Summary
Vaccines are among the greatest successes in the history of public health. However, past strategies for vaccine development are unlikely to succeed in the future against major global diseases such as AIDS, TB, and malaria. For such diseases, the correlates of protection are poorly defined and the pathogens evade immune detection and/or exhibit extensive genetic variability. Recent advances have heralded in a new era of vaccine discovery. However, translation of these advances into vaccines remains impeded by lack of understanding of key vaccinology principles in humans. We review these advances towards vaccine discovery and suggest that for accelerating successful vaccine development, new human immunology-based clinical research initiatives be implemented with the goal of elucidating and more effectively inducing vaccine-induced protective immune responses.
Introduction
At the end of the 18th century, Edward Jenner used cowpox-infected materials to immunize against smallpox and introduced the term “ vaccine” (1). A century later, Louis Pasteur developed methods for attenuation of bacteria (2) and Salmon and Smith developed methods for inactivation of micro-organisms (3). Together, these advances ushered in a new scientific era of vaccinology. Virus propagation in cell culture enabled the development of methods for attenuating viral vaccines (4), leading to a golden age of vaccine development in the second half of the 20th century with the development of several vaccines including polio, measles, mumps, and rubella (5-9). By the latter part of the 20th century, most of the vaccines that could be developed by direct mimicry of natural infection with live attenuated or killed/inactivated vaccines had been developed. New technologies, including protein conjugation to capsular polysaccharides, and the advent of methods to engineer recombinant DNA, led to the development of vaccines for prevention of bacterial pneumonia and meningitis, hepatitis B and the recent development of the human papillomavirus vaccine (10-12). Vaccines have now led to the eradication of smallpox, near eradication of polio, prevention of untold millions of deaths from infectious diseases each year, and are one of the most effective public health measures available (13). For example, prior to the introduction of the measles vaccine in the United States, incidence of measles peaked at nearly 900,000 cases per year, compared with an average of less than 100 cases of measles per year in recent years in the US (14). Similarly, using metrics to measure cost-effectiveness of vaccines such as disability adjusted life year (DALY), global vaccination for measles results in $17 per DALY, one of the most cost-effective health interventions in developing countries (15). Table 1 provides an overview of vaccine preventable diseases by currently licensed vaccines.
Table 1.
Major Global Infections Prevented by Vaccinesa
| Bacterial | Viral |
|---|---|
| Cholera | Adenovirus Based Diseases |
| Diphtheria | Hepatitis A |
| Haemophilus Influenza | Hepatitis B |
| Meningococcal Meningitis | Human Papillomavirus |
| Plague | Influenza |
| Pneumococcal Pneumonia | Japanese Encephalitis |
| Tetanus | Measles |
| Tuberculosis | Mumps |
| Typhoid Fever | Polio |
| Rabies | |
| Rotavirus Diarrhea | |
| Rubella | |
| Smallpox | |
| Tick Borne Encephalitis | |
| Varicella-Zoster | |
| Yellow Fever |
The level of efficacy for the vaccines noted above ranges in different populations and regions of the world.
There are several diseases, however, that cause significant global morbidity and mortality, for which vaccines do not currently exist (Table 2). In general, the viruses, bacteria, and parasites for which new vaccines are needed, are either much more complex in their pathogenesis, exhibit extensive variability, or have evolved immune evasion mechanisms to thwart the human immune system. For example, there are many cases such as influenza and dengue viruses for which immunologic memory induced by natural infection protects against reinfection by homologous serotypes but not by heterologous serotypes (16). Thus, minor changes in the outer glycoproteins from circulating strains of the influenza virus result in the need for annual immunizations against influenza. For viruses such as respiratory syncytial virus (RSV), reinfection with the same virus can occur, though disease is generally less severe with these sequential re-infections (17). For HIV, the hyper-variability of the virus coupled with its capacity to integrate in the host genome, results in the inability of the host to clear the infection (18). Lastly, for pathogens such as cytomegalovirus (CMV), herpes simplex and Mycobacterium tuberculosis, a carrier state is established with reactivation occurring in situations of immunosuppression (19). Clearly, new vaccine discovery and novel immunization paradigms will likely be required for successful vaccine development against HIV, Mycobacterium tuberculosis, Plasmodium falciparum, hepatitis C (HCV), and other challenging pathogens for which there currently are no licensed vaccines.
Table 2.
Major Global Diseases for which Vaccines do not Currently Exist
| Campylobacter |
| Chlamydia |
| Cytomegalovirus |
| Dengue |
| Epstein-Barr (Mononucleosis) |
| Helicobacter pylori- Gastrointestinal ulcers |
| Hepatitis C |
| Herpes Simplex |
| HIV |
| Influenza (Universal flu vaccine to replace need for annual flu vaccine) |
| Leishmaniasis |
| Malaria |
| Respiratory syncytial virus |
| Rhinovirus |
| Schistosomiasis |
| Shigella |
| Streptococcus Group A and B |
| Tuberculosis |
| Urinary tract infections |
| OTHER |
| Allergies; Autoimmune diseases; Cancersb |
HBV and HPV vaccines are effective in preventing liver and cervical cancers respectively
Recent technological advances in molecular genetics, molecular and cellular immunology, structural biology, bioinformatics, computational biology, nanotechnology, formulation technologies and systems biology have heralded in a new era in immunogen design, adjuvant discovery (i.e. agents that enhance immune responses, and immune monitoring). However, translation of these advances into successful vaccines remains significantly impeded by a lack of understanding of key vaccinology principles in humans. This includes the need for greater understanding of disease-specific mechanisms of protective immunity, immune evasion mechanisms, and strategies to drive the immune system towards preferred responses by immunization.
Though based on sound scientific principles, currently licensed vaccines have largely been developed empirically and protection by these vaccines is generally conferred by antigen-specific antibodies, which prevent or reduce infection (20). Viral neutralizing antibodies prevent virus replication by blocking virus binding and entry into cells. For hyper-variable viral pathogens such as HIV and hepatitis C, it is likely that broadly neutralizing antibodies (bnAbs), (i.e. those targeting highly conserved regions of these viruses), will be necessary for globally effective vaccines. Many bnAbs against HIV have now been identified from HIV+ subjects (21), but most of these bnAbs exhibit a high level of somatic hypermutation (22). Moreover, there is currently limited understanding of the optimal immunization strategies in humans necessary to drive the antibody maturation process from germline to a mature broadly neutralizing antibody (23). Strategies for focusing immune responses on protective epitopes and away from immunodominant decoy epitopes are also undefined, as are the strategies for induction of long-term memory responses. Moreover, most of what we know of human immunology has come from the study of blood, whereas the battle between pathogen and the immune system largely takes place in lymph nodes and other tissues. Finally, little is known of ways to overcome neonatal immaturity and immune senescence in the elderly, important for the development and optimal coverage of next generation vaccines. While small animal and nonhuman primate models serve a critical role in basic science and preclinical vaccine discovery, they are significantly limited when it comes to extrapolation in humans, in large part due to differences in immunogenetics, species-specificity of pathogens, and the resident microbiota that influences species-specific responses.
In this review, we discuss the technological advances that are fueling a new era in vaccine discovery, highlight advances in immune monitoring technologies and systems biology that offer significant potential for discovering new biomarkers of protective immunity and identify the limitations of animal models for screening and prioritizing human vaccines. Based on this analysis, we conclude that new human immunology-based clinical research initiatives with the goal of elucidating and more effectively inducing vaccine-induced protective immune responses, would greatly accelerate the development of next generation vaccines against major global killers such as AIDS, TB and malaria and provide a foundation for vaccine development against new and emerging diseases.
The 21st Century Technological Revolution Fueling a New Era in Vaccine Discovery
Effective vaccines work by eliciting effector and memory immune responses that confer protection against infection and disease. New technologies have recently been developed that are now fueling a revolution in vaccine discovery, which should help to address vaccine development challenges such as pathogen diversity and immune evasion. These technologies can be divided into three major categories related to antigen discovery, adjuvants and vaccine delivery, and deciphering human immune responses.
Antigen Discovery Technologies
The capacity to sequence whole genomes of microorganisms and utilize bioinformatics for design of vaccines is a relatively recent approach to antigen discovery and has been termed reverse vaccinology (24). The first pathogen for which reverse vaccinology was attempted was Meningococcus B, the cause of 50% of global meningococcal meningitis. With the genome of the bacterium sequenced, over 600 potential antigens were assessed for antigenicity, resulting in the identification of over 90 newly detected surface proteins, 30% of which could induce bactericidal antibodies. A subset of these antigens, when formulated as immunogens induced protective immunity in mice, provides proof-of-concept for ongoing clinical development (25). Reverse vaccinology has now been applied to many other bacterial pathogens as full genome sequencing has advanced, including group B streptococcus, group A streptococcus, Streptococcus pneumoniae, Staphylococcus aureus, and Chlamydia (26). Besides reverse vaccinology, genomic-based antigen discovery has been enhanced by new technologies enabling interrogation of the comprehensive antigenic repertoire using libraries of genetically expressed antigens and screening for immunogenicity of the proteins during infection, termed “ antigenome analysis” (27). In addition, advances in mass spectrometry have enabled direct testing of the presence and quantity of antigens on the surface of bacteria (28).
While these reverse vaccinology and antigenome technologies hold significant promise, particularly for identifying potential antigens for inclusion in vaccines, they remain limited in their capacity to predict which antigens are protective, as recently demonstrated by a Phase III trial of an experimental Staphylococcus aureus vaccine (29). Thus, greater efforts on understanding correlates of protection from human natural history studies, coupled with small, iterative clinical trials aimed at driving immune responses towards less immune dominant but protective epitopes, offers potential for greater utilization of these antigen discovery technologies.
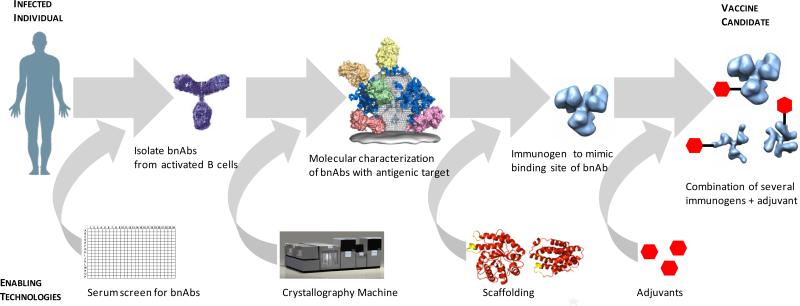
Antigen discovery technologies have also advanced for identification of antigens to be used in vaccine candidates in order to induce neutralizing antibody responses, and for the generation of T cell-based vaccines. For pathogens which are hypervariable such as HIV “reverse engineering of vaccines”, the concept that protective antigens could be identified through interrogation of the antibody repertoire from subjects infected with the pathogen (30) and formulated into effective vaccines is now being more fully exploited, due to recent technological advances in isolating monoclonal antibodies from memory B cells and plasma cells from infected patients, deep sequencing and bioinformatics. These efforts include: 1) Identification of subjects with broadly neutralizing antibody serum responses (21); 2) Identification of broadly neutralizing monoclonal antibodies (bnAbs) from such subjects by single-cell memory B cell techniques with or without antigen selection, and cloning the heavy and light chains into IgG vectors (31-35); 3) Determination of the structure of the binding sites of such bnAbs at the structural level using crystallographic methods (31-35); and, 4) Mimicking the epitopic binding sites of such bnAbs on carrier protein scaffolds or vectors to serve as the basis for immunogens to elicit such bnAbs (32, 33) (Figure 1). In addition to HIV, bnAbs against HCV (36) and influenza (37) are serving as templates for antigen designs for HCV and universal flu vaccines. The first proof-of-concept for reverse engineering of vaccines has now been achieved for respiratory syncytial virus (RSV), where computationally designed immunogens mimicking the binding site for an RSV neutralizing monoclonal antibody have successfully elicited RSV specific neutralizing antibodies in monkeys (38) .
Figure 1. Reverse engineering of vaccines.
The figure schematically depicts the identification of bnAbs; technological advances in high-throughput robotic crystallization platforms to enable greater precision in identifying the epitope targets for bnAbs; computational methods to design scaffolds mimicking the binding site of the bnAbs; and lastly greater understanding at the mechanistic level for selection of adjuvants to include in vaccine formulations for potentiating immune responses.
The technologies above highlight advances in vaccine discovery for which antibodies to the pathogen are the principal biomarker of vaccine efficacy. Similarly, there have been significant technological advances in the past decade with respect to antigen discovery and evaluation of cell mediated immune (CMI) responses. CMI responses play an integral role in controlling many acute and chronic diseases, provide critical helper signals for elicitation of antibody responses, and thus are important components of vaccine development strategies against some parasites, intracellular bacteria and some viral infections. Recent technological advances in CMI assays including ELISPOT and Intracellular Cytokine Staining (ICS), mass spectrometry coupled with epitope identification algorithms (39), tetrameric staining reagents (40), genomics and proteomics of more complex pathogens such as mycobacterium and plasmodium (41, 42), and advances in immune monitoring (see below) have combined to enable significant advances for T cell-based vaccine antigen discovery. For example, long peptides encompassing conserved regions of HIV have been identified which induce polyfunctional T cells in macaques with breadth superior to single gene vaccines (43). In addition, computational optimization methods have been applied to the design of “mosaic” proteins, assembled from fragments of natural viral genetic sequences, that provide diversity coverage comparable to that of thousands of separate peptides but are tractable for vaccines (44).
These technologies are beginning to be applied both for antigen identification and for assessment of vaccine efficacy. With respect to antigen identification, epitope specific algorithms aimed at identifying key cellular immunity epitopes have been assessed for vaccinia virus, to better understand the protective components of the efficacious smallpox vaccine. Antigens recognized by CD4+ responses differed significantly from those recognized by CD8+ responses, with such data now being applied to reverse vaccinology strategies for development of epitope specific vaccines (45, 46) .
Simian immunodeficiency virus (SIV) vaccine efficacy studies with heterologous adenovirus vectors compared in nonhuman primates the level of Gag-specific CMI responses with viral load. Greater control of infection correlated with numbers of Gag-specific epitope responses (47). More recently, using MHC typed monkeys to probe the efficacy of an SIV epitope-based vaccine, it was demonstrated that CMI responses to epitope-specific targets could control SIV infection in nonhuman primates (48). In contrast, studies with malaria immunogens demonstrated that heterologous vectors (chimp Adeno 63 + MVA) induced robust cellular immunity to specific blood stage antigens as measured by the magnitude of IFN-gamma ELISPOT responses, but had no effect on parasite growth rates in the blood (49) Similarly, a recently completed efficacy trial of MVA85A, a new tuberculosis vaccine in infants previously vaccinated with BCG, elicited modest CD4+ cellular immune responses as measured by ELISPOT and ICS assays, but failed to demonstrate efficacy against tuberculosis (50).
Adjuvants and Vaccine Vector Delivery Technologies
In the past decade, there have been significant advances in identification of signaling pathways and receptors of the innate immune system and a greater appreciation of the importance of innate immunity in influencing adaptive immune responses (51). Detection of microbes by the innate immune system is largely driven by pattern recognition receptors such as toll-like receptors (TLRs), that recognize common molecular structures found on microbes. In recent years, there has been a revolution in understanding of the receptors and molecules that drive innate immune responses (52-54), and this is now leading to the development and testing of novel vaccine adjuvants.
Adjuvants are an important component of vaccine formulations that potentiate immune responses via interaction with one or more TLRs, particularly important for those vaccines composed of proteins that are not highly immunogenic. For example, co-delivery of vaccine antigens with pattern recognition receptor agonists, may lead to enhanced immune responses to vaccines (55-57). This enables more mechanistic-based design of adjuvants, for potentiation of humoral and cellular immune responses. By extension, this should help to facilitate development of next generation adjuvants to help address vaccine challenges where immune responses may be compromised or less than optimal such as with immune senescence in the elderly. For example, adjuvant trials in individuals hyporesponive to hepatitis B vaccines showed that addition of CpG to the HBV vaccine enhanced the kinetics, magnitude and longevity of the seroprotective response (58). However, there is still a very limited database on improved efficacy in the elderly with adjuvanted vaccines, highlighting the need for additional studies in these populations. Adjuvants currently used in licensed vaccines include aluminum salts, oil in water emulsions, and virosomes and TLR4 agonists such as bacterially derived monophosphorylylipid A (MPL). In addition, there are a large number of adjuvants currently in development, aimed at boosting CD4+ helper T cell CD8+ cytotoxic T cell, and humoral immune responses (51) .
Many emerging and re-emerging pathogens, including those responsible for respiratory, gastrointestinal, and sexually transmitted diseases for which vaccines do not currently exist, initiate infection at a mucosal surface, suggesting that induction of mucosal immunity may be necessary or beneficial for prevention and control (59). Vector delivery systems designed to stimulate mucosal and systemic immunity have advanced in recent years, with some already in clinical trials including adenovirus, paramyxovirus, and some bacterial vectors (60). Viral vectors enable heterologous antigens to be delivered to antigen-processing pathways needed to stimulate Human Leukocyte Antigen (HLA)-class I restricted cytotoxic T cell responses, in addition to priming for effective humoral immune responses. Further, replicating viral vectors more closely mimic the attributes of efficacious live attenuated vaccines (61). However, one of the major challenges for vector delivery systems is the need to overcome pre-existing immunity specific to the vector. For example, adenovirus type 5 (Ad5) is a potent vector for induction of cell mediated immune responses, yet the majority of the world's population has been previously exposed to Ad5 or closely related adenoviruses thus limiting the potential of this vector for vaccine development, particularly for use in the developing world. Strategies to overcome pre-existing or nascent anti-vector immunity are now being assessed, including use of vectors against which humans have low seroprevalence, simian and canine vectors for which pre-existing immunity in human populations is negligible, prime-boost regimens and administration of vector-based vaccines via a mucosal route. Low seroprevalence adenoviruses, such as Ad26, Ad35 and chimpAd63, are currently in clinical trials and preclinical studies with cytomegalovirus vectors have demonstrated the capacity for overcoming anti-vector immunity (62).
Dendritic cells (DC) are antigen presenting cells that provide initial surveillance mechanisms for exposure to pathogens, whereupon they undergo rapid maturation, and migrate to lymph nodes where induction of immune responses occurs. DCs play a central role in the induction of vaccine mediated immune responses, by providing antigen-specific and co-stimulatory signals required for T cell activation. Increased understanding of the mechanisms of antigen presentation, guided by elucidation of dendritic cell subsets and their functional plasticity, offer additional opportunities for targeting and potentiation of immune responses by vaccines (63). Targeting dendritic cells in vivo via antibodies to specific DC cell surface receptors, is now being explored for vaccines (64). Some viral vectors, such as vesicular stomatitis virus (VSV), target dendritic cells, which may account for the potent immune responses when VSV is used as a vector (65).
Deciphering Human Immune Responses
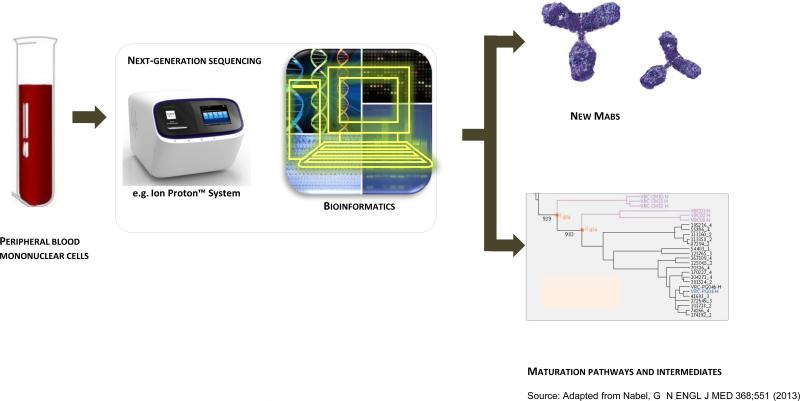
Along with advances in antigen discovery, adjuvant and vaccine vector delivery technologies, the past decade has seen a technological revolution in the capacity to analyze immune responses at the both the single cell and systems level, which offers tremendous potential for deciphering human immune responses to vaccines and identification of immune correlates of protection. Advances in systems biology, coupled with recent gains in understanding the pivotal role of innate immunity in augmenting adaptive immune responses, are now being applied to vaccine discovery in what is termed “systems vaccinology” (66). Technological advances with DNA microarrays and high through-put DNA sequencing, mass spectrometry powered proteomics, bioinformatics and computational methods enable data integration that serves as the basis of systems vaccinology (67). Initial studies on yellow fever vaccination in humans have identified early innate signatures that correlate with immunogenicity of the vaccines (68), and similar approaches are now being undertaken for influenza and malaria vaccine development. Interrogation of the antibody repertoire by deep sequencing and bioinformatics, has led to the identification of intermediates on the path to broadly neutralizing antibodies that is now helping to guide HIV vaccine development (69) (Figure 2). Concomitant with advances in systems biology approaches, immune monitoring technologies have advanced to enable new approaches to characterize and interrogate human immune responses (70). Multi-plexed flow cytometric and intracellular cytokine staining assays now allow analyses of cell phenotype, subset identity, activation and intracellular signaling status and effector activities (71-73), and single-cell gene expression analyses allow greater specificity in immune monitoring. Such monitoring should enable greater precision in assessments of correlates of protection for currently licensed vaccines, which will help guide next generation vaccine development. For example, induction of effector memory responses as determined by flow cytometric analysis is thought now to play an important role in the significant control of SIV infection conferred by CMV vector-based vaccine candidates (62). Combining these tools with other new methods for analysis of antigen-specific cells using multiplexed tetramer technologies, and highly sensitive microarray and sequencing technologies, will allow not only for assessment of responses in blood, but due to the sensitivity of these methods, may also provide insights into selected tissue level responses. This will enable future analyses to be made in the tissues, where the battle between pathogen and host largely takes place, rather than simply relying on measurements from peripheral blood.
Figure 2. Technological advances towards deciphering human immune responses.
This figure depicts technological advances in deep sequencing and bioinformatics which now allow identification of the antibodyome and tracing antibody development from germline through somatically hypermutated intermediates to the designated fully mature antibody Such tracing can help guide B cell lineage-based vaccine design (23).
Taken together, recent progress in technological development for antigen discovery, adjuvant and vector discovery and technologies aimed at deciphering human immune responses, provide the foundation for significant advances in vaccine discovery and their application towards accelerating vaccine development against the major global diseases for which vaccines do not currently exist.
The Importance of Studying Vaccines in Humans: Limitations of Current Animal Models
Despite best efforts, in vitro and small animal models do not effectively recapitulate the dynamics of human immune responses to vaccines. Mouse models, in particular the use of inbred mice, have been extremely effective as a tool for basic immunologists, yet have been largely unsuccessful as models for clinical application (74, 75). For example, inbred mice often have a number of homozygous recessive defects that alter the regulation of immune responses (76). Differences in pattern recognition receptors e.g. TLR9 expression, may account for significant differences between humans and rodents in response to microbial stimuli (77, 78). In addition, protocols in small animal models do not adequately reflect human vaccination studies. Many murine studies utilize intravenous or intraperitoneal injections, whereas human vaccines are generally administered intramuscularly or subcutaneously (79). Different routes of immunization, for example mucosal vs. intramuscular, can change patterns of recognition by dendritic cell subsets, leading to modifications in immune responses (80). In addition, dose and regimen of immunizations are often different between small animal studies and human clinical trials, which can affect the quality and quantity of priming, effector and memory responses.
As a result of these deficiencies, there is renewed interest in the use of “humanized” mice, which are usually immunodeficient mice that are reconstituted with human hematopoietic stem cells, peripheral blood mononuclear cells or tissue transplants. Although these models are potentially very promising, they have yet to be validated for predicting human immune responses to licensed vaccines, and human hematopoietic cells developed from stem cell transplantation in immunodeficient mice are not always phenotypically and functionally identical to those that develop in humans (81). Thus, such mice may not fully recapitulate human immune responses, particularly where T cell helper and other effector cell mechanisms are integral for optimization of vaccine-induced immune responses.
Although nonhuman primates have played an important role in the development of hepatitis B and other vaccines, there are additional limitations of nonhuman primate models with respect to human vaccine development, including differences in immunogenetics between macaques and humans, species specificity of some viral vectors being developed as vaccine candidates, and the impact of the microbiome of humans on vaccine evaluation. For example, HIV vaccine developers primarily rely on simian-immunodeficiency virus (SIV) replication in rhesus macaques, as a challenge model for assessing vaccine concepts, due to the limited replication of HIV in macaques (82). However, the hyper variability of HIV cannot readily be modeled by SIV, due to the limited numbers of referenced strains of SIV. Moreover, differences between major histocompatibility complex (MHC) restrictions in monkeys and HLA restrictions in human='s limits the assessment of epitopes for inclusion in vaccine candidates. In addition, vectors carrying HIV genes such as cytomegalovirus and replication competent Adenovirus type 4 are species-specific for humans.
The microbiome of humans consists of the plethora of viruses, bacteria, and parasites that infect and reside in our tissues, contributing a significant proportion of genetic information to our metagenome, and thus affecting susceptibility and resistance to disease, particularly inflammatory diseases such as type 1 diabetes, ulcerative colitis and Crohn's disease (83). The concept of microbiome is being applied to personalized medicine, but also should be viewed as a limitation when contemplating animal models for human vaccine development (84).
In summary, although murine models have provided important insights into basic immunology, and small animal models plus nonhuman primates have been important in the development of some currently licensed vaccines, there are significant limitations of these models in predicting human responses to vaccines. This was demonstrated yet again when SHIV protection studies in monkeys suggested that an Ad5-based HIV vaccine could suppress viral load and control disease, yet human efficacy trials failed to confirm these observations (85, 86). Thus, greater attention needs to be focused on immunogenicity studies in humans aimed at answering specific questions that impede vaccine development, and translating this information towards the development of next generation and more efficacious vaccines against globally important diseases.
Future Directions: Optimizing Protective Immune Responses in Humans
Although licensed vaccines continue to provide tremendous public health benefits, success rates in vaccine development are not optimal, and are even worse for the subset of complex pathogens for which viability and immune evasion mechanisms present additional challenges. For example, in one survey of over 200 vaccine development projects, success rates for vaccines was only 22% compared with 40 % for biopharmaceuticals (87). In our view, this high rate of failure for vaccines is directly related to: 1) Lack of information about mechanisms for protective immunity directly applicable to pathogen-specific vaccine development; and, 2) Lack of understanding of optimal strategies to elicit vaccine-induced protective immune responses in humans and thus inability to effectively predict which immunogens would have a greater chance for success in vaccine efficacy trials.
For the major global diseases for which vaccines do not currently exist (Table 2), identification of vaccine-induced protective immune responses will require greater understanding of pathogen-specific mechanism(s) of protective immunity, particularly from human natural history studies and in some cases from human challenge models when available, such as malaria. This is important in cases where natural infection by specific pathogens provide protection against subsequent exposure but also in cases where natural infection does not confer such protection. Thus, greater linkages of human disease-specific pathogenesis studies with applied vaccinology studies, will be central to accelerating next generation vaccine development.
Moreover, when one factors in the current lack of understanding in humans of how to fine control the antibody affinity maturation that is likely required to elicit broadly protective responses to highly antigenically variable pathogens; of how to focus immune responses on subdominant yet critical protective epitopes; and of how to elicit long-term central and effector memory responses, it's no surprise that vaccine success rates against complex pathogens such as HIV, mycobacteria, and plasmodium are worse than those against non-variable, acute infections. However, the confluence of recent technological advances for vaccine discovery, systems biology and immune monitoring, yields a tremendous opportunity for accelerating vaccine development. If harnessed effectively, they can lead to a greater understanding of disease-specific mechanisms of protective immunity, elucidate key principles of vaccinology and be used to optimize humoral and cellular protective immune responses. Application of this information across the spectrum of disease-specific vaccine development programs will likely shorten the timelines for successful development of new and improved vaccines.
Clinical trials of vaccines are currently driven by product-specific issues, as sponsors or vaccine developers have preferred to quickly advance candidates from Phase I/II safety immunogenicity trials, to Phase IIb/III efficacy trials, rather than prioritize vaccine discovery related issues. Unfortunately, for diseases such as HIV, TB, malaria and others, the gap in understanding how best to elicit the requisite humoral and cellular effector and memory responses in humans has slowed the pace of vaccine development. In the absence of a greater understanding of these key vaccinology principles in humans, the vaccine field is left to rely on large and expensive field trials to provide any guidance. Such trials can cost $50 million - $100 million or more, take several years to complete and often are undertaken with low probabilities of success. This is not to suggest that field trials should be abandoned. It is rather to suggest that in parallel, significant resources be directed towards conducting small, iterative, human clinical studies to address key questions currently impeding the development of next generation candidate vaccines.
In summary, successful development of vaccines against the major global diseases for which vaccines do not currently exist would be transformational for public health, with huge benefits across society. Historical paradigms of empirical product development efforts alone will unlikely be successful against major global killers that cause significant morbidity and mortality today. This is due in large part to the current lack of the understanding of mechanisms for protective immunity, immune evasion mechanisms, and immunization strategies on how best to elicit protective immune responses against such pathogens in humans to mimic the best attributes of successful licensed vaccines. To accelerate next generation vaccine development, we suggest that new human immunology-based clinical research initiatives be established, focused on addressing key questions impeding vaccine development, with the goal of elucidating and more effectively inducing vaccine-induced protective immune responses. Collectively, such a “Human Vaccines Project” holds the potential to greatly accelerate the development of next generation vaccines against major global killers such as AIDS, TB, malaria and other infectious diseases, enable more successful vaccine development against allergies, autoimmune diseases, and cancers, and provide a foundation for vaccine development against new and emerging diseases.
Acknowledgments
The authors acknowledge the assistance of Sandi Glass, Lisa Gieber, Olga Shmaidenko, Michelle Dees and Bill Hayes in the preparation of this manuscript.
REFERENCES
- 1.Jenner E. The Origin of the Vaccines Inoculation. Shury; London, UK: 1801. [Google Scholar]
- 2.Pasteur L. De l'attenuation du virus du Cholé ra des poules. CR Acad. Sci. Paris. 1880;91:673. [Google Scholar]
- 3.Salmon D, Smith T. On a new method of producing immunity from contagious diseases. American Vet Review. 1886;10:63. [Google Scholar]
- 4.Enders JF, Weller TH, Robbins FC. Cultivation of the Lansing Strain of Poliomyelitis Virus in Cultures of Various Human Embryonic Tissues. Science. 1949;109:85. doi: 10.1126/science.109.2822.85. [DOI] [PubMed] [Google Scholar]
- 5.Hilleman MR, Buynak EB, Weibel RE, Stokes J., Jr. Live, attenuated mumps-virus vaccine. N Engl J Med. 1968;278:227. doi: 10.1056/NEJM196802012780501. [DOI] [PubMed] [Google Scholar]
- 6.Plotkin SA, Farquhar JD, Katz M, Buser F. Attenuation of RA 27-3 rubella virus in WI-38 human diploid cells. Am J Dis Child. 1969;118:178. doi: 10.1001/archpedi.1969.02100040180004. [DOI] [PubMed] [Google Scholar]
- 7.Sabin AB, Hennessen WA, Winsser J. Studies on variants of poliomyelitis virus. I. Experimental segregation and properties of avirulent variants of three immunologic types. J Exp Med. 1954;99:551. doi: 10.1084/jem.99.6.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salk JE, et al. Formaldehyde treatment and safety testing of experimental poliomyelitis vaccines. Am J Public Health Nations Health. 1954;44:563. doi: 10.2105/ajph.44.5.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilleman MR, et al. Development and evaluation of the Moraten measles virus vaccine. JAMA. 1968;206:587. [PubMed] [Google Scholar]
- 10.Black S, et al. Efficacy, safety and immunogenicity of heptavalent pneumococcal conjugate vaccine in children. Northern California Kaiser Permanente Vaccine Study Center Group. Pediatr Infect Dis J. 2000;19:187. doi: 10.1097/00006454-200003000-00003. [DOI] [PubMed] [Google Scholar]
- 11.McNeil C. Who invented the VLP cervical cancer vaccines? J Natl Cancer Inst. 2006;98:433. doi: 10.1093/jnci/djj144. [DOI] [PubMed] [Google Scholar]
- 12.Schneerson R, Barrera O, Sutton A, Robbins JB. Preparation, characterization, and immunogenicity of Haemophilus influenzae type b polysaccharide-protein conjugates. J Exp Med. 1980;152:361. doi: 10.1084/jem.152.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Argenio DA, Wilson CB. A decade of vaccines: Integrating immunology and vaccinology for rational vaccine design. Immunity. 2010;33:437. doi: 10.1016/j.immuni.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Ehreth J. The value of vaccination: a global perspective. Vaccine. 2003;21:4105. doi: 10.1016/s0264-410x(03)00377-3. [DOI] [PubMed] [Google Scholar]
- 15.Miller MA, Hinman AR. In: Vaccines. Plotkin SA, Orenstein WA, Offit PA, editors. Saunders; Philadelphia: 2008. pp. 1594–1610. [Google Scholar]
- 16.Green S, Rothman A. Immunopathological mechanisms in dengue and dengue hemorrhagic fever. Curr Opin Infect Dis. 2006;19:429. doi: 10.1097/01.qco.0000244047.31135.fa. [DOI] [PubMed] [Google Scholar]
- 17.Sallusto F, Lanzavecchia A, Araki K, Ahmed R. From vaccines to memory and back. Immunity. 2010;33:451. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McElrath MJ, Haynes BF. Induction of immunity to human immunodeficiency virus type-1 by vaccination. Immunity. 2010;33:542. doi: 10.1016/j.immuni.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sacchettini JC, Rubin EJ, Freundlich JS. Drugs versus bugs: in pursuit of the persistent predator Mycobacterium tuberculosis. Nat Rev Microbiol. 2008;6:41. doi: 10.1038/nrmicro1816. [DOI] [PubMed] [Google Scholar]
- 20.Siegrist C-A. Vaccines. Sixth Edition W.B. Saunders; London: 2013. pp. 14–32. [Google Scholar]
- 21.Simek MD, et al. Human immunodeficiency virus type 1 elite neutralizers: Individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. Journal of Virology. 2009;83:7337. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burton DR, et al. A Blueprint for HIV Vaccine Discovery. Cell host & microbe. 2012;12:396. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol. 2012;30:423. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rappuoli R. Reverse vaccinology. Curr Opin Microbiol. 2000;3:445. doi: 10.1016/s1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- 25.Giuliani MM, et al. A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci U S A. 2006;103:10834. doi: 10.1073/pnas.0603940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sette A, Rappuoli R. Reverse vaccinology: developing vaccines in the era of genomics. Immunity. 2010;33:530. doi: 10.1016/j.immuni.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giefing C, et al. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J Exp Med. 2008;205:117. doi: 10.1084/jem.20071168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez-Ortega MJ, et al. Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat Biotechnol. 2006;24:191. doi: 10.1038/nbt1179. [DOI] [PubMed] [Google Scholar]
- 29.Daum RS. Vaccines. Sixth Edition W.B. Saunders; London: 2013. pp. 1161–1168. [Google Scholar]
- 30.Burton DR. Antibodies, viruses and vaccines. Nat Rev Immunol. 2002;2:706. doi: 10.1038/nri891. [DOI] [PubMed] [Google Scholar]
- 31.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou T, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Law M, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2008;14:25. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 37.Ekiert DC, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schief W. AIDS Vaccine. Suppl 2. Vol. 9. Boston, Massachusetts, USA: 2012. 2012. p. S03.04. [Google Scholar]
- 39.Zhang Q, et al. Immune epitope database analysis resource (IEDB-AR). Nucleic Acids Res. 2008;36:W513. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakker AH, Schumacher TN. MHC multimer technology: current status and future prospects. Curr Opin Immunol. 2005;17:428. doi: 10.1016/j.coi.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 41.Doolan DL, et al. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci U S A. 2003;100:9952. doi: 10.1073/pnas.1633254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMurry J, et al. Analyzing Mycobacterium tuberculosis proteomes for candidate vaccine epitopes. Tuberculosis (Edinb) 2005;85:95. doi: 10.1016/j.tube.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Rosario M, et al. Long peptides induce polyfunctional T cells against conserved regions of HIV-1 with superior breadth to single-gene vaccines in macaques. Eur J Immunol. 2010;40:1973. doi: 10.1002/eji.201040344. [DOI] [PubMed] [Google Scholar]
- 44.Fischer W, et al. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med. 2007;13:100. doi: 10.1038/nm1461. [DOI] [PubMed] [Google Scholar]
- 45.Moutaftsi M, et al. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol. 2006;24:817. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 46.Moutaftsi M, et al. Vaccinia virus-specific CD4+ T cell responses target a set of antigens largely distinct from those targeted by CD8+ T cell responses. J Immunol. 2007;178:6814. doi: 10.4049/jimmunol.178.11.6814. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, et al. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature. 2009;457:87. doi: 10.1038/nature07469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mudd PA, et al. Vaccine-induced CD8+ T cells control AIDS virus replication. Nature. 2012;491:129. doi: 10.1038/nature11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheehy SH, et al. ChAd63-MVA-vectored blood-stage malaria vaccines targeting MSP1 and AMA1: assessment of efficacy against mosquito bite challenge in humans. Mol Ther. 2012;20:2355. doi: 10.1038/mt.2012.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tameris MD, et al. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet. 2013 doi: 10.1016/S0140-6736(13)60177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coffman RL, Sher A, Seder RA. Vaccine adjuvants: putting innate immunity to work. Immunity. 2010;33:492. doi: 10.1016/j.immuni.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beutler BA. TLRs and innate immunity. Blood. 2009;113:1399. doi: 10.1182/blood-2008-07-019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 54.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 55.Huleatt JW, et al. Vaccination with recombinant fusion proteins incorporating Toll-like receptor ligands induces rapid cellular and humoral immunity. Vaccine. 2007;25:763. doi: 10.1016/j.vaccine.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 56.Tighe H, et al. Conjugation of immunostimulatory DNA to the short ragweed allergen amb a 1 enhances its immunogenicity and reduces its allergenicity. J Allergy Clin Immunol. 2000;106:124. doi: 10.1067/mai.2000.107927. [DOI] [PubMed] [Google Scholar]
- 57.Wille-Reece U, et al. HIV Gag protein conjugated to a Toll-like receptor 7/8 agonist improves the magnitude and quality of Th1 and CD8+ T cell responses in nonhuman primates. Proc Natl Acad Sci U S A. 2005;102:15190. doi: 10.1073/pnas.0507484102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cooper CL, Angel JB, Seguin I, Davis HL, Cameron DW. CPG 7909 adjuvant plus hepatitis B virus vaccination in HIV-infected adults achieves long-term seroprotection for up to 5 years. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2008;46:1310. doi: 10.1086/533467. [DOI] [PubMed] [Google Scholar]
- 59.Chen K, Cerutti A. Vaccination strategies to promote mucosal antibody responses. Immunity. 2010;33:479. doi: 10.1016/j.immuni.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu MA. Immunologic basis of vaccine vectors. Immunity. 2010;33:504. doi: 10.1016/j.immuni.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 61.Koff WC, et al. Replicating viral vectors as HIV vaccines Summary Report from IAVI Sponsored Satellite Symposium, International AIDS Society Conference. Biologicals. 2008 Jul;2236:2007, 277. doi: 10.1016/j.biologicals.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 62.Hansen SG, et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med. 2009;15:293. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Palucka K, Banchereau J, Mellman I. Designing vaccines based on biology of human dendritic cell subsets. Immunity. 2010;33:464. doi: 10.1016/j.immuni.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lai L, et al. Prevention of infection by a granulocyte-macrophage colony-stimulating factor co-expressing DNA/modified vaccinia Ankara simian immunodeficiency virus vaccine. J Infect Dis. 2011;204:164. doi: 10.1093/infdis/jir199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boudreau JE, et al. Recombinant vesicular stomatitis virus transduction of dendritic cells enhances their ability to prime innate and adaptive antitumor immunity. Mol Ther. 2009;17:1465. doi: 10.1038/mt.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity. 2010;33:516. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hyduke DR, Palsson BO. Towards genome-scale signalling network reconstructions. Nat Rev Genet. 2010;11:297. doi: 10.1038/nrg2750. [DOI] [PubMed] [Google Scholar]
- 68.Querec TD, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu X, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Germain RN. Vaccines and the future of human immunology. Immunity. 2010;33:441. doi: 10.1016/j.immuni.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 71.Bandura DR, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 72.Chattopadhyay PK, Hogerkorp CM, Roederer M. A chromatic explosion: the development and future of multiparameter flow cytometry. Immunology. 2008;125:441. doi: 10.1111/j.1365-2567.2008.02989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schulz KR, Danna EA, Krutzik PO, Nolan GP. Single-cell phospho-protein analysis by flow cytometry. Curr Protoc Immunol. 2007 doi: 10.1002/0471142735.im0817s78. Chapter 8, Unit 8 17. [DOI] [PubMed] [Google Scholar]
- 74.Davis MM. A prescription for human immunology. Immunity. 2008;29:835. doi: 10.1016/j.immuni.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seok J, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.von Herrath MG, Nepom GT. Lost in translation: barriers to implementing clinical immunotherapeutics for autoimmunity. J Exp Med. 2005;202:1159. doi: 10.1084/jem.20051224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Campbell JD, et al. CpG-containing immunostimulatory DNA sequences elicit TNF-alpha-dependent toxicity in rodents but not in humans. J Clin Invest. 2009;119:2564. doi: 10.1172/JCI38294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 79.Quintana-Murci L, Alcais A, Abel L, Casanova JL. Immunology in natura: clinical, epidemiological and evolutionary genetics of infectious diseases. Nat Immunol. 2007;8:1165. doi: 10.1038/ni1535. [DOI] [PubMed] [Google Scholar]
- 80.Klechevsky E, et al. Functional specializations of human epidermal Langerhans cells and CD14+ dermal dendritic cells. Immunity. 2008;29:497. doi: 10.1016/j.immuni.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ito R, Takahashi T, Katano I, Ito M. Current advances in humanized mouse models. Cell Mol Immunol. 2012 doi: 10.1038/cmi.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Song B, et al. Retrovirus restriction by TRIM5alpha variants from Old World and New World primates. J Virol. 2005;79:3930. doi: 10.1128/JVI.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Virgin HW, Todd JA. Metagenomics and personalized medicine. Cell. 2011;147:44. doi: 10.1016/j.cell.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 85.Watkins DI, Burton DR, Kallas EG, Moore JP, Koff WC. Nonhuman primate models and the failure of the Merck HIV-1 vaccine in humans. Nat Med. 2008;14:617. doi: 10.1038/nm.f.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Buchbinder SP, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Struck MM. Vaccine R&D success rates and development times. Nat Biotechnol. 1996;14:591. doi: 10.1038/nbt0596-591. [DOI] [PubMed] [Google Scholar]