Abstract
Background
Parkinson’s disease (PD) continues to be an important neurological disorder. It is caused by the loss of dopaminergic neurons in the substantia nigra. Dopamine, the neurotransmitter produced from dopaminergic neurons, is a major precursor of endogenous morphine. There are approximately 18 genes associated with PD; their roles have not yet been completely established. PARK2 is a gene that encodes for the protein parkin, and PINK1 is a gene that encodes for PTEN-induced putative kinase 1.
Material/Methods
Our objective was to determine if morphine treatment of HTB-11 cells affects the expression of PINK1 and PARK2. HTB-11 cells were treated with 10−7 M morphine for 2 h and a microarray analysis was conducted. To verify the microarray analysis, 3 Q-PCR trials were run using 10−6 M naloxone, morphine (10−7 M), or a naloxone/morphine mix.
Results
In both the microarray analysis and the Q-PCR analysis, PARK2 was up-regulated and PINK1 was down-regulated.
Conclusions
Morphine can affect the expression of PD-associated genes.
Keywords: Parkinson’s Disease, Morphine, PINK1, PARK2
Background
Parkinson’s disease was discovered in 1817 by James Parkinson, and continues to be an important disease [1]. PD is the second most common neurodegenerative disorder. It occurs in the substantia nigra located in the midbrain, and is due to the loss of dopaminergic neurons [1].
Many factors have been associated with this disease, and factors may differ between individuals. The factors that have been associated with this disease include combinations of environmental exposure, gene-environmental interactions, and polygenic inheritance [2]. Environmental factors show an association with an increased risk of developing PD. For example, agriculture-related exposure may increase a person’s risk due to the pesticides used in agricultural work as well as time spent in close proximity to pesticide-treated land [3]. Another risk factor for PD is aging [4]. Epidemiological studies have suggested that aging is the biggest risk factor in developing PD because aging also causes some loss of dopaminergic neurons, which is exacerbated in PD [4]. More than 1% of the population worldwide over the age of 65 years, and approximately 4% of the population over 85 years is diagnosed with PD. Younger people may also be diagnosed with PD, but this is rare [5].
There are many symptoms that are involved in this disorder, with the best known ones being cardinal motor signs such as: rigidity, tremor, bradykinesia (slowed movement), and postural instability [2]. Recent studies have shown that many patients experience a wide range of non-motor symptoms (NMS) such as mood disorders, loss of sense of smell, and constipation [6]. Approximately 21% of PD patients having non-motor features [6], which occur due to the continuous loss of dopaminergic cells located in the substantia nigra pars compacta [2]. This loss becomes drastic when around 80% of the striatal dopamine and about 50% of nigral neurons are lost [7]. The NMS that have been reported in PD patients include depression, autonomic insufficiency, olfactory deficits, sleep disturbance, anxiety, and psychosis [2].
There are currently many treatments available for PD patients, but there is no cure. The most popular treatment for PD is levodopa (L-dopa). L-Dopa increases the dopamine levels to make up for the loss of the dopaminergic neurons. This treatment is often complicated by adverse effects like dyskinesias and motor fluctuations [8]. Other treatments include anti-cholinergics, dopamine agonists, amantadine, and deep brain stimulation [2]. These treatments may not always work for all patients and some of them may eventually lose effectiveness in some patients.
No clear genetic mechanism has been identified for PD; however, rare causes and familial PD have been described. About 20% of PD patients report a family history of the disease [9]. The first major study that helped connect genetics to PD was the analysis of an Italian family throughout many generations [10]. In this study, the gene SNCA (PARK1) was discovered, demonstrating that there may be an autosomal dominant pattern for some forms of PD [10].
A number of genes have a role in PD, with mutations in about 6 different genes being recognized as leading to heritable forms of the disorder: SNCA, LRRK2, PARK2, DJ1, PINK1, and ATP13A2 [1,11]. However, these mutations are found in only 5% of individuals with PD, suggesting that there are other genes that promote this disease [12]. A total of 18 genes are known to have a link with PD, but their role is not fully understood [13].
For this investigation, we explored the role of 2 in PD: PARK2 and PINK1 (PARK6). PARK2 is a very large gene with a length of about 4kb and is found on the chromosome 6q25.2-q27 [14]. This gene specifically encodes a protein known as parkin, which has the length of 465 amino acids and belongs to the “ring between ring fingers” (RBR) family that pertains to E3 ubiquitin ligases [15]. PARK2 has also been found to be a tumor suppressor gene [14]. Previous studies showed that mutations in this gene cause approximately 50% of autosomal recessive juvenile Parkinsonism (ARJP) [1].
PINK1 is a gene that also causes early cases of PD, and was first identified in an Italian family on the chromosome 1p35-p36 [16]. PINK1 stands for (PTEN)-induced putative kinase 1 and has functions that aid the mitochondria, such as protecting it from damage [17]. This gene contains a mitochondrial targeting motif that maintains and helps regulate mitochondrial morphology and/or function [18].
With both of these genes playing a role in ARJP, previous studies have shown that the proteins for these genes are linked where the pathways connect, which is critical in maintaining mitochondrial integrity and function [19]. If the mitochondria is damaged and not functioning, the 2 proteins will work together to degrade the mitochondria. In addition, studies have shown that overexpression can compensate for the loss of PINK1 protein [19]. However, overexpression of PINK1 does not compensate for loss of parkin expression, suggesting that PINK1 has a function that is upstream in the pathway connected to parkin [19].
The mitochondrial DNA within the substantia nigra has a higher mutation rate than in any other region in the brain [19]. This increased amount of damage that occurs to the mtDNA is closely associated with sporadic Parkinson’s disease, which involves multiple changes in nerve cells, causing a considerable mitochondrial dysfunction [19].
This study investigated the effect of morphine on expression of PARK 2 and PINK 1. It has been suggested that there is a connection between morphine and dopamine. Dopamine, the neurotransmitter that is drastically low in Parkinson patients, is known to be a precursor of endogenous morphine [20]. Morphine and dopamine have an important role in reward processes in the brain, especially in drug abuse [21]. Most neurons have both opioid and dopamine receptors [21]. With the biological activities relating to endogenous morphine, and its interactions with dopamine, it has been suggested that the morphine signaling system is commonly dysregulated in PD [20].
It has been suggested that there is a link between low morphine concentrations in PD and the gene expression seen in PD. Thus, we examined the effect of morphine on gene expression of PINK1 and PARK2. These 2 genes were specifically chosen due to their shared biological pathway. To determine how morphine affects the gene expression on these genes, we measured their expression in HTB-11 cells treated with morphine for 2 hours. Changes in expression were assessed using microarray and quantitative PCR (Q-PCR) analysis. Microarray technique allows for a broad overview of how the genes are expressed due to the morphine treatment, while Q-PCR specifically analyzes the genes individually for a more quantitatively accurate measurement of expression. We aimed to determine the significance of morphine in Parkinson’s disease by altering the gene expression of 2 important associated genes.
Material and Methods
Cell culture
HTB-11 cells (ATCC) were grown in vitro in 10% FBS MEM media with Pen Strep. (Life Technologies) and were sub-cultured following standard protocol when they achieved 90% confluence.
For microarray analysis, nine wells with a seeding density of 0.3×106 of HTB-11 cells in 2 ml of media were each treated with either 10−7 M morphine sulfate or an equivalent volume of vehicle (phosphate buffered saline) as a control. The cells were placed in an incubator at 37°C and 5% CO2 for 2 h.
For Quantitative PCR (Q-PCR) analysis, HTB-11 cells in 2–6 welled plates with a seeding density of 0.3×106 were treated with either 20 μL of PBS, 10−7 M morphine sulfate, 10−6 M naloxone or pretreated for 10 min with naloxone (10−6 M) prior to morphine addition. The cells were then placed in an incubator at 37°C and 5% CO2 for 2h.
RNA isolation
After the 2hr treatment period, the media in the wells was aspirated, and RNA was isolated using the RNeasy Kit as per manufacturer’s instructions (QIAGEN). The cells were disrupted using a total of 600 μL of Buffer RLT. The purified RNA was eluted using 50 μL of RNase free water and stored at −70°C overnight.
The RNA samples for the microarray analysis were checked for quality and quantity using a RNA 6000 Nano Chip (Agilent Technologies). Samples were loaded into the chip along with the gel-Dye mix and RNA 6000 Nano marker following standard procedures.
The isolated RNA samples for the Q-PCR trials were also tested for quality and quantity using a Genequant 2 spectrophotometer.
cDNA synthesis
The volume of the isolated RNA samples was adjusted to give 2 μg of RNA in a final volume of 10 μL. The RNA was denatured in a 9700 thermocycler (Applied Biosystems) for 5 minutes at 95°C and the samples were placed on ice for 1 min. The reverse transcription reaction contained dNTP’s, 5× Buffer, DTT, Random primers, and RNase inhibitor (Invitrogen). The samples were placed at room temperature and then 1 μL of Reverse Transcriptase was added. Samples were placed into the thermocycler for 1 hour at 40°C and then 10 minutes at 65°C, afterwards being stored at −20°C.
Microarray analysis
Four control RNA samples and four morphine treated RNA samples were prepped for microarray analysis as per the Agilent Technologies standard Microarray protocol using the Agilent Low Input Linear Amplification kit. To prepare the labeling reaction the RNA samples was brought up to a final volume 11.5 μL. A cDNA Master Mix 5× First Strand Buffer, 0.1 M DTT, 10 mM dNTP mix, MMLV-RT and RNase Out (Agilent Quick Amp Kit) was prepared and a volume of 8.5 μL of cDNA Master Mix was then added to each of the 8 samples. The Transcription Master Mix contained 4× Transcription Buffer, 0.1 M DTT, NTP mix, 50% PEG, RNaseOut (Inhibitor), Inorganic pyrophosphatase, T7 RNA Polymerase and Cyanine 3-CTP.
Samples containing the amplified cRNA were purified using the Qiagen RNeasy standard procedure and kit. The cRNA samples were brought to a total volume of 100 μL using nuclease-free water. The RNA was eluted in a volume of 30 μL of RNase-free water and the purified samples and was placed on ice.
To prepare the hybridization samples, procedures were followed as per the Agilent Technologies standard Microarray protocol. Procedures followed the steps for a fragmentation mix for a 4×44K microarray and was prepared with a total volume of 55 μL which contained Cyanine 3-labeled, linearly amplified cRNA, 10× Blocking Agent, nuclease-free water, and 25× Fragmentation Buffer. To stop the fragmentation reaction, 55 μL of 2× GEx Hybridization Buffer HI-RPM was added to the 4×44K microarray format. Samples were loaded onto the array slides immediately and hybridized for 18 h at 65°C.
The hybridization samples were then washed following the Agilent Technologies standard microarray wash protocol. After the wash procedure, the microarray slides were loaded into the Agilent DNA microarray scanner 2505C (Agilent Technologies, Santa Clara, Ca) with each slide being scanned for 8 minutes.
Quantitative PCR analysis
A primary master mix with a final concentration of 1× containing 2× universal master mix, 20× detector set (Life Technologies) (primer and probe combined) (Thermo Scientific), and Nuclease-free H2O was used to amplify both the experimental genes PARK2 and PINK1 (Table 1), and the house keeping gene GAPDH using 3 μL of cDNA in a 96 well reaction plate (Applied Biosytems). The 96 well reaction plate was then placed into the 7500 Real Time PCR System (Applied Biosystems, Foster City, Ca), using cycles of 10 min. at 95°C, 15 s at 95°C and then for 1min at 60°C for 40 cycles. The real time PCR detector set consisted of a forward primer, reverse primer, and a Taqman probe (Life Technologies) which was designed to span an exon-exon boundary. Three trials of Q-PCR, using different cDNA samples each time, were run for both genes.
Table 1.
Experimental gene information for Q-PCR. Information of the two genes PINK1 and PARK2 for the quantitative PCR analysis.
| Gene | Q-PCR information |
|---|---|
| PINK1 | Assay ID from Applied Biosytems (Hs00260868_m1). The probe was centered on position 483 of the mRNA sequence (GenBank accession number NM_032409). The PCR product was 104 bp |
| PARK2 | Assay ID was Hs01038325_m1. The probe was centered on position 1221 of the mRNA sequence set forth by GenBank accession number NM_004562. The PCR product was 96 bp |
Results
Microarray analysis
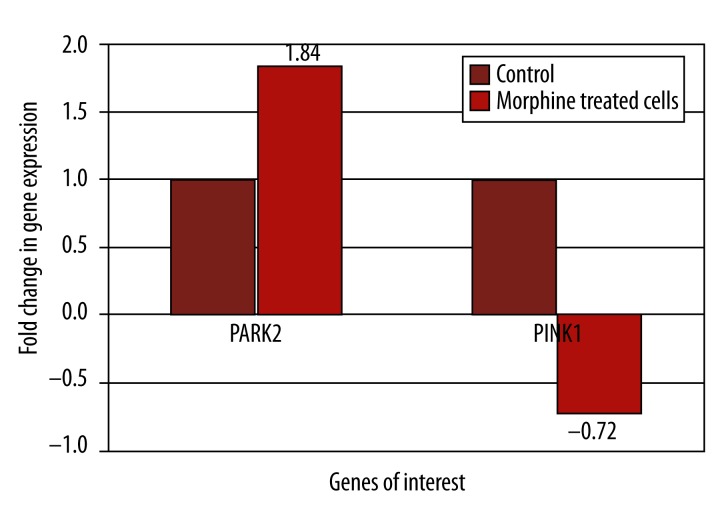
In the present study, HTB-11 cells were grown in vitro and treated with 10−7 M morphine for a 2 hour incubation period. HTB-11 cell gene expression was analyzed using microarray analysis and showed that the PARK2 gene had a 1.84 fold increase in expression, and the PINK1 gene expression decreased by 0.72 fold (Figure 1). The HTB-11 cells acting as a control for both genes had a normalized fold change value of 1.
Figure 1.
Microarray analysis of gene expression with 2 hour morphine treatment. Microarray analysis was conducted on HTB-11 cells which had been treated with a 10−7 M morphine. The gene expression levels of the control (untreated) cells were set at 1 for both genes to normalize the data. The expression of the genes PARK2 and PINK1 were specifically analyzed through the software program GeneSpring to calculate the fold changes.
Quantitative PCR
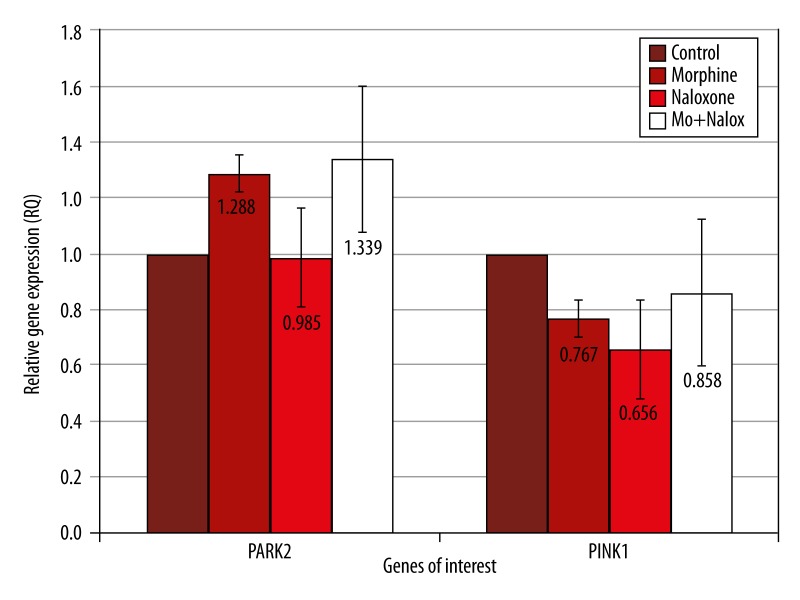
To validate the results of the microarray analysis, HTB-11 cells were then treated with either 10−7 M morphine, 10−6 M naloxone or a naloxone/morphine mix. Naloxone was used to verify that morphine was changing the expression since it has the function of blocking the morphine binding receptor. The PARK2 relative gene expressions had a mean value of 1.288 for the morphine treatment, which was an increased expression (Table 2, Figure 2). The naloxone treated cells for the PARK2 gene had a mean value, with no expression change, of 0.98. The morphine/naloxone mix expressed an averaged relative gene expression value of 1.339, which was an increase in gene expression (Table 2, Figure 2). The Q-PCR trials for the gene PINK1 expressed a slight decrease with a mean value of 0.767 for the morphine treated HTB-11 cells (Table 2, Figure 2). For the naloxone treated cells, PINK1 showed an average relative decreased gene expression of 0.656 (Table 2, Figure 2). The average gene expression value for the naloxone/morphine mix was calculated to be 0.858 for the three trials, which was a slight decrease in expression. (Table 2, Figure 2)
Table 2.
Averages of relative gene expression for quantitative PCR. The expression of PARK2 and PINK1 were determined in HTB-11 cells which were treated with 10−7 M morphine, 10−6 M naloxone, or a morphine/naloxone mix. Three trials of Q-PCR for each of the two genes, PARK2 and PINK1 were performed and the averages for each variable were calculated.
| Gene | Function | Morphine RQ avg. | Naloxone RQ avg. | Mo+Nalox RQ avg. |
|---|---|---|---|---|
| PARK2 | Makes protein, parkin. Parkin plays role in the cell machinery that breaks down unneeded proteins by tagging damaged and excess proteins with molecules called ubiquitin | 1.288 | 0.985 | 1.339 |
| PINK1 | Makes protein called PTEN (induced putative kinase 1) located in the mitochondria. Helps with cellular stress | 0.767 | 0.656 | 0.858 |
Figure 2.
Real time PCR analysis of PARK2 and PINK1 expression in HTB cells Treated with morphine and naloxone. HTB-11 cells were treated with 10−7 M morphine, 10−6 M naloxone, or a morphine/naloxone mix, and the expression of PINK1 and PARK2 were determined using Q-PCR analysis. The standard deviation error was calculated to show the variations in expression for the three trials and the controls were set at a value of 1 to normalize the data.
Discussion
We investigated how a 2 hour morphine treatment on HTB-11 cells affected the Parkinson’s associated genes PINK1 and PARK2. The present study demonstrates that morphine has varying effects on the gene expression for the Parkinson’s associated genes PINK1 and PARK2. All three PCR trials showed an up-regulation in PARK2 expression in the morphine treated HTB-11 cells verifying the microarray data. The HTB-11 cells treated with the morphine/naloxone mix also showed an up-regulation in PARK2 expression, the extent of which significantly varied for all three trials. This was not the expected results for this treatment because naloxone should have blocked the morphine from binding to its receptor and therefore no change in expression would occur. Accordingly, morphine might be acting via a different mechanism that is not influenced by the same receptor.
The Q-PCR results for the gene PINK1 showed that this gene was down-regulated 0.8 fold in the presence of morphine (Figure 2). This relates to the microarray analysis where the gene was also down-regulated due to morphine. Though this is the average, we did not find it to be statistically significant.
Previous studies have shown that overexpression of PARKIN can compensate for the loss of PINK1 expression [19]. Therefore, our results of the PARK2 being up-regulated, and the PINK1 gene being down-regulated are preliminary and subject to revision because we expect these genes work together in a pathway where they sense mitochondria that are in distress [19].
Mutations in the PARK2 and PINK1 genes have led to early onset cases of Parkinson’s disease [1]. Over 170 different mutations have been described for the gene PARK2 and have been known to lead to the loss of function for the protein Parkin [22]. PINK1 has also been known to have mutations which have been found to be both homozygous and heterozygous, both leading to reduction in enzymatic activity [22]. In the present study, morphine has shown to affect the expression for both of these genes. Mutations have been proven in previous studies to cause early onset of PD, but the heterozygous mutations that occur may also have a significant factor in the development of later onset PD as well [23].
The changes in gene expression that occurred in this study due to the morphine treatments can be associated with PD. Morphine has been known to be synthesized in animal tissues, and it also has been shown that it is present in the human immune cells as well [24–26]. The connection between PD and morphine is that dopamine, the neurotransmitter that is present in very low concentrations in PD patients due to the loss of dopaminergic neurons, has been shown to be a major precursor of morphine [20]. Dopamine is produced in the same pathway from its precursor L-DOPA and it has been shown that about 5% of the dopamine made from L-DOPA is converted into morphine [24]. This suggests that the pathways are linked and morphine levels will increase if dopamine levels increase. By using morphine in this study, we were able to verify the connection between morphine and expression of PARK2 and PINK1, two genes that have been implicated in PD.
Along with this knowledge, it has also been demonstrated that human neuroblastoma cells have the ability to synthesize morphine [27]. We chose human neuroblastoma cells for this study because they have dopaminergic markers. Since human neuroblastoma cells have the ability to synthesize morphine, it is possible that these cells are causing the PD associated genes to be expressed the way they did. Adding additional morphine to these cells as we did in this study would increase the overall morphine concentration in the cells.
This study was limited to a short term morphine exposure. Increasing the incubation time could give us a better understanding of how morphine affects these genes. The use of naloxone helped to verify if the changes that were occurring were due morphine acting on its receptor. Although we were able to verify the connection of morphine and the expression of these 2 genes, we determined that this does not mimic the behavior in PD because it has been suggested that PD patients have low concentrations of morphine.
As previously stated PARK2 is known to be a gene which encodes for the protein known as PARKIN and pertains to E3 ubiquitin ligases [15]. With this knowledge and the results presented in this study that PARK2 was up-regulated, it is possible that the removal of damaged proteins is increased due to morphine. We are only able to speculate what morphine is doing for the PARKIN protein (PARK2). Morphine may be restoring normal PARKIN signaling that may be lacking in a patient with low dopamine and therefore insufficient morphine. PARK2 increases were in line what we expect from morphine’s ability to enhance the proteasome system [28].
Conclusions
This study demonstrates that morphine affects the expression of Parkinson’s-associated genes in HTB-11 cells. Our results help elucidate the gene expression patterns that may occur in PD, as well as verifying the connection between morphine and dopamine when they are in the same pathway. To further test the gene expressions that occurred, treating different types of cells (e.g., dopaminergic neurons) would be a good approach as these neurons are directly associated with PD. By using dopaminergic neurons, we may be able to obtain a more accurate understanding of how morphine affects the expression of PD-associated genes.
Acknowledgements
This study was part of CS’s capstone research project at Stevenson University. Thank you to Dr. George B. Stefano for allowing CS to intern at the Neuroscience Research Institute and providing me with the necessary materials to complete the project. Thank you to Dr. Federico Casares for explaining and guiding CS through the computer software programs so that we were able to analyze the data. Finally, thank you to all of the Stevenson University faculty members that have made an impact on CS’s college career and driving CS towards becoming successful and staying determined.
Footnotes
Source of support: Departmental sources
References
- 1.Spratt DE, Martinez-Torres RJ, Noh YJ, et al. A molecular explanation for the recessive nature of parkin-linked Parkinson’s disease. Nat Commun. 2013;4:1983. doi: 10.1038/ncomms2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bekris LM, Mata IF, Zabetian CP. The genetics of Parkinson disease. J Geriatr Psychiatry Neurol. 2010;23:228–42. doi: 10.1177/0891988710383572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willis AW, Evanoff BA, Lian M, et al. Metal emissions and urban incident Parkinson disease: a community health study of Medicare beneficiaries by using geographic information systems. Am J Epidemiol. 2010;172:1357–63. doi: 10.1093/aje/kwq303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collier TJ, Kanaan NM, Kordower JH. Ageing as a primary risk factor for Parkinson’s disease: evidence from studies of non-human primates. Nat Rev Neurosci. 2011;12:359–66. doi: 10.1038/nrn3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Rijk MC, Launer LJ, Berger K, et al. Prevalence of Parkinson’s disease in Europe: A collaborative study of population-based cohorts. Neurologic Diseases in the Elderly Research Group. Neurology. 2000;54:S21–S23. [PubMed] [Google Scholar]
- 6.Khoo TK, Yarnall AJ, Duncan GW, et al. The spectrum of nonmotor symptoms in early Parkinson disease. Neurology. 2013;80:276–81. doi: 10.1212/WNL.0b013e31827deb74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fearnley JM, Lees AJ. Ageing and Parkinson’s disease: substantia nigra regional selectivity. Brain. 1991;114(Pt 5):2283–301. doi: 10.1093/brain/114.5.2283. [DOI] [PubMed] [Google Scholar]
- 8.Samii A, Nutt JG, Ransom BR. Parkinson’s disease. Lancet. 2004;363:1783–93. doi: 10.1016/S0140-6736(04)16305-8. [DOI] [PubMed] [Google Scholar]
- 9.Sellbach AN, Boyle RS, Silburn PA, Mellick GD. Parkinson’s disease and family history. Parkinsonism Relat Disord. 2006;12:399–409. doi: 10.1016/j.parkreldis.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Polymeropoulos MH, Higgins JJ, Golbe LI, et al. Mapping of a gene for Parkinson’s disease to chromosome 4q21-q23. Science. 1996;274:1197–99. doi: 10.1126/science.274.5290.1197. [DOI] [PubMed] [Google Scholar]
- 11.Lesage S, Brice A. Parkinson’s disease: from monogenic forms to genetic susceptibility factors. Hum Mol Genet. 2009;18:R48–59. doi: 10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- 12.Pankratz N, Beecham GW, DeStefano AL, et al. Meta-analysis of Parkinson’s disease: identification of a novel locus, RIT2. Ann Neurol. 2012;71:370–84. doi: 10.1002/ana.22687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrer MJ. Genetics of Parkinson disease: paradigm shifts and future prospects. Nat Rev Genet. 2006;7:306–18. doi: 10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
- 14.Kay DM, Stevens CF, Hamza TH, et al. A comprehensive analysis of deletions, multiplications, and copy number variations in PARK2. Neurology. 2010;75:1189–94. doi: 10.1212/WNL.0b013e3181f4d832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimura H, Hattori N, Kubo S, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–5. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 16.Healy DG, Abou-Sleiman PM, Gibson JM, et al. PINK1 (PARK6) associated Parkinson disease in Ireland. Neurology. 2004;63:1486–88. doi: 10.1212/01.wnl.0000142089.38301.8e. [DOI] [PubMed] [Google Scholar]
- 17.Bonifati V, Rohe CF, Breedveld GJ, et al. Early-onset parkinsonism associated with PINK1 mutations: frequency, genotypes, and phenotypes. Neurology. 2005;65:87–95. doi: 10.1212/01.wnl.0000167546.39375.82. [DOI] [PubMed] [Google Scholar]
- 18.Haque ME, Mount MP, Safarpour F, et al. Inactivation of Pink1 gene in vivo sensitizes dopamine-producing neurons to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) and can be rescued by autosomal recessive Parkinson disease genes, Parkin or DJ-1. J Biol Chem. 2012;287:23162–70. doi: 10.1074/jbc.M112.346437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narendra DP, Jin SM, Tanaka A, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stefano GB, Mantione KJ, Kralickova M, et al. Parkinson’s disease, L-DOPA, and endogenous morphine: a revisit. Med Sci Monit. 2012;18:RA133–37. doi: 10.12659/MSM.883259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shih YY, Chiang YC, Shyu BC, et al. Endogenous opioid-dopamine neurotransmission underlie negative CBV fMRI signals. Exp Neurol. 2012;234:382–88. doi: 10.1016/j.expneurol.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moura KC, Campos JM, de Rosso AL, et al. Genetic analysis of PARK2 and PINK1 genes in Brazilian patients with early-onset Parkinson’s disease. Dis Markers. 2013;35:181–85. doi: 10.1155/2013/597158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abou-Sleiman PM, Muqit MM, McDonald NQ, et al. A heterozygous effect for PINK1 mutations in Parkinson’s disease? Ann Neurol. 2006;60:414–19. doi: 10.1002/ana.20960. [DOI] [PubMed] [Google Scholar]
- 24.Zhu W, Mantione KJ, Shen L, Stefano GB. In vivo and in vitro L-DOPA and reticuline exposure increases ganglionic morphine levels. Med Sci Monit. 2005;11(5):MS1–5. [PubMed] [Google Scholar]
- 25.Weitz CJ, Lowney LI, Faull KF, et al. Morphine and codeine from mammalian brain. Proc Natl Acad Sci, USA. 1986;83:9784–88. doi: 10.1073/pnas.83.24.9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu W, Cadet P, Baggerman G, et al. Human white blood cells synthesize morphine: CYP2D6 modulation. J Immunol. 2005;175:7357–62. doi: 10.4049/jimmunol.175.11.7357. [DOI] [PubMed] [Google Scholar]
- 27.Boettcher C, Fellermeier M, Boettcher C, et al. How human neuroblastoma cells make morphine. Proc Natl Acad Sci USA. 2005;102:8495–500. doi: 10.1073/pnas.0503244102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rambhia S, Mantione KJ, Stefano GB, Cadet P. Morphine modulation of the ubiquitin-proteasome complex is neuroprotective. Med Sci Monit. 2005;11(11):BR386–96. [PubMed] [Google Scholar]