Abstract
Breast cancers with HER2 overexpression are sensitive to drugs targeting the receptor or its kinase activity. HER2-targeting drugs are initially effective against HER2-positive breast cancer, but resistance inevitably occurs. We previously found that nuclear factor kappa B is hyper-activated in a subset of HER-2 positive breast cancer cells and tissue specimens. In this study, we report that constitutively active NF-κB rendered HER2-positive cancer cells resistant to anti-HER2 drugs and cells selected for Lapatinib resistance up-regulated NF-κB. In both circumstances, cells were anti-apoptotic and grew rapidly as xenografts. Lapatinib-resistant cells were refractory to HER2 and NF-κB inhibitors alone but were sensitive to their combination, suggesting a novel therapeutic strategy. A subset of NF-κB-responsive genes was overexpressed in HER2-positive and triple-negative breast cancers, and patients with this NF-κB signature had poor clinical outcome. Anti-HER2 drug resistance may be a consequence of NF-κB activation, and selection for resistance results in NF-κB activation, suggesting this transcription factor is central to oncogenesis and drug resistance. Clinically, the combined targeting of HER2 and NF-κB suggests a potential treatment paradigm for patients who relapse after anti-HER2 therapy. Patients with these cancers may be treated by simultaneously suppressing HER2 signaling and NF-κB activation.
Keywords: Breast cancer, HER2-positive, duel therapy targets
Corresponding Author: Debajit K. Biswas Department of Cancer Biology Dana-Farber Cancer Institute 450 Brookline Avenue, Smith 958 Boston, MA 02215 debajit_biswas@dfci.harvard.edu Tel: 617-632-4684 FAX: 617-632-3709
INTRODUCTION
Diverse histopathology and clinical manifestations of breast cancer are linked to hormone and growth factor-mediated cellular and molecular pathways (1). For therapeutic and prognostic purposes, human breast cancers are sub-classified based on their hormone and growth factor receptor expression. One subclass is defined by amplification of the human epidermal growth factor 2 (HER2) protein, which is encoded by the ERBB2 gene. This protein, a member of the epidermal growth factor receptor (EGFR) family (2, 3) and lacks a ligand-binding domain; thus, its signals are propagated by dimerization with other ligand-bound EGFR family members to form a signaling complex (2, 4-6). HER2 kinase activation leads to the stimulation of downstream signaling, which is mediated by the mitogen-activated protein kinase (MAPK) and phosphatidylinositol-3-kinase (PI3K) pathways (4, 5, 7). Elevated HER2 protein expression magnifies its kinase activity, leading to a cellular dependence on HER2 signaling and sensitivity to HER2-targeted therapies (2, 8-12). HER2 inhibition is an effective treatment for patients with HER2-positive breast cancers. Trastuzumab, a humanized monoclonal antibody directed against the HER2 extracellular domain, has been used as first-line therapy for HER2-positive breast cancers. The supplementation of chemotherapy with trastuzumab increases the survival time of patients with metastatic HER2-positive disease, and its addition to standard treatment reduces the odds of recurrence by 50% (9, 13). Lapatinib, which is a highly effective small molecule inhibitor of HER2 tyrosine kinase activity (7), was first used in clinical trials in 2005 (5). Resistance to Lapatinib therapy has been reported to be common and may be due to activation of compensatory growth factor pathways (14).
In certain breast cancers, HER2 signaling leads to stimulation of the nuclear factor kappa B (NF-κB) transcription factor pathway (15, 16). The NF-κB transcription factor is a dimeric complex of rel family proteins that is sequestered in its basal state in the cytoplasm with inhibitor of NF-κB (IκB) proteins. Classical NF-κB activation is initiated by cell surface receptors responding to a variety of diverging stimulants, leading to signal propagation that ultimately activates the inhibitory kappa B kinase (IKK) complex (17). The IKK complex consists of two catalytic subunits, IKKα and IKKβ, and one regulatory subunit, IKKγ (also known as NF-κB essential modulator; NEMO)(11). This complex phosphorylates IκB, leading to its degradation and the release of NF-κB, allowing this transcription factor to translocate into the nucleus and transactivate target genes. The inhibition of IKK or HER2 by specific inhibitors blocks NF-κB activation (12). NF-κB activation was linked to human breast cancer in studies demonstrating the nuclear localization of NF-κB family proteins in breast cancer cell lines and patient tissue (18).
Mammary gland epithelial hyperplasia and ductal branching is increased in mice lacking the inhibitor of kappa B alpha (IκBα) protein, and development is impaired by NF-κB signaling pathway interference (19). Mice with MMTV-driven c-rel develop late-onset mammary carcinomas of various pathologies (20). The stimulation of EGFR family receptors with EGF or heregulin results in NF-κB activation in breast cancer cells, and inhibition of the IKK complex blocks NF-κB activation and xenograft outgrowth (12, 16). Active NF-κB signaling is present in estrogen receptor (ER)-negative breast cancers, including triple-negative and HER2-positive subtypes (16, 18), its activation is a natural apoptosis inhibitor, and the inhibition of NF-κB activation induces apoptosis in breast cancer cells, leading to tumor regression (21).
In this study, we examine the oncogenic role of HER2-induced NF-κB signaling in a clonal derivative (SKR6) of the HER2-positive, ER-negative human breast cancer cell line SKBR3. SKR6 cells expressing constitutively activated NF-κB are resistant to anti-HER2 drugs, and NF-κB is over activated in Lapatinib-resistant SKR6 cells. In both cell types with over activated NF-κB, apoptosis is blocked profoundly and both rapidly generate xenografts. A set of genes over-expressed in both cell types is identified as a consequence of NF-κB over-activation. The SKR6 cells expressing constitutively activated NF-κB and Lapatinib-resistant SKR6 cells express a common anti-apoptotic gene set that is also found in tumors from patients with poor outcome.
MATERIALS AND METHODS
Cell lines and nomenclature
The nomenclature of the SKBR3 (from ATCC) and derivatives are as follows: 1) SKR6: A clonal derivative of SKBR3 cells that was isolated by fluorescence-activated cell sorting (FACS) to enrich for elevated HER2 levels. 2) SKR6CA: SKR6 cells that were retrovirally transduced with constitutively active NF-κB relA/p65 (CAp65) (22). 3) SKR6 vector: SKR6 cells that were transduced with the pQCXIP empty retroviral vector and selected with puromycin. 4) SKR6LR: SKR6 cells that were treated with increasing Lapatinib concentrations from 0.2 to 5 μM for several months. Cells were maintained in RPMI 1640 supplemented with 10% FBS and antibiotics (rich medium). Minimal medium was RPMI 1640, phenol red-free supplemented with dextran-coated charcoal-treated FBS (Hyclone, Inc, Salt Lake City, UT). The resistant cells were subsequently maintained under standard culture conditions in the presence of 0.2 μM Lapatinib. The genetic identity of the SKBR3 and derivative cell lines was confirmed by DNA fingerprinting with small tandem repeats (STRs), which was performed in the Dana-Farber Cancer Institute Molecular Diagnostic Core facility. The cell lines were grown in rich or minimal medium as previously described (12).
Reagents
The heregulin (HRG) β1 and anti-actin antibodies were from purchased Sigma (St Louis, MO). Trastuzumab, the humanized anti-HER2 antibody (Herceptin; Genentech, San-Francisco, CA), was obtained from the Dana-Farber Cancer Institute pharmacy. The anti-HER2 and anti-IκBα antibodies were obtained from Cell Signaling Technologies (Billerica, MA). The anti-relA (p65) (sc-8008) antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Lapatinib was purchased from Fisher Scientific. The NEMO-binding domain (NBD) peptide (11, 23) was synthesized at the Dana-Farber Cancer Institute core facilities. Matrigel (HC) was obtained from BD Biosciences (Bedford, MA). The retroviral vector pQCXIP-CAp65 (22) was a generous gift from Drs. Harikrishna Nakshatri and Poornima Bhat-Nakshatri of the Department of Surgery, Indiana University School of Medicine, Indianapolis, IN.
NF-κB Activation Assay
The activity of NF-κB was determined in nuclear extracts by electrophoretic mobility shift assay (EMSA) as described (12,16). NF-κB activation was also determined by nuclear translocation of NF-κB p65 by immunofluorescence (16).
Xenograft growth
Approximately 3-6 × 106 cells were suspended in 0.25 ml of sterile PBS, thoroughly mixed with 0.25 ml Matrigel (BD Biosciences, Bedford, MA), and implanted under the dorsal skin of 6-week-old female nu/nu mice (NCR NUM Homozygous, Taconic, Hudson, NY). Tumor growth was monitored weekly by measuring its dimensions, and the tumor volume was calculated using the following formula: SS2 X LS X 0.41, where SS = short side and LS = long side (24).
FACS analysis of HER2 levels
Approximately 1 × 106 cells were incubated with humanized anti-HER2 antibody (Herceptin) followed by incubation with FITC-conjugated goat anti-human IGG. Cells incubated with secondary antibody alone served as a control for non-specific binding and auto-fluorescence signals from the FACS analysis.
Chromatin Immunoprecipitation (ChIP) assay
ChIP was performed as previously described (25). Briefly, approximately 5 × 106 SKBR-3 cells were seeded in hormone-depleted media. After three days, the cells were treated with 10 μM HRG in the absence and presence of 10 μM NBD for 40 min. The cells were cross-linked for 10 min at room temperature with 1% formaldehyde, which was then quenched with 0.125 M glycine for 5 min. The cells were washed in PBS and then lysed in 1% SDS lysis buffer and sonicated. The fragmented chromatin was immunoprecipitated overnight using an antibody directed against p65 (C-20, Santa Cruz). The DNA was then purified using the QIAquick PCR Purification Kit (Qiagen), and the samples were subjected to RT-PCR using the Power SYBR Green PCR Master Mix (Applied Biosystems). The following primers corresponding to NF-κB binding sites near known NF-κB targets were used for analysis: TRAF2: TGGAAAGTCCCTGAGAGGAG (forward) and ACAAAATCAAAGGCCACAGC (reverse), TNF: CCTGAACCAGCCACTTAACC (forward) and AGGTCAGGCCTTCTCTCACA (reverse), NFKB1: CTCGACGTCAGTGGGAATTT (forward) and GCGAAACCTCCTCTTCCTG (reverse), and NFKB2: CGCACAAACTCAAGATACGC (forward) and GAATCCCGAGCTTCTCAGTG (reverse).
Apoptosis analysis
Apoptosis was measured in cells and tissues using the Annexin V binding assay kit (Southern Biotech) and terminal deoxynucleotidyl transferase dUTP nick end-labeling (TUNEL) assay using the APOPTAG kit (Millipore Corporation) and FACS. Apoptosis was measured in paraffin sections of xenograft tissues using the APOPTAG assay kit, and the results were examined by microscopy and subjected to digital pathology.
Digital pathology of xenograft tissue
For digital pathology, slides were loaded onto a ScanScope (Aperio) and read in the X, Y, and Z-axes at different magnifications. Digital images were acquired using the ScanScope, and the images were automatically added to the Spectrum default data group.
Gene expression analysis
Total RNA was isolated from the SKBR3 derivative cell lines using a combination of TRIzol (Sigma) and the RNeasy Mini Kit (Qiagen). The RNA was processed and hybridized to Affymetrix Human Genome U133 2.0 Plus 2 microarrays. The expression data were normalized using the robust multichip average (RMA) method and analyzed with linear models for microarray data (Limma) using R to determine the significant differentially regulated genes with a p-value < 0.01. The common differentially expressed genes were then clustered using Pearson correlation with pair wise complete linkage in the hierarchical clustering method. For GO analysis, we used the Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatics resource (26, 27). The microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) repository under accession number GSE52707 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE52707).
Oncomine concepts map
The NF-κB-regulated, anti-apoptotic genes were compared with gene expression datasets derived from the tumors of patients with breast cancer using the Oncomine Concepts Map (Compendia Bioscience; https://www.oncomine.com). The patient datasets that had a significant association with the interrogated genes were then represented in a network using Cytoscape (28), which we used to generate node connections that represent associations with a p-value < 0.005 and an Odds Ratio > 2. The data in Figure 6 are derived from the following data sets: Bild et al. (29), Chin et al. (30), Esserman et al. (31), Gluck et al. (32), Hatzis et al. (33), Kao et al. (34), Korde et al. (35), Lu et al. (36), Minn et al. (37), Richardson et al. (38), Tabchy et al. (39), Waddell et al. (40), and The Cancer Genome Atlas (TCGA) (1;http://cancergenome.nih.gov/). Kaplan-Meier was performed to determine the survival of patients in the Kao dataset using the survival package in R version 2.15.2. The significance of the difference between the survival curves was calculated using the log-rank test.
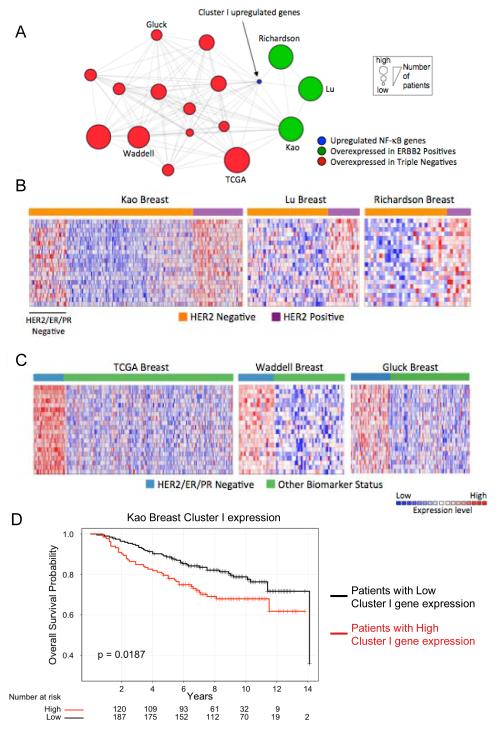
Figure 6.
Cluster I genes overexpressed in SKR6CA and SKR6LR are associated with HER2-positive and triple-negative human breast cancer. A, Oncomine Concepts Map analysis (https://www.oncomine.com). Cluster I genes were interrogated in publically available primary breast tumor gene expression datasets to determine significant overexpression in tumor subsets. These associations were represented in a network using Cytoscape (http://www.cytoscape.org). In this network, edges connect datasets that are significantly associated with cluster I genes (shown in blue). Significant associations were found with datasets from patients with triple-negative breast cancer (red circles) and those with HER2-positive breast cancer (green circles). Each node size is proportional to the number of patients demonstrating association in each dataset. B, Heatmaps of the Kao, Lu, and Richardson datasets (shown as green nodes in A) demonstrate the relative expression of the cluster I genes in HER2-positive versus HER2-negative patients (Materials and Methods for references). C) Heatmaps of Cluster I gene expression in the TCGA, Waddell, and Gluck breast datasets, representative of the HER2/ER/PR-negative datasets (Triple-negative, are shown as red nodes in A) demonstrate their relative expression in HER2/ER/PR-negative patients. D, Kaplan-Meier analysis of the probability of overall survival in HER2-positive and HER2/ER/PR-negative patients based on expression of Cluster I genes, dichotomized by the median gene expression into High and Low expression groups. The p-value was calculated with the log-rank test.
RESULTS
Functional assessment of NF-κB activation in parental SKBR3 and derivatives
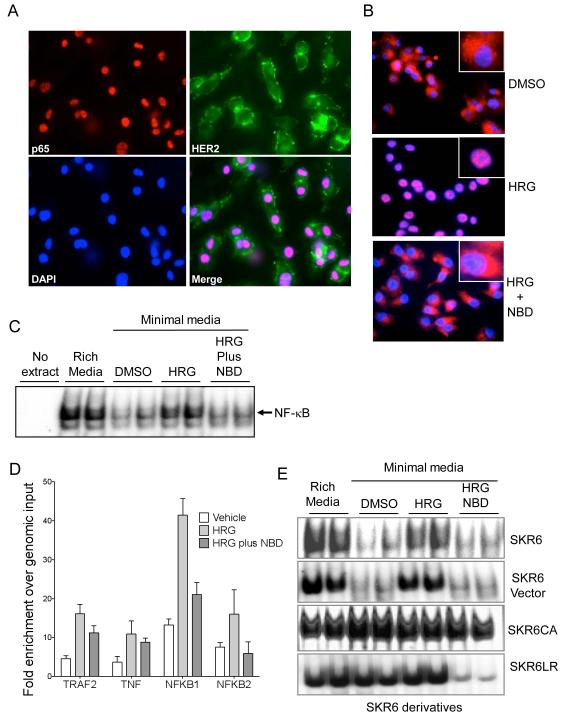
Nuclear localization of relA, the p65 NF-κB subunit, indicates activation of the NF-κB pathway. As expected, SKBR3 cells grown in rich medium harbored nuclear p65 and prominent HER2 membrane staining (Figure 1A). Culturing SKBR3 cells in hormone and growth factor-depleted media leads to the relocalization of p65 to the cytoplasm. Treatment with heregulin (HRG), a HER2 agonist, but not vehicle (dimethyl sulfoxide; DMSO) leads to the translocation of p65 into the nucleus (Figure 1B, top and middle panels). NF-κB activation was blocked by treatment with the NBD peptide, which inhibits the IKK complex, confirming NF-κB activation following HER2 stimulation by HRG (Figure 1B, bottom panel).
Figure 1.
Functional assessment of NF-κB activation in SKBR3 cells. A, Intracellular localization of NF-κB p65 and HER2 was determined by immunofluorescence. p65 (red, upper left) is found in the nucleus of SKBR3 cells growing exponentially in rich medium. Membranous staining of HER2 (green, upper right) reflects the cell surface localization of the receptor-like protein. Nuclei are stained with DAPI (blue, bottom left). The merged image of p65, HER2 and DAPI is shown (bottom right). B, Cells grown in minimal medium were treated with DMSO (vehicle, top panel), stimulated with 2nM HRG for 18 hours (middle panel), or HRG-stimulated cells were treated with 10μM NBD for 72h. NF-κB p65 is in red and nuclei were stained with DAPI. The inset shows a higher magnification of a single cell. C, The NF-κB DNA binding activity in 10μg of nuclear protein from parental SKBR3 cells cultured in the indicated conditions: Minimal medium stripped of growth factors, rich or complete media, minimal media supplemented with DMSO, minimal media plus 2nM HRG for 18 hours, or HRG plus 10 μg NBD for an additional 72 hours. NF-kB p65 DNA binding was assessed by EMSA. D, DNA Binding activity of indicated genes in SKR6 cells by chromatin immunoprecipitation assay (ChIP). TNF, tumor necrosis factor; TRAF2, TNF-receptor activating Factor 2; NFKB1, nuclear factor kappa B 1; and nuclear factor kappa B 2. E, NF-kB DNA binding activity by EMSA in nuclear proteins. 10μg of nuclear extracts from SKR6 and SKR6 Vector and 5μg of nuclear extracts from SKR6CA and SKR6LR cells were used. Each derivative was grown in rich medium, minimal medium, or minimal medium supplemented as in panel C.
Next, we examined the level of basal and activated NF-κB in SKBR3 cells by DNA binding activity using an electrophoretic mobility shift assay (EMSA). In agreement with the immunofluorescence results, we observed a NF-κB mobility shift in nuclear extracts from SKBR3 cells grown in rich media. The level of NF-κB DNA binding activity was reduced in cells grown in minimal media, and binding was elevated in the presence of HRG. This binding was reduced when cells were treated with HRG in the presence of NBD (Figure 1C).
We confirmed these data with a ChIP assay in which the level of NF-κB p65 binding was measured in the presence and absence of HRG plus and minus NBD. Increased p65 binding was found at NF-κB binding sites in NF-κB responsive genes in the presence of HER2 stimulation. Binding was reduced when cells were treated with HRG in the presence of NBD (Figure 1D). The ChIP data validate the EMSA results and demonstrate that NF-κB is under the control of HER2 signaling, and its activation is mediated via the IKK complex in the SKBR3 cells.
Different HER2 protein levels in SKBR3 cells may lead to different oncogenic potentials. Heterogeneity in non-clonal populations may obscure these differences. Thus, we sorted SKBR3 cells to select for a fraction expressing elevated HER2 levels and derived a single-cell clone from this fraction to acquire a population with stable and uniform elevated HER2 expression, which we designated SKR6 (Figure S1). The SKR6 clone was then transduced with a control virus or retrovirus encoding constitutively activated NF-κB p65 (Cap65) to create SKR6-Vector and SKR6CA cells, respectively. As expected, the SKR6CA cells demonstrated elevated and persistent NF-κB DNA binding activity under all culture conditions, demonstrating its constitutive activity. The SKR6 and SKR6-Vector cells exhibited regulated NF-κB activity that was similar to the parental SKBR3 cells. The stable Lapatinib-resistant cell clone SKR6LR, which was selected from SKR6 cells by long-term, stepwise Lapatinib treatment (0.25 – 5 μM), maintained elevated NF-κB activity in rich and minimal media containing vehicle or HRG. However, treatment with NBD blocked the elevated NF-κB activity in SKR6LR cells, suggesting that Lapatinib resistance involves a step upstream of the IKK complex (Figure 1E).
NF-κB activation is a mechanism of anti-HER2 drug resistance
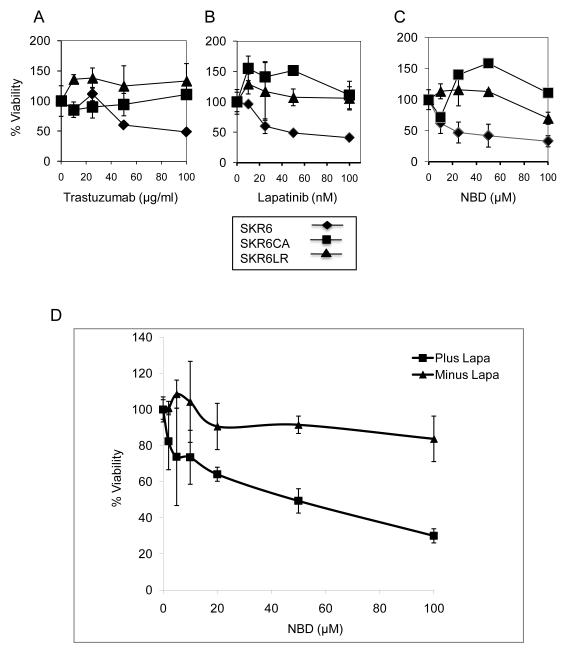
We analyzed the influence of trastuzumab, Lapatinib and NBD on the viability of the SKR6 and derivative cell lines. Parental SKR6 cells are sensitive to all three drugs, whereas SKR6CA and SKR6LR cells are resistant to these drugs (Figure 2, panels A – C). Treatment of SKR6LR cells with increasing doses of Lapatinib reduced the level of HER2 phosphorylation immediately downstream of HER2 activation in a dose-responsive fashion). The identical dose-response pattern was observed in SKR6 cells (Figure S2). This result demonstrates resistance to Lapatinib is not explained by the inability of the drug to inhibit the tyrosine kinase activity of the HER2 protein or by alteration of HER2 signaling at the initial step of HER2 phosphorylation.
Figure 2.
Sensitivity of SKR6 and derivative cells to anti-HER2 treatment or to the specific IKK inhibitor, NBD. SKR6, SKR6CA, and SKR6LR cells were grown in rich medium treated as indicated. A, Trastuzumab at the doses indicated. B, Lapatinib at the doses indicated. C, NBD at the doses indicated. Cells were treated for 72 hours and viability measured by the MTS assay. Error bars are one standard deviation, each assay was done in triplicate. D, SKR6LR cells were grown in the presence of DMSO only (Minus Lapa) or 500nM Lapatinib (Plus Lapa) with increasing concentrations of NBD (μM). The cell viability was measured by the MTS assay after 72 hours; error bars represent 1 standard deviation of triplicates.
Treatment with NBD alone leads to only a 17% reduction in cell viability in SKR6LR cells at a concentration of 100 uM; lower NBD concentrations had no effect (Figure 2C). In SKR6LR cells, Lapatinib at a concentration of 500 nM had no effect on cell viability. However, increasing concentrations of NBD in combination with a constant dose of Lapatinib (500 nM) led to a dose-dependent decrease in cell viability, which was consistently observed at NBD doses as low as 10 μM (Figure 2D). These results suggest a potential role for NF-κB inhibition combined with HER2 suppression in Lapatinib-resistant breast cancer.
NF-κB activity promotes xenograft growth and is anti-apoptotic in tumors
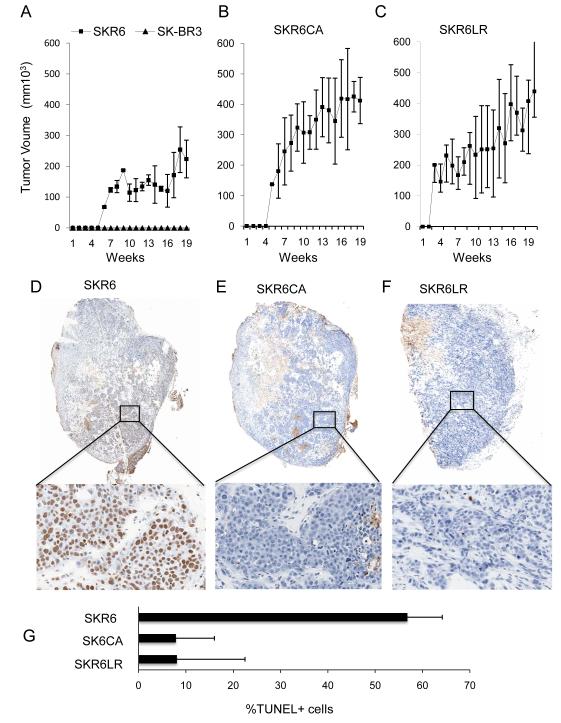
Parental SKBR3 cells and SKR6 cell derivatives were tested for their ability to grow as xenografts in nu/nu mice. While parental SKBR3 xenografts did not grow for up to 20 weeks, HER2-enriched SKR6 xenografts were palpable by 6 weeks with a moderate increase in tumor volume over 20 weeks (Figure 3A). In contrast, SKR6CA xenografts grew rapidly and reached the maximum size allowable by 20 weeks (Figure 3B). SKR6LR xenografts grew with comparable kinetics to SKR6CA xenografts, reaching maximal size by 20 weeks (Figure 3C). The SKR6, SKR6CA and SKR6LR cell lines formed high-grade tumors with large nuclei, prominent nucleoli, numerous mitotic cells, and large nests of dense tumor cells with central necrosis. Smaller nests of tumor cells with lower tumor cellularity and no necrosis were found in SKR6 xenografts (Figure S3).
Figure 3.
Xenograft growth and apoptotic fractions of SKBR3 and its derivatives. A, Growth of SKBR3 or its HER2-enhanced clonal derivative SKR6. B, SKR6CA. C, SKR6LR. 3×106 cells were suspended in 0.25ml PBS mixed with 0.25 ml of matrigel and implanted subcutaneously on the dorsal surface of nu/nu mice. Tumor volume was measured weekly; error bars are 1 standard deviation of the mean of 10 tumors. At 8-10 weeks, tumors from SKR6CA and SKR6LR cells, and at 17 weeks tumors from SKR6 were excised, formalin fixed and paraffin embedded. Sections were stained with Apop Tag™ for apoptotic nuclei. Representative images are shown at 1.5X magnification, and expanded to 20X to visualize the apoptotic fraction. D, Representative section from SKR6 xenografts. E, Representative section from SKR6CA xenografts. F, Representative section from SKR6LR xenografts. G, Mean apoptotic fractions from SKR6, SKR6CA and SKR6LR xenografts were determined by analysis of five independent sections from each of three tumors quantified by image analysis. Error bars are 1 standard deviation of the mean apoptotic fraction.
The enrichment of HER2 in SKR6 cells promoted tumor growth, and SKR6 cells contained elevated NF-κB activity compared with SKBR3 parental cells (Figure S4A). SKR6CA and SKR6LR cells and xenografts displayed very elevated NF-κB activity, which was much higher than that observed in SKR6 cells or xenografts (Figures S4A and S4B). A stepwise increase in HER2 levels in SKBR3, SKR6 and SKR6CA leads to elevated NF-κB activity, suggesting HER2 connects to the NF-κB transcriptional machinery. Constitutively active NF-κB leads directly to increased xenograft growth, tumor necrosis and HER2-directed drug resistance. Cells selected for Lapatinib resistance grew rapidly as xenografts and displayed levels of NF-κB activation equivalent to SKR6CA.
The fraction of apoptotic tumor cells measured by TUNEL staining in tumor sections was captured and quantified. Parental SKR6 xenografts demonstrated a mixed proportion of apoptotic and non-apoptotic cells (Figure 3D). In contrast, there was a dramatic absence of TUNEL-positive cells in tissue sections from SKR6CA and SKR6LR xenografts (Figures 3E and F). The absence of apoptosis correlates with the increase in NF-κB activity observed in SKR6CA and SKR6LR xenografts. In SKR6 tumors, approximately 60% of the cells in multiple tumors and a variety of sections from the same tumor were TUNEL positive. In contrast, SKR6CA and SKR6LR xenografts both displayed less than 10% TUNEL-positive cells (Figure 3G). The anti-apoptotic state of the SKR6LR cells was strikingly similar to that of the SKR6CA cells, implying that NF-κB activation is responsible for the anti-apoptotic state of tumors derived from Lapatinib-resistant cells.
Effect of combined HER2 and NF-κB inhibition on apoptosis in drug-resistant cells
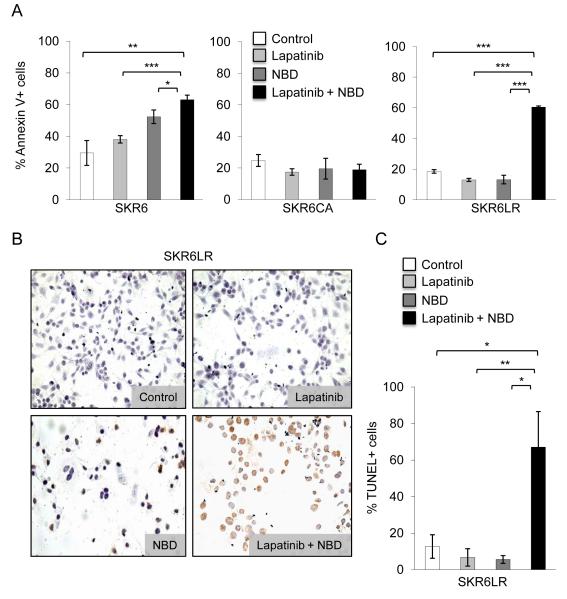
Because Lapatinib and NBD combined treatment retarded the growth of SKR6LR cells and overcame resistance, we examined this dual targeting strategy and its effect on apoptosis. We treated the SKBR3 derivative cells with vehicle, NBD, Lapatinib, and NBD plus Lapatinib and measured apoptosis using Annexin V binding and TUNEL assays. While we observed an increase in Annexin V binding in SKR6 cells in response to all treatments, cells treated with NBD plus Lapatinib demonstrated the highest level of apoptosis. As expected, SKR6CA cells maintained low apoptotic levels during all treatments (Figure 4A). In the SKR6LR cells, we observed little to no apoptosis in the presence of NBD or Lapatinib alone, whereas treatment with both compounds (10 μM NBD plus 100 nM Lapatinib) led to a dramatic increase in apoptosis (Figure 4A). In addition, TUNEL staining demonstrated massive apoptosis only in SKR6LR cells treated with combined HER2 and NF-κB inhibition (Figures 4B and C).
Figure 4.
Apoptotic fraction of the drug treated cultured cells. A, SKR6, SKR6CA, and SKR6LR cells were grown in minimal media in the presence of DMSO, Lapatinib (100nM), NBD (10μM) or Lapatinib (100nM) plus NBD (10μM) for 18h. Cells were stained with AnnexinV-FITC and propidium iodide and analyzed by FACS. Error bars represent 1 standard deviation of triplicate determinations. B, SKR6LR cells were grown in rich media in the presence of DMSO (control), Lapatinib (100nM), NBD (10μM) or Lapatinib (100nM) plus NBD (10μM) for 18h. Apoptosis was visualized using Apop Tag™ and displayed at 20X. C, Quantitation of the apoptotic fraction of SKR6LR measured by TUNEL-FACS using from cells treated as in B was measured by TUNELFACS analysis using APO BrdU™. Error bars represent 1 standard deviation of triplicate determinations.
Expression profiles of activated NF-κB and Lapatinib-resistant cell lines
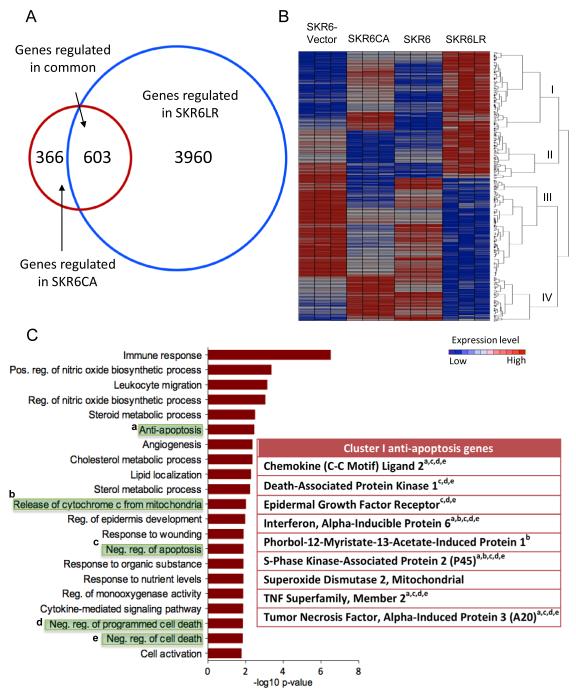
We performed expression analysis to interrogate genes that are differentially expressed in SKR6-Vector compared with SKR6CA cells and SKR6 compared with SKR6LR cells. We found 969 genes differentially expressed in SKR6CA compared with SKR6-Vector cells and 4,563 genes differentially expressed in SKR6LR compared with SKR6 cells with a two-fold difference and p < 0.01. Comparison of the genes regulated in each analysis revealed 603 genes in common (Figure 5A).
Figure 5.
Overlapping gene expression in SKR6CA and SKR6LR cells. A, Venn diagram showing the number of differentially expressed genes in SKR6CA vs. SKR6 Vector (red circle) and SKR6LR vs. SKR6 cells (blue circle). 603 genes were found in differentially expressed in both SKR6CA and SKR6LR. B, Hierarchical clustering diagram of the 603 differentially expressed genes in common. Four top clusters were identified: Cluster I, up-regulated in both SKR6CA and SKR6LR cells; Cluster II, down-regulated in SKR6CA and up-regulated in SKR6LR cells; Cluster III, down-regulated in SKR6CA and SKR6LR cells; Cluster IV, up-regulated in SKR6CA and down-regulated in SKR6LR cells. C, Gene ontology of Cluster I genes demonstrates enrichment for genes associated with apoptosis. The table lists genes found in the following GO categories (shaded in green): a, anti-apoptosis; b, release of cytochrome c from mitochondria; c, negative regulation of apoptosis; d, negative regulation of programmed cell death; e, negative regulation of death.
The 603 common genes were grouped by hierarchical clustering into four main clusters (Figure 5B). We next performed gene ontology (GO) analysis within each of the clusters to determine the biological processes influenced by the differentially expressed genes. Cluster I is comprised of genes up-regulated in SKR6CA and SKR6LR cells. This cluster is enriched for pathways associated with the negative regulation of programmed cell death and apoptosis in addition to genes associated with pathways regulated by NF-κB signaling (Figure 5C). GO analysis of genes in Clusters II – IV did not reveal strong associations with apoptosis or cell death (Figure S5). Because the relative overexpression of Cluster I genes is consistent with the loss of apoptosis observed in SKR6CA and SKR6LR cells and xenografts, we focused on these genes for further analysis.
Interrogation of human breast cancer datasets
Oncomine is a collection of gene expression datasets derived from patients with cancer and includes analysis tools (41). Using Oncomine, we performed a molecular concepts analysis of the list of Cluster I genes to determine whether these genes were also overexpressed in tumor samples derived from patients with breast cancer. We found that Cluster I genes were significantly overexpressed in tumors from patients with HER2-positive and triple-negative breast cancers (Figure 6A). Cluster I overexpression in HER2-positive breast cancer agrees with the findings reported in this study. Relatively elevated NF-κB activity is also observed in triple-negative breast cancer cell lines and tumors (16, 42). The overexpression of Cluster I genes in many triple-negative datasets from clinical samples suggests that NF-κB may be active in this subset as well.
Heatmaps demonstrate the relative difference in the expression of Cluster I genes in HER2-negative versus HER2-positive tumors. There was an overall relative down-regulation of expression of Cluster I genes in HER2-negative tumors compared with those that were HER2-positive (Figure 6B). The Kao breast cancer dataset contains gene expression data from 327 breast tumors from Taiwanese patients and is balanced for the content of HER2-positive, triple-negative and ER-positive cancers. In this dataset, the HER2-negative tumors that were also characterized as negative for ER and PR (triple-negative) exhibited overexpression of Cluster I genes (Figure 6B). These data are in agreement with the heatmaps that include datasets comparing Cluster I gene expression in tumors that are triple-negative versus those that possess some other biomarker status (Figure 6C).
Finally, we explored the association of Cluster I genes with patient outcome. Because treatments varied over time and across institutions contributing tumor expression data, we simply asked whether Cluster I genes were statistically associated with tumors from patients who suffered a poor outcome, which is defined as having a metastatic event, recurrence, or death within one to five years of diagnosis. Each dataset was analyzed separately. We found that Cluster I gene overexpression is commonly enriched in tumors from patients with poor outcome (Table 1). In addition, Kaplan-Meier analysis of patients with HER2-positive and triple-negative tumors in the Kao dataset demonstrates that those whose tumors contain high expression of Cluster I genes have worse overall survival than those with low expression of these genes (Figure 6D). We propose that the poor clinical outcome of patients with tumors enriched for overexpressed Cluster I genes are at least partially a consequence of NF-κB activation.
Table 1.
Cluster I genes associated with poor outcome
| Concept Name | No. of Overlap Genes |
P-Value | Q-Value |
|---|---|---|---|
| Metastatic Event at 3 Years(Kao et al.) | 72 | 3.1E-10 | 5.4E-08 |
| Metastatic Event at 3 Years (Bos et al.) | 64 | 3.1E-07 | 2.2E-05 |
| Dead at 3 Years (Kao et al.) | 63 | 6.8E-07 | 4.1E-05 |
| Metastatic Event at 3 Years (Symmans et al.) | 53 | 1.1E-06 | 6.1E-05 |
| Metastatic Event at 1 Year (Hatzis et al.) | 33 | 1.8E-06 | 9.3E-05 |
| Metastatic Event at 5 Years (Kao et al.) | 61 | 3.1E-06 | 1.4E-04 |
| Recurrence at 5 Years (Ma 2 et al.) | 54 | 7.3E-06 | 3.1E-04 |
| Dead at 5 Years (Boersma et al.) | 31 | 1.3E-05 | 5.0E-04 |
| Dead at 5 Years (Kao et al.) | 58 | 2.5E-05 | 8.5E-04 |
| Recurrence at 1 Year (Sorlie et al.) | 21 | 3.4E-05 | 1.0E-03 |
| Metastatic Event at 3 Years (vantVeer et al.) | 43 | 3.7E-05 | 1.0E-03 |
| Dead at 3 Years (Sorlie et al.) | 32 | 5.2E-05 | 2.0E-03 |
| Metastatic Event at 3 Years (Hatzis et al.) | 48 | 5.3E-05 | 2.0E-03 |
| Recurrence at 1 Year (Sorlie et al.) | 31 | 1.3E-04 | 3.0E-03 |
| Dead at 1 Year (Sorlie et al.) | 31 | 1.6E-04 | 4.0E-03 |
| Metastatic Event at 3 Years (Minn et al.) | 46 | 2.1E-04 | 5.0E-03 |
| Recurrence at 3 Years (Sorlie et al.) | 19 | 3.0E-04 | 6.0E-03 |
| Metastatic Event at 3 Years (Desmedt et al.) | 45 | 4.1E-04 | 8.0E-03 |
| Dead at 1 Year (Sorlie et al.) | 18 | 7.0E-04 | 1.2E-02 |
| Recurrence at 3 Years (Sorlie) | 18 | 7.0E-04 | 1.2E-02 |
| Recurrence at 3 Years (Loi et al.) | 52 | 9.8E-04 | 1.6E-02 |
| Metastatic Event at 1 Year (Desmedt et al.) | 26 | 1.0E-03 | 1.7E-02 |
| Recurrence at 3 Years (Desmedt et al.) | 26 | 1.0E-03 | 1.7E-02 |
| Metastatic Event at 5 Years (Minn et al.) | 9 | 2.0E-03 | 2.2E-02 |
| Metastatic Event at 5 Years (vantVeer et al.) | 37 | 2.0E-03 | 2.9E-02 |
| Dead at 3 Years (vandeVijver et al.) | 44 | 3.0E-03 | 3.6E-02 |
| Dead at 5 Years (Sotiriou et al.) | 25 | 3.0E-03 | 3.6E-02 |
| Metastatic Event at 5 Years (Chin et al.) | 49 | 3.0E-03 | 3.7E-02 |
| Dead at 3 Years (Esserman et al.) | 28 | 6.0E-03 | 5.8E-02 |
| Dead at 1 Year (Bild et al.) | 33 | 7.0E-03 | 6.3E-02 |
| Dead at 3 Years (Boersma et al.) | 40 | 7.0E-03 | 6.6E-02 |
| Metastatic Event at 5 Years (Bos et al.) | 48 | 7.0E-03 | 6.6E-02 |
DISCUSSION
The oncogenic potential of HER family receptors in breast cancer has been studied in great depth (20, 43). In most of these studies, genetic manipulations were employed to enhance HER family receptor-mediated signaling (12). The primary goal of this study was to explore connections between HER2 and NF-κB in HER2-positive breast cancer cells. We wanted to explore consequences of NF-κB activation, therapeutic opportunities for inhibitors, and potential pathways of resistance to HER2-directed drugs. In our previous studies, we included several other HER2 positive cell lines, but decided to explore a single HER2-positive line, SKBR3, and acquire several derivatives of these cells. In particular, we derived a homogeneous clone of SKBR3 selected for uniform HER2 expression, engineered a derivative of this clone with constitutively active NF-κB, and selected a line resistant to Lapatinib, a potent inhibitor of HER2 and EGFR. HER2 enrichment was achieved by FACS and subsequent cloning by serial dilution. Significantly, the enhanced HER2 expression in the cloned derivative, SKR6, was stable in nearly continuous passage for one year (Figure S1). This clone generated xenografts in immune-compromised mice readily, while the parental SKBR3 cell line acquired from ATCC for these studies was only weakly tumorigenic.
The HER2-enriched SKR6 cells also harbored elevated levels of NF-κB activity, measured by DNA binding when compared to the originating SKBR3 cell line (Figure S4). We previously demonstrated the heterodimeric NF-κB p50/p65 complex is the predominant NF-κB transcription complex bound to DNA in SKBR3 cells, and confirmed the presence of p65 by chromatin immunoprecipitation (ChIP) in these same cells (Figure 1D). Xenografts derived from SKR6 cells also contain considerable NF-κB DNA binding activity, which was greater than an equivalent extract from SKBR3 parental cells growing in culture (Figure S4). In cell lines, heregulin stimulation of HER2 signaling results in NF-κB activation, which is blocked entirely by the specific inhibition of IKK by the NBD peptide (Figure 1A). These observations are consistent with a role for NF-κB in the enhanced tumor-forming capability of SKR6 cells, which is likely a consequence of elevated HER2 protein levels in these cells. Furthermore, anti-HER2 drugs blocked NF-κB activation in SKBR3 cells, mirroring the effects of an IKK specific inhibitor (13). Signals that funnel through the IKK complex to activate NF-κB appear to be the predominant mechanism of connection between HER2 and NF-κB.
Our studies emphasize the importance of IKK in transmitting growth and anti-apoptotic signaling from HER2. We selected a derivative of SKR6 cells, SKR6LR, which is resistant to the dual HER2/EGFR tyrosine kinase inhibitor Lapatinib. Lapatinib is capable of completely blocking HER2 signaling in both SKR6 and SKR6LR cells, indicating that alternative growth pathways are responsible for the continued proliferation of SKR6LR in vitro, and for its enhanced tumorigenicity in vivo. However, the combination of HER2 blockade with the specific inhibition of IKK by the NBD peptide was capable of inhibiting the growth of SKR6LR cells (Figure 2D) and provoking massive apoptosis in these same Lapatinib resistant cells (Figure 4). Although SKR6LR cells are completely resistant to growth inhibition by Lapatinib, continued application of Lapatinib in combination with IKK inhibition is required for growth inhibition and the induction of apoptosis. The requirement for continued HER2 blockade in HER2-directed drug-resistant cancer cells is consistent with common clinical practice. New drugs given to patients with advanced HER2-positive breast cancer resistant to the HER2 inhibitor trastuzumab are added to, rather than substituted for, trastuzumab. Clinical trials with level 1 data demonstrate the superiority of continuing trastuzumab in combination rather than switching to an entirely new drug in metastatic breast cancer that has become resistant to trastuzumab (44).
This finding is consistent with activation of parallel growth signaling pathways in acquired resistance to HER2 blockade. If the blockade is lifted, cancer cells are again able to use HER2 signaling to promote growth. The concept of signal transduction along parallel growth factor pathways was recently suggested as a drug resistance mechanism (14). Overexpression of the receptor tyrosine kinase HGF/MET or amplification of the HGF/MET gene is commonly observed in gefitinib-resistant non-small cell lung cancer and trastuzumab-resistant metastastic breast cancer (45). Similarly, Azuma et al. proposed a mechanism that involves switching addiction from the HER2 to the FGF2 pathway, making cancer cells resistant to Lapatinib (46). Because many of these upstream parallel pathways may funnel growth signals through IKK, this downstream kinase emerges as a logical target for inhibition, particularly in cells resistant to inhibitors that act on upstream signaling targets.
Lapatinib is a small molecule inhibitor of EGFR and HER2-linked tyrosine kinases. Trastuzumab results in HER2 internalization and degradation, and may exert an immune-dependent mode of action as well. Both HER2 inhibitors are effective in a proportion of patients with HER2-positive breast cancer but result in treatment resistance in many patients who initially respond (46). The results of our study suggests that drug resistance to HER2 inhibition and the consequent recruitment of upstream receptor tyrosine kinase signaling may be overcome by blocking the NF-κB activation at the level of IKK while maintaining the inhibition of HER2. This strategy may be an alternative for patients who are resistant to anti-HER2 treatment.
Both HER2 and NF-κB are anti-apoptotic. Rapidly growing human cancers frequently contain a high fraction of apoptotic cells. Xenografts produced from SKR6 cells consistently harbor apoptotic fractions that exceed 50% (Figure 3). This is in striking contrast to xenografts generated from SKR6CA in which NF-κB is constitutively active and SKR6LR, which acquired high levels of NF-κB activation after selection for Lapatinib resistance. In xenografts from both of these cell lines, apoptotic fractions are consistently below 10% despite large areas of tumor necrosis (Figure 3 and Figure S3). These findings are consistent with the activation of NF-κB and the induction of anti-apoptotic gene products. Induction of NF-κB in tumors potentially is a powerful strategy that allows HER2-positive cancers to avoid apoptosis after inhibition of HER2 signaling and potentially after cytotoxic chemotherapy.
We examined the gene expression profiles in SKR6 derivatives. Expression of active NF-κB induced an anti-apoptotic gene program in both SKR6CA and SKR6LR cells (Figure 5C) and is likely responsible for the profound reduction in the apoptotic fraction observed in xenografts grown from these cells. To test whether the over-expressed anti-apoptotic genes found in Cluster I from Figure 5B are clinically relevant, we examined publicly available datasets and found that enrichment of Cluster I gene expression is predominantly observed in HER2-positive and triple-negative breast cancers. Estrogen receptor-positive cancers are quite distinct in their pattern of expression for Cluster 1 genes (Figure 5). We simply dichotomized the Cluster 1 signature around its mean and examined HER2-positive and triple-negative tumors from patients available in the Kao dataset (Figure 6) (33). Patients whose tumors expressed a high Cluster 1 signature had a poorer overall survival than those whose tumors expressed a lower Cluster 1 signature, and patients with a high Cluster 1 signature were more apt to having deleterious outcomes (Figure 6D and Table 1)
Our results demonstrate that NF-kB activation is downstream of HER2 signaling and potentially a mechanism for drug resistance and accelerated tumor growth. This concept is supported by engineering activated NF-κB and selecting lapatinib resistance, two independent strategies that both result in NF-κB hyper-activation. Activated NF-kB induces a gene expression profile that includes anti-apoptotic genes, and the anti-apoptotic state in tumors, whether a cause or consequence of drug resistance, results in poor patient prognosis (Figure S7). Recent reports suggest, in addition to its role in cell survival, death, inflammation, and immunity, NF-κB activation also regulates the renewal of breast cancer stem cells and plays a key role in chemoresistance (47). We propose breast cancers that are resistant to HER2-directed therapies may be susceptible to treatments that include NF-κB inhibition and continued HER2 suppression. Development of effective and clinically useful NF-kB inhibitors is a priority.
Supplementary Material
Acknowledgements
We are thankful to professors Arthur B. Pardee and Peter Sicinski of the Cancer Biology Department, Dana-Farber Cancer Institute for reading the manuscript. We are also thankful to Dr. Sabina Signoretti for her advice on the use and data analysis in ScanScope (Aperio) and Christine L. Unitt of the Brigham and Women’s Hospital Pathology Core Facility for her technical assistance. Authors are thankful to Drs. Harikrishna Nakshatri and Poornima Bhat-Nakshatri for providing the retroviral expression vector QCXIP-CAp65 and to Dr. Nicoli Birbuk for his input and data interpretation.
This manuscript is dedicated in the memory of the late Dr. Edward Fox, Director of Dana-Farber Cancer Institute Molecular Core facilities.
Funding Sources This study was supported by funds from Susan F. Smith Center for Women’s Cancers at Dana Farber Cancer Institute, by the Eileen and Mark Epstein Family Fund, and by funds from the NCI SPORE in Breast Cancer at Dana-Farber Cancer Institute (P50CA89303).
Footnotes
IMPLICATIONS The combination of an IKK inhibitor and anti-HER2 drugs may be a novel treatment strategy for drug-resistant human breast cancers.
Author contributions
Conception and design: Debajit. K. Biswas
Overall organization and supervision: James. D. Iglehart and Myles Brown
Experimental Methodology: Shannon T. Bailey, Penelope Miron, Yoon J. Choi, Debajit K. Biswas, Ruan Tian
Acquisition of data, analysis and interpretation: ShannonT.Bailey: Penelope Miron, Yoon J. Choi; Debajit K. Biswas, Ruan Tian; Alexander Miron; Bose Kochupurakkal; Gautham Maulik
Pathological analysis and Interpretation: Scott J. Rodick and Theresa Bowman
Technical assistance: Kathy M. Foley and Theresa Bowman.
The authors declare no conflict of interest
REFERENCES
- 1.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature. 2012 Oct 4;490(7418):61–70. doi: 10.1038/nature11412. PubMed PMID: 23000897. Pubmed Central PMCID: 3465532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nature reviews Cancer. 2009 Jul;9(7):463–75. doi: 10.1038/nrc2656. PubMed PMID: 19536107. Epub 2009/06/19. eng. [DOI] [PubMed] [Google Scholar]
- 3.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nature reviews Molecular cell biology. 2001 Feb;2(2):127–37. doi: 10.1038/35052073. PubMed PMID: 11252954. Epub 2001/03/17. eng. [DOI] [PubMed] [Google Scholar]
- 4.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nature reviews Cancer. 2005 May;5(5):341–54. doi: 10.1038/nrc1609. PubMed PMID: 15864276. [DOI] [PubMed] [Google Scholar]
- 5.Spector N, Xia W, El-Hariry I, Yarden Y, Bacus S. HER2 therapy. Small molecule HER-2 tyrosine kinase inhibitors. Breast Cancer Res. 2007 Mar 2;9(2):205. PubMed PMID: 17338834. [Google Scholar]
- 6.Yarden Y. Biology of HER2 and its importance in breast cancer. Oncology. 2001;61(Suppl 2):1–13. doi: 10.1159/000055396. PubMed PMID: 11694782. [DOI] [PubMed] [Google Scholar]
- 7.Yarden Y, Pines G. The ERBB network: at last, cancer therapy meets systems biology. Nature reviews Cancer. 2012 Aug;12(8):553–63. doi: 10.1038/nrc3309. PubMed PMID: 22785351. Epub 2012/07/13. eng. [DOI] [PubMed] [Google Scholar]
- 8.Heiser LM, Wang NJ, Talcott CL, Laderoute KR, Knapp M, Guan Y, et al. Integrated analysis of breast cancer cell lines reveals unique signaling pathways. Genome biology. 2009;10(3):R31. doi: 10.1186/gb-2009-10-3-r31. PubMed PMID: 19317917. Pubmed Central PMCID: 2691002. Epub 2009/03/26. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989 May 12;244(4905):707–12. doi: 10.1126/science.2470152. PubMed PMID: 2470152. Epub 1989/05/12. eng. [DOI] [PubMed] [Google Scholar]
- 10.Hortobagyi GN. Treatment of breast cancer. The New England journal of medicine. 1998 Oct 1;339(14):974–84. doi: 10.1056/NEJM199810013391407. PubMed PMID: 9753714. Epub 1998/10/01. eng. [DOI] [PubMed] [Google Scholar]
- 11.May MJ, D’Acquisto F, Madge LA, Glockner J, Pober JS, Ghosh S. Selective inhibition of NF-kappaB activation by a peptide that blocks the interaction of NEMO with the IkappaB kinase complex. Science. 2000 Sep 1;289(5484):1550–4. doi: 10.1126/science.289.5484.1550. PubMed PMID: 10968790. [DOI] [PubMed] [Google Scholar]
- 12.Singh S, Shi Q, Bailey ST, Palczewski MJ, Pardee AB, Iglehart JD, et al. Nuclear factor-kappaB activation: a molecular therapeutic target for estrogen receptor-negative and epidermal growth factor receptor family receptor-positive human breast cancer. Molecular cancer therapeutics. 2007 Jul;6(7):1973–82. doi: 10.1158/1535-7163.MCT-07-0063. PubMed PMID: 17620428. Epub 2007/07/11. eng. [DOI] [PubMed] [Google Scholar]
- 13.Baselga J, Cortes J, Kim SB, Im SA, Hegg R, Im YH, et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. The New England journal of medicine. 2012 Jan 12;366(2):109–19. doi: 10.1056/NEJMoa1113216. PubMed PMID: 22149875. Epub 2011/12/14. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012 Jul 26;487(7408):505–9. doi: 10.1038/nature11249. PubMed PMID: 22763448. Epub 2012/07/06. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS., Jr. Selective activation of NF-kappa B subunits in human breast cancer: potential roles for NF-kappa B2/p52 and for Bcl-3. Oncogene. 2000 Feb 24;19(9):1123–31. doi: 10.1038/sj.onc.1203412. PubMed PMID: 10713699. Epub 2000/03/14. eng. [DOI] [PubMed] [Google Scholar]
- 16.Biswas DK, Shi Q, Baily S, Strickland I, Ghosh S, Pardee AB, et al. NF-kappa B activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc Natl Acad Sci U S A. 2004 Jul 6;101(27):10137–42. doi: 10.1073/pnas.0403621101. PubMed PMID: 15220474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986 Aug 29;46(5):705–16. doi: 10.1016/0092-8674(86)90346-6. PubMed PMID: 3091258. [DOI] [PubMed] [Google Scholar]
- 18.Sovak MA, Bellas RE, Kim DW, Zanieski GJ, Rogers AE, Traish AM, et al. Aberrant nuclear factor-kappaB/Rel expression and the pathogenesis of breast cancer. The Journal of clinical investigation. 1997 Dec 15;100(12):2952–60. doi: 10.1172/JCI119848. PubMed PMID: 9399940. Pubmed Central PMCID: 508506. Epub 1998/01/31. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao Y, Karin M. NF-kappaB in mammary gland development and breast cancer. Journal of mammary gland biology and neoplasia. 2003 Apr;8(2):215–23. doi: 10.1023/a:1025905008934. PubMed PMID: 14635796. Epub 2003/11/26. eng. [DOI] [PubMed] [Google Scholar]
- 20.Romieu-Mourez R, Kim DW, Shin SM, Demicco EG, Landesman-Bollag E, Seldin DC, et al. Mouse mammary tumor virus c-rel transgenic mice develop mammary tumors. Molecular and cellular biology. 2003 Aug;23(16):5738–54. doi: 10.1128/MCB.23.16.5738-5754.2003. PubMed PMID: 12897145. Pubmed Central PMCID: 166341. Epub 2003/08/05. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biswas DK, Martin KJ, McAlister C, Cruz AP, Graner E, Dai SC, et al. Apoptosis caused by chemotherapeutic inhibition of nuclear factor-kappaB activation. Cancer Res. 2003 Jan 15;63(2):290–5. PubMed PMID: 12543776. [PubMed] [Google Scholar]
- 22.Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene. 2007 Feb 1;26(5):711–24. doi: 10.1038/sj.onc.1209808. PubMed PMID: 16862183. Epub 2006/07/25. eng. [DOI] [PubMed] [Google Scholar]
- 23.Gaurnier-Hausser A, Patel R, Baldwin AS, May MJ, Mason NJ. NEMO-binding domain peptide inhibits constitutive NF-kappaB activity and reduces tumor burden in a canine model of relapsed, refractory diffuse large B-cell lymphoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011 Jul 15;17(14):4661–71. doi: 10.1158/1078-0432.CCR-10-3310. PubMed PMID: 21610150. Pubmed Central PMCID: 3273413. Epub 2011/05/26. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grigorian M, Ambartsumian N, Lykkesfeldt AE, Bastholm L, Elling F, Georgiev G, et al. Effect of mts1 (S100A4) expression on the progression of human breast cancer cells. Int J Cancer. 1996 Sep 17;67(6):831–41. doi: 10.1002/(SICI)1097-0215(19960917)67:6<831::AID-IJC13>3.0.CO;2-4. PubMed PMID: 8824556. [DOI] [PubMed] [Google Scholar]
- 25.Bailey ST, Shin H, Westerling T, Liu XS, Brown M. Estrogen receptor prevents p53-dependent apoptosis in breast cancer. Proc Natl Acad Sci U S A. 2012 Oct 30;109(44):18060–5. doi: 10.1073/pnas.1018858109. PubMed PMID: 23077249. Pubmed Central PMCID: 3497783. Epub 2012/10/19. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang da W, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic acids research. 2007 Jul;35(Web Server issue):W169–75. doi: 10.1093/nar/gkm415. PubMed PMID: 17576678. Pubmed Central PMCID: 1933169. Epub 2007/06/20. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. PubMed PMID: 19131956. Epub 2009/01/10. eng. [DOI] [PubMed] [Google Scholar]
- 28.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011 Feb 1;27(3):431–2. doi: 10.1093/bioinformatics/btq675. PubMed PMID: 21149340. Pubmed Central PMCID: 3031041. Epub 2010/12/15. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006 Jan 19;439(7074):353–7. doi: 10.1038/nature04296. PubMed PMID: 16273092. Epub 2005/11/08. eng. [DOI] [PubMed] [Google Scholar]
- 30.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer cell. 2006 Dec;10(6):529–41. doi: 10.1016/j.ccr.2006.10.009. PubMed PMID: 17157792. Epub 2006/12/13. eng. [DOI] [PubMed] [Google Scholar]
- 31.Esserman LJ, Berry DA, Cheang MC, Yau C, Perou CM, Carey L, et al. Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657) Breast cancer research and treatment. 2012 Apr;132(3):1049–62. doi: 10.1007/s10549-011-1895-2. PubMed PMID: 22198468. Pubmed Central PMCID: 3332388. Epub 2011/12/27. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gluck S, Ross JS, Royce M, McKenna EF, Jr., Perou CM, Avisar E, et al. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine +/− trastuzumab. Breast cancer research and treatment. 2012 Apr;132(3):781–91. doi: 10.1007/s10549-011-1412-7. PubMed PMID: 21373875. Epub 2011/03/05. eng. [DOI] [PubMed] [Google Scholar]
- 33.Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA : the journal of the American Medical Association. 2011 May 11;305(18):1873–81. doi: 10.1001/jama.2011.593. PubMed PMID: 21558518. Epub 2011/05/12. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kao KJ, Chang KM, Hsu HC, Huang AT. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC cancer. 2011;11:143. doi: 10.1186/1471-2407-11-143. PubMed PMID: 21501481. Pubmed Central PMCID: 3094326. Epub 2011/04/20. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korde LA, Lusa L, McShane L, Lebowitz PF, Lukes L, Camphausen K, et al. Gene expression pathway analysis to predict response to neoadjuvant docetaxel and capecitabine for breast cancer. Breast cancer research and treatment. 2010 Feb;119(3):685–99. doi: 10.1007/s10549-009-0651-3. PubMed PMID: 20012355. Epub 2009/12/17. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu X, Lu X, Wang ZC, Iglehart JD, Zhang X, Richardson AL. Predicting features of breast cancer with gene expression patterns. Breast cancer research and treatment. 2008 Mar;108(2):191–201. doi: 10.1007/s10549-007-9596-6. PubMed PMID: 18297396. Epub 2008/02/26. eng. [DOI] [PubMed] [Google Scholar]
- 37.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005 Jul 28;436(7050):518–24. doi: 10.1038/nature03799. PubMed PMID: 16049480. Pubmed Central PMCID: 1283098. Epub 2005/07/29. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer cell. 2006 Feb;9(2):121–32. doi: 10.1016/j.ccr.2006.01.013. PubMed PMID: 16473279. Epub 2006/02/14. eng. [DOI] [PubMed] [Google Scholar]
- 39.Tabchy A, Valero V, Vidaurre T, Lluch A, Gomez H, Martin M, et al. Evaluation of a 30-gene paclitaxel, fluorouracil, doxorubicin, and cyclophosphamide chemotherapy response predictor in a multicenter randomized trial in breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010 Nov 1;16(21):5351–61. doi: 10.1158/1078-0432.CCR-10-1265. PubMed PMID: 20829329. Epub 2010/09/11. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waddell N, Cocciardi S, Johnson J, Healey S, Marsh A, Riley J, et al. Gene expression profiling of formalin-fixed, paraffin-embedded familial breast tumours using the whole genome-DASL assay. The Journal of pathology. 2010 Aug;221(4):452–61. doi: 10.1002/path.2728. PubMed PMID: 20593485. Epub 2010/07/02. eng. [DOI] [PubMed] [Google Scholar]
- 41.Rhodes DR, Kalyana-Sundaram S, Tomlins SA, Mahavisno V, Kasper N, Varambally R, et al. Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia. 2007 May;9(5):443–54. doi: 10.1593/neo.07292. PubMed PMID: 17534450. Pubmed Central PMCID: 1877973. Epub 2007/05/31. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaguchi N, Ito T, Azuma S, Ito E, Honma R, Yanagisawa Y, et al. Constitutive activation of nuclear factor-kappaB is preferentially involved in the proliferation of basal-like subtype breast cancer cell lines. Cancer science. 2009 Sep;100(9):1668–74. doi: 10.1111/j.1349-7006.2009.01228.x. PubMed PMID: 19538528. Epub 2009/06/23. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988 Jul 1;54(1):105–15. doi: 10.1016/0092-8674(88)90184-5. PubMed PMID: 2898299. Epub 1988/07/01. eng. [DOI] [PubMed] [Google Scholar]
- 44.Cardoso F, Costa A, Norton L, Cameron D, Cufer T, Fallowfield L, et al. 1st International consensus guidelines for advanced breast cancer (ABC 1) Breast. 2012 Jun;21(3):242–52. doi: 10.1016/j.breast.2012.03.003. PubMed PMID: 22425534. [DOI] [PubMed] [Google Scholar]
- 45.Corso S, Giordano S. Cell-autonomous and non-cell-autonomous mechanisms of HGF/MET-driven resistance to targeted therapies: from basic research to a clinical perspective. Cancer discovery. 2013 Sep;3(9):978–92. doi: 10.1158/2159-8290.CD-13-0040. PubMed PMID: 23901039. [DOI] [PubMed] [Google Scholar]
- 46.Azuma K, Tsurutani J, Sakai K, Kaneda H, Fujisaka Y, Takeda M, et al. Switching addictions between HER2 and FGFR2 in HER2-positive breast tumor cells: FGFR2 as a potential target for salvage after lapatinib failure. Biochemical and biophysical research communications. 2011 Apr 1;407(1):219–24. doi: 10.1016/j.bbrc.2011.03.002. PubMed PMID: 21377448. Epub 2011/03/08. eng. [DOI] [PubMed] [Google Scholar]
- 47.Shostak K, Chariot A. NF-kappaB, stem cells and breast cancer: the links get stronger. Breast Cancer Res. 2011;13(4):214. doi: 10.1186/bcr2886. PubMed PMID: 21867572. Pubmed Central PMCID: 3236328. Epub 2011/08/27. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.