Abstract
Oligodendrocytes—the myelin-forming cells of the central nervous system—can be regenerated during adulthood. In adults, new oligodendrocytes originate from oligodendrocyte progenitor cells (OPCs), but also from neural stem cells (NSCs). Although several factors supporting oligodendrocyte production have been characterized, the mechanisms underlying the generation of adult oligodendrocytes are largely unknown. Here we show that genetic inactivation of SIRT1, a protein deacetylase implicated in energy metabolism, increases the production of new OPCs in the adult mouse brain, in part by acting in NSCs. New OPCs produced following SIRT1 inactivation differentiate normally, generating fully myelinating oligodendrocytes. Remarkably, SIRT1 inactivation ameliorates remyelination and delays paralysis in mouse models of demyelinating injuries. SIRT1 inactivation leads to the upregulation of genes involved in cell metabolism and growth factor signalling, in particular PDGF receptor α (PDGFRα). Oligodendrocyte expansion following SIRT1 inactivation is mediated at least in part by AKT and p38 MAPK—signalling molecules downstream of PDGFRα. The identification of drug-targetable enzymes that regulate oligodendrocyte regeneration in adults could facilitate the development of therapies for demyelinating injuries and diseases, such as multiple sclerosis.
In the adult brain, oligodendrocytes can be generated from OPCs or from NSCs (refs 1–6). Factors involved in oligodendrocyte production have been identified6–11, but the molecular basis of adult oligodendrocyte generation is mostly undefined. Emerging evidence suggests a role for lysine deacetylation in oligodendrocyte differentiation12,13, raising the possibility that lysine deacetylation also affects OPC expansion. Several members of the sirtuin family (SIRTs) are NAD+-dependent deacetylases, which connect stress stimuli and energy metabolism to lysine deacetylation14, making them appealing candidates for mediating oligodendrocyte regeneration1,15. Although SIRT2—a cytoplasmic sirtuin—has been shown to inhibit postnatal oligodendrocyte differentiation13, the role of other NAD+-dependent deacetylases in oligodendrocyte differentiation or regeneration has not been examined.
SIRT1—a nuclear sirtuin—is upregulated in a mouse model of multiple sclerosis in regions of inflammation16, raising the possibility that the SIRT1 protein participates in the regulation of adult oligodendrocytes. SIRT1 has been extensively studied in the context of energy metabolism17–25, neurodegenerative diseases26–29 and memory30,31. SIRT1 is important in terminally differentiated cells, including neurons, in which it promotes survival and synaptic plasticity30–34. Emerging evidence suggests a role for SIRT1 in embryonic stem cells35–37, embryonic neural stem and progenitor cell fate determination16,38 and in other tissue-specific stem cells39,40. However, whether and how SIRT1 regulates the function of adult NSCs and more committed progenitors originating from these cells—in particular OPCs—has not been explored. Here we report that SIRT1 inactivation in adult NSCs leads to OPC expansion in vitro and in vivo, and improves demyelinating injuries in vivo. OPC expansion following SIRT1 inactivation is mediated at least in part by activation of AKT and p38 MAPK signalling.
RESULTS
SIRT1 is expressed in adult NSCs and neural progenitors
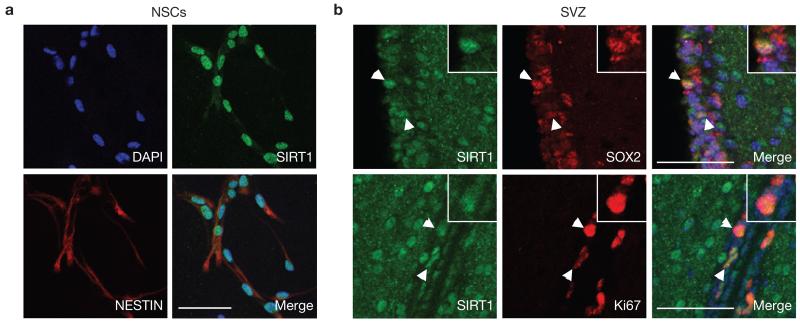
Immunostaining with an antibody against SIRT1 revealed that the SIRT1 protein is expressed in the nucleus of adult NSCs and neural progenitors in primary cultures (Fig. 1a and Supplementary Fig. S1a). In vivo, SIRT1 protein is expressed—albeit at low levels—in both adult NSC niches: the subventricular zone (SVZ) and the dentate gyrus of the hippocampus (Fig. 1b and Supplementary Fig. S1b–d). In these NSC niches, SIRT1 is expressed in a subset of cells positive for SOX2, a marker of neural stem/progenitor cells, and in all cells positive for the proliferation marker Ki67 (Fig. 1b and Supplementary Fig. S1b–d). SIRT1 is also expressed in mature neurons (Supplementary Fig. S1e), in some cells of the oligodendrocyte lineage (Supplementary Fig. S1f), but not in mature astrocytes (Supplementary Fig. S1e). Thus, although SIRT1 is not highly expressed in NSC niches in vivo, it is most consistently found in proliferating cells within these niches, suggesting that SIRT1 might play an important role in these proliferating cells.
Figure 1.
SIRT1 is expressed in adult NSCs and neural progenitors. (a) SIRT1 is expressed in the nuclei of NESTIN-positive cells in adult NSC and neural progenitor cultures. Immunocytochemistry on adherent monolayer cultures of adult NSCs and neural progenitors using antibodies against SIRT1 (green) and against the NSC/progenitor marker NESTIN (red). DAPI (blue) is a nuclear marker. (b) SIRT1 is expressed in NSCs and neural progenitors in the SVZ of the adult brain. Immunohistochemistry on brain sections of 8-week-old mice using antibodies against SIRT1 (green) and SOX2 (red, upper panel) or Ki67 (red, lower panel). White arrowheads show examples of double-stained cells. Insets show magnification of a cell positive for SIRT1 and SOX2 or Ki67. Merged images include DAPI to illustrate nuclei. Scale bars, 50 μm.
Inducible inactivation of SIRT1 in adult NSCs and neural progenitors expands the oligodendrocyte lineage
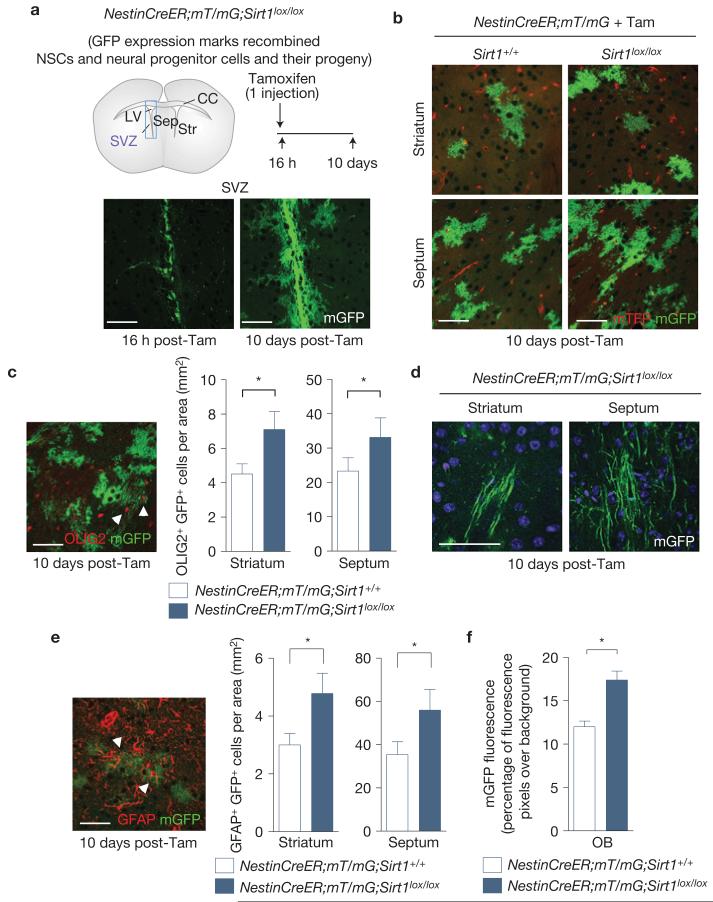
To determine the function of SIRT1 in adult NSCs and neural progenitors, we generated an inducible SIRT1 knockout mouse in which SIRT1 can be selectively inactivated in NSCs and neural progenitors in adults. To this end, we crossed mice in which exon 4 of the Sirt1 gene is flanked by loxP sites (Sirt1lox/lox) with NestinCreERT2–IRES–hPLAP transgenic mice41–44 (NestinCreER), which express a tamoxifen-inducible form of Cre (CreER) in adult NSCs and neural progenitors (Fig. 2a). We verified that the NestinCreERT2–IRES–hPLAP transgene was indeed expressed in the SVZ NSC niche, but not in other regions of the brain such as the striatum, septum and corpus collosum (Supplementary Fig. S2a). The protein product of the Sirt1 gene lacking exon 4 has no deacetylase activity and mice with germline homozygous deletion of exon 4 are phenotypically similar to Sirt1−/− mice45. To trace the progeny of adult NSCs and neural progenitors in which SIRT1 is inactivated, we crossed NestinCreER;Sirt1lox/lox mice with mice carrying a double fluorescent Cre reporter gene (mT/mG) in which Cre recombination leads to deletion of a membrane-targeted tomato fluorescent protein (mTFP) and expression of a membrane-targeted green fluorescent protein46 (mGFP; Fig. 2a and Supplementary Fig. S2e,f). We confirmed that the NestinCreER;Sirt1lox/lox mouse line allowed the specific deletion of the full-length SIRT1 protein in adult NSCs and neural progenitors when mice were injected with tamoxifen (Supplementary Fig. S2b,d), with no deletion from other brain regions (Supplementary Fig. S2c,d). We also verified that there was negligible Cre activity when mice were injected with vehicle (Supplementary Fig. S2e,f).
Figure 2.
Inducible inactivation of SIRT1 in adult NSCs and neural progenitors expands the oligodendrocyte lineage. (a) SIRT1 can be conditionally inactivated in adult NSCs and neural progenitors by tamoxifen injection of NestinCreER;mT/mG;Sirt1lox/lox mice. Upper left: schematic of a coronal mouse brain section. LV, lateral ventricle; SVZ, subventricular zone; Sep, septum; Str, striatum; CC, corpus callosum. Not drawn to scale. Upper right, adult mice were injected with tamoxifen once before analysis. Bottom, membrane-targeted green fluorescent protein (mGFP) serves as a reporter for Cre recombinase activity in NestinCreER;mT/mG;Sirt1lox/lox mice. (b) Progeny of adult NSCs and neural progenitors were observed in the striatum and septum of both NestinCreER;mT/mG;Sirt1lox/lox and NestinCreER;mT/mG control mice 10 days after tamoxifen injection. Concentrated areas of membrane-targeted tomato red fluorescent protein (mTFP) are blood vessels. (c) Inactivation of SIRT1 in adult NSCs and neural progenitors leads to greater production of OLIG2-positive NSC and neural progenitor progeny in the surrounding NSC niche. Mean±s.e.m. of 4 independent cohorts of 2 mice per genotype for each cohort, except one in which there were 3 control mice (n = 4). Two-tailed, paired Student’s t-test, striatum, *P =0.0294; septum, *P =0.0174. Arrowheads point to cells co-expressing mGFP and OLIG2. (d) mGFP-positive cells with morphologies characteristic of myelinating oligodendrocytes were observed in the striatum and septum of NestinCreER;mT/mG;Sirt1lox/lox brains. (e) Deletion of active SIRT1 in adult NSCs and neural progenitors leads to greater production of GFAP-positive NSC and neural progenitor progeny in the surrounding NSC niche. Mean±s.e.m. of 4 independent cohorts of 2 mice per genotype for each cohort, except one in which there were 3 control mice (n = 4). Two-tailed, paired Student’s t-test, striatum, *P = 0.0282; septum, *P = 0.0144. Arrowheads point to cells co-expressing mGFP and GFAP. (f) Deletion of active SIRT1 in adult NSCs and neural progenitors enhances the area in the olfactory bulb (OB) covered by mGFP fluorescence 10 days after tamoxifen injection. Mean±s.e.m. of n = 3 NestinCreER;mT/mG and n = 4 NestinCreER;mT/mG;Sirt1lox/lox mice. Two-tailed, unpaired Student’s t-test, *P = 0.01. Tam, tamoxifen. All scale bars, 50 μm.
To determine the consequences of the inducible inactivation of SIRT1 in adult NSCs and neural progenitors, we injected NestinCreER;mT/mG;Sirt1lox/lox and control NestinCreER;mT/mG mice with tamoxifen and analysed brain sections 16 h or 10 days later (Fig. 2a). Sixteen hours after tamoxifen injection, only the NSC niches (SVZ and dentate gyrus) began expressing mGFP (Fig. 2a), confirming that Cre-mediated deletion occurs specifically in adult brain regions containing NSCs and neural progenitors. In contrast, 10 days after tamoxifen injection, a larger number of mGFP-positive cells were observed in the SVZ (Fig. 2a) and mGFP-positive cells were also found in areas adjacent to the SVZ, including the striatum and septum (Fig. 2a,b), suggesting that cells generated from NSC niches have migrated to these regions.
The striatum and the septum are known to contain a relatively large population of OPCs that can give rise to mature oligodendrocytes6,47. We tested the possibility that SIRT1 inactivation might affect cells in the OPC lineage. Notably, the striatum and septum of NestinCreER;mT/mG;Sirt1lox/lox mice injected with tamoxifen contained a greater density of mGFP-positive cells when compared with control NestinCreER;mT/mG mice injected with tamoxifen (Fig. 2b,c). A proportion of these GFP-positive cells co-expressed OLIG2, a marker of the oligodendrocyte lineage (Fig. 2c), suggesting that SIRT1 inactivation in NSCs and neural progenitors increases the production of OPCs in the adult brain. This increase in cells of the oligodendrocyte lineage was not at the expense of other cell types because there was also an increase in the number of cells that are derived from NSCs and neural progenitors in vivo, such as astrocytes and olfactory bulb neuroblasts (Fig. 2e,f). Together, these observations indicate that SIRT1 inactivation in adult NSCs and neural progenitors enhances the generation of neural cell pools in the brain, including OPCs. Importantly, these genetic studies complement and extend viral lineage tracing studies5,6 and underscore that a substantial fraction of OPCs are generated from NSCs and neural progenitors under basal conditions in the adult wild-type brain, even in the absence of demyelinating injuries.
Brain-specific inactivation of SIRT1 increases the population of proliferating NSCs and OPCs
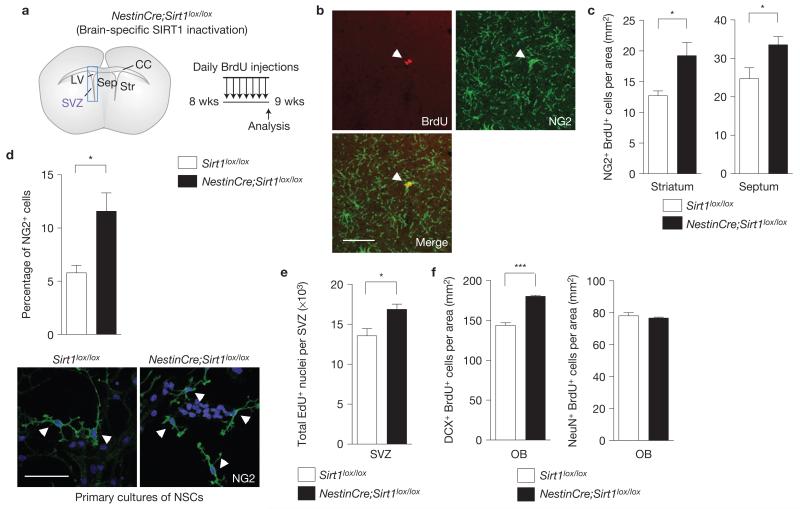
Chronic inactivation of SIRT1 in NSCs and neural progenitors from early development using NestinCre;Sirt1lox/lox mice (Fig. 3a and Supplementary Fig. S3a,b) also resulted in a greater number of proliferating OPCs in the adult striatum and septum when compared with control Sirt1lox/lox mice in vivo (Fig. 3b,c). These observations confirm that SIRT1 inactivation leads to the expansion of adult OPCs and indicate that OPC expansion is not due to a combined toxic effect of tamoxifen and SIRT1 inactivation. Primary cultures of adult NSCs and neural progenitors from NestinCre;Sirt1lox/lox mice exhibited a greater percentage of cells positive for NG2, an OPC marker (Fig. 3d and Supplementary Fig. S4a), further validating that SIRT1 inactivation results in an increase in the OPC pool. Moreover, NestinCre;Sirt1lox/lox mice also exhibited an increase in NSC and neural progenitor proliferation in vivo and in vitro (Fig. 3e and Supplementary Figs S3c,d and S4a,b), suggesting that the larger pool of NSCs and neural progenitors in the absence of SIRT1 may cause, at least in part, the expansion of OPCs. As could be expected from the larger NSC pool observed in NestinCre;Sirt1lox/lox mice, SIRT1 inactivation also resulted in an increased number of Doublecortin (DCX)-positive neuroblasts in the olfactory bulb (Fig. 3f and Supplementary Fig. S3e). However, SIRT1 inactivation did not significantly affect the number of more mature NeuN-positive neurons (Fig. 3f and Supplementary Fig. S3e), suggesting that newborn neurons may not survive in the absence of SIRT1. Furthermore, mice lacking active SIRT1 in the brain do not have proportionally larger brains than control mice or increased overall cell density in the brain (Supplementary Fig. S3f,g), indicative of a homeostatic control of the production of new cells under basal conditions. In the rest of this study, we focused on OPCs generated following SIRT1 inactivation because of the importance of these cells in myelination and the largely uncharacterized regulation of adult OPC generation.
Figure 3.
Brain-specific SIRT1 inactivation amplifies the proliferating OPC and NSC populations. (a) Mice lacking active SIRT1 specifically in NSCs and neural progenitors during embryonic development were generated by breeding mice in which exon 4 of Sirt1 is flanked by loxP sites (Sirt1lox/lox) with mice carrying a transgene in which Cre recombinase is expressed under the Nestin promoter (NestinCre). NestinCre;Sirt1lox/lox and control Sirt1lox / lox adult mice were injected with bromodeoxyuridine (BrdU) for 7 days to assess proliferation. SVZ, subventricular zone; LV, lateral ventricle; Sep, septum; Str, striatum; CC, corpus callosum. Schematic not drawn to scale. (b) Proliferating NG2-positive cells were observed in the striatum and septum of both NestinCre;Sirt1lox/lox and Sirt1lox/lox mice injected with BrdU for 7 days. Arrowheads indicate a double-labelled cell that is a dividing OPC. Scale bar, 50 μm. (c) The density of proliferating OPCs in the striatum and septum is greater in NestinCre;Sirt1lox/lox mice when compared with control mice. Mean±s.e.m. of n = 5 Sirt1lox/lox and n 4 = NestinCre;Sirt1lox/lox mice. Two-tailed, unpaired Student’s t-test; striatum, *P = 0.0139; septum, P = 0.041. (d) Absence of active SIRT1 increases the percentage of NG2-positive cells in proliferating NSC and neural progenitor cultures. Primary NSC and neural progenitor cultures from Sirt1lox/lox and NestinCre;Sirt1lox/lox 8-week-old mice were grown for a week in proliferation medium, and then immunostained for the OPC marker NG2. Arrowheads indicate NG2-positive cells. Mean±s.e.m. of 4 independent experiments. Two-tailed, unpaired Student’s t-test, *P = 0.0209. Scale bar, 50 μm. (e) Adult mice lacking SIRT1 in the nervous system have increased numbers of proliferating cells in the SVZ. Mice were injected with ethynyldeoxyuridine (EdU) for 2 h. Mean±s.e.m. of 4 mice per genotype. Two-tailed, unpaired Student’s t-test, *P = 0.0257. (f) The density of DCX-positive proliferating neuroblasts, but not NeuN-positive newly formed neurons, in the olfactory bulb (OB) granule cell layer is greater in NestinCre;Sirt1lox/lox mice when compared with control Sirt1lox/lox mice. Mice were injected with BrdU daily for 7 days before analysis. Mean±s.e.m. of 3 mice per genotype. Two-tailed, unpaired Student’s t-test, ***P = 0.0009.
SIRT1 inactivation does not alter the differentiation and myelination potential of oligodendrocytes
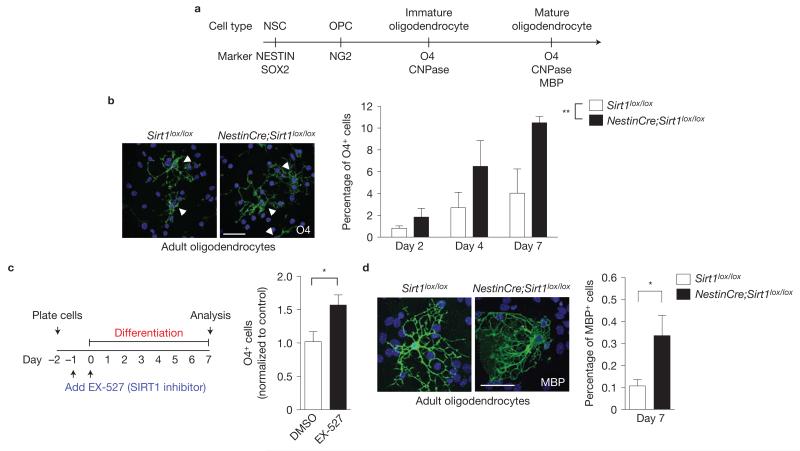
We investigated whether OPCs lacking active SIRT1 could generate fully differentiated oligodendrocytes. Importantly, inducible inactivation of SIRT1 in adult NSCs and neural progenitors on tamoxifen injection of NestinCreER;mT/mG;Sirt1lox/lox mice produced mGFP-positive cells in the striatum and septum that exhibited morphologies characteristic of myelinating oligodendrocytes (Fig. 2d), consistent with the notion that NSCs and neural progenitors lacking active SIRT1 can give rise to mature, fully myelinating oligodendrocytes. In culture, NSCs and neural progenitors from NestinCre;Sirt1lox/lox mice produced cells positive for the immature oligodendrocyte marker O4 when placed in differentiation conditions for 2, 4 and 7 days (Fig. 4a,b). In contrast, NSCs and neural progenitors from NestinCre;Sirt1lox/lox mice did not exhibit significant differences in the production of neurons and astrocytes on differentiation (Supplementary Fig. S4c), indicating that SIRT1 inactivation has a relatively specific effect on oligodendrocyte generation, at least in vitro. Interestingly, the SIRT1-specific inhibitor EX-527 also increased the production of O4-positive oligodendrocytes in NSC and neural progenitor cultures after 7 days of differentiation (Fig. 4c). OPCs lacking active SIRT1 could generate normal oligodendrocytes: immature oligodendrocytes that formed without active SIRT1 had morphologies similar to those from control cultures (Supplementary Fig. S4d). Furthermore, NestinCre;Sirt1lox/lox NSC and neural progenitor cultures also gave rise to cells expressing myelin basic protein (MBP), a marker for mature, myelinating oligodendrocytes (Fig. 4a,d). Finally, postnatal OPCs isolated from NestinCre;Sirt1lox/lox mice had normal morphologies and did not exhibit a change in differentiation ability in vitro (Supplementary Fig. S5). Overall, these findings indicate that SIRT1 inactivation results in an increase in the generation of normal myelinating oligodendrocytes both in vivo and in culture.
Figure 4.
SIRT1 inactivation does not alter the differentiation and myelination potential of oligodendrocytes. (a) Markers for specific cell types during differentiation of NSCs into mature oligodendrocytes. (b) Absence of active SIRT1 increases the percentage of O4-positive cells in differentiated adult NSC and neural progenitor cultures. Primary NSC and neural progenitor cultures from Sirt1lox/lox and NestinCre;Sirt1lox/lox 8-week-old mice were differentiated by growth factor removal and addition of 1% fetal bovine serum (FBS) after the first passage and immunostained after 2, 4 and 7 days for the oligodendrocyte marker O4. Images are representative of O4-positive cells at day 7 of differentiation. Arrowheads indicate individual O4-positive cells. Mean±s.e.m. of 5 independent experiments. Two-way ANOVA analysis: both genotype and differentiation day are significantly different, **P = 0.0055 and P = 0.0029, respectively. No interaction. Bonferroni post-test: day 7, P < 0.05. (c) SIRT1 inhibition enhances production of oligodendrocytes from NSCs and neural progenitor cells. Proliferating NSCs and neural progenitors were incubated with the SIRT1-specific inhibitor EX-527 (5 μM) or dimethylsulphoxide (DMSO) vehicle for 24 h. Cells were then switched to differentiation media (1% FBS) containing the same concentration of compound or vehicle and grown for a further 48 h. Cells were allowed to differentiate without added compound for 5 more days before staining for the oligodendrocyte marker O4. Mean±s.e.m. of 3 independent experiments. One-tailed, unpaired, Student’s t-test, *P < 0.05. (d) Absence of active SIRT1 increases the percentage of MBP-positive cells in differentiated adult NSC and neural progenitor cultures. Primary NSC and neural progenitor cultures from Sirt1lox/lox and NestinCre;Sirt1lox/lox 8-week-old mice were differentiated for 7 days by growth factor removal and addition of 1% FBS after the first passage and immunostained 7 days for the mature oligodendrocyte marker MBP. Images are representative of MBP-positive cells at day 7 of differentiation. Mean±s.e.m. of 6 independent experiments. Two-tailed, unpaired Student’s t-test, *P = 0.0398. Scale bars, 50 μm (b,d).
SIRT1 inactivation enhances remyelination upon demyelinating injuries and delays onset of paralysis in a chronic model of demyelination
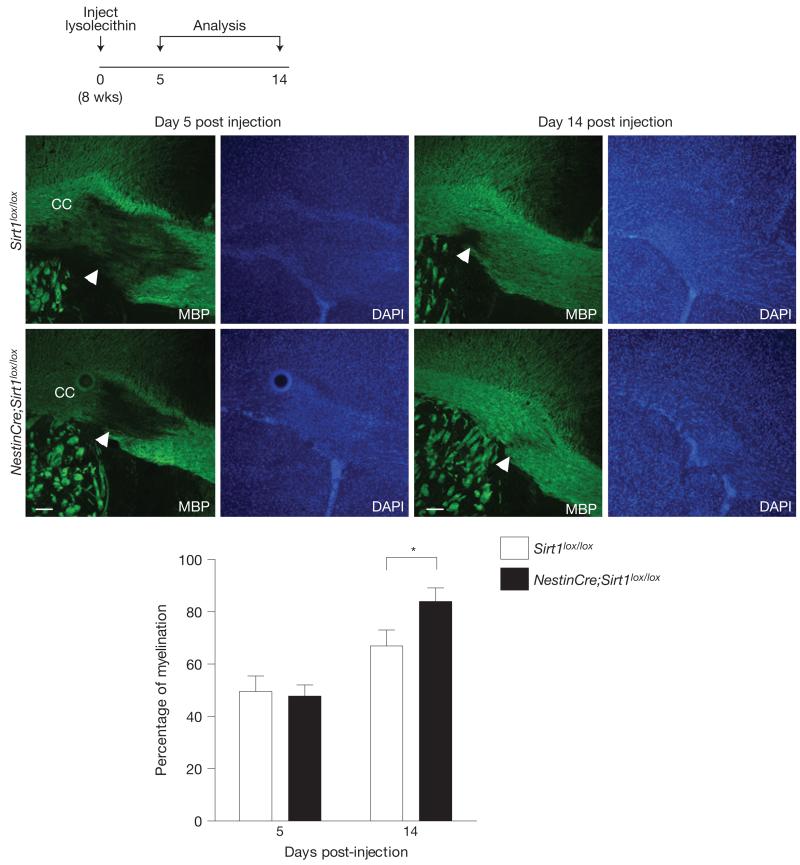
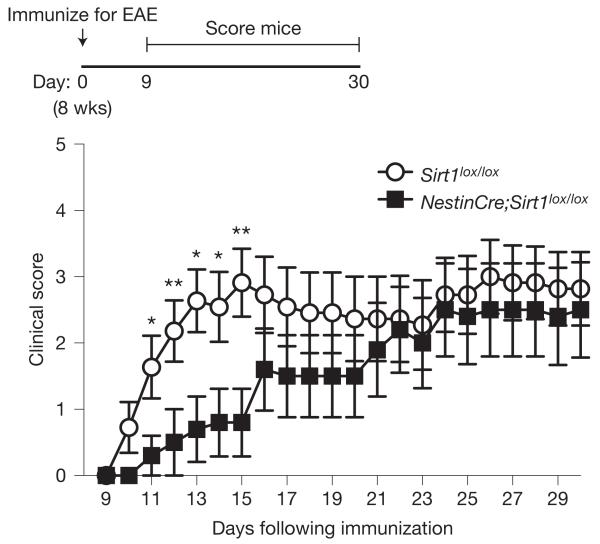
The production of oligodendrocytes could be particularly helpful in demyelination diseases or injuries. We first investigated the remyelination ability of mice lacking active SIRT1 following lysolecithin-induced demyelination of the corpus callosum. We found that 14 days after focal injection of lysolecithin into the corpus callosum, mice lacking active SIRT1 in the brain exhibited higher levels of remyelination in the region of injection when compared with control mice (Fig. 5). These results indicate that OPCs lacking SIRT1 can form normal oligodendrocytes following injury in the adult brain. We then examined whether SIRT1 inactivation in the brain affected the phenotype of mice with chronic experimental autoimmune encephalomyelitis (EAE), an established mouse model of multiple sclerosis48–50. Remarkably, NestinCre;Sirt1lox/lox mice were initially protected against the onset of paralysis in MOG35–55-induced chronic EAE when compared with control Sirt1lox/lox mice (Fig. 6). These data are compatible with the possibility that proliferating OPCs in NestinCre;Sirt1lox/lox brains might help remyelinate EAE-induced demyelinating lesions, although SIRT1 inactivation may also minimize axonal damage or reduce astrogliosis. Together, these data indicate that SIRT1 inactivation in the nervous system enhances the rate of remyelination on demyelinating injury and minimizes the severity of clinical symptoms in a mouse model of multiple sclerosis.
Figure 5.
SIRT1 inactivation enhances remyelination in the corpus callosum after lysolecithin-induced demyelination. Mice lacking active SIRT1 in the nervous system exhibit enhanced remyelination after focal lysolecithin-induced demyelination in the corpus callosum (CC). Sirt1lox/lox and NestinCre;Sirt1lox/lox 8-week-old mice were stereotactically injected with 1% lysolecithin in the corpus callosum and allowed to recover for 5 or 14 days. Levels of myelination were blindly quantified in the region of injection by coverage of MBP staining over background. One-tailed, unpaired Student’s t-test, *P = 0.028. Mean±s.e.m. of n = 3 Sirt1lox/lox and n = 5 NestinCre;Sirt1lox/lox mice for day 5 and n = 10 Sirt1lox/lox and n = 8 NestinCre;Sirt1lox/lox mice for day 14. Arrowheads indicate demyelination areas. Scale bars, 50 μm.
Figure 6.
SIRT1 inactivation delays onset of paralysis in chronic EAE. Mice lacking active SIRT1 in the nervous system are initially protected against the onset of paralysis in EAE. Mice were assessed for clinical signs of EAE daily after immunization with MOG35–55 peptide, Freund’s adjuvant and pertussis toxin. Score of 0, no paralysis; 1, loss of tail tone; 2, hindlimb weakness; 3, hindlimb paralysis; 4, hindlimb and forelimb paralysis; 5, moribund or dead. Mean±s.e.m. of n = 11 Sirt1lox/lox and n = 10 NestinCre;Sirt1lox/lox mice. Mann–Whitney U test, *P < 0.05, **P < 0.01.
SIRT1 inactivation in adult NSCs and neural progenitors leads to the upregulation of genes involved in metabolism, protein translation and growth factor signalling
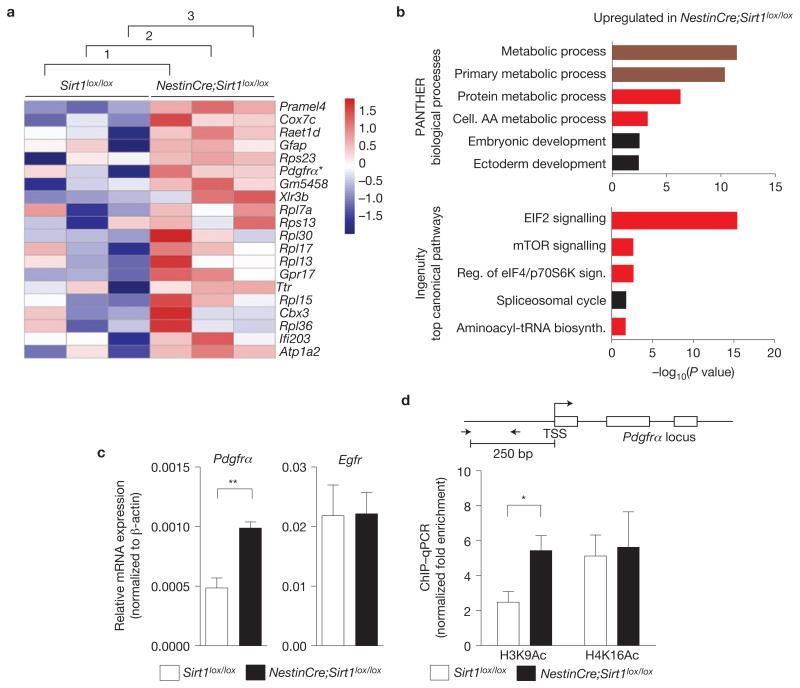
To gain insight into the molecular mechanisms underlying the expansion of OPCs in the absence of SIRT1, we assessed the genome-wide gene expression profile of primary NSC and neural progenitors from adult NestinCre;Sirt1lox/lox mice when compared with control Sirt1lox/lox cells using microarrays. Unbiased rank product analysis of the microarrays revealed a set of genes significantly upregulated in NestinCre;Sirt1lox/lox NSCs and neural progenitors when compared with control cells (Fig. 7a and Supplementary Table S1). A significant proportion of the genes upregulated in the absence of active SIRT1 encoded proteins with functions related to growth factor signalling (for example, platelet-derived growth factor receptor α (Pdgfrα)), cell metabolism (for example, cytochrome c oxidase subunit VIIc (Cox7c), pyruvate dehydrogenase kinase, isoenzyme 1 (Pdk1), acyl-coA dehydrogenase, long chain (Acadl)) and protein translation (for example, ribosomal protein large and small subunits (Rpl13, Rpl15, Rpl21, Rps2 and Rps8); Fig. 7a,b and Supplementary Table S1). These findings raise the possibility that SIRT1 inactivation affects growth factor signalling, cellular metabolism and protein translation, which in turn may impact OPC expansion. These data are consistent with the observation that oligodendrocytes have a greater metabolic rate when compared with other neural cells1,51. In contrast, genes downregulated in the absence of active SIRT1 were involved in neurogenesis and neuronal function (Supplementary Fig. S6 and Table S1), suggesting that neuronal function may be altered in the absence of SIRT1, as has been observed in previous studies30,31.
Figure 7.
SIRT1 inactivation leads to the upregulation of genes involved in metabolism, protein translation and growth factor signalling. (a) Genes upregulated in NestinCre;Sirt1lox/lox NSCs and neural progenitors compared with Sirt1lox/lox control NSCs and neural progenitors. Whole-genome microarray data from three independent biological replicates of proliferating NestinCre;Sirt1lox/lox and Sirt1lox/lox NSCs and neural progenitor isolated from 8-week-old mice. Heat map represents a subset of genes upregulated in the absence of active SIRT1 that was selected on the basis of a false discovery rate of less than 0.001%. Numbers 1–3 designate independent biological experiments. Colours represent the Z score of the robust multi-array analysis-normalized expression level for each gene (red is high expression; blue is low expression). Asterisk highlights Pdgfrα, a gene important for OPC proliferation and maintenance9,10,52,53. (b) Genes upregulated following SIRT1 inactivation are significantly over-represented in biological processes related to metabolism and ribosomal biology. Genes differentially regulated by SIRT1 were selected on the basis of a false discovery rate of less than 1% and analysed using PANTHER and Ingenuity Pathway Analysis (IPA). Canonical pathways significantly over-represented in the genes upregulated following SIRT1 inactivation are presented. Upper graph shows 6 of the top 7 PANTHER biological processes. Lower graph shows the top 5 IPA processes. Dark red colour highlights categories related to metabolism and bright red colour highlights categories related to protein synthesis and metabolism. AA, amino acid. (c) Pdgfrα mRNA expression is greater in proliferating NSCs and neural progenitors lacking active SIRT1 than in control cells. RT–qPCR on total RNA from primary NSC and neural progenitor cultures from NestinCre;Sirt1lox / lox and Sirt1lox/lox 8-week-old mice with primers to Pdgfrα and Egfr. Expression of Pdgfrα and Egfr was normalized to β-actin. Mean±s.e.m. of 3 independent experiments. Two-tailed, unpaired Student’s t-test, **P = 0.0068. (d) Increased H3K9 acetylation at the Pdgfrα promoter in NSCs and neural progenitors lacking active SIRT1. ChIP-qPCR on NSCs and neural progenitors from Sirt1lox / lox and NestinCre;Sirt1lox/lox 8-week-old mice with antibodies against H3K9Ac or H4K16Ac with primers to the Pdgfrα promoter. Data are presented as fold enrichment at the Pdgfrα promoter normalized to that at the promoter of HoxA10, a silent gene in these cells. Mean±s.e.m. of 3 independent experiments. Two-tailed, unpaired Student’s t-test, *P = 0.0491. TSS, transcriptional start site. The Pdgfrα locus is not drawn to scale.
To identify SIRT1-regulated pathways that could mediate the effect of SIRT1 on OPC expansion, we focused on the top upregulated genes in NestinCre;Sirt1lox/lox NSCs and neural progenitors (Fig. 7a). Among these top upregulated genes is Pdgfrα, a known regulator of OPC proliferation and function9,10,52,53, raising the possibility that SIRT1 may regulate OPCs by affecting PDGFRα signalling. We confirmed that SIRT1 inactivation results in increased Pdgfrα expression in NSCs and neural progenitors by reverse transcription followed by quantitative PCR (RT–qPCR; Fig. 7c). This effect was relatively specific to Pdgfrα as lack of active SIRT1 did not affect the expression of genes encoding other tyrosine kinase receptors (Egfr, TrkC and Fgfr1; Fig. 7c and Supplementary Fig. S7a). These results suggest that SIRT1 normally serves to repress Pdgfrα expression in NSCs and neural progenitors.
The increase in Pdgfrα messenger RNA levels could be an indirect consequence of SIRT1 deletion. To determine whether SIRT1 inactivation affects Pdgfrα expression in a direct manner, we examined the chromatin state at the Pdgfrα gene locus following SIRT1 deletion. SIRT1 has been shown to deacetylate acetylated Lys 9 of histone 3 (H3K9Ac) and acetylated Lys 16 of histone H4 (H4K16Ac; ref. 54). Chromatin immunoprecipitation (ChIP) showed a twofold enrichment of H3K9Ac at the promoter of Pdgfrα in cultured adult NSCs and neural progenitors from NestinCre;Sirt1lox/lox mice when compared with NSCs and neural progenitors from control mice (Fig. 7d). In contrast, SIRT1 inactivation in NSCs and neural progenitors did not affect H4K16Ac or global H3K9 acetylation (Fig. 7d and Supplementary Fig. S7b,c). This finding suggests that SIRT1 normally catalyses H3K9 deacetylation at the Pdgfrα promoter, thereby repressing the transcription of Pdgfrα, although it is possible that SIRT1 also deacetylates transcription factors that bind in the regulatory regions of the Pdgfrα gene. These findings are also consistent with the notion that SIRT1 inactivation directly leads to increased production of PDGFRα, which could in turn help support the expansion of OPCs.
Inhibition of p38MAPK and AKT signalling reduces the production of oligodendrocytes from NSCs and neural progenitors lacking active SIRT1
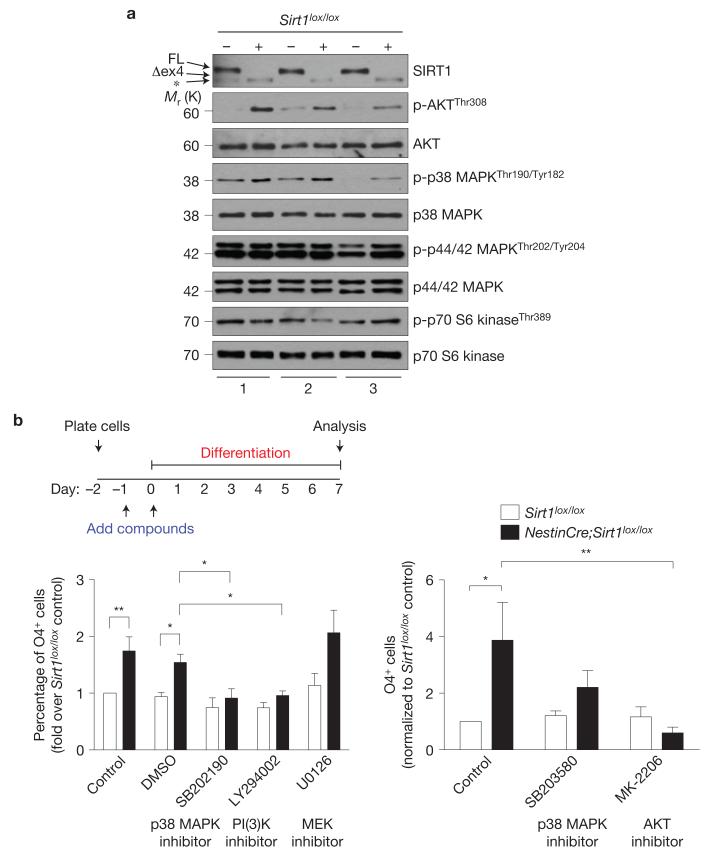
We examined whether PDGFRα signalling could mediate the SIRT1-dependent increase in oligodendrocyte number in NSC and neural progenitor cultures. PDGFRα stimulation is known to elicit a number of signalling pathways, including p38 MAPK, p44/p42 MAPK, AKT and mTOR-S6K (refs 55–57). Immunoblotting with phospho-specific antibodies against the active forms of these protein kinases or their substrates revealed that loss of active SIRT1 leads to increased phospho-p38 MAPK and phospho-AKT levels in adult NSCs and neural progenitors in vitro (Fig. 8a and Supplementary Fig. S8). In contrast, SIRT1 inactivation did not result in consistent changes in the phosphorylation of p44/42 MAPK or p70 S6 kinase, a target of mTOR (Fig. 8a and Supplementary Fig. S8). These results indicate that lack of active SIRT1 leads to p38 MAPK and AKT pathway activation.
Figure 8.
Inhibition of p38 MAPK and AKT signalling reduces the production of oligodendrocytes from NSCs and neural progenitors lacking active SIRT1. (a) Increased AKT and p38 MAPK phosphorylation in NSCs and neural progenitors lacking active SIRT1. Immunoblots of lysates from 3 independent cultures of NSC and neural progenitors isolated from Sirt1lox / lox and NestinCre;Sirt1lox / lox adult mice. Antibodies specific against phosphorylated forms of AKT, p38 MAPK, p44/42 MAPK and p70 S6 kinase were used. Antibodies specific against total proteins were used as loading controls. FL, full-length SIRT1; Δex4, truncated inactive SIRT1 protein generated from the Sirt1 gene that lacks exon 4. *, protein that is distinct from the truncated SIRT1 protein and probably represents a splice variant of SIRT1 (see Supplementary Fig. S2d). (b) Activity of signalling kinases p38 MAPK and AKT is important for oligodendrocyte generation from NSCs and neural progenitors. Proliferating Sirt1lox / lox and NestinCre;Sirt1lox/lox NSCs and neural progenitors were incubated with p38 MAPK inhibitor SB202190 (10 μM) or SB203580 (1 μM), PI(3)K inhibitor LY294002 (10 μM), AKT inhibitor MK-2206 (1 μm), MEK inhibitor U0126 (10 μm) or dimethylsulphoxide (DMSO) vehicle for 24 h. Cells were then switched to differentiation medium (1% FBS) containing the same concentration of compounds or vehicle and grown for a further 48 h. Cells were allowed to differentiate without added compounds for 5 more days, with complete medium changes every other day. Cells were stained for the oligodendrocyte marker O4. Left panel: mean±s.e.m. of the following number of independent experiments, n = 9 for control and dimethylsulphoxide; n = 5 for SB202190; n = 6 for LY294002; and n = 4 for U0126. Two-way ANOVA, compound, P < 0.0001; genotype, P < 0.0001; interaction, P = 0.0165; Bonferroni post-tests, *P < 005, **P < 0.01. Right panel: mean ± s.e.m. of 4 independent experiments. Two-way ANOVA, compound, P = 0.0684; genotype, P = 0.0438, interaction, P = 0.0416; Bonferroni post-tests, *P < 0.05, **P < 0.01. = Uncropped images of blots are shown in Supplementary Fig. S8.
We investigated whether the p38 MAPK and AKT pathways were required for adult OPC expansion due to SIRT1 inactivation. Inhibition of either p38 MAPK signalling with the inhibitor SB202190 or phosphatidylinositol-3-OH kinase (PI(3)K) signalling with the inhibitor LY294002 significantly decreased the production of cells positive for the oligodendrocyte marker O4 in NestinCre;Sirt1lox/lox cultures after induction of differentiation (Fig. 8b). An independent inhibitor of p38 MAPK (SB203580) and a direct inhibitor of AKT (MK-2206) also reduced the formation of oligodendrocytes in NestinCre;Sirt1lox/lox cultures after induction of differentiation (Fig. 8b), reinforcing the notion that the p38 MAPK and AKT signalling pathways are critical for the expansion of the oligodendrocyte lineage. In contrast, inhibition of the p44/42 MAPK pathway with U0126 compound had no effect on the production of O4-positive cells following SIRT1 inactivation, indicating that p44/p42 MAPK activity is dispensable for OPC proliferation/maintenance in these cultures (Fig. 8b). These results suggest that SIRT1 inactivation promotes the expansion of the OPC pool at least in part by leading to the activation of the p38 MAPK and AKT signalling pathways. We propose that SIRT1 inactivation in NSCs and neural progenitors leads to increased H3K9Ac at the Pdgfrα promoter, which would lead to elevated Pdgfrα expression, thereby allowing the activation of the AKT and p38 MAPK pathways. As both signalling pathways have been shown to regulate postnatal OPC proliferation/survival55,58, the activation of the AKT and p38 MAPK pathways following SIRT1 inactivation could help support the expansion of OPCs from adult NSCs. OPCs generated following SIRT1 inactivation retain their capacity to terminally differentiate into myelinating oligodendrocytes, which raises the possibility that manipulating SIRT1 expression or activity or downstream PDGFRα–p38 MAPK and AKT pathways could be potentially used in regenerative medicine to enhance the generation of oligodendrocytes in vitro or in vivo.
DISCUSSION
Here we show that the conditional deletion of a gene in adult NSCs and neural progenitors influences OPC production in adults. Our results also identify a new role for the protein deacetylase SIRT1 in normally limiting the expansion of OPCs as well as several cell types from SVZ NSCs and neural progenitors. Importantly, lack of active SIRT1 in adult NSCs and neural progenitors increases the pool of proliferating OPCs while maintaining their differentiation capacity, which could provide an effective approach to generate more oligodendrocytes to fight myelinating injury and diseases, including multiple sclerosis. Consistent with this possibility, SIRT1 inactivation ameliorates remyelination following demyelination and delays signs of paralysis in MOG35–55-induced chronic EAE, a mouse model for multiple sclerosis48–50. Delayed paralysis may be due to enhanced remyelination in the brain and/or the spinal cord following SIRT1 inactivation, although loss of active SIRT1 in the nervous system might also diminish astrogliosis, minimize axonal damage and/or alter blood–brain barrier dynamics. Our findings contrast with the observation that SIRT1 inactivation in the whole body worsens EAE symptoms59, probably owing to the requirement of SIRT1 for T-cell tolerance. Thus, harnessing the benefits of SIRT1 inactivation for therapeutic purposes will probably require specific and transient targeting of the nervous system.
We also show that SIRT1 regulates OPC expansion at least in part by affecting signalling molecules downstream of PDGFRα—AKT and p38 MAPK—that have previously been implicated in OPC proliferation/survival at postnatal stages55,58. We demonstrate that in adult NSCs and neural progenitors, signalling through AKT and p38 MAPK is critical for generating the oligodendrocyte lineage following SIRT1 inactivation, which may provide further strategies for manipulating the OPC pool in the adult brain. OPCs lacking active SIRT1 are able to give rise to mature oligodendrocytes, suggesting that increased PDGFRα signalling is transient. Our findings also suggest that SIRT1 affects Pdgfrα expression through direct action as a histone deacetylase at the Pdgfrα promoter. However, we cannot exclude other mechanisms for SIRT1 action. The changes in genes involved in cell metabolism and protein translation following SIRT1 inactivation may also mediate SIRT1 function on oligodendrocyte production, in particular as oligodendrocytes are highly metabolically active1,51.
The ability of SIRT1 to repress the production of OPCs from NSCs and neural progenitors without affecting oligodendrocyte differentiation contrasts with that of another sirtuin family member SIRT2, which acts to limit differentiation and maturation of oligodendrocytes from postnatal OPCs (ref. 13). The role of SIRT1 in OPC expansion also contrasts with that of HDAC1/2, histone deacetylases that promote oligodendrocyte lineage progression by allowing oligodendrocyte-specific gene expression12. Thus, SIRT1 may act in concert with other lysine deacetylases to coordinate oligodendrocyte expansion and maturation. There exist relatively specific chemical inhibitors for SIRT1, SIRT2 and HDAC1/2 enzymes60–62, and we observed that the SIRT1-specific inhibitor EX-527 increases the production of oligodendrocytes from differentiating NSCs and neural progenitor cells in vitro. Therefore, combined inactivation of SIRT1 together with SIRT2 or HDAC1/2 may be particularly helpful for generating large pools of mature oligodendrocytes for therapeutic purposes.
Conversely, the role we identified for SIRT1 in normally limiting proliferation of NSCs and neural progenitors and the production of OPCs may also be important to prevent tumour formation63. Indeed, SIRT1 protein expression is downregulated in oligodendrogliomas and glioblastomas64. Chronic downregulation of SIRT1 in actively dividing OPCs, which have been proposed to be the cell of origin for oligodendrogliomas65–69, could increase the risk for deleterious mutations. Together, these results suggest that SIRT1 has functions in multiple tissues and that manipulating SIRT1 activity for therapeutic purposes may require cell type specificity. The function of SIRT1 to limit cell expansion may be beneficial to limit cancer, but may be detrimental for normal renewal and homeostasis during ageing or degenerative disease. This may explain why SIRT1-overexpressing mice have positive effects on metabolism at midlife, but do not exhibit extension of maximal lifespan24,70. As SIRT1 inactivation is predicted to have both beneficial and detrimental effects, transient pulses of SIRT1 inactivation may be particularly helpful to generate new NSCs and neural progenitors and their progeny to fight brain injury without inducing long-term detrimental consequences. Together, our findings have important ramifications for the development of cellular or pharmacological therapies for myelin injuries and demyelinating diseases, such as multiple sclerosis.
METHODS
Animals
Sirt1+/− and Sirt1lox / lox mice in FVB/N and 129/Sv backgrounds, respectively, were generously provided by F. Alt (Harvard University, USA). NestinCre mice in a C57BL/6 background were purchased from Jackson Laboratories and were crossed with female Sirt1lox / lox mice owing to inheritance of the transgene through the male germline. mT/mG mice46 were also obtained from Jackson Laboratories and maintained on a 129/Sv background. NestinCreERT2–IRES–hPLAP (abbreviated NestinCreER) mice expressing CreERT2 under the control of the rat Nestin promoter and second intron as a transgene were generated in the S.J.B. laboratory and maintained on a predominantly 129/Sv background41,42,44. All mice used for in vivo experiments were male. Primary cultures of NSC/progenitors were derived from a mixture of male and female mice (sex-matched between genotypes). Mice were maintained in the Stanford University Animal Facility. All care and procedures were in accordance with the Guide for the Care and Use of Laboratory Animals (NHHS Publication No. (NIH) 85-23).
Antibodies
AKT, Santa Cruz #sc-1618; β-actin, Novus Biologicals #NB600-501; BrdU, AbD Serotec #OBT0030CX; CNPase, Santa Cruz #sc-30158 (immunoblot), Millipore #MAB326 (immunocytochemistry); DCX, Santa Cruz #sc-8066; GalC, B.A.B. laboratory; GFAP, rat Calbiochem #345860, guinea pig Advanced Immunochemicals #031223; H3, Abcam #1791; H3K9Ac, Millipore #07-352; H4, Abcam #31827; H4K16Ac, Millipore #07-329 (immunoblot), Santa Cruz #sc-8662-R (ChIP); IgG, Invitrogen #02-6102; Ki67, BD Pharmingen #550609; MBP, Santa Cruz #sc-13914; NESTIN, BD Pharmingen #556309; NeuN, Millipore #MAB377; NG2, Millipore #AB5320; O4, B.A.B. laboratory; OLIG2, Millipore #AB9610; p-AKT, CST #9275; p-p38 MAPK, Biosource #44-684G; p38 MAPK, CST #9212; p-p44/42 MAPK, CST #4370; p44/42 MAPK, CST #9102; p-p70 S6 kinase, CST #9206; p70 S6 kinase, CST #2708; SIRT1, Upstate/Millipore #07-131; SOX2, Santa Cruz #sc-17320; Thy1.2, Serotec #MCA02R; Tuj1, Covance #PRB-435P.
Fluorescence-conjugated secondary antibodies were obtained from Jackson ImmunoResearch and Molecular Probes.
EAE
Induction of MOG35–55 chronic EAE was performed as described previously71. Mice were scored by blinded investigators using the following guidelines: 0, no paralysis; 1, loss of tail tone; 2, hindlimb weakness; 3, hindlimb paralysis; 4, hindlimb and forelimb paralysis; 5, moribund or dead. The Mann–Whitney U-test was used for statistical analysis.
In vivo demyelination
Eight-week-old mice were anaesthetized with ketamine/xylazine solution (10 mg ml−1 ketamine; 1 mg ml−1 xylazine) and placed into a stereotactic injection apparatus. A hole was drilled in the skull to inject 1 μlof 1% lysolecithin solution (l-α-lysophosphatidylcholine) into the corpus callosum (0.5 mm anterior–posterior, 1.0 mm medial–lateral, −2.1 mm dorsal–ventral relative to bregma) using a 33-gauge Hamilton needle. Myelination was assessed by quantifying the coverage of fluorescent MBP staining in the injection region over background using Adobe Photoshop.
Tamoxifen treatment
A solution of 20 mg ml−1 tamoxifen (Sigma) was prepared in an ethanol/corn oil (1:9) mixture. Eight-week-old mice were injected intraperitoneally once daily for 3 days with vehicle or tamoxifen (0.225 mg g−1 mouse). A 20-day retention period was observed before NSC/tissue isolation. For fate analysis, 8–10-week-old mice were injected once with vehicle or tamoxifen (0.133 mg g−1 mouse) and perfused after 10 days. To quantify the density of mGFP-positive cells, 30 μm sections from bregma 0 mm to +1.15 mm were counted for mGFP-positive cells and normalized to the area. To assess the percentage of GFP-positive cells of a particular cell fate, Z-stacks containing the SVZ were assessed for cells that were positive for both GFP and the marker of interest.
In vivo proliferation and fate analysis
To quantify proliferation, 10-week-old mice were injected with EdU (50 mg kg−1—Invitrogen) intraperitoneally for 2 h. The Invitrogen Click-It EdU Cell Proliferation Assay kit was used on every sixth coronal section (30 μm) from bregma −1.65 mm to +1.15 mm for the SVZ and from bregma −3.60 to −1.35 mm for the dentate gyrus according to manufacturer’s instructions. Sections were counted for EdU-positive nuclei in the dentate gyrus granule cell layer (GCL) and subgranular zone (at the hilus/GCL border) and in the SVZ. The total number of EdU-positive nuclei per SVZ was estimated by multiplying the total number of EdU-positive nuclei by 6. The number of EdU-positive nuclei in the dentate gyrus was normalized to the GCL volume.
To assess the number and fate of cells near the SVZ, BrdU (50 mg kg−1—Calbiochem or Sigma) was injected intraperitoneally into 8-week-old male mice daily for 7 days before perfusing on the eighth day. Coronal brain sections (40 μm) were stained for BrdU and NG2 as described below. To quantify the density of proliferating cells, sections from bregma 0 mm to +1.15 mm were counted for BrdU-positive cells in the striatum and septum and normalized to the area. To quantify the percentage of BrdU-positive cells of a particular fate, Z-stacks of sections were assessed for cells that were positive for both BrdU and the cell fate marker of interest.
Immunohistochemistry
Brains were processed for immunohistochemistry as described previously72. Heat-induced antigen retrieval was performed except in the case of NG2. Antibodies were diluted as follows: SIRT1 1:1,000, OLIG2 1:500, NG2 1:500, NeuN 1:600, GFAP 1:500, BrdU 1:400, Ki67 1:200, SOX2 1:200, DCX 1:200, MBP 1:500. BrdU staining was performed as described previously72. Immunostaining for alkaline phosphatase activity was performed with the Vector Blue Alkaline Phosphatase Substrate Kit III (#SK-5300 Vector Laboratories).
NSC and neural progenitor cultures
Isolation and propagation of adult mouse NSCs/progenitors were performed as described previously72.
Postnatal OPC cultures
OPCs were isolated and cultured from postnatal day 7–8 mouse pups as described previously73. OPCs were cultured at 2,000 cells ml−1 in proliferation media (containing CNTF, PDGF-AA, NT-3) for 5 days or cultured at 10,000 cells ml−1 in proliferation media for 2 days and then placed in differentiation media (containing CNTF and T3 without PDGF-AA and NT-3) for 3 days. Antibodies were used at the following dilutions: NG2 1:400, CNPase: 1:100, MBP: 1:500, secondary 1:1,000. For EdU incorporation assays, proliferating and differentiating OPCs were incubated with 2.5 μM EdU for 4 h and 24 h, respectively, before staining using the Invitrogen Click-iT EdU staining kit.
Differentiation of NSCs and neural progenitors
Primary NSCs/progenitors were dissociated and plated onto poly-d-lysine-coated coverslips overnight (50,000 cells ml−1). The following day, proliferation/self-renewal medium was replaced by differentiation medium: NeuroBasal-A medium with PSQ, serum-free B27 supplement without vitamin A, and 1% FBS. Differentiation medium was changed every other day. To examine the effect of kinase inhibitors on NSC/progenitor differentiation, primary NSCs/progenitors were dissociated and plated overnight (25,000 cells ml−1). The following day, half of the medium was replaced by fresh proliferation/self-renewal media (1× growth factors) containing 2× vehicle or compounds. Compounds were used at the following final concentrations: U0126 monoethanolate (Sigma #U120; 10 μM), LY294002 (EMD #440202; 10 μM), SB202190 (Sigma #S7067; 10 μM), EX-527 (BioVision; 5 μM), SB203580 (Selleck Chemicals; 1 μM), MK-2206 (Selleck Chemicals; 1 μM). Doses were chosen on the basis of published literature as well as the concentrations known to target these enzymes specifically. Twenty-four hours later, medium was replaced by differentiation medium containing 1× dimethylsulphoxide or compounds. Thereafter, medium was changed completely with differentiation medium (without vehicle or compounds) every other day.
Immunocytochemistry
Cells were fixed with 4% PFA/2% sucrose, permeabilized with PBS + 0.4% Triton X-100, and stained with antibodies diluted in 5% goat serum and 7.5% BSA in 1× PBS at the following concentrations: GFAP 1:1,000, MBP 1:500, NESTIN 1:200, SIRT1 1:1,000, Tuj1 1:1,000, fluorescence-conjugated secondary antibody 1:500. For O4 staining, unfixed cells were incubated with the antibody against O4 (1:1 in 10% goat serum/1% BSA/0.1M l-lysine) before fixation and secondary antibody staining. Coverslips were mounted using Vectashield solution containing DAPI (Vector Laboratories).
ChIP
NSCs and neural progenitors from 8- to 10-week-old mice were collected at early passage 3 and processed for ChIP as described previously72. Standard curves were determined for each primer set using a serial dilution of input chromatin and then used to calculate fold enrichment over the negative control gene HoxA10 (not expressed in NSCs and neural progenitors).
Primers
Pdgfrα forward, 5′-TTGTTGAAGTCTGGGGGTTG-3′; Pdgfrα reverse, 5′-TCCCTAGGACTCCTTGCTCTC-3′; HoxA10 forward, 5′-ATGTCAGCCAGAAAGGGCTA-3′; HoxA10 reverse, 5′-ATGAGCGAGTCGACCAAAAA-3′.
RT–qPCR
Total RNA from NSCs and neural progenitors was collected and purified using the Qiagen RNeasy kit and processed for RT–qPCR as described previously72.
Primers
Pdgfrα forward, 5′-TCCATGCTAGACTCAGAAGTCA-3′; Pdgfrα reverse, 5′-TCCCGGTGGACACAATTTTTC-3′; Egfr forward, 5′-GCCATCCTGTCCAACTATGG-3′; Egfr reverse, 5′-GATGGGGTTGTTGCTGAATC-3′; Fgfr1 forward, 5′-CGCAGACTTTGGCTTAGCTC-3′; Fgfr1 reverse, 5′-GGTGTGTGTAGATCCGGTCA-3′; TrkC forward, 5′-GCCAGAGCCTTTACTGCATC-3′; TrkC reverse, 5′-TGGCTCACACTGATCTCTGG-3′; β-actin forward, 5′-TGTTACCAACTGGGACGACA-3′; β-actin reverse, 5′-CTCTCAGCTGTGGTGGTGAA-3′.
Microarray
RNA from proliferating adult NSCs and neural progenitors at passage 2 was collected and purified using the Qiagen RNeasy kit. Samples were processed by the Stanford University Protein and Nucleic Acid Facility using Affymetrix mouse GeneChip 1.0 ST array according to the manufacturer’s protocol. Background adjustment and normalization was performed with robust multi-array analysis. Z-score normalization was used to generate the expression heat maps. The Rank Products method74 (RankProd package from Bioconductor) was used to compare gene expression between NestinCre;Sirt1lox / lox and Sirt1lox / lox controls cells. Genes upregulated in NestinCre;Sirt1lox / lox samples using a false discovery rate of less than 1% were analysed using PANTHER (http://www.pantherdb.org/), Ingenuity Pathway Analysis (Ingenuity Systems, http://david.abcc.ncifcrf.gov/).
Immunoblotting
Immunoblotting was performed on NSCs and neural progenitor or tissue extracts as described previously72. A 6 or 8% gel was used to resolve full-length SIRT1 from truncated Δex4 SIRT1.
Image analysis
Fluorescent images were blindly taken with either a Zeiss epifluorescence microscope or a Zeiss LSM 510 confocal microscope. Images from randomly chosen fields distributed across the coverslip or brain section (2–4 sections per brain) were analysed with Zeiss AxioVision 4, Metamorph or LSM Image Browser. Alternatively, when positively stained cells were unevenly distributed throughout the coverslip, the total number of positively stained cells on each coverslip was counted by eye using a ×20 objective. This number was divided by an estimate of the total nuclei on the coverslip (estimated by counting 3–5 random fields of DAPI staining).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5. Data are presented as mean±s.e.m. unless otherwise noted. When two groups were compared, a two-tailed unpaired Student’s t-test was used unless otherwise noted. Two-way analysis of variance (ANOVA) followed by Bonferroni post-tests was used to analyse experiments with two variables.
Supplementary Material
ACKNOWLEDGEMENTS
We thank F. Alt for the generous gift of the Sirt1+/− and Sirt1lox/lox mice. We thank T. Palmer, T. Rando, and T. Wyss-Coray for helpful suggestions. We thank M. Winslow and J. Sage for critical review of the manuscript and discussion of the experiments. We are grateful to B. Benayoun and A. Morgan for their advice on the microarray analysis. We thank members of the A.B. laboratory for their invaluable discussion of the experiments and manuscript, in particular D. Leeman, J. Lim and A. Webb. This work was supported by NIH/NIA grants (R01 AG026648 and P01 AG036695), a California Institute for Regenerative Medicine grant, a Brain Tumour Society grant, an AFAR grant, the Glenn Foundation for Medical Research (A.B.) and the National Multiple Sclerosis Society (J.C.D., A.I. and B.A.B.—RG4059A8). V.A.R. was supported by an NSF graduate fellowship and an NINDS/NRSA graduate fellowship (5F31NS064600). E.A.P. was supported by an NSF graduate fellowship and an NIA/NRSA graduate fellowship (F31AG043232). L.S. and P.P.H were supported by the NIH (R01 NS055997), the National Multiple Sclerosis Society and the Guthy–Jackson Charitable Foundation. S.J.B. was supported by NIH P01CA096832. L.M.L.C. was supported by the Canadian Institutes of Health Research.
Footnotes
Note: Supplementary Information is available in the online version of the paper
AUTHOR CONTRIBUTIONS
V.A.R. conceived and planned the study with the help of A.B. V.A.R. performed the experiments and wrote the paper with the help of A.B. P.P.H. and V.A.R. designed and performed the EAE experiments (Fig. 6) under the supervision of L.S. J.O.B. performed and analysed the RT–qPCR experiments (Fig. 7c and Supplementary Fig. S7a). D.U. helped with microarray analysis (Fig. 7a and Supplementary Fig. S6a and Table S1). The postnatal OPC experiments (Supplementary Fig. S5) were conceived and planned by J.C.D. and performed by A.I. under the supervision of B.A.B. E.A.P. performed the analysis of global histone acetylation (Supplementary Fig. S7b,c). L.M.L.C. and S.J.B. generated and characterized the NestinCreER mice. All authors discussed the results and commented on the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Accession numbers. The primary accession number for raw microarray data is GSE36997.
References
- 1.McTigue DM, Tripathi RB. The life, death, and replacement of oligodendrocytes in the adult CNS. J. Neurochem. 2008;107:1–19. doi: 10.1111/j.1471-4159.2008.05570.x. [DOI] [PubMed] [Google Scholar]
- 2.Franklin RJ, Ffrench-Constant C. Remyelination in the CNS: from biology to therapy. Nat. Rev. Neurosci. 2008;9:839–855. doi: 10.1038/nrn2480. [DOI] [PubMed] [Google Scholar]
- 3.Picard-Riera N, et al. Experimental autoimmune encephalomyelitis mobilizes neural progenitors from the subventricular zone to undergo oligodendrogenesis in adult mice. Proc. Natl Acad. Sci. USA. 2002;99:13211–13216. doi: 10.1073/pnas.192314199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nait-Oumesmar B, et al. Progenitor cells of the adult mouse subventricular zone proliferate, migrate and differentiate into oligodendrocytes after demyelination. Eur. J. Neurosci. 1999;11:4357–4366. doi: 10.1046/j.1460-9568.1999.00873.x. [DOI] [PubMed] [Google Scholar]
- 5.Menn B, et al. Origin of oligodendrocytes in the subventricular zone of the adult brain. J. Neurosci. 2006;26:7907–7918. doi: 10.1523/JNEUROSCI.1299-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez-Perez O, Romero-Rodriguez R, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Epidermal growth factor induces the progeny of subventricular zone type B cells to migrate and differentiate into oligodendrocytes. Stem Cells. 2009;27:2032–2043. doi: 10.1002/stem.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, et al. Notch1 signaling plays a role in regulating precursor differentiation during CNS remyelination. Proc. Natl Acad. Sci. USA. 2009;106:19162–19167. doi: 10.1073/pnas.0902834106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu QR, et al. Sonic hedgehog-regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- 9.Richardson WD, Pringle N, Mosley MJ, Westermark B, Dubois-Dalcq M. A role for platelet-derived growth factor in normal gliogenesis in the central nervous system. Cell. 1988;53:309–319. doi: 10.1016/0092-8674(88)90392-3. [DOI] [PubMed] [Google Scholar]
- 10.Woodruff RH, Fruttiger M, Richardson WD, Franklin RJ. Platelet-derived growth factor regulates oligodendrocyte progenitor numbers in adult CNS and their response following CNS demyelination. Mol. Cell. Neurosci. 2004;25:252–262. doi: 10.1016/j.mcn.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 11.Jablonska B, et al. Chordin-induced lineage plasticity of adult SVZ neuroblasts after demyelination. Nat. Neurosci. 2010;13:541–550. doi: 10.1038/nn.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen S, et al. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat. Neurosci. 2008;11:1024–1034. doi: 10.1038/nn.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, et al. Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating α-tubulin. J. Neurosci. 2007;27:2606–2616. doi: 10.1523/JNEUROSCI.4181-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guarente L. The logic linking protein acetylation and metabolism. Cell Metab. 2011;14:151–153. doi: 10.1016/j.cmet.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Smith J, Ladi E, Mayer-Proschel M, Noble M. Redox state is a central modulator of the balance between self-renewal and differentiation in a dividing glial precursor cell. Proc. Natl Acad. Sci. USA. 2000;97:10032–10037. doi: 10.1073/pnas.170209797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prozorovski T, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat. Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 17.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 18.Satoh A, et al. SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J. Neurosci. 2010;30:10220–10232. doi: 10.1523/JNEUROSCI.1385-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen DE, Supinski AM, Bonkowski MS, Donmez G, Guarente LP. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 2009;23:2812–2817. doi: 10.1101/gad.1839209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc. Natl Acad. Sci. USA. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feige JN, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 22.Banks AS, et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun C, et al. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. 2007;6:307–319. doi: 10.1016/j.cmet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Bordone L, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 25.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 26.Jeong H, et al. Sirt1 mediates neuroprotection from mutant huntingtin by activation of the TORC1 and CREB transcriptional pathway. Nat. Med. 2011;18:519–165. doi: 10.1038/nm.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses β-amyloid production by activating the α-secretase gene ADAM10. Cell. 2010;142:320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Donmez G, et al. SIRT1 Protects against α-synuclein aggregation by activating molecular chaperones. J. Neurosci. 2012;32:124–132. doi: 10.1523/JNEUROSCI.3442-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Kim D, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao J, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michan S, et al. SIRT1 is essential for normal cognitive function and synaptic plasticity. J. Neurosci. 2010;30:9695–9707. doi: 10.1523/JNEUROSCI.0027-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 33.Shindler KS, Ventura E, Rex TS, Elliott P, Rostami A. SIRT1 activation confers neuroprotection in experimental optic neuritis. Invest. Ophthalmol. Vis. Sci. 2007;48:3602–3609. doi: 10.1167/iovs.07-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo W, et al. Sirt1 overexpression in neurons promotes neurite outgrowth and cell survival through inhibition of the mTOR signaling. J. Neurosci. Res. 2011;89:1723–1736. doi: 10.1002/jnr.22725. [DOI] [PubMed] [Google Scholar]
- 35.Han MK, et al. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calvanese V, et al. Sirtuin 1 regulation of developmental genes during differentiation of stem cells. Proc. Natl Acad. Sci. USA. 2010;107:13736–13741. doi: 10.1073/pnas.1001399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saunders LR, et al. miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging. 2010;2:415–431. doi: 10.18632/aging.100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hisahara S, et al. Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proc. Natl Acad. Sci. USA. 2008;105:15599–15604. doi: 10.1073/pnas.0800612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fulco M, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 40.Ou X, et al. SIRT1 deficiency compromises mouse embryonic stem cell hematopoietic differentiation, and embryonic and adult hematopoiesis in the mouse. Blood. 2011;117:440–450. doi: 10.1182/blood-2010-03-273011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cicero SA, et al. Cells previously identified as retinal stem cells are pigmented ciliary epithelial cells. Proc. Natl Acad. Sci. USA. 2009;106:6685–6690. doi: 10.1073/pnas.0901596106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavado A, Lagutin OV, Chow LM, Baker SJ, Oliver G. Prox1 is required for granule cell maturation and intermediate progenitor maintenance during brain neurogenesis. PLoS Biol. 2010;8:e1000460. doi: 10.1371/journal.pbio.1000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lavado A, Oliver G. Six3 is required for ependymal cell maturation. Development. 2011;138:5291–5300. doi: 10.1242/dev.067470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu G, et al. Pten deletion causes mTorc1-dependent ectopic neuroblast differentiation without causing uniform migration defects. Development. 2012;139:3422–3431. doi: 10.1242/dev.083154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng HL, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl Acad. Sci. USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 47.Dawson MR, Polito A, Levine JM, Reynolds R. NG2-expressing glial progenitor cells: an abundant and widespread population of cycling cells in the adult rat CNS. Mol. Cell. Neurosci. 2003;24:476–488. doi: 10.1016/s1044-7431(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 48.Olitsky PK, Yager RH. Experimental disseminated encephalomyelitis in white mice. J. Exp. Med. 1949;90:213–224. doi: 10.1084/jem.90.3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levine S, Sowinski R. Experimental allergic encephalomyelitis in inbred and outbred mice. J. Immunol. 1973;110:139–143. [PubMed] [Google Scholar]
- 50.Mendel I, Kerlero de Rosbo N, Ben-Nun A. A myelin oligodendrocyte glycoprotein peptide induces typical chronic experimental autoimmune encephalomyelitis in H-2b mice: fine specificity and T cell receptor V β expression of encephalitogenic T cells. Eur. J. Immunol. 1995;25:1951–1959. doi: 10.1002/eji.1830250723. [DOI] [PubMed] [Google Scholar]
- 51.Connor JR, Menzies SL. Relationship of iron to oligodendrocytes and myelination. Glia. 1996;17:83–93. doi: 10.1002/(SICI)1098-1136(199606)17:2<83::AID-GLIA1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 52.Noble M, Murray K, Stroobant P, Waterfield MD, Riddle P. Platelet-derived growth factor promotes division and motility and inhibits premature differentiation of the oligodendrocyte/type-2 astrocyte progenitor cell. Nature. 1988;333:560–562. doi: 10.1038/333560a0. [DOI] [PubMed] [Google Scholar]
- 53.Shi J, Marinovich A, Barres BA. Purification and characterization of adult oligodendrocyte precursor cells from the rat optic nerve. J. Neurosci. 1998;18:4627–4636. doi: 10.1523/JNEUROSCI.18-12-04627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 55.Baron W, Metz B, Bansal R, Hoekstra D, de Vries H. PDGF and FGF-2 signaling in oligodendrocyte progenitor cells: regulation of proliferation and differentiation by multiple intracellular signaling pathways. Mol. Cell. Neurosci. 2000;15:314–329. doi: 10.1006/mcne.1999.0827. [DOI] [PubMed] [Google Scholar]
- 56.Chew LJ, Coley W, Cheng Y, Gallo V. Mechanisms of regulation of oligodendrocyte development by p38 mitogen-activated protein kinase. J. Neurosci. 2010;30:11011–11027. doi: 10.1523/JNEUROSCI.2546-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Franke TF, et al. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 58.Flores AI, et al. Akt-mediated survival of oligodendrocytes induced by neuregulins. J. Neurosci. 2000;20:7622–7630. doi: 10.1523/JNEUROSCI.20-20-07622.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J, et al. The type III histone deacetylase Sirt1 is essential for maintenance of T cell tolerance in mice. J. Clin. Invest. 2009;119:3048–3058. doi: 10.1172/JCI38902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Solomon JM, et al. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol. Cell. Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Outeiro TF, et al. Sirtuin 2 inhibitors rescue α-synuclein-mediated toxicity in models of Parkinson’s disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 62.Park JH, et al. Class I histone deacetylase-selective novel synthetic inhibitors potently inhibit human tumor proliferation. Clin. Cancer Res. 2004;10:5271–5281. doi: 10.1158/1078-0432.CCR-03-0709. [DOI] [PubMed] [Google Scholar]
- 63.Fang Y, Nicholl MB. Sirtuin 1 in malignant transformation: friend or foe? Cancer Lett. 2011;306:10–14. doi: 10.1016/j.canlet.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 64.Lages E, et al. MicroRNA and target protein patterns reveal physiopathological features of glioma subtypes. PLoS ONE. 2011;6:e20600. doi: 10.1371/journal.pone.0020600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sugiarto S, et al. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer Cell. 2011;20:328–340. doi: 10.1016/j.ccr.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Persson AI, et al. Non-stem cell origin for oligodendroglioma. Cancer Cell. 2010;18:669–682. doi: 10.1016/j.ccr.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu C, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lindberg N, Kastemar M, Olofsson T, Smits A, Uhrbom L. Oligodendrocyte progenitor cells can act as cell of origin for experimental glioma. Oncogene. 2009;28:2266–2275. doi: 10.1038/onc.2009.76. [DOI] [PubMed] [Google Scholar]
- 69.Lu QR, et al. Oligodendrocyte lineage genes (OLIG) as molecular markers for human glial brain tumors. Proc. Natl Acad. Sci. USA. 2001;98:10851–10856. doi: 10.1073/pnas.181340798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herranz D, et al. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat. Commun. 2010;1:3. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grant JL, et al. Reversal of paralysis and reduced inflammation from peripheral administration of β-amyloid in TH1 and TH17 versions of experimental autoimmune encephalomyelitis. Sci. Transl. Med. 2012;4:145ra105. doi: 10.1126/scitranslmed.3004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Renault VM, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. Functional genomic analysis of oligodendrocyte differentiation. J. Neurosci. 2006;26:10967–10983. doi: 10.1523/JNEUROSCI.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.