Summary
Genetic diversity at the human β-globin locus has been implicated as a modifier of sickle cell anaemia (SCA) severity. However, haplotypes defined by restriction fragment length polymorphism sites across the β-globin locus have not been consistently associated with clinical phenotypes. To define the genetic structure at the β-globin locus more thoroughly, we performed high-density single nucleotide polymorphism (SNP) mapping in 820 children who were homozygous for the sickle cell mutation (HbSS). Genotyping results revealed very high linkage disequilibrium across a large region spanning the locus control region and the HBB (β-globin gene) cluster. We identified three predominant haplotypes accounting for 96% of the βS-carrying chromosomes in this population that could be distinguished using a minimal set of common SNPs. Consistent with previous studies, fetal haemoglobin level was significantly associated with βS-haplotypes. After controlling for covariates, an association was detected between haplotype and rate of hospitalization for acute chest syndrome (ACS) (incidence rate ratio 0.51, 95% confidence interval 0.29–0.89) but not incidence rate of vaso-occlusive pain or presence of silent cerebral infarct (SCI). Our results suggest that these SNP-defined βS-haplotypes may be associated with ACS, but not pain or SCI in a study population of children with SCA.
Keywords: Sickle cell anaemia, acute chest syndrome, β-globin, genetic analysis, haplotype
Introduction
Sickle haemoglobinopathies are Mendelian disorders caused by mutation in the haemoglobin β-chain gene (HBB) and characterized by considerable phenotypic heterogeneity. Even among individuals with sickle cell anaemia (SCA), which is the result of homozygosity for a single base pair substitution (HBB: p.Glu7Val; rs334), there is a dramatic range in the incidence of clinical manifestations (Steinberg and Hebbel 1983). Deoxygenated sickle haemoglobin (HbS), produced from aberrant βS-globin chains, form long inflexible polymers that result in the characteristic red blood cell sickling and haemolysis (Ferrone and Nagel 2001). Despite being a monogenic disease, the variable severity has only partially been explained by environmental and genetic factors. A better understanding of the pleiotropic effects that result from HbS could lead to targeted treatment and intervention strategies.
The two best-described genetic modifiers of SCA phenotype are α-thalassaemia and fetal haemoglobin (HbF) concentration (Driss et al 2009, Lettre 2012, Nagel and Steinberg 2001, Steinberg and Sebastiani 2012). The co-presence of α+-thalassaemia, typically resulting from 3.7 kb deletions in the α-globin (HBA1/2) genes, decreases haemolysis in individuals with SCA as a result of the decreased intracellular concentration of HbS available for polymerization (Steinberg and Embury 1986). Similarly, the beneficial effect of increased HbF level on many complications of SCA is tied to decreasing HbS concentration and inhibiting its polymerization under low oxygen conditions (Akinsheye et al 2011, Goldberg et al 1977, Poillon et al 1993). HbF level is heritable, and regulation of its expression has been mapped so far to three regions of the genome, including cis effects at the β-globin cluster, although many of the responsible functional elements remain unknown (Dover et al 1981, Garner et al 2000, Lettre et al 2008).
The β-globin cluster, which consists of a locus control region (LCR) upstream of the epsilon (HBE1), gamma-G (HBG2), gamma-A (HBG1), delta (HBD), and beta (HBB) globin genes, is under complex genetic regulation. The β locus globins are expressed differentially during development according to their order on the chromosome, switching in predominance from embryo to fetus and again after birth (Fritsch et al 1980, Hardison 2001, Wilber et al 2011). Classically, βS-gene (HBBs) cluster haplotypes have been mapped to geographic regions by thirteen or fewer restriction fragment length polymorphisms (RFLPs) that create or destroy DNA cleavage sites across the greater than 70 kilobase β-globin locus (Antonarakis et al 1985, Powars 1991). In descending order, the Benin, Central African Republic (CAR), and Senegal haplotypes are the most common reported in the Americas with significant chromosomal heterozygosity found in the United States (Hattori et al 1986, Sarnaik and Ballas 2001, Schroeder et al 1989, Steinberg et al 1995, Taylor et al 2008).
These RFLP-defined haplotypes at the βS locus have also been studied for their association with HbF and for a role in modulating disease severity; however, consistent associations have not been observed, perhaps due in part to unresolved genetic complexity of this region, small sample size, and limited inclusion of other clinical and genetic factors (Liu et al 2009, Powars et al 1994, Rieder et al 1991, Sarnaik and Ballas 2001, Schroeder et al 1989, Steinberg et al 1995, Taylor et al 2008). For example, Liu et al (2009) recently demonstrated that five commonly used RFLP sites at the β-globin locus are not equally informative in haplotype-tagging analyses and identified multiple single nucleotide polymorphisms (SNPs) within the HindIII restriction site near the HBG2 gene that produce identical digestion patterns, suggesting that these markers may not be sufficient to reveal the genomic diversity in this region. However, in spite of the unique regulation of the β-globin locus, the identification of hundreds of SNPs distributed densely across this large genomic region, and the ever-increasing efficiency of SNP genotyping assays, few other efforts have been reported using high-throughput genotyping techniques to fine-map chromosomes carrying the HbS mutation.
To define βS-haplotypes more accurately and determine their role in modifying clinical outcomes, we performed high-density SNP mapping of the β-globin locus in 820 Black or African American individuals who were homozygous for the sickle cell mutation (HbSS; HBB E6V) in the Silent Cerebral Infarct Transfusion (SIT) Trial. Using results from 131 common variants, we examined the linkage disequilibrium (LD) structure of the region spanning the LCR and the β-globin cluster and defined βS-haplotypes that could be distinguished using a minimal set of common SNPs. Finally, we tested the hypothesis that. after adjusting for clinical and genetic modifiers, SNP-defined βS-haplotypes were associated with sickle cell disease morbidity indicated by attenuated incidence rates of pain, acute chest syndrome (ACS), or silent cerebral infarcts (SCI).
Materials and Methods
Study population
Samples were selected from the SIT Trial (Casella et al 2010), registered at www.ClinicalTrials.gov (NCT00072761). Briefly, patients with an established relationship with a haematologist were enrolled at clinical sites across North America and Europe between 2004 and 2010. All participants were children aged 5 to 14 years, with haemoglobin (Hb) genotypes of SS or Sβ0–thalassaemia. Patients with a history of regular transfusion therapy or hydroxycarbamide therapy were excluded from the SIT Trial study. The current genetic analysis was further restricted to only those individuals carrying two copies of haemoglobin S (HbSS) with SCA. Among first-degree relatives participating in the study, only the first enrolled sibling with DNA available for genotyping was included. Finally, participants with missing data on rate of hospitalization for either vaso-occlusive pain or ACS were excluded. In total, 820 SIT Trial Black or African American children were available for haplotype analysis (Supplemental Figure 1).
Clinical definitions
Hospitalizations were recorded retrospectively at enrollment; patients were not receiving hydroxycarbamide or blood transfusion therapy when these data were collected. All vaso-occlusive pain episodes and ACS episodes that required hospitalization over a 3-year period prior to enrollment were recorded for each patient (Casella et al 2010). Pain events were defined locally as episodes that could not be attributed to causes other than sickle cell disease and required hospitalization and treatment with opiates. Local site coordinators verified these episodes for the patients in the highest 10% for rates of painful events. ACS was defined locally, based on a commonly accepted set of criteria that included fever, increased respiratory rate, decreased oxygen saturation, a new radiodensity of chest radiograph, and pneumonia (Castro et al 1994, Vichinsky et al 2000, Vichinsky et al 1997). ACS events occurring during hospitalization for vaso-occlusive crisis were classified as ACS, and local site coordinators verified events for the patients in the highest 10% for rates of ACS.
Children with asthma were identified by a positive response from a parent or legal guardian to the question, “Does the patient currently carry a diagnosis of asthma?” The use of asthma medication was also recorded. When a diagnosis of asthma was made and no asthma medication was recorded in the SIT Trial data base, the diagnosis was reconfirmed with a review of the medical records by the site coordinator using confirmation criteria of any hospital admissions, Emergency Department visits, or medications (Advair, Flovent, Montelukast) for asthma. Similarly, if the patient was recorded as having prescriptions for inhaled corticosteroids, bronchodilators, or a cysteinyl leucotriene receptor antagonist, but the parent did not state that the child had asthma, the site coordinator was required to recheck the medical records for a diagnosis of asthma (An et al 2011).
Additionally, results of SCI screenings were available for 605 of the SIT Trial participants included in the present analysis. SCI was defined by an area of abnormal magnetic resonance imaging (MRI) signal intensity on fluid attenuated inversion recovery T2-weighted images in a child with no prior history or physical findings of a focal neurological deficit as ascertained by a study neurologist’s physical examination (Casella et al 2010, Thangarajh et al 2012, Vendt et al 2009).
Genotyping
DNA for this study was isolated from Epstein-Barr virus (EBV)-transformed lymphoblast cells established from lymphocytes isolated from the blood of patients enrolled in the SIT Trial. High density SNP mapping was performed with two overlapping panels of probes designed to known variants in the 85 kilobase (kb) region containing β-globin locus (GenBank: U01317.1) on the short arm of chromosome 11. Samples were genotyped using the Illumina Infinium HumanOmni1-Quad BeadChip, a custom GoldenGate panel, or both according to manufacturer’s protocol (Illumina, Inc., San Diego, CA).
For the HumanOmni1-Quad BeadChip, cluster plots for assays on chromosome 11 from position 5,233,697 to 5,321,686 (Genome Reference Consortium Human Build 37) were manually reviewed using GenomeStudio (Illumina), and genotype calls for 98 high quality SNPs were exported for subsequent analyses. Similarly, GenomeStudio (Illumina) was used to review cluster plots and confirm genotype calls for SNPs run on the GoldenGate platform, and 55 high quality SNPs from the same 85 kb region containing the β-globin locus were selected for further analyses, including 22 SNPs present on the Illumina Omni1 BeadChip. SNPs with greater than 4% failed genotype calls were excluded from analysis, and the average genotyping call rate for each panel was >99%. Fifteen percent of samples were typed with both the HumanOmni1-Quad BeadChip and the custom GoldenGate panel, and genotype calls were over 99.5% concordant for the overlapping SNPs successfully genotyped on both platforms.
Haplotype analysis
Linkage disequilibrium at the β-globin locus among individuals with SCA was visualized using Haploview Version 4.2 (Barrett et al 2005) for common SNPs with minor allele frequencies >5%. Individual haplotypes were estimated with PHASE Version 2.1 (Stephens et al 2001) using 5 cross-validated SNPs spanning the β-globin cluster: rs11036351, rs4320977, rs16912210, rs2855039, and rs7482144. The three most common resulting five-SNP haplotypes were named H1, H2, and H3 in order of their prevalence in this population.
Statistical analysis
All other statistical analyses were conducted using SAS Version 9.3 (SAS Institute, Cary, NC). The distribution of demographic and clinical characteristics [age at enrollment, sex, asthma diagnosis, steady state white blood count, steady state haemoglobin, steady state reticulocytes, total bilirubin, HBA1/2 deletion status, and heme oxygenase-1 (HMOX1) promoter repeat class was calculated for the entire study population (n=820). Steady state levels were defined at a single time point at routine visit when the patients were clinically well. For children with more than one bilirubin measurement, the highest value during the 3-year period was used for the analysis. HMOX1 (GT) n promoter repeat alleles were classified according to number of dinucleotide repeats as short (S) with ≤ 25 repeats and long (L) with > 25 repeats (Bean et al 2012).
For association tests, individuals were grouped by haplotype with the most prevalent group, H1,H1, serving as the referent group for comparison to those carrying H2 or H3 alleles. Sixty-three of the 820 samples (7.7%) carried rare recombinant haplotypes with allele frequencies of ≤ 1% and were excluded from further analysis. In regression modelling, H1, H2 heterozygotes were grouped with H2,H2 homozygotes, and H1,H3 heterozygotes were grouped with H3,H3 homozygotes. However, 24 heterozygous H2,H3 individuals (24/820; 2.9%) could not be unambiguously grouped with either the H2- or H3-carrying category and were therefore also excluded from analysis. After these exclusions, 733 SIT Trial participants were available for testing for association with laboratory values and clinical outcomes (Supplemental Figure 1). Mean steady-state haemoglobin was slightly elevated (p<0.05) in the excluded group (84 ± 11 g/l) compared to the analysed group (81 ± 10 g/l). However, there were no differences in average age, gender, asthma, steady-state white blood count, steady-state reticulocytes, total bilirubin, HBA1/2 deletion status, or HMOX1 promoter genotype distribution between these two groups (data not shown).
A subset of SIT Trial participants with haplotypes available for analysis had an HbF assessment performed at a central laboratory at enrollment (n=479). For these samples, mean HbF percentage was calculated for the haplotype pairs and differences were assessed using the Kruskal-Wallis test for non-normal data. Multiple linear regression models were used to evaluate the association between haplotype group and HbF (log transformed). The initial models included all demographic and clinical characteristics as covariates; only those factors associated with a ≥10% change in effect estimate (age and gender) were retained in the final model. There was no difference in mean age, sex, race, asthma, white blood cell count, haemoglobin, reticulocytes, total bilirubin, α-globin genotype distribution, or HMOX1 (GT) n genotype distribution between individuals with and without HbF assessment at the central laboratory (data not shown).
Negative binomial regression models with a scale parameter estimated by maximum likelihood were used to estimate the incidence of hospitalization for ACS and pain episodes according to haplotype group. The aforementioned criterion was used to determine which covariates to include in the final model. Thus, the ACS model included asthma, gender, HBA1/2 deletion status, and HMOX1 promoter repeat class as covariates. Similarly, the pain model included age, steady state haemoglobin, reticulocytes, α HBA1/2 deletion status, and HMOX1 promoter repeat class. Finally, the association between SCI and haplotype was estimated using log-binomial regression with HBA1/2 deletion status and HMOX1 promoter repeat class included as covariates.
Results
SIT Trial participants
The demographic, clinical, and genetic characteristics of the 820 SIT Trial participants available for the present analyses are presented in Table I. Participants included or excluded in subsequent analyses are summarized in Supplemental Figure 1.
Table I.
Demographic, clinical, and genotypic characteristics of 820 Black or African American SIT Trial participants.
| Characteristic* | Mean (SD) or % |
|---|---|
| Age at registration (years) | 8.9 (2.5) |
| Male (%) | 52.3 |
| Ever had asthma (%) | 23.4 |
| Steady-state white blood count (×109/l) | 12.7 (5.5) |
| Steady-state haemoglobin (g/l) | 81 (11) |
| Steady-state reticulocytes | 12.2 (5.3) |
| Total bilirubin (µmol/l) | 65 (46.2) |
| α-globin genotype (%) | |
| αα/αα | 58.3 |
| −α/αα | 36.3 |
| −α/− α | 5.4 |
| HMOX1 promoter (GT)n genotype** (%) | |
| L/L | 73.1 |
| S/L | 23.2 |
| S/S | 3.7 |
Steady-state levels defined at a single routine clinical well visit.
(GT)n allele classes defined by number of repeats: S ≤ 25 and L > 25.
Linkage disequilibrium across the β-Globin locus
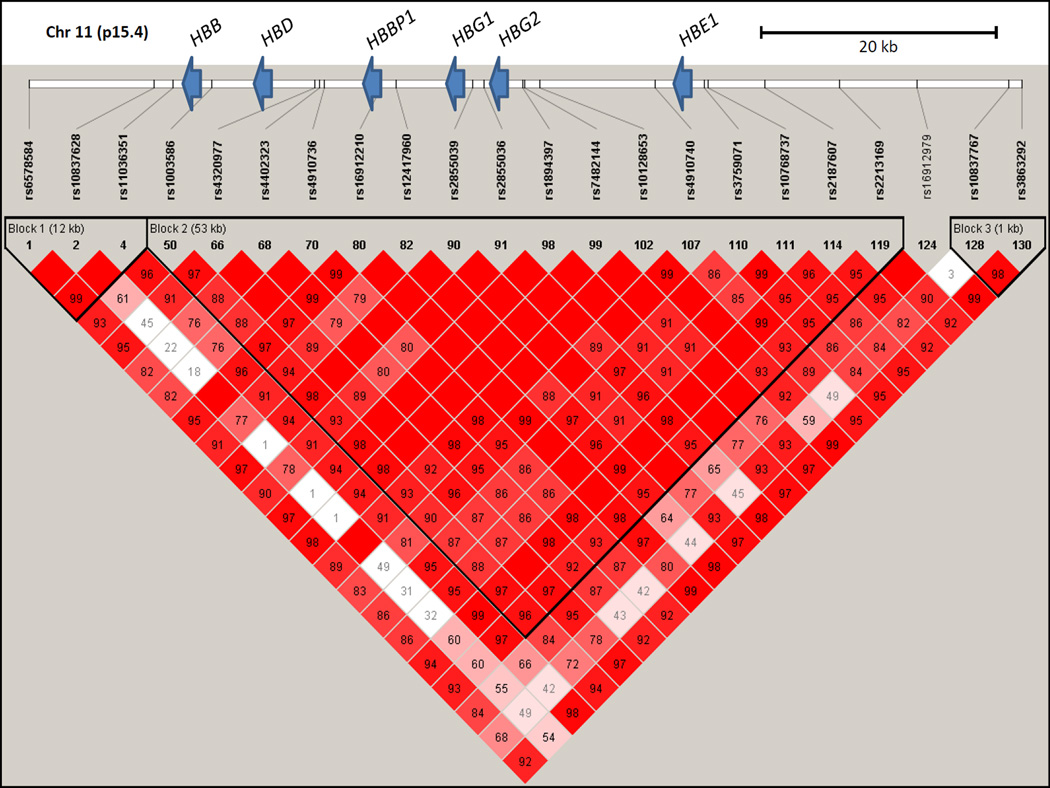
To explore genomic structure in individuals with SCA thoroughly, high-density genotyping was successfully performed for 131 SNPs across the 85kb β-globin region that includes the LCR and the HBE1, HBG2, HBG1, HBD, and HBB genes. The average genotyping call rate was >99%, and nearly one-half (64/131) of the SNPs were very rare or monomorphic with minor allele frequencies of <0.001 in this cohort (Supplemental Table I). Analysis of common markers with >5% minor allele frequencies revealed a pattern of very high LD across the entire region (Figure 1).
Figure 1.
Linkage disequilibrium across the β-globin locus in SIT Trial participants with sickle cell anaemia. Haploview Version 4.2 visualization of LD (D′) is shown for 22 SNPs with minor allele frequencies >5% genotyped on the HumanOmni1-Quad BeadChip. The calculated D′ for pairwise comparisons between SNPs is displayed in each box as the decimal value between 0 and 99, and blank boxes represent complete LD (D′ = 1). The relative positions of the β-globin cluster genes compared to the SNPs genotyped along chromosome 11p15.4 are indicated by blue arrows.
PHASE analysis using a minimal set of five SNPs spanning the β-globin cluster identified a total of twelve haplotypes in this population. The three most common haplotypes accounted for 96% of the predicted haplotypes and were named here as H1, H2, and H3 in order of their prevalence in this population (Table II). The nine remaining haplotypes had allele frequencies of ≤1%, and individuals carrying at least one of these “other” rare haplotypes were excluded from further analysis (Table II). The relationship between classical haplotypes and SNP-defined β-globin haplotypes is summarized in Supplemental Table II.
Table II.
Summary of single nucleotide polymorphisms (SNPs) and haplotypes present in SIT Trial participants.
| Number | Position* | SNP | Alleles | MAF** |
| 1 | chr11:5246000 | rs11036351 | [C/T] | T (0.32) |
| 2 | chr11:5258162 | rs4320977 | [A/G] | G (0.23) |
| 3 | chr11:5263853 | rs16912210 | [A/G] | G (0.21) |
| 4 | chr11:5271671 | rs2855039 | [A/G] | A (0.10) |
| 5 | chr11:5276169 | rs7482144 | [A/G] | A (0.09) |
| Haplotype | Genotype*** | Frequency | ||
| H1 | CAAGG | 0.665 | ||
| H2 | TGGGG | 0.202 | ||
| H3 | TAAAA | 0.094 | ||
| Other | 0.040 | |||
Genome Reference Consortium GRCh37 chromosomal coordinates.
Observed minor allele frequency.
Haplotype estimated with PHASE Version 2.1 using 5 SNPs spanning the HBB cluster. The three most common resulting five-SNP haplotypes were named H1, H2, and H3 in order of their prevalence in this population.
Haplotype association with fetal haemoglobin (HbF)
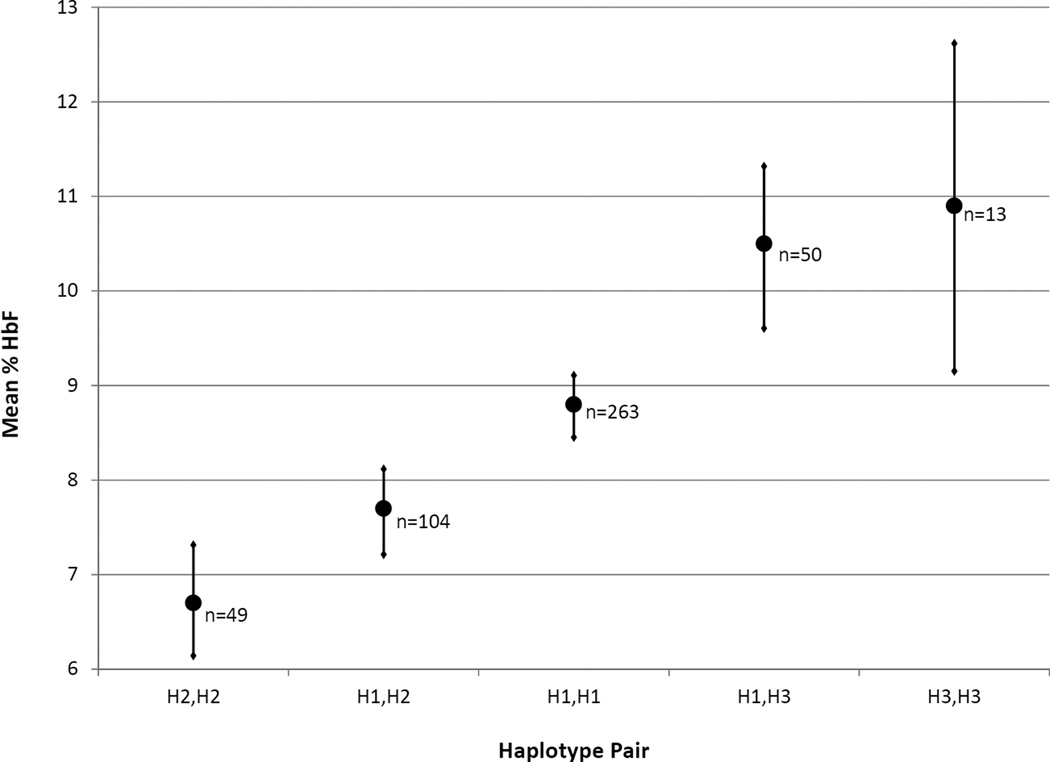
HbF assessment was performed at a central laboratory for a subset of SIT Trial participants at enrollment (Casella et al 2010). Mean HbF level in these participants was associated with βS-globin H1, H2 and H3 haplotype groups (Figure 2). Individuals carrying two of the most common haplotype, H1, had an intermediate HbF level and served as the referent group. Carrying one or two copies of H2 was significantly associated with decreased HbF (p=0.01). In contrast, carrying one or two copies of H3 was significantly associated with increased HbF (p=0.05). Heterozygous H2,H3 individuals (n=24) could not be exclusively assigned to either the H2 or H3 carrying groups and were therefore excluded from analysis, as described above.
Figure 2.
Fetal haemoglobin (HbF) levels by haplotype group in SIT Trial participants (n=479). Mean HbF (filled circle) and standard error (horizontal bar) is shown with number of individuals (n) in each haplotype pair group indicated. For statistical analyses, H2,H2 and H1,H2 were grouped together, and H3,H3 and H1,H3 were grouped together. Kruskal-Wallis non-parametric testing demonstrated a significant difference in the sample means (p=0.006). To test the association with haplotype, HbF was log transformed and multiple linear regression models were adjusted for age and gender. Carrying one or two copies of H2 was associated with a decreased HbF (7.4%) compared to the 8.8% level in the H1,H1 reference group (95% confidence interval [CI]: 0.72–0.98; p=0.01). Conversely, carrying one or two copies of H3 was associated with increased HbF (10.5%) compared to the H1,H1 reference group (95% CI: 1.02–1.39; p=0.05).
Vaso-occlusive episodes and haplotypes
The association between haplotype group and hospitalization for ACS, hospitalization for pain, or SCI was tested. The rates of clinical complications in individuals carrying one or two H2 haplotypes, associated with decreased HbF, and individuals with one or two H3 haplotypes, associated with increased HbF, were compared to rates in the H1,H1 homozygous reference group. Individuals carrying one or two copies of the H3 haplotype, associated with increased HbF, had a significantly lower rate of hospitalization for ACS after correcting for asthma, gender, HBA1/2 genotype, and HMOX1 promoter repeat class (p=0.02). The rate of ACS in individuals with one or two copies of the H2 haplotype was similar to the homozygous H1 reference group (Table III). The rate of hospitalization for vaso-occlusive pain was somewhat higher among those carrying one or two copies of the H3 haplotype (0.71; n=99) compared to those carrying one or two copies of the H2 haplotype (0.62; n=224) and the homozygous H1,H1 group (0.60; n=410), although this difference did not achieve statistical significance in crude or adjusted modelling. Similarly, we found no evidence for an association with the incidence of SCI in individuals carrying one or two copies of H3 (31.7%, n=79) or those with one or two copies of H2 (27.5%, n=189) compared to the H1,H1 homozygous reference group (34.1%, n=337).
Table III.
Rate of hospitalization for Acute Chest syndrome (ACS) in β-globin haplotype groups in SIT Trial participants.
| Haplotype | N | ACS rate | IRR* (95% CI) unadjusted |
p-value | IRR (95% CI) adjusted** |
p-value |
|---|---|---|---|---|---|---|
| H1,H2 / H2,H2 | 224 | 0.16 | 1.13 (0.84–1.52) | 0.42 | 1.04 (0.76–1.43) | 0.79 |
| H1,H1 | 410 | 0.15 | REF | REF | ||
| H1,H3 / H3,H3 | 99 | 0.09 | 0.66 (0.41–1.06) | 0.09 | 0.51 (0.29–0.89) | 0.02 |
Incidence rate ratio (95% confidence interval).
Adjusted for asthma, gender, HBA1/2 deletion status, and HMOX1 promoter repeat class.
Discussion
SCA, the most common inherited sickle haemoglobinopathy, is a monogenic disorder with striking clinical variability (Steinberg and Hebbel 1983). Two factors, α-thalassaemia and HbF concentration, have long been recognized to influence disease severity (Altay et al 1981, Edington and Lehmann 1955, Watson 1948). However, much of the variability in the severity of the disease remains unexplained, continuing to complicate efforts to identify individuals at high-risk of vaso-occlusive crises before significant organ damage accumulates. Attempts to determine the role of genetic variation at the β-globin locus itself in modifying the severity of clinical outcomes of SCA have been largely restricted to studies of haplotypes defined by a limited number of variants or RFLP sites in and near the genes in the β-globin locus (Powars 1991, Rieder et al 1991, Schroeder et al 1989, Steinberg et al 1995). The HBB cluster is under unique developmental regulation, and additional genetic complexity in the region not resolved by classical RFLP analysis may contribute to inconsistent results of βS-haplotype studies (Hardison 2001, Liu et al 2009). In the present analysis, we sought use high-density SNP mapping to define βS-haplotypes more accurately and determine their role in modifying clinical phenotypes in a large study of children with SCA.
Previous reports of high-density genomic analysis of the HBB cluster were performed in small numbers of individuals with HbSS and did not consider disease severity (Hanchard et al 2007, Liu et al 2009). In the present study of over 800 individuals homozygous for the sickle mutation, we used high density SNP mapping to perform a detailed analysis of this region in individuals participating in a clinical trial. We observed a striking pattern of very high LD across a large region of the βS-globin locus, supporting the idea that βS-haplotypes are conserved over long genomic distances due to selective pressure (Hanchard et al 2007, Liu et al 2009). In spite of typing over one hundred SNPs at the β-globin locus, we identified three predominant haplotypes that accounted for the majority of the βS-chromosomes in this population, similar to previous RFLP studies that report Benin, CAR, and Senegal as the most prevalent haplotypes in populations in the United States (Hattori et al 1986, Powars 1991, Schroeder et al 1989).
HbF level is partly controlled by genetic variation at the HBB cluster, although the exact responsible functional cis elements remain largely unknown (Dover et al 1981, Galarneau et al 2010, Lettre et al 2008, Nagel et al 1985, Nagel et al 1987, Steinberg 2005). We found a clear connection between HbF level and SNP-defined βS-haplotypes in children with SCA. Individuals homozygous for the most common haplotype, H1, had an intermediate mean HbF level, whereas carrying H2 was significantly associated with decreased HbF and carrying H3 was significantly associated with increased HbF. These results were consistent with previous studies that report intermediate HbF levels associated with the Benin haplotype, lowest HbF levels associated with the CAR haplotype, and highest HbF levels associated with the Senegal haplotype (Antonarakis et al 1984, Hattori et al 1986, Powars 1991).
Similar to studies that report no correlation between RFLP-defined HBBs cluster haplotypes and acute clinical outcomes or cerebrovascular accidents, we did not find an association between SNP-defined βS haplotypes and incidence rate of hospitalization for pain or prevalence of SCI (de Montalembert et al 1993, Flanagan et al 2011, Rieder et al 1991). However, we were able to detect a significant association between SNP-defined βS-haplotypes and ACS in children with SCA. The H3 haplotype was protective in this population, and individuals with at least one H3 haplotype had approximately half as many hospitalizations for ACS per year compared to those that did not carry this haplotype. Only after controlling for covariates (asthma, gender, HBA1/2 genotype, and HMOX1 promoter repeat class) associated with the incidence of ACS was thus association with the haplotype detected.
Our results may not fully address the significance of variation at the HBBS cluster on long-term prognosis of disease severity. Although we have a large number of individuals, including both heterozygotes and homozygotes for each of the predominant SNP-defined haplotypes, our study population is limited to children in whom severe end-organ failure may not yet be evident. Additionally, individuals at risk for the most severe SCA may be under-represented in this study, because enrollment was restricted to those not previously on regular transfusion or hydroxycarbamide therapy (Casella et al 2010). Despite these limitations, this cohort has previously been used to replicate other established associations, such as asthma with the incidence of vaso-occlusive pain episodes and ACS (An et al 2011), as well as document new associations, such as HMOX1 promoter polymorphisms and the incidence of ACS (Bean et al 2012) .
In this study, we thoroughly explored genetic variation in the HBB cluster, performing high density SNP genotyping and haplotype analysis in a large number of chromosomes carrying the HBB E6V (HbS) mutation. We observed a remarkable pattern of high LD and a small number of predominant βS-haplotypes that could be easily determined in future studies by genotyping a small number of common SNPs reported herein. We identified a close relationship between SNP-defined haplotype and mean HbF level, and detected an association between βS-haplotypes and rate of hospitalization for ACS. Our results suggest that these SNP-defined βS-haplotypes, in combination with covariates, can be used to delineate clinical heterogeneity in a large study of Black or African American children with SCA as measured by acute events. Future rigorous studies are needed to validate the predictive value of these markers with additional clinical outcomes and in more severely affected and older individuals.
Supplementary Material
Acknowledgements
The authors would like to thank the families and children with SCD who were participants, the SIT Trial study coordinators, and both past and present laboratory members in the Division of Blood Disorders (CDC) and Division of Pediatric Hematology (JHU). Support for this study included RO1-HL079937 (NHLBI) and Burroughs Wellcome Foundation for M.R.D., U01-NS-042804 (NINDS) for E.B.C., 1U54HL090515 (NHLBI) and U54 HL090515, RO1 HL091759 and M01-RR00052 (CTSA) for J.F.C.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. Dr. Casella has received an honorarium and travel expenses in the past and presently receives salary support through Johns Hopkins for providing consultative advice to Adventrx Pharmaceuticals regarding a proposed clinical trial of an agent for treating vaso-occlusive crisis in sickle cell disease.
Footnotes
Authorship
C.J.B. contributed to designing the genotyping experiments, analysing the data and writing the manuscript; S.L.B. analysed the data and contributed to writing the manuscript; A.B.P., N.G., and P.B. contributed to analysing and interpreting the data; G.Y. and M.E.P. contributed to performing the genotyping; W.C.H. contributed to designing the experiments and writing the manuscript; and as part of the SIT Biologic Repository, M.R.D., J.F.C, E.B.C., and J.K. contributed to collection of key reagents and data, designing the experiments and writing the manuscript.
Conflict of interest disclosure
The remaining authors declare no competing financial interests.
References
- Akinsheye I, Alsultan A, Solovieff N, Ngo D, Baldwin CT, Sebastiani P, Chui DH, Steinberg MH. Fetal hemoglobin in sickle cell anemia. Blood. 2011;118:19–27. doi: 10.1182/blood-2011-03-325258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altay C, Gravely ME, Joseph BR, Williams DF. alpha-thalassemia-2 and the variability of hematological values in children with sickle cell anemia. Pediatric Research. 1981;15:1093–1096. doi: 10.1203/00006450-198108000-00004. [DOI] [PubMed] [Google Scholar]
- An P, Barron-Casella EA, Strunk RC, Hamilton RG, Casella JF, DeBaun MR. Elevation of IgE in children with sickle cell disease is associated with doctor diagnosis of asthma and increased morbidity. The Journal of Allergy and Clinical Immunology. 2011;127:1440–1446. doi: 10.1016/j.jaci.2010.12.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonarakis SE, Boehm CD, Serjeant GR, Theisen CE, Dover GJ, Kazazian HH., Jr Origin of the beta S-globin gene in blacks: the contribution of recurrent mutation or gene conversion or both. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:853–856. doi: 10.1073/pnas.81.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonarakis SE, Kazazian HH, Jr, Orkin SH. DNA polymorphism and molecular pathology of the human globin gene clusters. Human Genetics. 1985;69:1–14. doi: 10.1007/BF00295521. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bean CJ, Boulet SL, Ellingsen D, Pyle ME, Barron-Casella EA, Casella JF, Payne AB, Driggers J, Trau HA, Yang G, Jones K, Ofori-Acquah SF, Hooper WC, DeBaun MR. Heme oxygenase-1 gene promoter polymorphism is associated with reduced incidence of acute chest syndrome among children with sickle cell disease. Blood. 2012;120:3822–3828. doi: 10.1182/blood-2011-06-361642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casella JF, King AA, Barton B, White DA, Noetzel MJ, Ichord RN, Terrill C, Hirtz D, McKinstry RC, Strouse JJ, Howard TH, Coates TD, Minniti CP, Campbell AD, Vendt BA, Lehmann H, Debaun MR. Design of the silent cerebral infarct transfusion (SIT) trial. Journal of Pediatric Hematology/Oncology. 2010;27:69–89. doi: 10.3109/08880010903360367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro O, Brambilla DJ, Thorington B, Reindorf CA, Scott RB, Gillette P, Vera JC, Levy PS. The acute chest syndrome in sickle cell disease: incidence and risk factors. The Cooperative Study of Sickle Cell Disease. Blood. 1994;84:643–649. [PubMed] [Google Scholar]
- de Montalembert M, Maier-Redelsperger M, Girot R, Belloy M, Vilmer E, Ducrocq R, Guidal C, Elion J. Beta-globin gene cluster haplotype and alpha-thalassemia do not correlate with the acute clinical manifestations of sickle cell disease in children. Blood. 1993;82:2595–2596. [PubMed] [Google Scholar]
- Dover GJ, Boyer SH, Pembrey ME. F-cell production in sickle cell anemia: regulation by genes linked to beta-hemoglobin locus. Science. 1981;211:1441–1444. doi: 10.1126/science.6162200. [DOI] [PubMed] [Google Scholar]
- Driss A, Asare KO, Hibbert JM, Gee BE, Adamkiewicz TV, Stiles JK. Sickle Cell Disease in the Post Genomic Era: A Monogenic Disease with a Polygenic Phenotype. Genomics Insights. 2009;2009:23–48. [PMC free article] [PubMed] [Google Scholar]
- Edington GM, Lehmann H. Expression of the sickle-cell gene in Africa. British Medical Journal. 1955;1:1308–1311. doi: 10.1136/bmj.1.4925.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrone F, Nagel RL. Polymer structure and polymerization of dexoyhemoglobin S. In: Steinberg MH, Forget BG, Higgs DR, Nagel RL, editors. Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management. Boston, USA: Cambridge University Press; 2001. [Google Scholar]
- Flanagan JM, Frohlich DM, Howard TA, Schultz WH, Driscoll C, Nagasubramanian R, Mortier NA, Kimble AC, Aygun B, Adams RJ, Helms RW, Ware RE. Genetic predictors for stroke in children with sickle cell anemia. Blood. 2011;117:6681–6684. doi: 10.1182/blood-2011-01-332205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch EF, Lawn RM, Maniatis T. Molecular cloning and characterization of the human beta-like globin gene cluster. Cell. 1980;19:959–972. doi: 10.1016/0092-8674(80)90087-2. [DOI] [PubMed] [Google Scholar]
- Galarneau G, Palmer CD, Sankaran VG, Orkin SH, Hirschhorn JN, Lettre G. Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nature Genetics. 2010;42:1049–1051. doi: 10.1038/ng.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner C, Tatu T, Reittie JE, Littlewood T, Darley J, Cervino S, Farrall M, Kelly P, Spector TD, Thein SL. Genetic influences on F cells and other hematologic variables: a twin heritability study. Blood. 2000;95:342–346. [PubMed] [Google Scholar]
- Goldberg MA, Husson MA, Bunn HF. Participation of hemoglobins A and F in polymerization of sickle hemoglobin. Journal of Biological Chemistry. 1977;252:3414–3421. [PubMed] [Google Scholar]
- Hanchard N, Elzein A, Trafford C, Rockett K, Pinder M, Jallow M, Harding R, Kwiatkowski D, McKenzie C. Classical sickle beta-globin haplotypes exhibit a high degree of long-range haplotype similarity in African and Afro-Caribbean populations. BMC Genetics. 2007;8:52. doi: 10.1186/1471-2156-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison R. Organization, evolution, and regulation of the globin genes. In: Steinberg MH, Forget BG, Higgs DR, Nagel RL, editors. Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management. Boston, USA: Cambridge University Press; 2001. [Google Scholar]
- Hattori Y, Kutlar F, Kutlar A, McKie VC, Huisman TH. Haplotypes of beta S chromosomes among patients with sickle cell anemia from Georgia. Hemoglobin. 1986;10:623–642. doi: 10.3109/03630268609036566. [DOI] [PubMed] [Google Scholar]
- Lettre G. The Search for Genetic Modifiers of Disease Severity in the beta-Hemoglobinopathies. Cold Spring Harbor Perspectives in Medicine. 2012;2 doi: 10.1101/cshperspect.a015032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G, Sankaran VG, Bezerra MA, Araujo AS, Uda M, Sanna S, Cao A, Schlessinger D, Costa FF, Hirschhorn JN, Orkin SH. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11869–11874. doi: 10.1073/pnas.0804799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Muralidhar S, Singh M, Sylvan C, Kalra IS, Quinn CT, Onyekwere OC, Pace BS. High-density SNP genotyping to define beta-globin locus haplotypes. Blood Cells, Molecules & Diseases. 2009;42:16–24. doi: 10.1016/j.bcmd.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel RL, Steinberg MH. Role of epistatic (modifier) genes in the modulation of the phenotypic diversity of sickle cell anemia. Pediatric Pathology & Molecular Medicine. 2001;20:123–136. [PubMed] [Google Scholar]
- Nagel RL, Fabry ME, Pagnier J, Zohoun I, Wajcman H, Baudin V, Labie D. Hematologically and genetically distinct forms of sickle cell anemia in Africa. The Senegal type and the Benin type. New England Journal of Medicine. 1985;312:880–884. doi: 10.1056/NEJM198504043121403. [DOI] [PubMed] [Google Scholar]
- Nagel RL, Rao SK, Dunda-Belkhodja O, Connolly MM, Fabry ME, Georges A, Krishnamoorthy R, Labie D. The hematologic characteristics of sickle cell anemia bearing the Bantu haplotype: the relationship between G gamma and HbF level. Blood. 1987;69:1026–1030. [PubMed] [Google Scholar]
- Poillon WN, Kim BC, Rodgers GP, Noguchi CT, Schechter AN. Sparing effect of hemoglobin F and hemoglobin A2 on the polymerization of hemoglobin S at physiologic ligand saturations. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:5039–5043. doi: 10.1073/pnas.90.11.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powars DR. Beta s-gene-cluster haplotypes in sickle cell anemia. Clinical and hematologic features. Hematology/Oncology Clinics of North America. 1991;5:475–493. [PubMed] [Google Scholar]
- Powars DR, Meiselman HJ, Fisher TC, Hiti A, Johnson C. Beta-S gene cluster haplotypes modulate hematologic and hemorheologic expression in sickle cell anemia. Use in predicting clinical severity. The American Journal of Pediatric Hematology/Oncology. 1994;16:55–61. [PubMed] [Google Scholar]
- Rieder RF, Safaya S, Gillette P, Fryd S, Hsu H, Adams JG, 3rd, Steinberg MH. Effect of beta-globin gene cluster haplotype on the hematological and clinical features of sickle cell anemia. American Journal of Hematology. 1991;36:184–189. doi: 10.1002/ajh.2830360305. [DOI] [PubMed] [Google Scholar]
- Sarnaik SA, Ballas SK. Molecular characteristics of pediatric patients with sickle cell anemia and stroke. American Journal of Hematology. 2001;67:179–182. doi: 10.1002/ajh.1103. [DOI] [PubMed] [Google Scholar]
- Schroeder WA, Powars DR, Kay LM, Chan LS, Huynh V, Shelton JB, Shelton JR. Beta-cluster haplotypes, alpha-gene status, and hematological data from SS, SC, and S-beta-thalassemia patients in southern California. Hemoglobin. 1989;13:325–353. doi: 10.3109/03630268909003397. [DOI] [PubMed] [Google Scholar]
- Steinberg MH. Predicting clinical severity in sickle cell anaemia. British Journal of Haematology. 2005;129:465–481. doi: 10.1111/j.1365-2141.2005.05411.x. [DOI] [PubMed] [Google Scholar]
- Steinberg MH, Embury SH. Alpha-thalassemia in blacks: genetic and clinical aspects and interactions with the sickle hemoglobin gene. Blood. 1986;68:985–990. [PubMed] [Google Scholar]
- Steinberg MH, Hebbel RP. Clinical diversity of sickle cell anemia: genetic and cellular modulation of disease severity. American Journal of Hematology. 1983;14:405–416. doi: 10.1002/ajh.2830140412. [DOI] [PubMed] [Google Scholar]
- Steinberg MH, Sebastiani P. Genetic modifiers of sickle cell disease. American Journal of Hematology. 2012;87:795–803. doi: 10.1002/ajh.23232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg MH, Hsu H, Nagel RL, Milner PF, Adams JG, Benjamin L, Fryd S, Gillette P, Gilman J, Josifovska O, Hellman-Erlingsson S, Safaya S, Huey L, Rieder RF. Gender and haplotype effects upon hematological manifestations of adult sickle cell anemia. American Journal of Hematology. 1995;48:175–181. doi: 10.1002/ajh.2830480307. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JGt, Ackah D, Cobb C, Orr N, Percy MJ, Sachdev V, Machado R, Castro O, Kato GJ, Chanock SJ, Gladwin MT. Mutations and polymorphisms in hemoglobin genes and the risk of pulmonary hypertension and death in sickle cell disease. American Journal of Hematology. 2008;83:6–14. doi: 10.1002/ajh.21035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangarajh M, Yang G, Fuchs D, Ponisio MR, McKinstry RC, Jaju A, Noetzel MJ, Casella JF, Barron-Casella E, Hooper WC, Boulet SL, Bean CJ, Pyle ME, Payne AB, Driggers J, Trau HA, Vendt BA, Rodeghier M, Debaun MR. Magnetic resonance angiography-defined intracranial vasculopathy is associated with silent cerebral infarcts and glucose-6-phosphate dehydrogenase mutation in children with sickle cell anaemia. British Journal of Haematology. 2012;159:352–359. doi: 10.1111/bjh.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendt BA, McKinstry RC, Ball WS, Kraut MA, Prior FW, Barton B, Casella JF, DeBaun MR. Silent Cerebral Infarct Transfusion (SIT) trial imaging core: application of novel imaging information technology for rapid and central review of MRI of the brain. Journal of Digital Imaging. 2009;22:326–343. doi: 10.1007/s10278-008-9114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vichinsky EP, Styles LA, Colangelo LH, Wright EC, Castro O, Nickerson B. Acute chest syndrome in sickle cell disease: clinical presentation and course. Cooperative Study of Sickle Cell Disease. Blood. 1997;89:1787–1792. [PubMed] [Google Scholar]
- Vichinsky EP, Neumayr LD, Earles AN, Williams R, Lennette ET, Dean D, Nickerson B, Orringer E, McKie V, Bellevue R, Daeschner C, Manci EA. Causes and outcomes of the acute chest syndrome in sickle cell disease. National Acute Chest Syndrome Study Group. New England Journal of Medicine. 2000;342:1855–1865. doi: 10.1056/NEJM200006223422502. [DOI] [PubMed] [Google Scholar]
- Watson J. The significance of the paucity of sickle cells in newborn Negro infants. The American Journal of the Medical Sciences. 1948;215:419–423. doi: 10.1097/00000441-194804000-00008. [DOI] [PubMed] [Google Scholar]
- Wilber A, Nienhuis AW, Persons DA. Transcriptional regulation of fetal to adult hemoglobin switching: new therapeutic opportunities. Blood. 2011;117:3945–3953. doi: 10.1182/blood-2010-11-316893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.