative colitis in the United States. Clin Gastroenterol Hepatol. 2007;5:1424–1429. doi: 10.1016/j.cgh.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369:1627–1640. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- 4.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 5.Fava F, Danese S. Intestinal microbiota in inflammatory bowel disease: friend of foe? World J Gastroenterol. 2011;17:557–566. doi: 10.3748/wjg.v17.i5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Packey CD, Sartor RB. Interplay of commensal and pathogenic bacteria, genetic mutations, and immunoregulatory defects in the pathogenesis of inflammatory bowel diseases. J Intern Med. 2008;263:597–606. doi: 10.1111/j.1365-2796.2008.01962.x. [DOI] [PubMed] [Google Scholar]
- 7.Sartor RB. Mechanisms of disease: pathogenesis of Crohn’s disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 8.Sartor RB. Pathogenesis and immune mechanisms of chronic inflammatory bowel diseases. Am J Gastroenterol. 1997;92:5S–11S. [PubMed] [Google Scholar]
- 9.Sydora BC, Tavernini MM, Wessler A, Jewell LD, Fedorak RN. Lack of interleukin-10 leads to intestinal inflammation, independent of the time at which luminal microbial colonization occurs. Inflamm Bowel Dis. 2003;9:87–97. doi: 10.1097/00054725-200303000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Aranda R, Sydora BC, McAllister PL, Binder SW, Yang HY, Targan SR, Kronenberg M. Analysis of intestinal lymphocytes in mouse colitis mediated by transfer of CD4+, CD45RBhigh T cells to SCID recipients. J Immunol. 1997;158:3464–3473. [PubMed] [Google Scholar]
- 11.Onderdonk AB, Franklin ML, Cisneros RL. Production of experimental ulcerative colitis in gnotobiotic guinea pigs with simplified microflora. Infect Immun. 1981;32:225–231. doi: 10.1128/iai.32.1.225-231.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karlinger K, Gyorke T, Mako E, Mester A, Tarjan Z. The epidemiology and the pathogenesis of inflammatory bowel disease. Eur J Radiol. 2000;35:154–167. doi: 10.1016/s0720-048x(00)00238-2. [DOI] [PubMed] [Google Scholar]
- 13.Issa M, Vijayapal A, Graham MB, Beaulieu DB, Otterson MF, Lundeen S, Skaros S, Weber LR, Komorowski RA, Knox JF, Emmons J, Bajaj JS, Binion DG. Impact of Clostridium difficile on inflammatory bowel disease. Clin Gastroenterol Hepatol. 2007;5:345–351. doi: 10.1016/j.cgh.2006.12.028. [DOI] [PubMed] [Google Scholar]
- 14.Willing B, Halfvarson J, Dicksved J, Rosenquist M, Jarnerot G, Engstrand L, Tysk C, Jansson JK. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 15.Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR, Timmis KN, Schreiber S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 17.Erdman SE, Poutahidis T, Tomczak M, Rogers AB, Cormier K, Plank B, Horwitz BH, Fox JG. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2-deficient mice. Am J Pathol. 2003;162:691–702. doi: 10.1016/S0002-9440(10)63863-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox JG, Ge Z, Whary MT, Erdman SE, Horwitz BH. Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Mucosal Immunol. 2011;4:22–30. doi: 10.1038/mi.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erdman SE, Rao VP, Poutahidis T, Rogers AB, Taylor CL, Jackson EA, Ge Z, Lee CW, Schauer DB, Wogan GN, Tannenbaum SR, Fox JG. Nitric oxide and TNF-alpha trigger colonic inflammation and carcinogenesis in Helicobacter hepaticus-infected, Rag2-deficient mice. Proc Natl Acad Sci U S A. 2009;106:1027–1032. doi: 10.1073/pnas.0812347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schicho R, Nazyrova A, Shaykhutdinov R, Duggan G, Vogel HJ, Storr M. Quantitative metabolomic profiling of serum and urine in DSS-induced ulcerative colitis of mice by (1)H NMR spectroscopy. J Proteome Res. 2010;9:6265–6273. doi: 10.1021/pr100547y. [DOI] [PubMed] [Google Scholar]
- 21.Jansson J, Willing B, Lucio M, Fekete A, Dicksved J, Halfvarson J, Tysk C, Schmitt-Kopplin P. Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS One. 2009;4:e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murdoch TB, Fu H, MacFarlane S, Sydora BC, Fedorak RN, Slupsky CM. Urinary metabolic profiles of inflammatory bowel disease in interleukin-10 gene-deficient mice. Anal Chem. 2008;80:5524–5531. doi: 10.1021/ac8005236. [DOI] [PubMed] [Google Scholar]
- 23.Williams HR, Cox IJ, Walker DG, North BV, Patel VM, Marshall SE, Jewell DP, Ghosh S, Thomas HJ, Teare JP, Jakobovits S, Zeki S, Welsh KI, Taylor-Robinson SD, Orchard TR. Characterization of inflammatory bowel disease with urinary metabolic profiling. Am J Gastroenterol. 2009;104:1435–1444. doi: 10.1038/ajg.2009.175. [DOI] [PubMed] [Google Scholar]
- 24.Bjerrum JT, Nielsen OH, Hao F, Tang H, Nicholson JK, Wang Y, Olsen J. Metabonomics in ulcerative colitis: diagnostics, biomarker identification, and insight into the pathophysiology. J Proteome Res. 2010;9:954–962. doi: 10.1021/pr9008223. [DOI] [PubMed] [Google Scholar]
- 25.Le GG, Noor SO, Ridgway K, Scovell L, Jamieson C, Johnson IT, Colquhoun IJ, Kemsley EK, Narbad A. Metabolomics of Fecal Extracts Detects Altered Metabolic Activity of Gut Microbiota in Ulcerative Colitis and Irritable Bowel Syndrome. J Proteome Res. 2011 doi: 10.1021/pr2003598. [DOI] [PubMed] [Google Scholar]
- 26.Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, Dedon PC. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci U S A. 2012;109:E1820–E1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wegrzyn JL, Bark SJ, Funkelstein L, Mosier C, Yap A, Kazemi-Esfarjani P, La Spada AR, Sigurdson C, O’Connor DT, Hook V. Proteomics of dense core secretory vesicles reveal distinct protein categories for secretion of neuroeffectors for cell-cell communication. J Proteome Res. 2010;9:5002–5024. doi: 10.1021/pr1003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q, Tang N, Brock JW, Mottaz HM, Ames JM, Baynes JW, Smith RD, Metz TO. Enrichment and analysis of nonenzymatically glycated peptides: boronate affinity chromatography coupled with electron-transfer dissociation mass spectrometry. J Proteome Res. 2007;6:2323–2330. doi: 10.1021/pr070112q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Fox JG, Whary MT, Yan L, Shames B, Zhao Z. SCID/NCr mice naturally infected with Helicobacter hepaticus develop progressive hepatitis, proliferative typhlitis, and colitis. Infect Immun. 1998;66:5477–5484. doi: 10.1128/iai.66.11.5477-5484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young SN, Anderson GM, Gauthier S, Purdy WC. The origin of indoleacetic acid and indolepropionic acid in rat and human cerebrospinal fluid. J Neurochem. 1980;34:1087–1092. doi: 10.1111/j.1471-4159.1980.tb09944.x. [DOI] [PubMed] [Google Scholar]
- 31.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noga AA, Stead LM, Zhao Y, Brosnan ME, Brosnan JT, Vance DE. Plasma homocysteine is regulated by phospholipid methylation. J Biol Chem. 2003;278:5952–5955. doi: 10.1074/jbc.M212194200. [DOI] [PubMed] [Google Scholar]
- 33.Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ge Z, Feng Y, Taylor NS, Ohtani M, Polz MF, Schauer DB, Fox JG. Colonization dynamics of altered Schaedler flora is influenced by gender, aging, and Helicobacter hepaticus infection in the intestines of Swiss Webster mice. Appl Environ Microbiol. 2006;72:5100–5103. doi: 10.1128/AEM.01934-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whary MT, Danon SJ, Feng Y, Ge Z, Sundina N, Ng V, Taylor NS, Rogers AB, Fox JG. Rapid onset of ulcerative typhlocolitis in B6.129P2-IL10tm1Cgn (IL-10−/−) mice infected with Helicobacter trogontum is associated with decreased colonization by altered Schaedler’s flora. Infect Immun. 2006;74:6615–6623. doi: 10.1128/IAI.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams HR, Cox IJ, Walker DG, Cobbold JF, Taylor-Robinson SD, Marshall SE, Orchard TR. Differences in gut microbial metabolism are responsible for reduced hippurate synthesis in Crohn’s disease. BMC Gastroenterol. 2010;10:108–116. doi: 10.1186/1471-230X-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atkinson C, Frankenfeld CL, Lampe JW. Gut bacterial metabolism of the soy isoflavone daidzein: exploring the relevance to human health. Exp Biol Med (Maywood ) 2005;230:155–170. doi: 10.1177/153537020523000302. [DOI] [PubMed] [Google Scholar]
- 38.Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu P, Yetukuri L, Islam S, Felin J, Perkins R, Boren J, Oresic M, Backhed F. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res. 2010;51:1101–1112. doi: 10.1194/jlr.M002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winterkamp S, Weidenhiller M, Otte P, Stolper J, Schwab D, Hahn EG, Raithel M. Urinary excretion of N-methylhistamine as a marker of disease activity in inflammatory bowel disease. Am J Gastroenterol. 2002;97:3071–3077. doi: 10.1111/j.1572-0241.2002.07028.x. [DOI] [PubMed] [Google Scholar]
- 40.Selley ML. A metabolic link between S-adenosylhomocysteine and polyunsaturated fatty acid metabolism in Alzheimer’s disease. Neurobiol Aging. 2007;28:1834–1839. doi: 10.1016/j.neurobiolaging.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Tehlivets O. Homocysteine as a risk factor for atherosclerosis: is its conversion to s-adenosyl-L-homocysteine the key to deregulated lipid metabolism? J Lipids. 2011;2011:702853. doi: 10.1155/2011/702853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malanovic N, Streith I, Wolinski H, Rechberger G, Kohlwein SD, Tehlivets O. S-adenosyl-L-homocysteine hydrolase, key enzyme of methylation metabolism, regulates phosphatidylcholine synthesis and triacylglycerol homeostasis in yeast: implications for homocysteine as a risk factor of atherosclerosis. J Biol Chem. 2008;283:23989–23999. doi: 10.1074/jbc.M800830200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Musso G, Gambino R, Cassader M. Obesity, diabetes, and gut microbiota: the hygiene hypothesis expanded? Diabetes Care. 2010;33:2277–2284. doi: 10.2337/dc10-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venema K. Role of gut microbiota in the control of energy and carbohydrate metabolism. Curr Opin Clin Nutr Metab Care. 2010;13:432–438. doi: 10.1097/MCO.0b013e32833a8b60. [DOI] [PubMed] [Google Scholar]
- 45.Ooi M, Nishiumi S, Yoshie T, Shiomi Y, Kohashi M, Fukunaga K, Nakamura S, Matsumoto T, Hatano N, Shinohara M, Irino Y, Takenawa T, Azuma T, Yoshida M. GC/MS-based profiling of amino acids and TCA cycle-related molecules in ulcerative colitis. Inflamm Res. 2011;60:831–840. doi: 10.1007/s00011-011-0340-7. [DOI] [PubMed] [Google Scholar]
- 46.Schoultz I, Soderholm JD, McKay DM. Is metabolic stress a common denominator in inflammatory bowel disease? Inflamm Bowel Dis. 2010;17:2008–2018. doi: 10.1002/ibd.21556. [DOI] [PubMed] [Google Scholar]
- 47.Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kominsky DJ, Campbell EL, Colgan SP. Metabolic shifts in immunity and inflammation. J Immunol. 2010;184:4062–4068. doi: 10.4049/jimmunol.0903002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borregaard N, Herlin T. Energy metabolism of human neutrophils during phagocytosis. J Clin Invest. 1982;70:550–557. doi: 10.1172/JCI110647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 51.Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340:448–454. doi: 10.1056/NEJM199902113400607. [DOI] [PubMed] [Google Scholar]
- 52.Koomen JM, Li D, Xiao LC, Liu TC, Coombes KR, Abbruzzese J, Kobayashi R. Direct tandem mass spectrometry reveals limitations in protein profiling experiments for plasma biomarker discovery. J Proteome Res. 2005;4:972–981. doi: 10.1021/pr050046x. [DOI] [PubMed] [Google Scholar]
- 53.Villanueva J, Shaffer DR, Philip J, Chaparro CA, Erdjument-Bromage H, Olshen AB, Fleisher M, Lilja H, Brogi E, Boyd J, Sanchez-Carbayo M, Holland EC, Cordon-Cardo C, Scher HI, Tempst P. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest. 2006;116:271–284. doi: 10.1172/JCI26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Villanueva J, Nazarian A, Lawlor K, Tempst P. Monitoring peptidase activities in complex proteomes by MALDI-TOF mass spectrometry. Nat Protoc. 2009;4:1167–1183. doi: 10.1038/nprot.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hawkins JV, Emmel EL, Feuer JJ, Nedelman MA, Harvey CJ, Klein HJ, Rozmiarek H, Kennedy AR, Lichtenstein GR, Billings PC. Protease activity in a hapten-induced model of ulcerative colitis in rats. Dig Dis Sci. 1997;42:1969–1980. doi: 10.1023/a:1018887832465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.