Abstract
Obesity is known to be influenced by a number of genes, including the β3 subunit of G protein (GNB3), β3-adrenergic receptor (ADRB3), uncoupling protein 2 (UCP2), and peroxisome proliferator activated receptor gamma (PPARγ). The single nucleotide polymorphisms (SNPs) of the above genes, such as GNB3-C825T, ADRB3-Trp64Arg, UCP2-3′UTR 45 bp del/ins, and PPARγ-Pro12Ala, are associated with obesity and body mass index. The present study evaluates the impact of Bofutsushosan, a traditional Eastern Asian herbal medicine with known anti-obesity properties, on obese subjects according to the presence of the above-mentioned SNPs. Upon randomization, the volunteers were allocated to receive Bofutsushosan (n=55) or placebo (n=56) treatments for 8 weeks. Following the treatment schedule, significant reductions in total cholesterol and significant improvement in the Korean version of obesity-related quality of life scale were seen in the Bofutsushosan-treated group, but not in placebo. Bofutsushosan exerted significant anti-obesity effects on a number of parameters in the carriers of the GNB3-825T allele, but only on waist circumference in the GNB3-C/C homozygote. Significant anti-obesity impact of Bofutsushosan was also seen on a number of obesity-indices in both ADRB3-Arg64 carriers and ADRB3-Trp64 homozygotes, as well as in UCP2-D/D carriers, but not in UCP2-D/I+I/I variants. The effect of Bofutsushosan was more pronounced in PPARγ-Pro/Pro genotype compared to PPARγ-Pro/Ala variants. Thus, the results revealed differential responses of the subjects to the anti-obesity effects of Bofutsushosan treatment according to the polymorphism of the vital obesity-related genes. Our study provides new insight into individualized clinical applications of Bofutsushosan for obesity.
Key Words: : Bofutsushosan, obesity, single nucleotide polymorphism, clinical trial
Introduction
Obesity is one of the most alarming global public health problems, contributing to type 2 diabetes, hypertension and cardiovascular diseases, many forms of cancer, and death. Obesity is influenced by genetic and environmental factors and their interactions. Many studies have focused on the associations between specific genes and obesity phenotypes, with 426 findings of positive associations with 127 candidate genes.1 The most significant observation is that 22 genes are each supported by at least five positive studies.2 Accordingly, GNB3, ADRB3, PPARγ, and UCP2 have been found to be encoded by genes associated with obesity and body composition.2
The pharmacogenomics of obesity is a new and challenging area of research, which could be a valuable tool to understand why some patients being treated for obesity respond to pharmacological and/or behavioral interventions, while others do not (cited by Irizarry et al.).3 A single nucleotide polymorphism (SNP) is a variation in a single base pair in the DNA sequence of a particular gene, such as insertion, deletion, or substitution of a base, and represents the most common form of genetic variation. For the past few years, SNPs have been used extensively to explore obesity candidate gene regions, selected on functional and/or positional basis. Accordingly, GNB3-C825T, ADRB3-Trp64Arg, UCP2-3′UTR 45 bp del/ins, and PPARγ-Pro12Ala polymorphisms have been demonstrated to be SNPs that are associated with obesity and body mass index (BMI).1,4–18
Bofutsushosan, a traditional Eastern Asian herbal medicinal formulation that contains 18 crude drugs, is an effective agent for treating various disease states, especially obesity and metabolic disorders. Previous animal studies have demonstrated the anti-obesity effects of Bofutsushosan.19 Experimental results with rodent models have suggested that Bofutsushosan prevents adipogenesis in white adipocytes through the modulation of gene expression,20 and attenuates the weight and size gains of white adipose tissue (WAT), along with upregulation of uncoupling protein 1 (UCP1) mRNA in WAT.21 Additionally, a clinical trial on obese Japanese women concluded that Bofutsushosan could be a useful herbal medicine in treating obesity with impaired glucose tolerance.22 In our previous study on obese Korean volunteers where we evaluated the impact of Bofutsushosan on the Asian type obesity pattern, it was revealed that this herbal formulation significantly decreased body weight, BMI, waist circumference (WC), body fat mass, total cholesterol (T-Chol), and high-density lipoprotein cholesterol (HDL-Chol) in the subjects categorized under “liver Qi stagnation” pattern and WC in the subjects diagnosed with “indigestion” pattern.23
Although there have been a few clinical reports concerning Bofutsushosan treatment, to the best of our knowledge, no studies have been conducted to address the influence of genetic variations of GNB3, ADRB3, UCP2, and PPARγ simultaneously on Bofutsushosan response in obese subjects. Genetic polymorphisms greatly account for the interindividual differences in the responses to drug treatment for a particular disease, as well as for adverse drug reactions. For instance, type 2 diabetic patients with the Pro12Ala genotype in the PPAR2 gene exhibited better therapeutic response to rosiglitazone (a thiazolidinedione class of antidiabetic drug) than did by patients with the Pro12Pro genotype.24 In another study, a change in systolic blood pressure in hypertensive patients treated with irbesartan (an angiotensin II receptor antagonist, which is used primarily for the treatment of hypertension) was found to be related to the ApoA-IV A1449G, ApoA-V C31455T, and ApoB C711T SNPs of the apolipoprotein gene family.25 The presence of the C-allele of the LDLR C16730T SNP of low-density lipoprotein receptor gene was found to be associated with decreased systolic blood pressure in hypertensive patients treated with atenolol (a selective β1 receptor antagonist, which is used mainly for the treatment of hypertension and angina).25 Furthermore, a clinical study demonstrated that overweight and obese participants with LS or SS but not LL genotype for serotonin transporter (5-HTT ) gene had greater weight loss with sibutramine (an inhibitor of the reuptake of the neurotransmitters norepinephrine and serotonin, which was used before in the management of obesity) treatment compared to placebo.26 Moreover, it has been revealed that the XmnI and BCL11A SNPs may help to predict the response to hydroxyurea (an antineoplastic drug, which can also reactivate fetal hemoglobin production and thereby is frequently used to treat β-thalassemia) in Iranian β-thalassemia patients.27 It has been suggested that the presence of the XmnI T/T genotype or the BCL11A rs766432 C allele correlates strongly with response to hydroxyurea.27
The above observations prompted us to find whether any relationship exists between the genetic polymorphisms of GNB3-C825T, ADRB3-Trp64Arg, UCP2-3′UTR 45 bp del/ins, and PPARγ-Pro12Ala and the anti-obesity impact of Bofutsushosan. This study, which represents the extension of our previous work, was conducted on obese Korean subjects.23
Subjects and Methods
Subjects
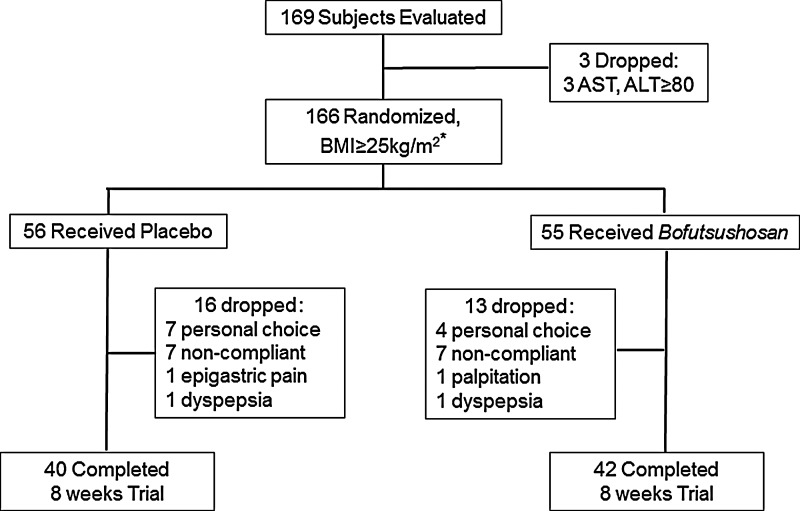
The clinical study presented here was part of the trial conducted by us in Dongguk University Oriental Hospital, Korea, to evaluate the efficacy and adverse effects of Bofutsushosan and Boiogito according to the oriental obesity pattern.23 Briefly, a total of 166 obese subjects were enrolled following the initial screening of 169 volunteers who responded to advertisements in local newspapers or the Web site of Dongguk University Oriental Hospital. The volunteers were recruited between August 2009 and September 2010, and the study was completed in November, 2010. A total of 56 subjects were randomly assigned to the placebo group and 55 subjects to the Bofutsushosan group (Fig. 1).
FIG. 1.
Flow chart of the participation history of the subjects. *Following randomization, 55 subjects (who were not part of the present study) received Boiogito treatment (for other study) that is not shown in this figure.
To qualify for this study, subjects had to be ethnically Korean, in general good physical condition, a nonsmoker, 18–65 years of age, with a BMI ≥25 kg/m2, and with a body weight that had remained stable within±3 kg during the previous 3 months. The basic aim and principles of the study, possible adverse effects of the treatment, as well as the right to withdraw from the study were thoroughly explained to all screened subjects. The subjects were then requested to read the informed consent forms carefully and sign the forms if they agreed to participate in the study. A physician interviewed and scrutinized the general health conditions of the volunteers. Exclusion criteria included: malignancy, heart, kidney, or liver diseases, history of alanine aminotransferase (ALT) or aspartate aminotransferase (AST) enzyme activities >2.5 times the normal upper limits, or level of creatinine >2.0 mg/dL. Subjects receiving medications that may affect the body composition and metabolism, and subjects who had displayed a hypersensitive reaction to a drug were also excluded. Furthermore, pregnancy at any time during the study was also among the exclusion criteria. The detailed inclusion and exclusion criteria of the present study are summarized in Table 1. The medical, scientific, and ethical aspects of the study were approved by the Institutional Review Board of Dongguk University Oriental Hospital.
Table 1.
Study Selection Criteria
| Inclusion criteria | Exclusion criteria |
|---|---|
| 1. 18–65 years of age 2. BMI ≥25 kg/m2 3. Stable body weight (≤±3 kg) in the preceding 3 months 4. Sedentary individuals 5. Otherwise healthy nonsmokers 6. Subjects signed informed consent forms |
1. Subjects with significant history or currently suffering from cardiovascular disease, hepatic or renal dysfunctions, cancer • ALT, AST >2.5 times normal range • Creatinine >2.0 mg/dL • Clinically significant abnormal ECG 2. Patient taking drugs that could interfere with the study 3. Known to have allergic responses or sensitivity to any of the medication 4. Pregnancy |
Study design
The study was a randomized, double-blind, placebo-controlled, 8-week trial. Following their recruitment, the subjects were randomized with the use of a table of random numbers compiled by an independent statistician. The active or Bofutsushosan group received Bofutsushosan capsules (12 capsules a day, each containing 237 mg of Bofutsushosan dry extract; Hanpoong Pharm. Co., Seoul, Korea), whereas the placebo group received capsules (12 capsules a day, 250 mg) filled with corn starch. The Bofutsushosan formulation is summarized in Table 2. The above formulation and dose of Bofutsushosan was selected on the basis of an earlier clinical study on Korean obese subjects performed for 12 weeks,28 which revealed a marked impact on body weight, WC, and BMI of the subjects.
Table 2.
The Composition of Bofutsushosan Dry Extract
| Crude drug | Content (mg)a |
|---|---|
| Zingiberis Rhizoma Recens | 16.7 |
| Glycyrrhizae Radix | 83.3 |
| Scutellariae Radix | 83.3 |
| Gardeniae Fructus | 50.0 |
| Gypsum Fibrosum | 83.3 |
| Saposhnikoviae Radix | 50.0 |
| Rhei Rhizoma | 62.5 |
| Natrii Sulfas | 31.3 |
| Platycodi Radix | 83.3 |
| Atractylodis marcrocephalae Rhizoma | 83.3 |
| Schizonepetae Herba | 50.0 |
| Chuanxiong Rhizoma | 50.0 |
| Angelicae Gigantis Radix | 50.0 |
| Menthae Herba | 50.0 |
| Ephedrae Herba | 50.0 |
| Forsythiae Fructus | 50.0 |
| Talcum | 125 |
| Paeoniae Radix Alba | 50.0 |
The amount of each crude drug required to prepare 237 mg of Bofutsushosan dry extract.
The randomization code was completely double-blinded to the research investigators and participants. The code was transferred to Hanpoong Pharm. Co. where Bofutsushosan and placebo capsules were manufactured and labeled according to the code, which was not revealed until the completion of the clinical trial. A serial number was marked on the surface of the box containing Bofutsushosan or placebo capsules, and the research pharmacist distributed those capsules to the participants. The capsules were administered orally three times a day, four capsules at one time. At each visit to the hospital, the capsules for a 2-week dose regimen were provided, and the subjects were asked to revisit once every second week during the 8-week period of the study. The subjects were also asked to return the capsules that they did not take for any reason, and this parameter was counted as a means of determining their adherence to the dose regimen. If final compliance to the treatment schedule was <80%, the data for that subject were excluded due to the violation of the protocol of the clinical trial. The randomization code was unblinded only after all the clinical and laboratory data collection was completed.
Subjects were educated on the dietary guidelines and were asked to consume 20–25 kcal/day per kg body weight, and to note down their total dietary intake within the entire 24 h period just before the day of the visit. Upon each visit, their dietary intake was assessed during an interview and with reference to the 24 h dietary recall notes. The data were recorded in the case report forms. The subjects were also asked to maintain their current lifestyle, without any new exercise program.
Measurements
The efficacy of treatment was assessed by measuring the changes in body weight, BMI, WC, body fat percent, body fat mass, resting metabolic rate (RMR), and serum clinical parameters, and by completion of the Korean version of obesity-related quality of life (KOQOL) scale. The safety of the treatment was evaluated by measuring the changes in cardiovascular parameters (blood pressure, heart rate, and electrocardiogram), liver enzymatic activities, and kidney function parameters, as well as by assessing the self-reported symptoms of the subjects. The genotypic analyses of the subjects to determine the mutation of particular genes were conducted before starting the treatment schedule.
The subjects' blood pressure was measured at every visit following a subject rest period of more than 10 min using a mercury sphygmomanometer (Sankei, Tokyo, Japan). The pulse rate was measured from the radial artery while the subject was sitting after resting for more than 10 min. The electrocardiogram of each subject was recorded and analyzed in a dedicated test room.
Anthropometric assessments
The height of each subject was measured to the nearest 0.1 cm at their first visit. Body weight was measured to the nearest 0.1 kg using a model GM1000 electro body weight scale (Neo GM Tech, Incheon, Korea) at their first visit and every second week of the study. WC was determined while each subject was in an upright posture by measuring the circumference of the midsection between the lowest costal margin and the highest iliac crest (3 cm top of the anterior superior iliac spine). The same skilled examiner verified the intravariability of the measurements, and the mean of three consecutive measurements was recorded.
Determination of body fat parameters
Body fat percent and body fat mass of the subjects were measured at the first visit and at weeks 4 and 8 using bioelectrical impedance (BIA) with an Inbody 720 unit (Biospace, Seoul, Korea).
Assessment of RMR
The RMR of the subjects were measured at the first visit, and at weeks 4 and 8 using a MedGem closed circuit indirect calorimeter (Health Tech, Golden, CO, USA). The measurement was performed following a 30 min rest.
Measurement of serum clinical parameters
The serum biochemical parameters were assessed at baseline and at the end of the study. Levels of glucose and lipid parameters were measured from blood samples collected following a 12 h fast. All serum clinical parameters were analyzed as described previously.9
Evaluation of KOQOL scale
Each subject completed a questionnaire to evaluate the KOQOL scale as described previously.29 The questionnaire is relatively simple to complete, but yields data that are highly reliable and accurate. For this measurement, each subject answered 15 questions on six different areas (mental and social health, physical health, job and domestic duty, daily life, sexual relations, and food). Based on the response, the KOQOL scale was determined. This evaluation was performed before and after the treatment schedule.
Genotype analyses
Genomic DNA was isolated from whole blood by proteinase K digestion using a QIAmp DNA blood Mini Kit (Qiagen, Valencia, CA, USA) following the manufacturer's instructions. The purity and concentration of DNA in the samples were determined spectrophotometrically. Subsequently, polymerase chain reaction (PCR) was performed to amplify the genomic DNA fragment representing a particular gene polymorphism. The amplification reactions were carried out using a commercial ready-to-use PCR reagent (AccuPower® PCR PreMix; Bioneer, Daejeon, Korea) in a total reaction volume of 20 μL containing PCR PreMix, 1 μL of cDNA, and specific primers (10 pmol for each). The sequences of the primers (Cosmogenetech, Seoul, Korea) are depicted in Table 3. The optimized annealing temperatures of the primers for PCR reactions were as follows: 60°C (C825T and Trp64Arg), 58°C (UCP2 del/ins), and 54°C (Pro12Ala). The PCR amplification conditions were as follows: a prior incubation step at 94°C for 10 min followed by 35 cycles of amplification encompassing denaturation at 94°C for 60 sec, annealing at the corresponding optimized temperature for 45 sec, extension at 72°C for 60 sec, and a final extension at 72°C for 10 min. The resultant PCR products were then either kept intact (for UCP2 del/ins) or digested with restriction enzymes BseD1 (for C825T; Fermentas, Thermo Fisher Scientific, Waltham, MA, USA), Mva1 (for Trp64Arg; Takara Bio, Shiga, Japan), or Hha1 (for Pro12Ala; Takara Bio). Finally, all the products were subjected to electrophoresis in a 1.5% agarose gel. The resulting genotype-specific band patterns were as follows: C825T: T/T, single band of 268 bp; C/T, three bands of 268, 152, and 116 bp; C/C, two bands of 152 and 116 bp; Trp64Arg, Trp64 homozygote: five bands of 99, 62, 30, 12, and 7 bp; Trp64/Arg64 heterozygote (also termed “Arg64 carrier” in this study), six bands of 161, 99, 62, 30, 12, and 7 bp (notably, the 30, 12, and 7 bp fragments were too small to be resolved on the gel); Arg64 homozygote, expected to be represented by four bands of 161, 30, 12, and 7 bp (there were no subjects who were homozygous for Arg64); UCP2 del/ins: D/D, a single band of 457 bp; D/I, two bands of 457 bp and 502 bp; I/I, a single band of 502 bp; Pro12Ala: Pro/Pro, a single band of 154 bp; Pro/Ala, three bands of 154, 132, and 22 bp; Ala/Ala: expected to be represented by two bands of 132 and 22 bp (there were no subjects who were homozygous for Ala).
Table 3.
Primer Sequences Used for Single Nucleotide Polymorphisms Analyses
| SNPs | Forward primer (5′-3′) | Reverse primer (5′-3′) | Ref. |
|---|---|---|---|
| C825T GNB3 | TGA CCC ACT TGC CAC CCG TGC | GCA GCA GCC AGG GCT GGC | 31 |
| Trp64Arg ADRB3 | CGC CCA ATA CCG CCA ACAC | CCA CCA GGA GTC CCA TCA CC | 17,32 |
| UCP2 del/ins | CAG TGA GGG AAG TGG GAG G | GGG GCA GGA CGA AGA TTC | 33 |
| Pro12Ala PPARγ2 | TCT GGG AGA TTC TCC TAT TGG | GTG GAA GAC AAC TAC AAG AG | 34 |
Statistical analyses
The data are expressed as mean±SD. All statistical analyses were carried out using SAS v9.1 (SAS Institute, Cary, NC, USA). Statistical evaluation was performed by the intention to treat (ITT) approach to ensure unbiased comparisons among the experimental groups, and the last observation carried forward (LOCF) approach was used to impute values for the missing data.
Independent t tests were employed to determine the level of significance for the following: (1) the differences between the placebo and Bofutsushosan groups (a) in the general characteristics of the subjects at baseline (Table 4) and (b) in the changes in general and safety outcome variables of the subjects after intervention (Tables 5 and 11, respectively); (2) the difference in the change of a given outcome parameter between the allelic variants for ADRB3, UCP2, and PPAR-γ2 genes in Bofutsushosan-treated subjects (Tables 7–9). One-way analysis of variance (ANOVA) was conducted to determine the level of significance in the difference in the change of a given outcome variable among the SNPs of GNB3 (Table 6). A paired t test was carried out for the comparison between pre- and post-treatment measurements of outcome variables under a particular evaluation (Tables 5–9 and 11). Differences were considered significant at P<.05.
Table 4.
Baseline Characteristics of the Subjects
| Bofutsushosan (n=55) | Placebo (n=56) | P valuea | |
|---|---|---|---|
| Age, years | 41.56±8.62 | 39.21±10.12 | .191 |
| Height, cm | 160.5±6.79 | 161.79±7.85 | .357 |
| Body weight, kg | 74.53±10.98 | 76.86±11.7 | .279 |
| WC, cm | 97.63±7.45 | 98.99±7.81 | .380 |
| BMI, kg/m2 | 29.72±6.17 | 29.28±3.11 | .640 |
| Body fat, % | 38.33±4.98 | 39.02±5.39 | .489 |
| Body fat mass, kg | 28.61±6.14 | 29.87±6.83 | .309 |
| RMR, kcal/d | 1396.73±383.55 | 1453.39±335.02 | .414 |
| Fasting BST, mg/dL | 99±9.33 | 98.36±9.04 | .713 |
| T-Chol, mg/dL | 207.69±39.36 | 191.84±40.12 | .038* |
| HDL, mg/dL | 55.27±14.7 | 50.29±11.32 | .047* |
| TG, mg/dL | 127.35±58.62 | 123.02±67.02 | .718 |
| KOQOL | 33.51±7.6 | 34.52±8.56 | .513 |
Bofutsushosan vs. placebo by independent t test.
P<.05.
WC, waist circumference; BMI, body mass index; RMR, resting metabolic rate; BST, blood sugar test; T-Chol, total cholesterol; HDL, high-density lipoprotein; TG, triglyceride; KOQOL, Korean version of obesity-related quality of life.
Table 5.
Changes in the Outcome Variables of the Subjects After Intervention
| Bofutsushosan (n=55) | Placebo (n=56) | P valuea | |
|---|---|---|---|
| Body weight, kg | |||
| Week 8 – baseline | −0.86±1.73 | −1.05±1.66 | .622 |
| P valueb | .0005* | .0001* | |
| WC, cm | |||
| Week 8 – baseline | −1.79±2.93 | −1.84±2.68 | .442 |
| P valueb | .0003* | .0001* | |
| BMI, kg/m2 | |||
| Week 8—baseline | −0.32±0.66 | −0.41±0.59 | .334 |
| P valueb | .0008* | .0001* | |
| Body fat percent | |||
| Week 8 – baseline | −0.67±1.6 | −0.68±1.26 | .742 |
| P valueb | .0096* | .0014* | |
| Body fat mass, kg | |||
| Week 8 – baseline | −0.97±1.65 | −0.89±1.25 | .401 |
| P valueb | .0005* | .0001* | |
| RMR, kcal/d | |||
| Week 8 – baseline | −11.19±451.4 | −12.2±495.24 | .911 |
| P valueb | .8731 | .8755 | |
| Fasting BST, mg/dL | |||
| Week 8 – baseline | 0.14±8.16 | 0.32±5.5 | .783 |
| P valueb | .9102 | .7138 | |
| T-Chol, mg/dLc | |||
| Week 8 – baseline | −11.45±27.39 | −3.85±19.65 | .586 |
| P valueb | .0098* | .2165 | |
| HDL, mg/dLc | |||
| Week 8 – baseline | −2.81±8.24 | 1.07±7.68 | .157 |
| P valueb | .0327* | .3762 | |
| TG, mg/dL | |||
| Week 8 – baseline | −7.43±59.61 | −15.34±59.73 | .366 |
| P valueb | .4239 | .1079 | |
| KOQOL | |||
| Week 8 – baseline | −2.69±7.1 | −1.31±14.92 | .382 |
| P valueb | .0184* | .5727 | |
| SBP, mmHg | |||
| Week 8 – baseline | −1.9±11.43 | −4.76±11.29 | .782 |
| P valueb | .2864 | .0102* | |
| DBP, mmHg | |||
| Week 8 – baseline | −1.24±9.73 | −3.76±11.87 | .265 |
| P valueb | .4145 | .0494* | |
| PR, bpm | |||
| Week 8 – baseline | −2.67±8.1 | −0.27±9.96 | .343 |
| P valueb | .0389* | .8640 | |
Bofutsushosan vs. placebo (by independent t test).
Pre- vs. post-treatment within group (by paired t test).
The treatment effects on this parameter were adjusted using general linear model.
P<.05.
SBP, systolic blood pressure; DBP, diastolic blood pressure; PR, pulse rate.
Table 11.
Changes in the Safety Outcome Variables of the Subjects
| Bofutsushosan (n=55) | Placebo (n=56) | P valuea | |
|---|---|---|---|
| ALT at baseline, IU/L | 22.36±16.86 | 22±17.16 | |
| Week 8 – baseline | −3.57±8.72 | −1.88±8.89 | .732 |
| P valueb | .0113* | 0.1838 | |
| AST at baseline, IU/L | 21.31±10.38 | 19.66±7.69 | |
| Week 8 – baseline | −2.14±5.94 | −1.17±5.5 | .573 |
| P valueb | .0243* | .1801 | |
| BUN at baseline, mg/dL | 14.02±4.15 | 12.99±3.53 | |
| Week 8 – baseline | −0.09±3.43 | 0.05±3.18 | .181 |
| P valueb | .8688 | .9223 | |
| Creatinine at baseline, mg/dL | 0.69±0.13 | 0.67±0.1 | |
| Week 8 – baseline | 0±0.08 | −0.01±0.08 | .305 |
| P valueb | .7476 | .49 |
Bofutsushosan vs. placebo (by independent t test).
Pre- vs. post-treatment within group (by paired t test).
P<.05.
ALT, alanine aminotransferase; AST, aspartate aminotransferase; BUN, blood urine nitrogen.
Table 7.
Changes in the Outcome Variables of the Subjects According to ADRB3 Polymorphism
| Bofutsushosan | ||||
|---|---|---|---|---|
| Genotypea | Pre | Post | P valueb | |
| Body weight, kg | Arg64 | 75.16±13.45 | 73.97±12.34 | .009* |
| Trp64 | 74.69±9.32 | 73.99±9.74 | .0219* | |
| BMI, kg/m2 | Arg64 | 29.53±3.65 | 29.1±3.46 | .0157* |
| Trp64 | 30.06±7.54 | 29.79±7.64 | .0222* | |
| WC, cm | Arg64 | 96.66±7.63 | 94.79±7.46 | .2446 |
| Trp64 | 98.84±7.09 | 95.67±6.38 | .0001* | |
| Body fat % | Arg64 | 39.63±5.59 | 39.06±5.73 | .6331 |
| Trp64 | 37.69±4.55 | 36.29±4.48 | .001* | |
| Body fat mass, kg | Arg64 | 29.9±7.7 | 28.47±6.97 | .1078 |
| Trp64 | 28.13±4.86 | 26.63±4.16 | .0007* | |
| RMR, kcal/d | Arg64 | 1363.33±390.02 | 1402.78±313.23 | .5657 |
| Trp64 | 1448.44±374.08 | 1417.08±320.67 | .4641 | |
| TG, mg/dL | Arg64 | 128.67±60.88 | 124±65.82 | .3917 |
| Trp64 | 125.81±59.69 | 100.83±41.34 | .0671 | |
| T-Chol, mg/dL | Arg64 | 203.33±32.47 | 191.67±28.09 | .015* |
| Trp64 | 208.41±43.39 | 199.33±38.93 | .1235 | |
| HDL-Chol, mg/dL | Arg64 | 53.9±13.69 | 52.61±10.69 | .0484* |
| Trp64 | 56.69±15.69 | 55.75±14.1 | .2849 | |
| Fasting BST, mg/dL | Arg64 | 98.38±8.29 | 95.22±5.78 | .1183† |
| Trp64 | 99.63±10.24 | 100.13±13.1 | .196 | |
| SBP, mmHg | Arg64 | 118.43±13.79 | 117.83±11.23 | .7191 |
| Trp64 | 120.28±11.85 | 118.58±15.04 | .2931 | |
| DBP, mmHg | Arg64 | 73.52±10.69 | 76.17±8.97 | .1892 |
| Trp64 | 77.53±7.38 | 74.17±9.56 | .0206*† | |
| PR, bpm | Arg64 | 77.33±9.03 | 73.67±8.07 | .0528 |
| Trp64 | 75.91±9.49 | 74.13±9.12 | .323 | |
Distribution (number of patients): Arg64, 21; Trp64, 32; unidentified, 2.
Pre- vs. post-treatment (by paired t test).
P<.05.
Significant difference (P<.05) between the allelic variants (by independent t test).
Table 8.
Changes in the Outcome Variables of the Subjects According to UCP2 Polymorphism
| Bofutsushosan | ||||
|---|---|---|---|---|
| Genotypea | Pre | Post | P valueb | |
| Body weight, kg | D/D | 74.88±10.08 | 73.92±10.02 | .0018* |
| D/I+I/I | 74.87±13.5 | 74.15±12.74 | .1291 | |
| BMI, kg/m2 | D/D | 30.24±7.16 | 29.88±7.24 | .0027* |
| D/I+I/I | 28.84±2.85 | 28.6±2.64 | .1464 | |
| WC, cm | D/D | 97.83±7.28 | 94.47±7.17 | <.0001*† |
| D/I+I/I | 98.33±7.64 | 97.14±5.69 | .9738 | |
| Body fat % | D/D | 38.47±5.11 | 37.4±5.2 | .0099* |
| D/I+I/I | 38.43±5 | 37.65±5.34 | .5913 | |
| Body fat mass, kg | D/D | 28.94±6.54 | 27.56±6.27 | .0011* |
| D/I+I/I | 28.55±5.17 | 27.12±3.6 | .2127 | |
| RMR, kcal/d | D/D | 1449.21±325.62 | 1399.66±318.97 | .4825 |
| D/I+I/I | 1327.33±492.03 | 1436.15±312.85 | .3643 | |
| TG, mg/dL | D/D | 131.05±63.19 | 120.38±56.36 | .7846 |
| D/I+I/I | 116.53±49.76 | 89.31±41.7 | .122 | |
| T-Chol, mg/dL | D/D | 206.89±39.65 | 197.62±36.04 | .0421* |
| D/I+I/I | 205.13±39.25 | 192.54±31.99 | .1113 | |
| HDL-Chol, mg/dL | D/D | 55.13±15.87 | 52.59±12.07 | .014* |
| D/I+I/I | 56.73±12.31 | 58.46±13.63 | .7338 | |
| Fasting BST, mg/dL | D/D | 98.29±9.9 | 98.21±12.39 | .4004 |
| D/I+I/I | 101.27±8.13 | 97.62±6.21 | .265 | |
| SBP, mmHg | D/D | 119.03±12.55 | 118.69±14.27 | .42 |
| D/I+I/I | 120.87±12.92 | 117.31±11.69 | .5079† | |
| DBP, mmHg | D/D | 75.5±9.24 | 75.52±8.6 | .5645 |
| D/I+I/I | 77.07±8.42 | 73.92±10.87 | .5807 | |
| PR, bpm | D/D | 77.13±9.78 | 74.86±9.55 | .0861 |
| D/I+I/I | 74.8±7.78 | 71.85±5.68 | .2798 | |
Distribution (number of patients): D/D, 38; D/I, 13; I/I, 2; unidentified, 2.
Pre- vs. post-treatment (by paired t test).
P<.05.
Significant difference (P<.05) between the allelic variants (by independent t test).
Table 9.
Changes in the Outcome Variables of the Subjects According to PPAR-γ2 Polymorphism
| Bofutsushosan | ||||
|---|---|---|---|---|
| Genotypea | Pre | Post | P valueb | |
| Body weight, kg | Pro/Pro | 74.74±11.18 | 73.83±10.83 | .0007* |
| Pro/Ala | 72.42±9.48 | 72.04±9.84 | .4971 | |
| BMI, kg/m2 | Pro/Pro | 29.84±6.38 | 29.51±6.42 | .0011* |
| Pro/Ala | 28.46±3.57 | 28.3±3.63 | .4997 | |
| WC, cm | Pro/Pro | 97.41±7.48 | 95.33±7 | .0007* |
| Pro/Ala | 99.9±7.55 | 94.83±3.88 | .2529 | |
| Body fat percent | Pro/Pro | 38.46±4.92 | 37.65±5.3 | .0108* |
| Pro/Ala | 36.96±6.01 | 35.27±2.71 | .6304 | |
| Body fat mass, kg | Pro/Pro | 28.75±5.97 | 27.66±5.65 | .0006* |
| Pro/Ala | 27.2±8.36 | 24.27±2.6 | .5254 | |
| RMR, kcal/d | Pro/Pro | 1405.2±393.65 | 1424.1±317.29 | .9526 |
| Pro/Ala | 1312±279.05 | 1240±240 | .7511 | |
| TG, mg/dL | Pro/Pro | 122.5±52.27 | 112.59±55.28 | .8844† |
| Pro/Ala | 175.8±98.47 | 187±16 | .1888 | |
| T-Chol, mg/dL | Pro/Pro | 206.28±39.46 | 195.72±35.42 | .0284* |
| Pro/Ala | 221.8±39.59 | 200.33±24.01 | .2013 | |
| HDL-Chol, mg/dL | Pro/Pro | 54.64±14.94 | 53.13±12.06 | .0101* |
| Pro/Ala | 61.6±11.22 | 71±10.15 | .0488*† | |
| Fasting BST, mg/dL | Pro/Pro | 98.9±9.71 | 98.03±11.13 | .8444 |
| Pro/Ala | 100±4.18 | 98±5.29 | .849 | |
| SBP, mmHg | Pro/Pro | 119.12±12.39 | 118.44±12.8 | .5772 |
| Pro/Ala | 122.4±16.18 | 116±23.64 | .2415 | |
| DBP, mmHg | Pro/Pro | 75.46±9.36 | 75.85±8.45 | .8583 |
| Pro/Ala | 76.8±4.32 | 64.33±14.64 | .2049† | |
| PR, bpm | Pro/Pro | 76.76±9.09 | 74.28±8.77 | .053* |
| Pro/Ala | 71.6±10.16 | 69.33±3.79 | .5352 | |
Distribution (number of patients): Pro/Ala, 5; Pro/Pro, 50.
Pre- vs. post-treatment (by paired t test).
P<.05.
Significant difference (P<.05) between the allelic variants (by independent t test).
Table 6.
Changes in the Outcome Variables of the Subjects According to GNB3 Polymorphism
| Bofutsushosan | ||||
|---|---|---|---|---|
| Genotypea | Pre | Post | P valueb | |
| Body weight, kg | C/C | 70.53±8.77 | 69.33±7.6 | .12 |
| C/T | 75.33±13.11 | 74.16±12.58 | .0147* | |
| T/T | 74.75±9.04 | 73.82±9.26 | .0705 | |
| BMI, kg/m2 | C/C | 27.52±1.17 | 27.1±0.66 | .1486 |
| C/T | 29.59±3.16 | 29.14±2.99 | .0155* | |
| T/T | 28.12±2.38 | 27.79±2.43 | .077 | |
| WC, cm | C/C | 95.8±8.38 | 93.07±6.4 | .0488* |
| C/T | 97.49±6.99 | 95.9±7.24 | .0206* | |
| T/T | 96.91±7.31 | 95.19±6.33 | .0581 | |
| Body fat percent | C/C | 36.22±6.36 | 35.75±6.68 | .5142 |
| C/T | 39.2±4.95 | 38.64±5.18 | .1148 | |
| T/T | 37±3.74 | 36.02±4.06 | .0433* | |
| Body fat mass, kg | C/C | 25.28±4.03 | 24.5±3.93 | .251 |
| C/T | 29.52±6.64 | 28.54±5.93 | .015* | |
| T/T | 27.7±5.04 | 26.63±5.09 | .0325* | |
| RMR, kcal/d | C/C | 1318.33±351.53 | 1448.33±217.39 | .4527 |
| C/T | 1434.17±445.61 | 1382.5±354.71 | .6238 | |
| T/T | 1450±340.69 | 1449.17±279.56 | .9938 | |
| TG, mg/dL | C/C | 129.33±56.22 | 171.33±92.78 | .2938 |
| C/T | 122±56.33 | 108.04±38.32 | .2592 | |
| T/T | 105±42.93 | 85.92±33.11 | .0462* | |
| T-Chol, mg/dL | C/C | 187.33±20.29 | 195.83±34.14 | .6638 |
| C/T | 214.04±46.27 | 197.46±39.34 | .0003* | |
| T/T | 204.5±39.73 | 193.33±25.8 | .2085 | |
| HDL-Chol, mg/dL | C/C | 48.33±10.21 | 50.17±7.25 | .2257 |
| C/T | 57.08±14.06 | 52.83±12.17 | .023* | |
| T/T | 61.92±19.13 | 59.67±14.93 | .398 | |
| Fasting BST, mg/dL | C/C | 101±6.99 | 99.5±10.63 | .6539 |
| C/T | 96.5±8.67 | 96.13±6.73 | .7912 | |
| T/T | 99.08±8.66 | 101.08±16.36 | .5351 | |
| SBP, mmHg | C/C | 115±1 2.98 | 112.17±13.69 | .6535 |
| C/T | 120.33±11.92 | 119.79±13.08 | .7927 | |
| T/T | 122.42±14.85 | 118.25±14.16 | .295 | |
| DBP, mmHg | C/C | 71.67±8.66 | 75±12.07 | .4765 |
| C/T | 75.75±10.18 | 76.79±8.52 | .5608 | |
| T/T | 79.58±5.73 | 71.5±9 | .007*† | |
| PR, bpm | C/C | 80.67±14.51 | 76±10.04 | .3493 |
| C/T | 76.21±8.71 | 73.83±9.11 | .1687 | |
| T/T | 75.33±8.69 | 73.08±7.22 | .275 | |
Distribution (number of patients): C/C, 7; C/T, 28; T/T, 18; unidentified, 2.
Pre- vs. post-treatment (by paired t test).
P<.05.
Significant difference (P<.05) among the allelic variants (by one-way ANOVA).
Results
Participation history and general characteristics of the subjects at baseline
There were no differences in the general characteristics of the subjects in the two groups at baseline (Table 4) except for T-Chol and HDL-Chol. Accordingly, a general linear model was applied to adjust the treatment effects on the above two cholesterol parameters. During the study, 29 subjects dropped out because of personal choice, noncompliance, or adverse effects of the treatments. Accordingly, 82 subjects completed the study, 42 in the Bofutsushosan group, and 40 in the placebo group (Fig. 1).
Effects of treatments on clinical outcome
Following completion of the study, body weight, WC, BMI, body fat percent, and body fat mass were significantly reduced in both Bofutsushosan and placebo groups, although the changes were not significant between groups (Table 5). However, the levels of T-Chol and HDL-Chol after intervention were found to be significantly lower than those at baseline in the Bofutsushosan group (P=.01 and .03, respectively) but not in the placebo group. Moreover, following the treatment schedule, the KOQOL score was not changed significantly in the placebo group, but it was significantly improved in the Bofutsushosan group (P=.02). Interestingly, after intervention, systolic blood pressure (SBP) and diastolic blood pressure (DBP) values were reduced in both the placebo and Bofutsushosan groups, but only significantly in the former group (P=.01 and .049 for SBP and DBP, respectively). However, the changes in those two parameters did not differ significantly between the groups. In contrast, following intervention, the pulse rate of the Bofutsushosan group but not of the placebo group was significantly reduced compared to the baseline.
Impact of GNB3-C825T polymorphism on the response to Bofutsushosan treatment
After the completion of the trial, the following C825T polymorphism-specific changes in the clinical outcomes were evident in the Bofutsushosan group (Table 6). In the C/C genotype, WC was significantly reduced, whereas in the C/T genotype, body weight (P=.015), BMI (P=.015), WC (P=.02), body fat mass (P=.015), T-Chol (P=.0003), and HDL-Chol (P=.02) were significantly decreased. On the other hand, in the T/T genotype, body fat percent (P=.04), body fat mass (P=.03), TG (P=.046), and DBP (P=.007) were significantly reduced.
Impact of ADRB3-Trp64Arg polymorphism on the response to Bofutsushosan treatment
After the completion of the study, the following Trp64Arg polymorphism-specific changes in the clinical outcomes were noted in the Bofutsushosan group (Table 7). In Arg64 carrier, body weight (P=.009), BMI (P=.016), T-Chol (P=.015), and HDL-Chol (P=.048) were significantly reduced. In the Trp64 homozygote, body weight (P=.02), BMI (P=.02), WC (P=.001), body fat percent (P=.001), body fat mass (P=.007), and DBP (P=.0206) were significantly decreased. Notably, a significant depletion in fasting glucose was noticed in the Arg64 carrier when the change in this parameter due to Bofutsushosan treatment was compared between Arg64 carrier and Trp64 homozygote.
Impact of UCP2-del/ins polymorphism on the response to Bofutsushosan treatment
Only a very small number of subjects (n=2) were I/I carriers in the Bofutsushosan group. Accordingly, the clinical outcome variables of the I/I genotypic group was combined with that of D/I genotypic group to evaluate the impact of Bofutsushosan on I allele carriers.
After the completion of the study, following del/ins polymorphism-specific changes in the clinical outcomes were observed in the Bofutsushosan group (Table 8). In the D/D genotype, body weight (P=.002), BMI (P=.003), body fat percent (P=.01), body fat mass (P=.001), T-Chol (P=.04), and HDL-Chol (P=.01) were significantly reduced. On the other hand, in the D/I+I/I genotype, no significant difference in any of the outcome variables was evident. Notably, the degree of depletion in WC in the D/D genotype was also found to be significantly greater than that in the D/I+I/I genotype. On the other hand, the decrease in SBP in the D/I+I/I genotype was significantly greater compared to that in the D/D genotype.
Impact of Pro12Ala polymorphism of PPARγ2 on the response to Bofutsushosan treatment
After the completion of the study, the following Pro12Ala polymorphism-specific changes in the outcomes were observed (Table 9). In the Pro/Pro genotype, body weight (P=.0007), BMI (P=.001), WC (P=.0007), body fat percent (P=.01), body fat mass (P=.0006), T-Chol (P=.03), and HDL-Chol (P=.01) were significantly decreased, while in the Pro/Ala genotype, HDL-Chol was significantly increased (P=.0488). Notably, significant depletion in TG and DBP was noticed in the Pro/Pro and Pro/Ala genotypes, respectively, when the changes in these two parameters were compared between the genotypes.
Adverse effects of the treatments
The adverse effects of the treatments on the subjects are summarized in Table 10. During the entire period of our study, a total of 19 adverse events were reported by 15 subjects. The most prevalent symptom was gastrointestinal problems such as dyspepsia and epigastric pain. In the Bofutsushosan group, 15 adverse cases included dyspepsia and epigastric pain (n=7), headache (n=2), diarrhea (n=3), nausea and vomiting (n=2), and palpitations (n=1). In the placebo group, four adverse cases included dyspepsia and epigastric pain (n=3) and headache (n=1). Notably, although there were no significant adverse effects that might greatly affect subject health, four subjects were dropped from the trial: two from the Bofutsushosan group (one case of palpitations and one case of dyspepsia) and two from the placebo group (one case of epigastric pain and one case of dyspepsia; Fig. 1).
Table 10.
Adverse Events Reported in the Study
| Treatment group | Dyspepsia, epigastric pain | Headache | Diarrhea | Nausea, vomiting | Insomnia | Palpitations | Total |
|---|---|---|---|---|---|---|---|
| Bofutsushosan (n=55) | 7 | 2 | 3 | 2 | 1 | 15 | |
| Placebo (n=56) | 3 | 1 | 4 | ||||
| Total | 10 | 3 | 3 | 2 | 1 | 19 |
Safety evaluation
The evaluation of the safety of the proposed treatments was based on the examination of liver- and kidney-related clinical indices of the subjects through blood test at both baseline and the end of the study. The outcome of this analysis is depicted in Table 11. Accordingly, Bofutsushosan treatment significantly decreased the activity of both ALT (P=.01) and AST (P=.02), whereas those two parameters were not significantly altered in the placebo group. However, the changes in ALT and AST activities in the Bofutsushosan group did not differ significantly from that in the placebo group. Notably, the changes in the level of blood urea nitrogen and creatinine were insignificant in both placebo and Bofutsushosan groups, and the differences between the groups were also insignificant.
Discussion
The present 8-week, randomized, placebo-controlled, double-blind clinical trial evaluated the anti-obesity impact of Bofutsushosan, as well as the association between the polymorphisms in obesity-related genes and the response of the subjects to Bofutsushosan treatment.
Both Bofutsushosan and placebo treatment resulted in a significant reduction in body weight, WC, BMI, body fat percent, and body fat mass. Although there were no significant differences in the above parameters between the placebo and Bofutsushosan groups, the data still revealed the following. First, in the Bofutsushosan group, there was a significant reduction in the level of T-Chol, but not in the placebo group. Second, following the completion of treatment schedule, the KOQOL score was significantly improved in the Bofutsushosan group, but not in the placebo group. Thus, our findings are in keeping with earlier reports that have substantiated the anti-obesity effect of Bofutsushosan.19–22
Notably, after completion of the trial, significant reductions in both SBP and DBP were observed in the placebo group, but not in the Bofutsushosan group, although the differences in those changes between the groups were insignificant. So far, no detailed studies have been conducted to evaluate the impact of Bofutsushosan treatment on the blood pressure of obese subjects. Indeed, changes in blood pressure after placebo administration have been seen in the placebo groups of a number of placebo-controlled trials.30,31 It has been suggested that certain conditions that may influence the clinical BP outcome are psychological factors such as the medical environment or the relationship between the patient and the physician,30 including verbal suggestions.31 It has also been opined that the verbal suggestions-induced fall of SBP is specifically mediated by autonomic nervous system efferents that are involved in the regulation of blood pressure, although the precise mechanism behind this is yet to be understood.31 Accordingly, without further detailed studies, no definite conclusion can be drawn from our study on the observed placebo effects on the subjects' blood pressure.
The family of genes responsible for obesity includes those expressing heterotrimetric G proteins, which are the vital components of the intracellular signaling pathway and which have a central role in adipogenesis. A common C→T substitution at nucleotide 825 of the gene encoding the β3 subunit of the G protein, commonly known as GNB3 C825T polymorphism, is associated with a number of pathophysiological conditions including obesity.32,33 It has been revealed that BMI, waist girth, hip girth, subscapular skinfold thickness, triceps skinfold thickness, and body fat percent are significantly higher in subjects with the T/T genotype compared to subjects with the C/T or C/C genotypes.32,33
Bofutsushosan treatment produced significant anti-obesity effects on a number of obesity-related clinical parameters in the carriers of the GNB3 825T allele, whereas in C/C homozygotes, the same treatment produced significant anti-obesity impact only on WC. Thus, our results demonstrate a more targeted anti-obesity impact of Bofutsushosan on the T allele, which is a clinically important finding. These findings are also comparable to an earlier study,26 where the 825(T/T+C/T) genotype, but not the 825C/C genotype, was associated with greater weight loss upon treatment with sibutramine, a weight-reducing drug that inhibits presynaptic reuptake of serotonin and norepinephrine, thereby causing accumulation of these neutrotransmitters in between the synapses. Accordingly, we could not exclude the possibility of a similar scenario in Bofutsushosan treatment, as ephedrine, the active ingredient of Ephedra in Bofutsushosan, is an amphetamine-type central nervous system stimulant that augments the release of norepinephrine from its storage vesicles.34 However, since Bofutsushosan is a multiherbal formulation rather than a single entity, no definite conclusion can be made on this postulation. Future in-depth studies (trials and observational studies) are needed to ascertain the mode of drug–gene interaction between Bofutsushosan and the C825T polymorphism.
ADRB3, which is a G protein coupled receptor, plays an important role in the regulation of thermogenesis and lipolysis in adipose tissue, as well as delivery of free fatty acids into the portal vein. A missense mutation in the codon 64 of ADRB3 that leads to the replacement of tryptophan by arginine (Trp64Arg) in the first intracellular loop of the receptor is associated with abdominal obesity and insulin resistance, as well as early development of type 2 diabetes.35,36 Our study revealed that the anti-obesity effects of Bofutsushosan are directed differentially among the Arg64 and TRP64 carriers of ADRB3 gene. This is relevant to an earlier clinical report that elucidated the effects of TRP64Arg polymorphism on multiple-dose rosiglitazone response in patients suffering from type 2 diabetes,37 where, although the Arg64 genotype had greater attenuated serum TG and lesser attenuated LDL-Chol, as well as smaller enhancement of adiponectin levels compared with the TRP64 carriers, there were no significant differences in most diabetes-related clinical parameters between these two genotypes. Additionally, in an earlier clinical study, no difference in the β1/β2/β3 AR-agonist isoproterenol– or ADRB3-specific agonist CGP-12117–induced lipolysis was seen among the omental fat tissues harvested from the Trp64/TRp64, TRP64/Arg64, or Arg64/Arg64 variants.38 Taking all this into consideration, we hypothesize that the differential anti-obesity effects of Bofutsushosan among the Trp64 and Arg64 carriers in our study might be attributable to the active ingredient ephedrine, as this compound is a nonspecific adrenergic agonist and, accordingly, its mode of action in the subjects might be differentially affected by a base mutational change of only one member of the adrenoceptor family. However, further detailed studies are required to understand the mode of drug–gene interaction between Bofutsushosan and Trp64Arg polymorphism.
Uncoupling protein 2 (UCP2), which is ubiquitously expressed in adult human tissues, plays an important role in energy balance, body weight regulation, and thermoregulation. A 45 bp del/ins polymorphism in the 3′-untranslated region (UTR) of exon 8 of the UCP2 gene39 is one of the common UCP2 gene variants in humans. It has been reported that the 3′ del/ins polymorphism in the UCP2 gene is associated with obesity and BMI in different populations.1 The I/I genotype was shown to be associated with increased BMI in South Indian women.40 It has been hypothesized that D/I polymorphism may influence UCP2 mRNA stability, which, in turn, could affect the expression of this protein and determine body weight by modulating energy expenditure and thermogenesis.40 Any impairment in UCP2 mRNA stability could compromise a subject's ability to remove excess calories through thermogenesis, particularly in a person who is inclined to obesity from other genetic or environmental influences.40 This hypothesis is partially supported by the observation of decreased UCP2 mRNA levels in the visceral fat of obese, but not lean, subjects.41
In the present study, Bofutsushosan treatment resulted in significant improvement in a number of obesity-related clinical parameters in the subjects with the UCP2 D/D genotype, while the same treatment did not produce significant change in any of the studied parameters in the UCP2 D/I+I/I variants. Thus, our results suggest that the anti-obesity effects of Bofutsushosan on the subjects are specifically disposed to the D/D genotype of 3′UTR UCP2 polymorphism. Notably, previous studies have revealed that β3-adrenergic agonists could induce the expression of UCP2 gene in a number of tissues,42,43 and perhaps thereby exert a thermogenic anti-obesity effect. The above findings suggest that in our study, probably ephedrine in Bofutsushosan, an indirect β3-adrenergic agonist, might induce UCP2 gene expression. However, this event could not be presently translated into the synthesis of UCP2 mRNA in D/I or I/I, but not in D/D carriers, most likely because of the destabilization of the newly produced UCP2 transcript in the former two genotypes. Accordingly, Bofutsushosan treatment might result in an augmented production of UCP2 protein in D/D carriers, thereby exerting a more pronounced anti-obesity impact on the D/D genotype compared to the other two UCP2 SNP variants studied.
The nuclear hormone receptor, PPARγ, is a transcriptional factor that plays a major role in adipogenesis,44 as well as in the induction of transcription of specific anti-obesity target genes.45 A number of genetic polymorphisms in the PPARγ gene have been discovered. Among them, Pro12Ala variant, a missense mutation, is most prevalent in the PPARγ2 isoform involving a C→G substitution at nucleotide 34 resulting in the replacement of a proline by alanine at residue 12 of the PPARγ2 protein.46 Functionally, the Ala variant of PPARγ2 has been demonstrated to have a reduced affinity for the response element and a lower capacity for activating target genes.47 Several studies have aimed to find a relationship between the Pro12Ala polymorphism and metabolic syndrome, but the results have been equivocal. However, an extensive meta-analysis using data from 30 independent studies with a total number of 19,136 subjects revealed that BMI was significantly higher in Ala allele carriers compared with Pro allele homozygotes.17 In female Korean subjects, body weight, BMI, and waist-to-hip ratio were significantly higher in Ala allele carriers than in noncarriers,48 consistent with the results of the above-mentioned meta-analysis.17 In our study, the Ala/Ala genotype was absent among the subjects, and the results presented here are derived from the Pro/Pro and Pro/Ala variants, which revealed that the anti-obesity impact of Bofutsushosan was more pronounced in the wild type Pro/Pro genotype than in mutated Pro/Ala variant where the functionality of PPARγ2 is thought to be impaired.47 This suggests a possible interaction between PPARγ and the ingredients of Bofutsushosan, especially Ephedra, although no definite conclusion can be made on this postulation. However, our hypothesis is in keeping with earlier studies,49,50 suggesting a cross-talk of the PPARγ pathway with signaling cascade of cAMP, the cellular content of which can be modulated by Ephedra.
Gastrointestinal problems, which were the most common adverse events reported in our study, were noticed in both the placebo and Bofutsushosan groups, but predominantly in the latter group. It has been opined that Bofutsushosan treatment may cause or aggravate anorexia, epigastric distress, nausea, vomiting, abdominal pain, soft feces, and diarrhea in the subjects.51 Indeed, in a clinical trial, 9 out of 127 obese patients receiving Bofutsushosan treatment experienced side effects, including diarrhea, abdominal pain, and abdominal discomfort.52 In another clinical study on obese Japanese females, the Bofutsushosan-treated group showed more improvement in constipation compared to the placebo group.22 However, in a preclinical study using a mouse model, diarrhea was found to be associated with Bofutsushosan treatment in the control (nonobese) group, but not in the disease (obese and diabetic) group.19 Moreover, repeated oral administration of Bofutsushosan in Sprague Dawley rats for 13 weeks did not produce any adverse effects in the animals in either sex up to and including doses of 2000 mg/kg/ per day.53 Thus, taking the above into consideration, no definite conclusion can be drawn on the possibility that gastrointestinal problems are a consequence of Bofutsushosan treatment. Furthermore, our safety assessment revealed that Bofutsushosan treatment did not produce any toxic or adverse impact on hepatic or renal function of the subjects. In our opinion, Bofutsushosan and/or certain subject-specific intrinsic factors as well as diet status might play an important role in contributing the gastrointestinal disorders in the volunteers. Furthermore, we cannot exclude the possible contribution of the SNPs to the adverse response to Bofutsushosan treatment. Many adverse drug reactions can be associated with SNPs in genes that regulate aspects of drug disposition. The members of these genes encompass drug metabolizing enzymes, which include phase 1 enzymes such as cytochrome P450 superfamily, phase 2 enzymes such as microsomal epoxide hydrolase, and receptors and transporters such as multidrug resistance protein 1 (MDR1) P-glycoprotein. Further studies are needed to investigate whether SNPs of the above genes and those we studied may contribute to the adverse response to Bofutsushosan treatment.
In conclusion, differential responses of the obesity indices to Bofutsushosan treatment were revealed according to the GNB3-C825T, ADRB3-Trp64Arg, UCP2-3′UTR 45 bp ins/del, and PPARγ-Pro12Ala SNPs of the subjects. Our results provide new insights into the clinical efficacy of Bofutsushosan.
Acknowledgments
This study was supported by a grant from the Traditional Korean Medicine R&D Project, Ministry for Health & Welfare & Family Affairs, Republic of Korea (B090014). The authors also thankfully acknowledge the Editor-in-Chief, Journal of Oriental Rehabilitation Medicine, for granting permission to reproduce some of the data from our earlier publication in the above journal.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Yang W, Kelly T, He J: Genetic epidemiology of obesity. Epidemiol Rev 2007;29:49–61 [DOI] [PubMed] [Google Scholar]
- 2.Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Pérusse L, Bouchard C: The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14:529–644 [DOI] [PubMed] [Google Scholar]
- 3.Irizarry K, Hu G, Wong ML, Licinio J, Lee CJ: Single nucleotide polymorphism identification in candidate gene systems of obesity. Pharmacogenomics J 2001;1:193–203 [DOI] [PubMed] [Google Scholar]
- 4.Klenke S, Kussmann M, Siffert W: The GNB3 C825T polymorphism as a pharmacogenetic marker in the treatment of hypertension, obesity, and depression. Pharmacogenet Genomics 2011;21:594–606 [DOI] [PubMed] [Google Scholar]
- 5.Hsiao TJ, Hwang Y, Liu CH, Chang HM, Lin E: Association of the C825T polymorphism in the GNB3 gene with obesity and metabolic phenotypes in a Taiwanese population. Genes Nutr 2013;8:137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oizumi T, Daimon M, Saitoh T, Kameda W, Yamaguchi H, Ohnuma H, Igarashi M, Eguchi H, Manaka H, Tominaga M, Kato T: Genotype Arg/Arg, but not Trp/Arg, of the Trp64Arg polymorphism of the beta(3)-adrenergic receptor is associated with type 2 diabetes and obesity in a large Japanese sample. Diabetes Care 2001;24:1579–1583 [DOI] [PubMed] [Google Scholar]
- 7.Widen E, Lehto M, Kanninen T, Walston J, Shuldiner AR, Groop LC: Association of a polymorphism in the beta 3-adrenergic-receptor gene with features of the insulin resistance syndrome in Finns. N Engl J Med 1995;333:348–351 [DOI] [PubMed] [Google Scholar]
- 8.Mitchell BD, Blangero J, Comuzzie AG, Almasy LA, Shuldiner AR, Silver K, Stern MP, MacCluer JW, Hixson JE: A paired sibling analysis of the beta-3 adrenergic receptor and obesity in Mexican Americans. J Clin Invest 1998;101:584–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malik SG, Saraswati MR, Suastika K, Trimarsanto H, Oktavianthi S, Sudoyo H: Association of beta3-adrenergic receptor (ADRB3) Trp64Arg gene polymorphism with obesity and metabolic syndrome in the Balinese: a pilot study. BMC Res Notes 2011;4:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee YH, Kim W, Yu BC, Park BL, Kim LH, Shin HD: Association of the ins/del polymorphisms of uncoupling protein 2 (UCP2) with BMI in a Korean population. Biochem Biophys Res Commun 2008;371:767–771 [DOI] [PubMed] [Google Scholar]
- 11.Yanovski JA, Diament AL, Sovik KN, Nguyen TT, Li H, Sebring NG, Warden CH: Associations between uncoupling protein 2, body composition, and resting energy expenditure in lean and obese African American, white, and Asian children. Am J Clin Nutr 2000;71:1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cassell PG, Neverova M, Janmohamed S, Uwakwe N, Qureshi A, McCarthy MI, Saker PJ, Albon L, Kopelman P, Noonan K, Easlick J, Ramachandran A, Snehalatha C, Pecqueur C, Ricquier D, Warden C, Hitman GA: An uncoupling protein 2 gene variant is associated with a raised body mass index but not Type II diabetes. Diabetologia 1999;42:688–692 [DOI] [PubMed] [Google Scholar]
- 13.Evans D, Minouchehr S, Hagemann G, Mann WA, Wendt D, Wolf A, Beisiegel U: Frequency of and interaction between polymorphisms in the beta3-adrenergic receptor and in uncoupling proteins 1 and 2 and obesity in Germans. Int J Obes Relat Metab Disord 2000;24:1239–1245 [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Chu WS, Lu T, Hasstedt SJ, Kern PA, Elbein SC: Uncoupling protein-2 polymorphisms in type 2 diabetes, obesity, and insulin secretion. Am J Physiol Endocrinol Metab 2004;286:E1–E7 [DOI] [PubMed] [Google Scholar]
- 15.Walder K, Norman RA, Hanson RL, Schrauwen P, Neverova M, Jenkinson CP, Easlick J, Warden CH, Pecqueur C, Raimbault S, Ricquier D, Silver MH, Shuldiner AR, Solanes G, Lowell BB, Chung WK, Leibel RL, Pratley R, Ravussin E: Association between uncoupling protein polymorphisms (UCP2-UCP3) and energy metabolism/obesity in Pima Indians. Hum Mol Genet 1998;7:1431–1435 [DOI] [PubMed] [Google Scholar]
- 16.Mattevi VS, Zembrzuski VM, Hutz MH: Effects of a PPARG gene variant on obesity characteristics in Brazil. Braz J Med Biol Res 2007;40:927–932 [DOI] [PubMed] [Google Scholar]
- 17.Masud S, Ye S: Effect of the peroxisome proliferator activated receptor-gamma gene Pro12Ala variant on body mass index: a meta-analysis. J Med Genet 2003;40:773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dedoussis GV, Vidra N, Butler J, Papoutsakis C, Yannakoulia M, Hirschhorn JN, Lyon HN: Peroxisome proliferator-activated receptor-gamma (PPARgamma) Pro12Ala polymorphism and risk for pediatric obesity. Clin Chem Lab Med 2009;47:1047–1050 [DOI] [PubMed] [Google Scholar]
- 19.Shimada T, Kudo T, Akase T, Aburada M: Preventive effects of Bofutsushosan on obesity and various metabolic disorders. Biol Pharm Bull 2008;31:1362–1367 [DOI] [PubMed] [Google Scholar]
- 20.Yamakawa J, Ishigaki Y, Takano F, Takahashi T, Yoshida J, Moriya J, Takata T, Tatsuno T, Sasaki K, Ohta T, Takegami T, Yoshizaki F: The Kampo medicines Orengedokuto, Bofutsushosan and Boiogito have different activities to regulate gene expressions in differentiated rat white adipocytes: comprehensive analysis of genetic profiles. Biol Pharm Bull 2008;31:2083–2089 [DOI] [PubMed] [Google Scholar]
- 21.Akagiri S, Naito Y, Ichikawa H, Mizushima K, Takagi T, Handa O, Kokura S, Yoshikawa T: Bofutsushosan, an oriental herbal medicine, attenuates the weight gain of white adipose tissue and the increased size of adipocytes associated with the increase in their expression of uncoupling protein 1 in high-fat diet-fed male KK/Ta mice. J Clin Biochem Nutr 2008;42:158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hioki C, Yoshimoto K, Yoshida T: Efficacy of bofu-tsusho-san, an oriental herbal medicine, in obese Japanese women with impaired glucose tolerance. Clin Exp Pharmacol Physiol 2004;31:614–619 [DOI] [PubMed] [Google Scholar]
- 23.Park JH, Lee MJ, Kim HJ, Hong SW, Lee DK, Yoo JW, Choi SM, Moon JS, Lim CY, Lee JB: Efficacy and adverse events of bangpungtongseong-san (bofutsusho-san) and bangkihwangki-tang (boiogiot-tang) by oriental obesity pattern identification on obese subjects: randomized, double blind, placebo-controlled trial. J Oriental Rehab Med 2011;21:265–278 [Google Scholar]
- 24.Kang ES, Park SY, Kim HJ, Kim CS, Ahn CW, Cha BS, Lim SK, Nam CM, Lee HC: Effects of Pro12Ala polymorphism of peroxisome proliferator-activated receptor gamma2 gene on rosiglitazone response in type 2 diabetes. Clin Pharmacol Ther 2005;78:202–208 [DOI] [PubMed] [Google Scholar]
- 25.Liljedahl U, Lind L, Kurland L, Berglund L, Kahan T, Syvänen AC: Single nucleotide polymorphisms in the apolipoprotein B and low density lipoprotein receptor genes affect response to antihypertensive treatment. BMC Cardiovasc Disord 2004;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grudell AB, Sweetser S, Camilleri M, Eckert DJ, Vazquez-Roque MI, Carlson PJ, Burton DD, Braddock AE, Clark MM, Graszer KM, Kalsy SA, Zinsmeister AR; A controlled pharmacogenetic trial of sibutramine on weight loss and body composition in obese or overweight adults. Gastroenterology 2008;135:1142–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banan M, Bayat H, Azarkeivan A, Mohammadparast S, Kamali K, Farashi S, Bayat N, Khani MH, Neishabury M, Najmabadi H: The XmnI and BCL11A single nucleotide polymorphisms may help predict hydroxyurea response in Iranian β-thalassemia patients. Hemoglobin 2012;36:371–380 [DOI] [PubMed] [Google Scholar]
- 28.Shin DH, Cho GH, Lee H, Moon MK., Kang DG, Yun YG, Park DS, Juhng SK, Lee HS: Clinical study of Bangpoongtongsungsan on body weight change in subjects with obesity. Korean J Oriental Med Prescrip 2008;16:133–144 [Google Scholar]
- 29.Park HS, Sun WS, Ou SW, Lee KY, Kim BS, Han JH, Kim SM, Lee HR, Yu BY, Lee KM, Suh YS, Nam YD, Park YW, Shin HC, Lee JK: Development of Korean version of obesity-related quality of life scale. Kor J Obes 2003;12:280–293 [Google Scholar]
- 30.Asmar R, Safar M, Queneau P: Evaluation of the placebo effect and reproducibility of blood pressure measurement in hypertension. Am J Hypertens 2001;14:546–552 [DOI] [PubMed] [Google Scholar]
- 31.Meissner K: The placebo effect and the autonomic nervous system: evidence for an intimate relationship. Phil Trans R Soc B 2011;366:1808–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stefan N, Stumvoll M, Machicao F, Koch M, Häring HU, Fritsche A: C825T polymorphism of the G protein beta3 subunit is associated with obesity but not with insulin sensitivity. Obes Res 2004;12:679–683 [DOI] [PubMed] [Google Scholar]
- 33.Hegele RA, Anderson C, Young TK, Connelly PW: G-protein beta3 subunit gene splice variant and body fat distribution in Nunavut Inuit. Genome Res 1999;9:972–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothman RB, Baumann MH, Dersch CM, Romero DV, Rice KC, Carroll FI, Partilla JS: Amphetamine-type central nervous system stimulants release norepinephrine more potently than they release dopamine and serotonin. Synapse 2001;39:32–41 [DOI] [PubMed] [Google Scholar]
- 35.Walston J, Silver K, Bogardus C, Knowler WC, Celi FS, Austin S, Manning B, Strosberg AD, Stern MP, Raben N, John D. Sorkin JD, Roth J, Shuldiner AR: Time of onset of non-insulin-dependent diabetes mellitus and genetic variation in the beta 3-adrenergic-receptor gene. N Engl J Med 1995;333:343–347 [DOI] [PubMed] [Google Scholar]
- 36.Kadowaki H, Yasuda K, Iwamoto K, Otabe S, Shimokawa K, Silver K, Walston J, Yoshinaga H, Kosaka K, Yamada N, Saito Y, Hagura R, Akanuma Y, Shuldiner A, Yazaki Y, Kadowaki T: A mutation in the beta 3-adrenergic receptor gene is associated with obesity and hyperinsulinemia in Japanese subjects. Biochem Biophys Res Commun 1995;215:555–560 [DOI] [PubMed] [Google Scholar]
- 37.Yang M, Huang Q, Wu J, Yin JY, Sun H, Liu HL, Zhou HH, Liu ZQ: Effects of UCP2-866 G/A and ADRB3 Trp64Arg on rosiglitazone response in Chinese patients with type 2 diabetes. Br J Clin Pharmacol 2009;68:14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Umekawa T, Yoshida T, Sakane N, Kogure A, Kondo M, Honjyo H: Trp64Arg mutation of beta3-adrenoceptor gene deteriorates lipolysis induced by beta3-adrenoceptor agonist in human omental adipocytes. Diabetes 1999;48:117–120 [DOI] [PubMed] [Google Scholar]
- 39.Dalgaard LT, Pedersen O: Uncoupling proteins: functional characteristics and role in the pathogenesis of obesity and type II diabetes. Diabetologia 2001;44:946–965 [DOI] [PubMed] [Google Scholar]
- 40.Cassell PG, Neverova M, Janmohamed S, Uwakwe N, Qureshi A, McCarthy MI, Saker PJ, Albon L, Kopelman P, Noonan K, Easlick J, Ramachandran A, Snehalatha C, Pecqueur C, Ricquier D, Warden C, Hitman GA: An uncoupling protein 2 gene variant is associated with a raised body mass index but not type II diabetes. Diabetologia 1999;42:688–692 [DOI] [PubMed] [Google Scholar]
- 41.Oberkofler H, Liu YM, Esterbauer H, Hell E, Krempler F, Patsch W: Uncoupling protein-2 gene: reduced mRNA expression in intraperitoneal adipose tissue of obese humans. Diabetologia 1998;41:940–946 [DOI] [PubMed] [Google Scholar]
- 42.Gong DW, He Y, Karas M, Reitman M: Uncoupling protein-3 is a mediator of thermogenesis regulated by thyroid hormone, beta3-adrenergic agonists, and leptin. J Biol Chem 1997;272:24129–24132 [DOI] [PubMed] [Google Scholar]
- 43.Boss O, Samec S, Dulloo A, Seydoux J, Muzzin P, Giacobino JP: Tissue-dependent upregulation of rat uncoupling protein-2 expression in response to fasting or cold. FEBS Lett 1997;412:111–114 [DOI] [PubMed] [Google Scholar]
- 44.Lee CH, Olson P, Evans RM: Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology 2003;144:2201–2207 [DOI] [PubMed] [Google Scholar]
- 45.Houtkooper RH, Auwerx J: Obesity: new life for antidiabetic drugs. Nature 2010;466:443–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yen CJ, Beamer BA, Negri C, Silver K, Brown KA, Yarnall DP, Burns DK, Roth J, Shuldiner AR: Molecular scanning of the human peroxisome proliferator activated receptor gamma (hPPAR gamma) gene in diabetic Caucasians: identification of a Pro12Ala PPAR gamma 2 missense mutation. Biochem Biophys Res Commun 1997;241:270–274 [DOI] [PubMed] [Google Scholar]
- 47.Deeb SS, Fajas L, Nemoto M, Pihlajamäki J, Mykkänen L, Kuusisto J, Laakso M, Fujimoto W, Auwerx J: A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet 1998;20:284–287 [DOI] [PubMed] [Google Scholar]
- 48.Kim KS, Choi SM, Shin SU, Yang HS, Yoon Y: Effects of peroxisome proliferator-activated receptor-gamma 2 Pro12Ala polymorphism on body fat distribution in female Korean subjects. Metabolism 2004;53:1538–1543 [DOI] [PubMed] [Google Scholar]
- 49.Sell H, Berger JP, Samson P, Castriota G, Lalonde J, Deshaies Y, Richard D: Peroxisome proliferator-activated receptor gamma agonism increases the capacity for sympathetically mediated thermogenesis in lean and ob/ob mice. Endocrinology 2004;145:3925–3934 [DOI] [PubMed] [Google Scholar]
- 50.Bogacka I, Gettys TW, de Jonge L, Nguyen T, Smith JM, Xie H, Greenway F, Smith SR: The effect of beta-adrenergic and peroxisome proliferator-activated receptor-gamma stimulation on target genes related to lipid metabolism in human subcutaneous adipose tissue. Diabetes Care 2007;30:1179–1186 [DOI] [PubMed] [Google Scholar]
- 51.Yamakawa J, Moriya J, Takeuchi K, Nakatou M, Motoo Y, Kobayashi J: Significance of kampo, Japanese traditional medicine, in the treatment of obesity: basic and clinical evidence. Evid Based Complement Alternat Med 2013;2013:943075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Itoh T, Senda A, Inoue H, Saitoh Y, Kagami M, Matsubara F, Aoyagi H: The effect of Bofutsushosan on weight reduction in humans. Kampo Med 2005;56:933–939 [Google Scholar]
- 53.Lee MY, Shin IS, Seo CS, Kim JH, Han SR, Shin HK: Subchronic oral toxicity studies of the traditional herbal formula Bangpungtongseong-san in Crl:CD (SD) rats. J Ethnopharmacol 2012;144:720–725 [DOI] [PubMed] [Google Scholar]