Abstract
Background
Correlation of data within electronic health records is necessary for implementation of various clinical decision support functions, including patient summarization. A key type of correlation is linking medications to clinical problems; while some databases of problem-medication links are available, they are not robust and depend on problems and medications being encoded in particular terminologies. Crowdsourcing represents one approach to generating robust knowledge bases across a variety of terminologies, but more sophisticated approaches are necessary to improve accuracy and reduce manual data review requirements.
Objective
We sought to develop and evaluate a clinician reputation metric to facilitate the identification of appropriate problem-medication pairs through crowdsourcing without requiring extensive manual review.
Approach
We retrieved medications from our clinical data warehouse that had been prescribed and manually linked to one or more problems by clinicians during e-prescribing between June 1, 2010 and May 31, 2011. We identified measures likely to be associated with the percentage of accurate problem-medication links made by clinicians. Using logistic regression, we created a metric for identifying clinicians who had made greater than or equal to 95% appropriate links. We evaluated the accuracy of the approach by comparing links made by those physicians identified as having appropriate links to a previously manually validated subset of problem-medication pairs.
Results
Of 867 clinicians who asserted a total of 237,748 problem-medication links during the study period, 125 had a reputation metric that predicted the percentage of appropriate links greater than or equal to 95%. These clinicians asserted a total of 2464 linked problem-medication pairs (983 distinct pairs). Compared to a previously validated set of problem-medication pairs, the reputation metric achieved a specificity of 99.5% and marginally improved the sensitivity of previously described knowledge bases.
Conclusion
A reputation metric may be a valuable measure for identifying high quality clinician-entered, crowdsourced data.
Keywords: Electronic health records, Crowdsourcing, Knowledge bases, Medical records, Problem-oriented
1. Introduction
Electronic health records (EHRs) contain vast amounts of data of many types, including medications, laboratory test results, problems, allergies, notes, visits, and health maintenance items. The volume of information is often overwhelming to clinicians and can lead to inefficiencies in patient care [1–4]. Methods for summarizing patient information are required to better organize patient data, which can lead to more effective medical decision making. Developing such summaries requires knowledge about the relationships between the EHR elements [5–7]. Many prior research efforts have described methods for generating this knowledge using standard terminologies [8–10], association-rule mining [11–14], and literature mining [15–17], although each has disadvantages with respect to generalizability, accuracy, and completeness. Crowdsourcing represents a new approach for generating knowledge about relationships between clinical data types that takes advantage of required manual linking by clinicians of these types, such as medications and problems, during e-ordering that overcomes many limitations of traditional approaches [18]. Initial attempts utilizing this approach showed promise, but there was room for improvement in determining the accuracy of the clinical knowledge [18]. To more accurately classify links, we explored the inclusion of a clinician reputation metric, hypothesizing that such a metric would correlate with the percentage of links made by the clinician that were appropriate.
2. Background
2.1. Clinical summarization
At present, most EHRs present clinical data to providers organized by data type or date [5]. With increasing EHR implementations and growing amounts of patient data, such presentations can hinder point-of-care information retrieval and decision making, leading to clinician dissatisfaction, poor adoption, and substandard patient care [1–5]. Problem-oriented EHRs, or clinical summaries, which organize patient data by relevant clinical problems, make up one approach to overcoming these challenges, but few EHRs have effectively implemented such capabilities [6, 7]. One potential cause of low implementation is the limited availability of computable knowledge about the relationships between data elements that is required to develop these summaries.
2.2. Problem-medication knowledge bases
Knowledge bases composed of problem-medication pairs are an important component of clinical summarization. They can also be utilized within EHRs in a variety of other ways, in addition to summarization, such as improving medication reconciliation by grouping together all medications used to treat a particular condition, facilitating order entry by enabling order by indication, and improving the specificity of clinical decision support by enabling different medication dose ranges based on patient condition. However, current procedures for constructing such knowledge bases have significant limitations. The use of standard terminologies or commercially available resources comprises one method, though development of such resources is difficult and expensive, often requiring substantial maintenance [8–10]. Data mining methods are also common but can be hard to execute and may be biased to only include common links [11–13]. Given the drawbacks of these existing methods, new approaches to developing problem-medication knowledge bases are necessary.
2.3. Crowdsourcing
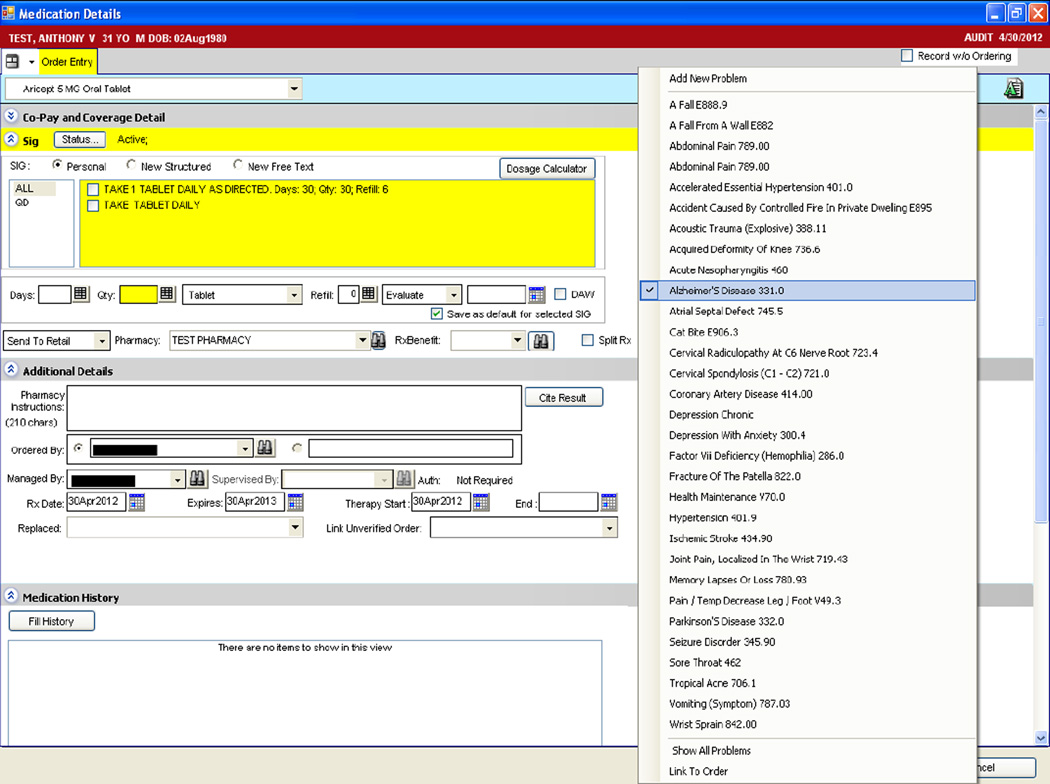
Crowdsourcing is defined as outsourcing a task to a group or community of people [19, 20]. This method has been used in various settings to generate large knowledge bases, such as encyclopedias [21]; drug discovery resources [22]; disease treatment, symptom, progression, and outcome data [23, 24]; and SNOMEDCT subsets [25]. In recent work, we have applied the crowdsourcing methodology to create a problem-medication knowledge base, which can facilitate the generation of clinical summaries and drive clinical decision support [18]. Fig. 1 depicts an example EHR screen through which clinicians e-prescribe medications (e.g., Aricept 5 MG Oral Tablet) and manually link the medication to the patient’s indicating problem (e.g., Alzheimer’s Disease). In our crowdsourcing research application, clinician EHR users represent the community, and generating problem-medication pairs for inclusion in the knowledge base represents the task.
Fig. 1.
Example screen showing problem manually linked to medication during e-prescribing.
Crowdsourcing relies on user input, and the quality of the resulting knowledge depends on correct data collected from the users. In our problem-medication pair application, clinicians may select an incorrect problem for linking due to poor usability, missing problem list entries, or carelessness. As a result, some metrics for evaluating the accuracy of the input for inclusion in a final knowledge base are required. Initial attempts to identify appropriate problem-medication links obtained through crowdsourcing approaches utilized link frequency (i.e., the number of times a problem and medication were manually linked by a provider) and link ratio (i.e., the number of times a co-occurring problem and medication were manually linked by a provider) [18]. However, these measures did not adequately determine the accuracy of all problem-medication pairs, indicating a need for additional metrics for evaluating crowdsourced data.
2.4. Reputation metrics
One method for determining data accuracy utilizes reputation metrics for evaluating user-generated content, such as e-commerce transactions [26], product reviews [27], and e-news or forum comments [28]. Several metrics for evaluating user-generated content have been reported. One approach evaluated feedback on content when a gold standard is not available, generating a reputation metric by comparing an individual’s response to others’ responses and disseminating ratings to encourage honest, appropriate responses [29]. A later approach expanded these methods, exploring various approaches for identifying true ratings from an aggregated data set [30]. Similarly, an evaluation of product reviews from Amazon.com showed that reviews with a high proportion of helpful votes had a higher impact on sales than those with a low proportion of helpful votes, demonstrating that user-generated content is frequently trusted by other users of a system [31].
More recently, reputation metrics have been applied to evaluating individuals who contribute to crowdsourced knowledge. One group of researchers described reputation and expertise as characteristics of a worker’s profile in a taxonomy of quality control in crowdsourcing [32]. In related work, the same authors developed a model for reputation management in crowdsourcing systems; however, like the metrics most frequently described in e-commerce settings, the model requires evaluation of workers by other workers [33]. Another approach used a consensus ratio for evaluating the accuracy of user-submitted map routes, measuring the ratio of agreements and disagreements between users; however, no evaluation of the metric was reported [34]. We hypothesized that these methods could be adapted to evaluate and identify appropriate problem-medication pairs, where clinicians are the users and problem-medication pairs are the user-generated content.
In this study, we developed and validated a clinician reputation metric to evaluate the accuracy of links between medications and problems asserted by clinicians in an EHR during e-prescribing. We hypothesized that the computed reputation metric for a clinician would positively correlate with the appropriateness of the problem-medication pairs that he or she had linked.
3. Methods
3.1. Study setting
We conducted the study at a large, multi-specialty, ambulatory academic practice that provides medical care for adults, adolescents, and children throughout the Houston community. Clinicians utilized Allscripts Enterprise Electronic Health Record (v11.1.7; Chicago, IL) to maintain patient notes and problem lists, order and view results of laboratory tests, and prescribe medications. Clinicians are required to manually link medications to an indication within the patient’s clinical problem list for all medications ordered through e-prescribing (Fig. 1). However, medications listed in the EHR not added through e-prescribing do not require selection of an indicated problem.
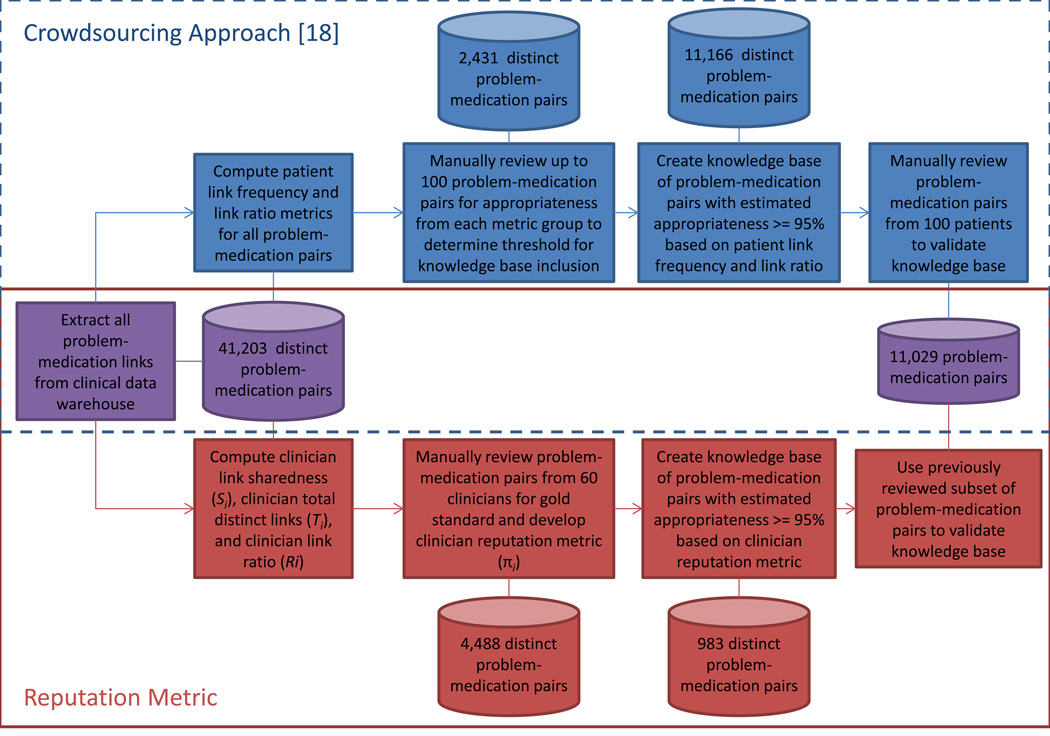
Fig. 2 depicts an overview of the methods for developing the reputation metric. We first retrieved medications from our clinical data warehouse that had been prescribed and linked to one or more problems by clinicians between June 1, 2010 and May 31, 2011. We excluded problem entries with an ICD-9 V code (e.g., V70.0 – Normal Routine History and Physical), as these concepts are for supplementary classification of factors and are not clinical problems, though they are frequently added to the problem list for billing purposes.
Fig. 2.
Flowchart of methods for developing and evaluating the crowdsourcing approach and reputation metric.
3.2. Development of a reputation metric
Our prior analysis suggested that the clinician linking of medications to problems is not always done accurately [18]. We found many situations where clinicians did not link a medication to a problem or linked medications to an unrelated problem. Further, the accuracy of linking differed among clinicians. As such, we sought to develop a reputation metric that would identify those clinicians who are likely to make accurate links. To develop the reputation metric, we included variables based on previous computational reputation metric literature and experience with real-world determinations of clinician reputation that were likely to predict whether links asserted by a clinician were appropriate. Based on our prior research and reputation metrics developed in other domains, we explored three contributors to the reputation metric: clinician link sharedness, clinician total distinct links, and clinician link ratio. We computed the values of each for all clinicians in the clinical data warehouse.
3.2.1. Clinician link sharedness
We first explored the variable most similar to that utilized in previous reputation metrics, where responses shared by other users were most likely to be appropriate [29, 30]. We calculated clinician link sharedness as the proportion of links asserted by a given clinician that were also asserted by another clinician. For example, a clinician who had linked 100 distinct problem-medication pairs, 80 of which were also linked by one or more other clinicians, would have a clinician link sharedness value of 80%.
Clinician link sharedness, Sx, is represented in Eq. (1), where Lx is the set of all links made by clinician x and Lx, is the set of all links made by clinicians other than clinician x:
| (1) |
3.2.2. Clinician total distinct links
We also hypothesized that clinicians who had asserted more links, and therefore had more experience linking medications and problems within the EHR, were likely to make more appropriate links compared to clinicians who had asserted very few links. We calculated clinician total distinct links as the number of unique problem-medication pairs linked by a given clinician. Clinician total distinct links, Tx, is represented in Eq. (2), where Dx is the set of distinct problem-medication pairs linked by clinician x.
| (2) |
3.2.3. Clinician link ratio
Our prior crowdsourcing study found that the “link ratio”, the proportion of patients receiving a particular medication and with a particular problem for which a link between the medication and problem has been manually asserted, was a predictor of accurate linking [18]. As such, we developed a similar metric at the clinician level which we called the clinician link ratio. We calculated the clinician link ratio for a given clinician by averaging, for each distinct problem-medication pair linked by the clinician, the proportion of links asserted by the clinician for all scenarios in which the clinician had the opportunity to link the problem and medication (i.e., the clinician prescribed the medication and the problem existed on the patient’s problem list).
The clinician link ratio, Rx, is represented in Eq. (3), where Lx is the set of all links made by clinician x, Dx, is the set of distinct problem-medication pairs linked by clinician x, and Px is the set of co-occurring patient problem and medication pairs which clinician x had the opportunity to link (i.e., the union of the Cartesian products of patients’ problem lists and prescribed medications across all patients for whom a clinician had prescribed a medication).
| (3) |
Consider an example clinician who had two patients with a medication order for metformin and diabetes on the problem list. In this scenario, Px contains both pairs of metformin and diabetes. If the clinician only linked the metformin to diabetes for one patient, the link ratio for the metformin-diabetes pair ( for p = metformin-diabetes) would be 50%; if the clinician linked metformin to diabetes for both patients, the link ratio would be 100%. The clinician link ratio would simply be the average of the link ratios for all problem-medication pairs linked by the clinician; a clinician with two problem-medication pairs linked with link ratios of 50% and 100% would be 75%.
3.3. Determination of clinician link appropriateness
Because it was not possible to review all links made by clinicians to determine the gold standard for appropriateness, we randomly selected 60 clinicians who had asserted at least one link during the study period to include in our analyses. We weighted our selection to include more clinicians with higher percentage of shared links, as these were most common; we included 10 clinicians with a percentage of shared links less than 25%, 10 clinicians with a percentage greater than or equal to 25% and less than 50%, 20 clinicians with a percentage greater than or equal to 50% and less than 75%, and 20 clinicians with a percentage greater than or equal to 75%. Two study investigators with medical training (SF, DR) reviewed the 4488 distinct problem-medication pairs linked by the 60 clinicians to determine whether it was appropriate (i.e., the medication was clinically relevant to use in the treatment or management of the problem). The reviewers first evaluated a set of 100 overlapping pairs each and discussed any disagreements to reach consensus. They then reviewed independently an additional 100 overlapping pairs to allow for evaluation of inter-rater reliability using the kappa statistic, then each reviewer evaluated half of the remaining pairs (2144 pairs each) asserted by the selected clinicians.
For further analyses, we created a binary outcome variable for each clinician, where clinicians having a link appropriateness percentage greater than or equal to 95% (i.e., 95% of links asserted by clinicians were determined to be appropriate by the reviewers) had an appropriateness outcome of one, and clinicians having a link appropriateness percentage less than 95% had an appropriateness outcome of zero. We selected the threshold of 95% to remain consistent with our previous study [18].
To determine whether the three variables in the reputation metric were associated with clinician link appropriateness for the 60 randomly selected clinicians, we first assessed each component in separate univariable analyses using logistic regression with the binary clinician link appropriateness variable as the dependent variable. We then included the components found to have a p-value less than 0.25 in univariable analyses in exploratory multivariable logistic regression analyses. We tested for multi-collinearity to ensure that the three components were not highly correlated. For the components represented as proportions, we reported odds ratios for a 0.1 unit increase. We applied the resulting model to all 867 clinicians to identify those predicted to have a binary appropriateness outcome of one (i.e., a clinician link appropriateness percentage greater than or equal to 95%), selecting a probability cutoff that maximized specificity for the logistic regression model. All analyses were performed using SAS 9.2.
3.4. Evaluation of the reputation metric
We included in our final reputation metric knowledge base all pairs linked by clinicians predicted to have a binary appropriateness outcome of one (i.e., clinician link appropriateness percentage greater than or equal to 95%). To evaluate the accuracy of the resulting reputation metric knowledge base, we repeated the evaluation of our previously described crowdsourcing approach, where we compared links automatically generated by the knowledge base to manual links (i.e., problem-medication pairs linked by clinicians during e-prescribing) [18]. Briefly, we reviewed all potential problem-medication pairs for 100 randomly selected patients, then we determined the sensitivity and specificity of the knowledge base at identifying links between the pairs. We also combined the manual links, crowdsourcing, and reputation metric approaches by including the pair in the combined knowledge base if any single approach included the pair, and we compared the resulting measures.
4. Results
4.1. Reputation metric development
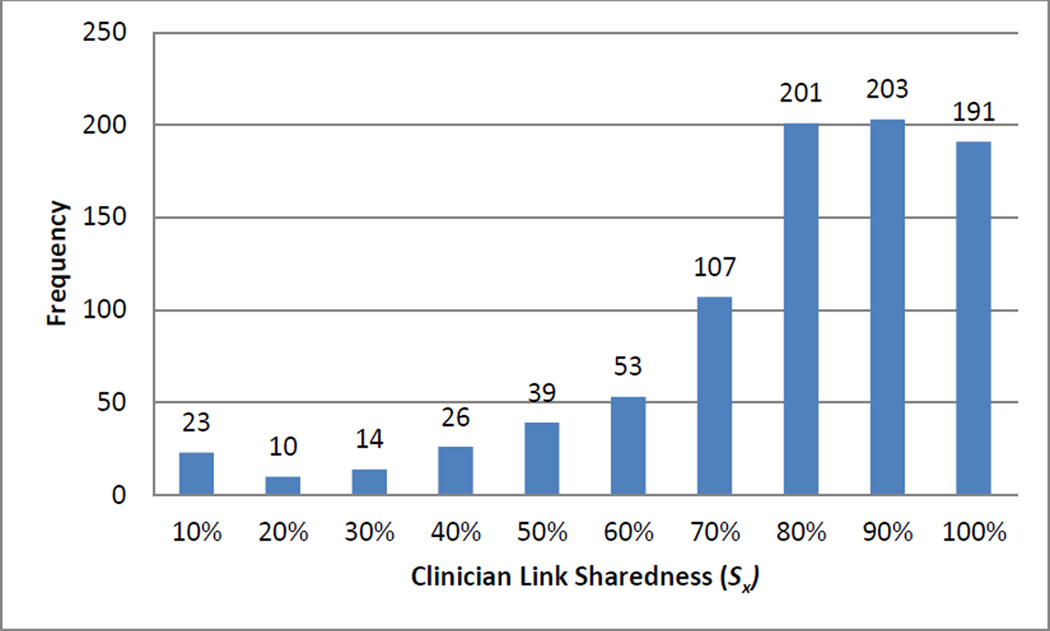
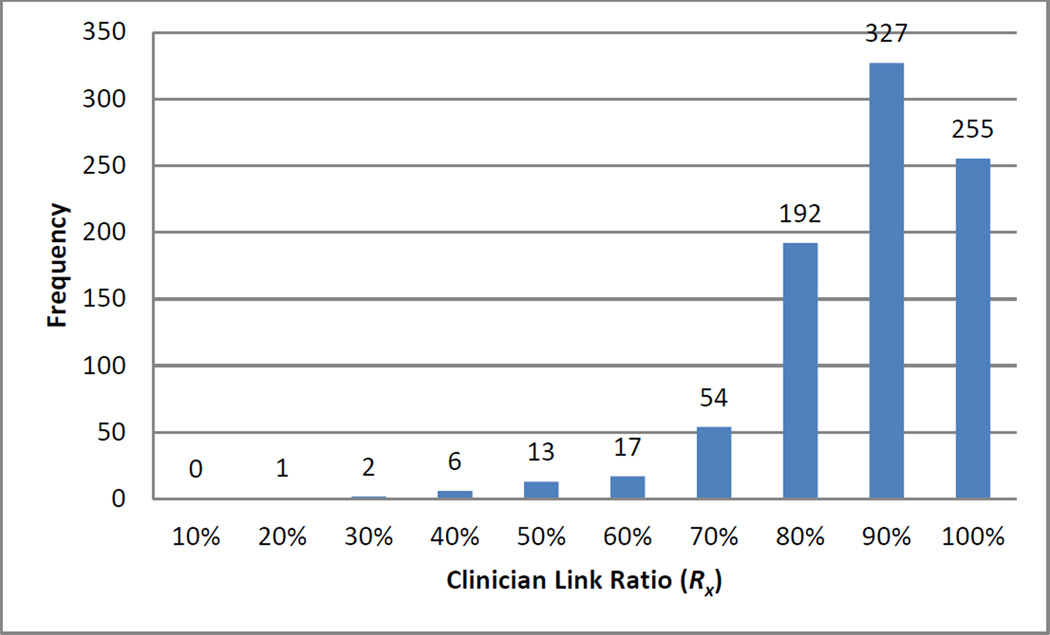
We calculated the three reputation metric components for 867 clinicians who had asserted a problem-medication link during the one-year study period. Combined, the clinicians asserted 237,748 total links (40,680 distinct problem-medication pairs). Figs. 3–5 depict the distributions for clinician link sharedness (Sx), clinician total distinct links (Tx), and the clinician link ratio (Rx) respectively for all clinicians.
Fig. 3.
Distribution of clinician link sharedness (Sx).
Fig. 5.
Distribution of clinician link ratio (Rx).
4.2. Clinician link appropriateness
The subset of 60 randomly selected clinicians included in the logistic regression analysis asserted a total of 4878 links, including 4488 distinct problem-medication pairs. The reviewers agreed on appropriateness for 98% of the 100 overlapping pairs (kappa = 0.79). Of the 4488 distinct pairs that were evaluated, 4063 (90.6%) were appropriate (91.2% of total links). Of the 60 clinicians, 27 (45%) had a link appropriateness percentage greater than or equal to 95% and therefore had a binary appropriateness outcome of one.
Clinician link sharedness (Sx), clinician total distinct links (Tx), and the clinician link ratio (Rx) all achieved significance (p < 0.25) in univariable analyses to be included in the multivariable analysis (p = 0.009, p = 0.2, p = 0.04 respectively). In the multivariable logistic regression analysis, clinician link sharedness (Sx), remained significant (OR = 1.28, p = 0.013); clinician total distinct links (Tx) and the clinician link ratio (Rx) were not significant (OR = 0.996, p = 0.3; OR = 1.60, p = 0.097 respectively).
We selected a predicted probability threshold of 0.7 from the logistic regression to achieve a model specificity of 94% for classifying the binary clinician link appropriateness outcome (i.e., the clinician having greater than or equal to 95% appropriate links). The resulting function is represented in the following equation.
| (4) |
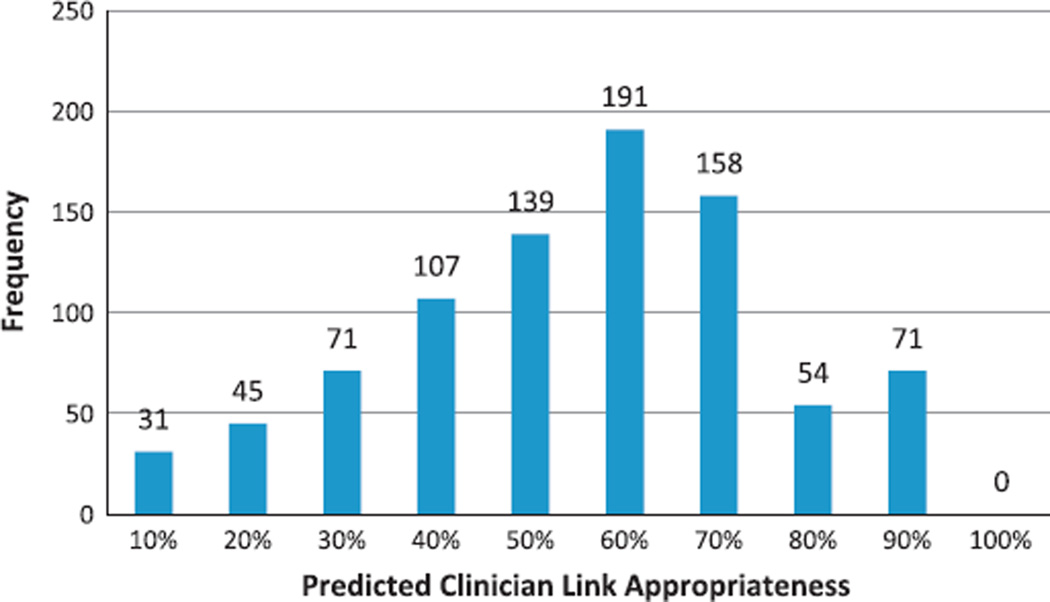
We applied the resulting function from the logistic regression to the 867 total clinicians who had asserted a link during the study period; 125 met the threshold, having a predicted probability of clinician link appropriateness greater than 70%. Problem-medication pairs linked by these clinicians totaled 2464 (983 distinct pairs). Fig. 6 depicts the distribution of predicted clinician link appropriateness.
Fig. 6.
Distribution of predicted clinician link appropriateness.
4.3. Reputation metric evaluation
Finally, we evaluated the knowledge base of 983 identified pairs using an alternate set of 11,029 problem-medication pairs from 100 randomly selected patients that were previously evaluated for appropriateness through manual review [18]. Our previously developed crowdsourcing knowledge base had a sensitivity of 56.2% and a specificity of 98.0%, and, when combined with manual links made by clinicians during e-prescribing, had a sensitivity of 65.8% and a specificity of 97.9% [18]. The reputation metric knowledge base alone had a sensitivity of 16.1% and a specificity of 99.5%, and, when combined with manual links, had a sensitivity of 49.3% and a specificity of 99.2%. When the reputation metric knowledge base was combined with the crowdsourcing knowledge base, the sensitivity was 56.6% and the specificity was 97.9%, and when combined with both the manual linking and crowdsourcing knowledge base, the sensitivity was 66.3% and the specificity was 97.8% (Table 1).
Table 1.
Comparison of reputation metric and crowdsourced knowledge bases to expert problem-medication pair review.
| Expert review |
Total | ||
|---|---|---|---|
| Positive | Negative | ||
| Reputation metric | |||
| Positive | 117 | 48 | 165 |
| Negative | 609 | 10,255 | 10,864 |
| Total | 726 | 10,303 | 11,029 |
| Sensitivity | Specificity | ||
| 16.1% | 99.5% | ||
| Reputation metric + manual links | |||
| Positive | 358 | 87 | 445 |
| Negative | 368 | 10,216 | 10,584 |
| Total | 726 | 10,303 | 11,029 |
| Sensitivity | Specificity | ||
| 49.3% | 99.2% | ||
| Reputation metric + crowdsourcing | |||
| Positive | 411 | 214 | 625 |
| Negative | 315 | 10,089 | 10,404 |
| Total | 726 | 10,303 | 11,029 |
| Sensitivity | Specificity | ||
| 56.6% | 97.9% | ||
| Reputation metric + crowdsourcing + manual links | |||
| Positive | 481 | 231 | 712 |
| Negative | 245 | 10,072 | 10,317 |
| Total | 726 | 10,303 | 11,029 |
| Sensitivity | Specificity | ||
| 66.3% | 97.8% | ||
5. Discussion
5.1. Significance
We expanded our previous crowdsourcing methodology, developing and evaluating a novel reputation metric for identifying appropriately linked problem-medication pairs. The reputation metric successfully facilitated identification of appropriately linked problem-medication pairs, outperforming our previously described crowdsourcing approach in specificity (99.5% compared to 98.0%) and, when combined with the previous approaches, marginally improving the sensitivity (66.3% compared to 65.8%) and nearly maintaining specificity (97.8% compared to 97.9%). Approaches for evaluating knowledge obtained through crowdsourcing approaches is important, as the data quality can be low if users input inaccurate data. While our application of crowdsourcing depends on well-trained clinicians who are capable of correctly linking clinical problems and prescribed medications, sociotechnical issues, such as a difficult user interface, mismatched workflow, or missing problem data, or inadequate training on the technology, may prevent correct input. Use of a reputation metric may reduce the need for extensive manual reviews, costly resources, or other computational approaches to generating knowledge bases.
5.2. Comparison to other approaches
This is the first study, to our knowledge, to develop and evaluate a reputation metric for generating knowledge bases of clinical data. Prior studies have described alternate methods for developing this knowledge, but each has disadvantages, as noted in our previous crowdsourcing work [18]. The reputation metric aims to overcome some of the disadvantages in the initial crowdsourcing work by improving the accuracy of pairs identified through the approach. Given the high specificity of the reputation metric, we believe the approach is successful.
Our work is based on a number of prior studies that describe approaches similar to the reputation metric [29–34]. However, unlike the reputation metrics described previously, which require some form of user feedback or voting on the input, our reputation metric can be computed without additional human effort. Further, these metrics have not all been evaluated for accuracy in identifying high quality input, so it is difficult to compare the approaches.
5.3. Limitations
Our study had some limitations. First, we selected a clinician appropriateness percentage of 95% and a predictive specificity of 94%, so our methods were likely to incorrectly classify some links. Because we erred in favor of specificity, our method had a low sensitivity, and it will be necessary to combine these approaches with other metrics to generate a complete and accurate knowledge base. Further effort is necessary to identify methods for effectively combining the approaches. Because of the extensive efforts required to manually determine the appropriateness of links, and therefore the percentage of clinician link appropriateness, we had a small sample size in developing our logistic regression model. Finally, because our study included only links generated by clinicians at a single study site, although the methods can be adopted by other institutions, our resulting knowledge base may not be generalizable to other settings. Further studies that utilize sources containing clinician links from multiple institutions are necessary to create more generalizable knowledge bases.
5.4. Future work
These findings have a number of implications for future research. As we have discussed previously, development of problem-medication knowledge bases can help inform problem-oriented summary screens, which may improve clinical care. The crowdsourcing methodology, including the reputation metric for analysis, which increases the accuracy of the resulting knowledge bases, can be used to generate knowledge bases for other data types that can be linked to problems, such as laboratory results and procedures. Finally, the clinician reputation metric can be used to evaluate other forms of user input within EHRs, such as alert overrides, helping informatics personnel to identify and improve poorly performing clinical decision support.
6. Conclusion
The clinician reputation metric achieved a high specificity when used to identify appropriate problem-medication pairs generated through the crowdsourcing methodology. This metric may be a valuable measure for evaluating clinician-entered EHR data.
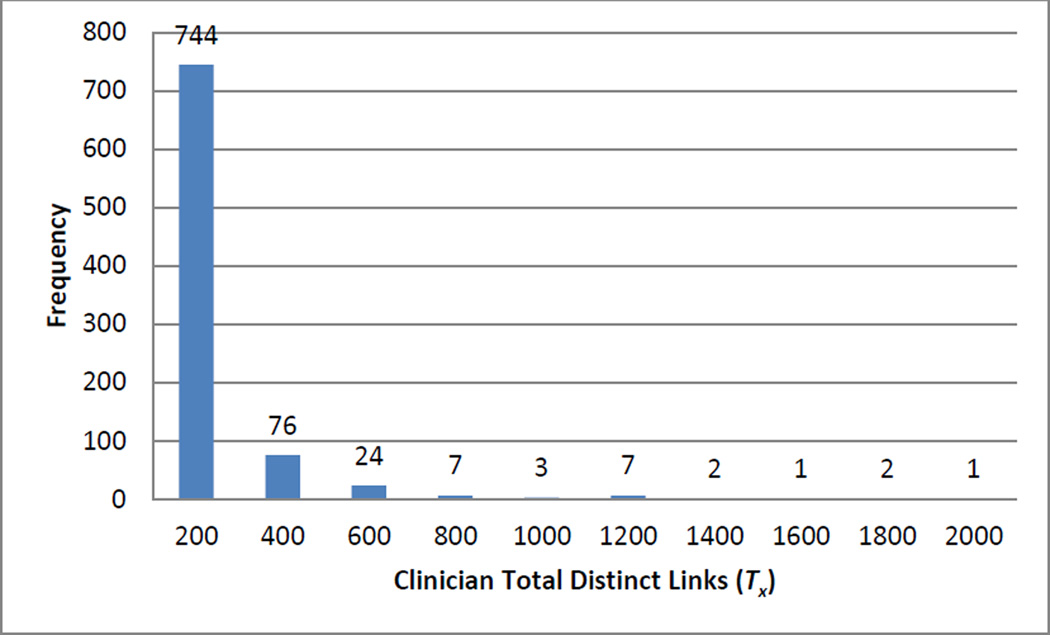
Fig. 4.
Distribution of clinician total distinct links (Tx).
Acknowledgments
This project was supported in part by a UTHealth Young Clinical and Translational Sciences Investigator Award (KL2 TR 000370-06A1), Contract No. 10510592 for Patient-Centered Cognitive Support under the Strategic Health IT Advanced Research Projects Program (SHARP) from the Office of the National Coordinator for Health Information Technology, and NCRR Grant 3UL1RR024148.
Contributor Information
Allison B. McCoy, Email: amccoy1@tulane.edu.
Adam Wright, Email: awright5@partners.org.
Deevakar Rogith, Email: deevakar.rogith@uth.tmc.edu.
Safa Fathiamini, Email: safa.fathiamini@uth.tmc.edu.
Allison J. Ottenbacher, Email: allison.ottenbacher@nih.gov.
Dean F. Sittig, Email: dean.f.sittig@uth.tmc.edu.
References
- 1.Koppel R, Metlay JP, Cohen A, Abaluck B, Localio AR, Kimmel SE, et al. Role of computerized physician order entry systems in facilitating medication errors. JAMA. 2005;293(10):1197–1203. doi: 10.1001/jama.293.10.1197. [DOI] [PubMed] [Google Scholar]
- 2.Ash JS, Sittig DF, Poon EG, Guappone K, Campbell E, Dykstra RH. The extent and importance of unintended consequences related to computerized provider order entry. J Am Med Inform Assoc. 2007;14(4):415–423. doi: 10.1197/jamia.M2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sittig DF, Singh H. Rights and responsibilities of users of electronic health records. CMAJ. 2012;184(13):1479–1483. doi: 10.1503/cmaj.111599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Health IT and Patient Safety: Building Safer Systems for Better Care. The National Academies Press; 2012. Committee on Patient Safety and Health Information Technology, Institute of Medicine. [PubMed] [Google Scholar]
- 5.Laxmisan A, McCoy AB, Wright A, Sittig DF. Clinical summarization capabilities of commercially-available and internally-developed electronic health records. Appl Clin Inform. 2012;3(1):80–93. doi: 10.4338/ACI-2011-11-RA-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feblowitz JC, Wright A, Singh H, Samal L, Sittig DF. Summarization of clinical information: a conceptual model. J Biomed Inform. 2011;44(4):688–699. doi: 10.1016/j.jbi.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Weed LL. Medical records that guide and teach. N Engl J Med. 1968;278(12):652–657. doi: 10.1056/NEJM196803212781204. [DOI] [PubMed] [Google Scholar]
- 8.Carter JS, Brown SH, Erlbaum MS, Gregg W, Elkin PL, Speroff T, et al. Initializing the VA medication reference terminology using UMLS metathesaurus cooccurrences. Proc AMIA Symp. 2002:116–120. [PMC free article] [PubMed] [Google Scholar]
- 9.Elkin PL, Carter JS, Nabar M, Tuttle M, Lincoln M, Brown SH. Drug knowledge expressed as computable semantic triples. Stud Health Technol Inform. 2011;166:38–47. [PubMed] [Google Scholar]
- 10.McCoy AB, Wright A, Laxmisan A, Singh H, Sittig DF. A prototype knowledge base and SMART app to facilitate organization of patient medications by clinical problems. AMIA Annu Symp Proc. 2011;2011:888–894. [PMC free article] [PubMed] [Google Scholar]
- 11.Wright A, Chen ES, Maloney FL. An automated technique for identifying associations between medications, laboratory results and problems. J Biomed Inform. 2010;43(6):891–901. doi: 10.1016/j.jbi.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Brown SH, Miller RA, Camp HN, Guise DA, Walker HK. Empirical derivation of an electronic clinically useful problem statement system. Ann Intern Med. 1999;131(2):117–126. doi: 10.7326/0003-4819-131-2-199907200-00008. [DOI] [PubMed] [Google Scholar]
- 13.Zeng Q, Cimino JJ, Zou KH. Providing concept-oriented views for clinical data using a knowledge-based system: an evaluation. J Am Med Inform Assoc. 2002;9(3):294–305. doi: 10.1197/jamia.M1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright A, McCoy A, Henkin S, Flaherty M, Sittig D. Validation of an association rule mining-based method to infer associations between medications and problems. Appl Clin Inform. 2013;4(1):100–109. doi: 10.4338/ACI-2012-12-RA-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen ES, Hripcsak G, Xu H, Markatou M, Friedman C. Automated acquisition of disease drug knowledge from biomedical and clinical documents: an initial study. J Am Med Inform Assoc. 2008;15(1):87–98. doi: 10.1197/jamia.M2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kilicoglu H, Fiszman M, Rodriguez A, Shin D, Ripple A, Rindflesch TC. Semantic MEDLINE: A web application for managing the results of PubMed searches. 2008:69–76. [Google Scholar]
- 17.Duke JD, Friedlin J. ADESSA: a real-time decision support service for delivery of semantically coded adverse drug event data. AMIA Annu Symp Proc. 2010;2010:177–181. [PMC free article] [PubMed] [Google Scholar]
- 18.McCoy AB, Wright A, Laxmisan A, Ottosen MJ, McCoy JA, Butten D, et al. Development and evaluation of a crowdsourcing methodology for knowledge base construction: identifying relationships between clinical problems and medications. J Am Med Inform Assoc. 2012;19(5):713–718. doi: 10.1136/amiajnl-2012-000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tapscott D. Wikinomics: how mass collaboration changes everything. New York: Portfolio; 2006. [Google Scholar]
- 20.Howe J. The rise of crowdsourcing. Wired Magazine. 2006;14(6):1–4. [Google Scholar]
- 21.Giles J. Internet encyclopaedias go head to head. Nature. 2005;438(7070):900–901. doi: 10.1038/438900a. [DOI] [PubMed] [Google Scholar]
- 22.Ekins S, Williams AJ. Reaching out to collaborators: crowdsourcing for pharmaceutical research. Pharm Res. 2010;27(3):393–395. doi: 10.1007/s11095-010-0059-0. [DOI] [PubMed] [Google Scholar]
- 23.Hughes S, Cohen D. Can online consumers contribute to drug knowledge? A mixed-methods comparison of consumer-generated and professionally controlled psychotropic medication information on the internet. J Med Internet Res. 2011;13(3) doi: 10.2196/jmir.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brownstein CA, Brownstein JS, Williams DS, 3rd, Wicks P, Heywood JA. The power of social networking in medicine. Nat Biotechnol. 2009;27(10):888–890. doi: 10.1038/nbt1009-888. [DOI] [PubMed] [Google Scholar]
- 25.Parry DT, Tsung-Chun Tsai. Crowdsourcing techniques to create a fuzzy subset of SNOMED CT for semantic tagging of medical documents. 2010 IEEE International Conference on Fuzzy Systems (FUZZ) IEEE. 2010:1–8. [Google Scholar]
- 26. [cited 05.09.13];Establishing a good eBay reputation: Buyer or Seller|eBay [Internet] < http://www.ebay.com/gds/Establishing-a-good-eBay-reputation-Buyer-or-Seller/10000000001338209/g.html>. [Google Scholar]
- 27. [cited 05.09.13];Amazon.com Help: about customer ratings [Internet] < http://www.amazon.com/gp/help/customer/display.html/ref=hp_left_sib?ie=UTF8&nodeId=200791020>. [Google Scholar]
- 28. [cited 05.09.13];FAQ – slashdot [Internet] < http://slashdot.org/faq>. [Google Scholar]
- 29.Miller N, Resnick P, Zeckhauser R. Eliciting informative feedback: the peerprediction method. Manag Sci. 2005;51(9):1359–1373. [Google Scholar]
- 30.McGlohon M, Glance N, Reiter Z. Star quality: aggregating reviews to rank products and merchants. 2010 [Google Scholar]
- 31.Chen P-Y, Dhanasobhon S, Smith MD. All reviews are not created equal: the disaggregate impact of reviews and reviewers at amazon.com. [cited 29.09.11];SSRN eLibrary [Internet] 2008 < http://ssrn.com/abstract=918083>. [Google Scholar]
- 32.Allahbakhsh M, Benatallah B, Ignjatovic A, Motahari-Nezhad HR, Bertino E, Dustdar S. Quality control in crowdsourcing systems: issues and directions. IEEE Internet Comput. 2013;17(2):76–81. [Google Scholar]
- 33.Allahbakhsh M, Ignjatovic A, Benatallah B, Beheshti S-M-R, Foo N, Bertino E. An analytic approach to people evaluation in crowdsourcing systems [Internet] Report No.:1211.3200. 2012 Nov; http://arxiv.org/abs/1211.3200. [Google Scholar]
- 34.Stranders R, Ramchurn SD, Shi B, Jennings NR. CollabMap: augmenting maps using the wisdom of crowds. [cited 30.07.13];Human computation [Internet] 2011 < http://www.aaai.org/ocs/index.php/WS/AAAIW11/paper/viewPDFInterstitial/3815/4247>. [Google Scholar]