Abstract
The transcription factor peroxisome proliferator-activated receptor (PPAR)-δ promotes oligodendrocyte differentiation and myelin formation in vitro and is prevalent throughout the brain and spinal cord. Its expression after injury, however, has not been examined. Thus, we used a spinal contusion model to examine the spatiotemporal expression of PPAR-δ in naïve and injured spinal cords from adult rats. As previously reported, PPAR-δ was expressed by neurons and oligodendrocytes in uninjured spinal cords; PPAR-δ was also detected in NG2 cells (potential oligodendrocyte progenitors) within the white matter and gray matter. After spinal cord injury (SCI), PPAR-δ mRNA and protein were present early and increased over time. Overall PPAR-δ+ cell numbers declined at 1 day post injury (dpi), likely reflecting neuron loss, and then rose through 14 dpi. A large proportion of NG2 cells expressed PPAR-δ after SCI, especially along lesion borders. PPAR-δ+ NG2 cell numbers were significantly higher than naive by 7 dpi and remained elevated through at least 28 dpi. PPAR-δ+ oligodendrocyte numbers declined at 1 dpi and then increased over time such that >20% of oligodendrocytes expressed PPAR-δ after SCI compared with ~10% in uninjured tissue. The most prominent increase in PPAR-δ+ oligodendrocytes was along lesion borders where at least a portion of newly generated oligodendrocytes (bromode-oxyuridine +) were PPAR-δ+. Consistent with its role in cellular differentiation, the early rise in PPAR-δ+ NG2 cells followed by an increase in new PPAR-δ+ oligodendrocytes suggests that this transcription factor may be involved in the robust oligodendrogenesis detected previously along SCI lesion borders.
INDEXING TERMS: PPAR-δ, differentiation, spinal cord injury, proliferation, cell genesis, myelin
Peroxisome proliferator-activated receptors (PPARs) were first discovered as molecules activated by carcinogenic drugs that led to marked production of liver peroxisomes (Issemann and Green, 1990). These molecules belong to the nuclear hormone receptor family and function as ligand-activated transcription factors (Qi et al., 2000). Three isoforms of PPAR have been identified to date: α, δ (also called β), and γ, all of which form heterodimers with retinoid X receptors (Qi et al., 2000; Hihi et al., 2002). PPARs are widely distributed throughout the body and are primarily known for their metabolic functions. For instance, PPAR-α and PPAR-γ are involved in lipid and glucose metabolism, respectively. PPAR-δ is the most abundant PPAR isoform in the body and recent studies demonstrate its involvement in muscle fatty acid oxidation (Wang et al., 2004), skin wound healing (Tan et al., 2007), and embryonic development (Nadra et al., 2006). In addition, it appears to play a global role in cellular differentiation, as has been shown for keratinocytes, endothelial cells, colonic epithelial cells, and adipocytes (Matsusue et al., 2004; Kim et al., 2006; Muller-Brusselbach et al., 2007).
Within the central nervous system (CNS), PPAR-δ is known to be expressed by oligodendrocytes and neurons (Woods et al., 2003; Benani et al., 2004; Hall et al., 2008). Its functions in the adult CNS are not entirely clear, but several reports implicate a role for PPAR-δ in myelination and oligodendrocyte development. For instance, PPAR-δ expression peaks during late development coinciding with the onset of myelination (Braissant and Wahli, 1998) and, accordingly, PPAR-δ knock-out mice display reduced myelination of the corpus callosum (Peters et al., 2000). Other work showed that PPAR-δ stimulates myelin protein expression and differentiation of oligodendrocyte lineage cells in vitro (Granneman et al., 1998; Saluja et al., 2001). The sustained expression of PPAR-δ in the adult CNS suggests that it contributes to normal physiological functions of oligodendrocytes (and neurons).
Because of its myelinogenic actions, PPAR-δ may also play an important role in CNS pathologies involving oligo-dendrocyte loss and/or demyelination. Spinal cord injury (SCI) is one such condition. Oligodendrocytes are rapidly lost at the injury site and oligodendrocyte apoptosis occurs for at least 2 weeks post injury (Crowe et al., 1997; Grossman et al., 2001; Zai and Wrathall, 2005). This dramatic loss in oligodendrocytes is thought to contribute to the vulnerability and abnormal conduction properties of axons after injury. Interestingly, recent reports demonstrate that oligodendrocyte generation occurs concomitant with oligodendrocyte apoptosis within the traumatized spinal cord and that new oligodendrocytes survive for at least 6 weeks after SCI (Zai and Wrathall, 2005; Horky et al., 2006; Rabchevsky et al., 2007; Tripathi and McTigue, 2007). Oligodendrogenesis and NG2+ oligodendrocyte progenitor cell accumulation is especially prominent during the first 14 days post injury (dpi) in tissue bordering the lesions (McTigue et al., 2001; Tripathi and McTigue, 2007). Determining which molecules regulate progenitor cell proliferation and differentiation in this region will provide insight into an environment that clearly supports spontaneous adult oligodendrogenesis. As described above, PPAR-δ is a potential candidate if it is expressed at the correct time and location post injury. However, no studies to date have examined PPAR-δ expression in injured CNS tissue. Hence, we characterized the spatio-temporal expression pattern and cellular distribution of PPAR-δ after spinal contusion in rats. Specifically, we determined the cells of origin and compared expression in the lesion borders with that in outlying spared tissue along the pial border.
Collectively, the data show that PPAR-δ+ cells increase significantly after SCI and remain elevated for several weeks. Many of these PPAR-δ+ cells are NG2 progenitors at 7 dpi and new oligodendrocytes at 14 dpi, suggesting that PPAR-δ is present in cells as they differentiate from a progenitor stage to a more mature oligodendrocyte. Most of these PPAR-δ+ cells are located along the lesion borders, revealing that PPAR-δ is a viable candidate for influencing glial responses and differentiation in the reactive gliogenic zone bordering spinal contusion lesions.
MATERIALS AND METHODS
Spinal cord injury
Adult female Sprague-Dawley rats (205–222 g; Harlan, Houston, TX) were anesthetized with ketamine (80 mg/kg, i.p) and xylazine (10 mg/kg, i.p.), and a dorsal laminectomy was performed at the T8 vertebral level. Rats then received a moderate spinal contusion injury using the Infinite Horizons device (Precision Systems and Instrumentation, Lexington, KY) with a preset force of 150 kDynes (actual forces were 150–167 kDynes). The muscles over-lying the spinal cord were then sutured and the skin was closed by using surgical clips. Animals were given 5 ml of saline and placed into warm recovery cages. Postsurgical care included 5 days of treatment with antibiotics (gentomicin, 5 mg/kg) and saline to maintain hydration, and twice-a-day manual bladder expression until spontaneous voiding returned. All procedures conformed to NIH and The Ohio State University animal care guidelines.
Quantitative real-time PCR analysis
Naïve and injured rats (n=4–5/group) at 7, 14, and 28 dpi were anesthetized and perfused with cold diethylene pyrocarbonate (DEPC)-treated phosphate-buffered saline (PBS; pH 7.4). Spinal cords were excised (4 mm) centered on the epicenter and homogenized in 1 ml of Trizol (Invitrogen, Carlsbad, CA). RNA was extracted by using a standard chloroform/phenol protocol. Known concentrations of purified RNA were used for cDNA synthesis. RNA was reverse-transcribed by using SuperScript II and random primers (Invitrogen) to obtain cDNA. Quantitative real-time polymerase chain reaction (PCR) analysis was conducted by using specific primers designed for rat tissue. The primer sequences for PPAR-δ are forward primer 5′-GGCTGAAGAAGACGGAGAGTGA′3′ and reverse primer 5′-TGCGTGCAGCCTTAGTACATG-3′ and for 18s ribosomal RNA they are forward primer 5′-TCCGGAACTGAGGCCATGAT-3′ and reverse primer 5′-TTTCGCTCTGGTCCGTCTTG-3′ The primer sequences were confirmed by BLAST analysis for highly similar sequences against known databases. A 96-well plate was set up by using 10 ng of cDNA, 500 nM of forward and reverse primer, and SYBR green master mix (Applied Biosystems, Foster City, CA) in a 20-µl reaction volume. Each sample was run in triplicate and SYBR green fluorescence was detected by Applied Biosystems 7300 (Ririe et al., 1997). A standard curve was generated for each gene by using a cDNA dilution series, and melting point analysis was performed to confirm a single amplification product. Relative PPAR-δ to 18s gene expression was quantitated by using the comparative CT method (Schmittgen et al., 2008).
Western blot analysis
Injured and naïve rats (n=2/group) at 1, 7, 14, and 28 dpi were anesthetized and perfused with cold 0.1 M PBS. Spinal cords (4 mm) centered on the epicenters were collected and homogenized in T-PERS buffer (pH 7.6; Pierce, Rockford, IL), EDTA (1X), and protease inhibitor (1X). Detailed blotting methods have been described previously (Tripathi and Mc-Tigue, 2008). Briefly, homogenates were centrifuged and supernatants were used to measure protein concentrations. Next, 10 µg of protein was loaded and resolved by using a 4–12% Bis-Tris Nupage gel (Invitrogen) and then transferred to a nitrocellulose membrane. The membrane was blocked in 5% milk in PBS (0.1 M) for 1 hour and then incubated at 4°C overnight on a shaker in the appropriate primary antibody. The next day the membrane was washed 4–5 times with 0.5% Tween in PBS and placed in an appropriate secondary anti-body for 2 hours at room temperature on a shaker. Next the membrane was washed and developed by using the Super Signal West pico chemiluminescent kit (Pierce) for 5 minutes. The membrane was exposed to autoradiographic X-ray films (ISC BioExpress, Kaysville, UT) and signal was detected. The primary antibodies used were rabbit polyclonal PPAR-δ (1:1,000; Cayman Chemicals, Ann Arbor, MI) and monoclonal α-tubulin (1:2,500; Abcam, Cambridge, MA), which was used as a loading control.
To confirm the specificity of PPAR-δ antibody, 1 µg of human recombinant protein of PPAR-α (cat. no. P1048; Protein One, Bethesda, MA), PPAR-δ (cat. no. 10007451; Cayman Chemicals), and PPAR-γ (cat. no. 10009987; Cayman Chemicals) were subjected to gel separation and detected as described.
Bromodeoxyuridine (BrdU) administration
Solution was made fresh daily by dissolving BrdU (Roche Diagnostics, Indianapolis, IN) in warmed sterile saline (20 mg/ml). Rats were given a single injection of BrdU (50 mg/kg, i.p.) daily through the first 7 dpi.
Immunohistochemistry
Naïve and injured rats (n=4/group) at 1, 3, 7, 14 or 28 dpi were anesthetized with an overdose of ketamine/xylazine (1.5X surgery dose) and intracardially perfused with 0.1 M PBS followed by 4% paraformaldehyde. Spinal cords were isolated and postfixed for 2 hours and then transferred to 0.2 M phosphate buffer (PB) overnight. Spinal tissue was cryopreserved in 30% sucrose for 48 hours, after which cords were blocked into 1-cm pieces centered on the injury epicenter, frozen in OCT (Electron Microscopy Sciences, Hatfield, PA), and then sectioned at 10 µm on a cryostat. Lesion epicenters were identified as sections containing the least amount of white matter sparing by using standard immunohistochemistry protocols to label myelin and axons (Tripathi and McTigue, 2007).
Double-labeling for PPAR-δ and NG2 was performed by first blocking with BP+ (4% bovine serum albumin [BSA]/0.1% Triton X-100) and then overlaying sections with a PPAR-δ-specific rabbit polyclonal antibody (1:1,500, Cayman Chemicals) and a monoclonal anti-NG2 antibody (1: 100; US Biological, Swamscott, MA) overnight. The next day, sections were incubated with biotinylated goat anti-rabbit antibody (1:2,000; Vector, Burlingame, CA) and a biotinylated mouse antibody (1:800; Vector) in BP+ for 1 hour and then treated with 6% hydrogen peroxide in methanol. Sections were next incubated with Elite Avidin Biotin Conjugate (ABC; Vector) and then visualized with 3, 3′-diaminobenzamide (DAB; Vector) substrate for PPAR-δ antibody and SG substrate (Vector) for NG2 antibody.
PPAR-δ expression in oligodendrocytes was visualized with immunofluorescence by blocking sections as above and then incubating them overnight with anti-PPAR-δ (1: 2,000) and the monoclonal CC1 antibody (1:2,000, Abcam), which labels oligodendrocyte cell bodies. The next day the sections were overlaid for 30 minutes with the secondary antibodies Alexa Fluor 546 and Alexa Fluor 488 (1:1,000; Molecular Probes, Eugene, OR) for PPAR-δ and CC1, respectively. Cell nuclei were counterstained with Draq5 (1:3,000, Biostatus Limited, Leicestershire, UK). PPAR-δ expression in neurons was examined by using a monoclonal NeuN antibody (1:5,000, Chemicon, Billerica, MA) as described above.
Triple-label immunofluorescence was used to label PPAR-δ in new oligodendrocytes that were generated after SCI. Sections were labeled for PPAR-δ and Alexa Fluor anti-rabbit 546 and then blocked for 30 minutes followed by incubation with the CC1 antibody for 2 hours at room temperature. Next, sections were overlaid with Alexa Fluor anti-mouse 633 (1: 1,000) for 30 minutes before incubating with 2 N HCl at 37°C for 30 minutes. Sections were then blocked for 1 hour as above and incubated overnight with monoclonal BrdU anti-body (1:100; Developmental Studies Hybridoma Bank, Iowa City, IA). Secondary Alexa Fluor anti-mouse 488 was applied the next day to visualize BrdU. All slides were coverslipped with either Permount (Fisher Scientific, Fair Lawn, NJ) for DAB sections or Immu-mount (Thermo Scientific, Pittsburgh, PA) for fluorescent sections.
All protocols included controls in which the primary or secondary antibodies were omitted. These controls consistently revealed no labeling. Digital plates were constructed by using Adobe Photoshop (Adobe Systems, San Jose, CA); brightness and contrast were enhanced when necessary on microscopic images to reproduce immunolabeling as viewed through the microscope. The high red Draq5 signal was converted to blue by using the confocal software.
Antibody characterization
PPAR-δ
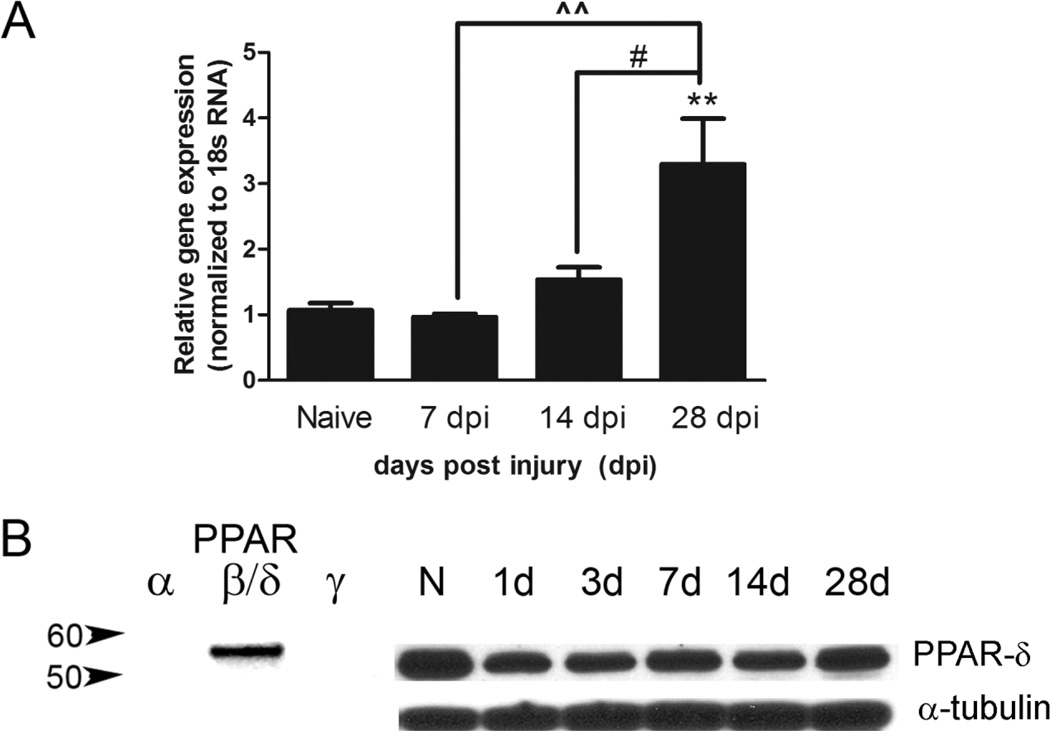
The PPAR-δ antibody used was raised against human PPAR-δ amino acids 39–54 (SSSYTDLSRSSSPPSL) and recognizes human PPAR-δ antigen. By using Western blots on rat CNS tissue, we observed that this antibody recognized a single band at ~50 kDa (Fig. 1B), which is the expected size for PPAR-δ as reported by the manufacturer using protein isolated from human cortical samples (Cayman Chemicals). Confirmation of antibody specificity was examined by performing Western blots with purified recombinant PPAR-α, δ, and γ proteins. This revealed a single band at the expected weight for the PPAR-δ isoform and an absence of bands for the other two isoforms (Fig. 1B). The immunohistochemical labeling pattern is nuclear and consistent with reports demonstrating nuclear PPAR-δ expression in neurons and oligodendrocytes in rodent CNS tissue (Woods et al., 2003). A second antibody toward PPAR-δ against the amino acids 1–29 (MEQPQEETPEAREEEKEEVA MGDGAPELN) and a positive control (COS cell lysates transfected with PPAR-δ vector) was generously given to us by Dr. Jeffery Peters (The Pennsylvania State University, University Park, PA); the second antibody showed a similar band size as the Cayman PPAR-δ antibody on rat SCI tissue, and the Cayman antibody labeled the positive control protein (data not shown). In addition, work conducted by Han et al. (2008) revealed decreased PPAR-δ expression with PPAR-δ-specific siRNA versus control siRNA when probed with the PPAR-δ antibody (Cayman Chemicals), supporting its specificity.
Figure 1.
Peroxisome proliferator-activated receptor-δ (PPAR-δ) levels increase chronically after SCI. A: Quantitative real-time PCR for PPAR-δ was performed on homogenates of injured spinal cords at different days post injury (dpi) and normalized to the control gene 18s ribosomal RNA. A significant increase in PPAR-δ mRNA was detected at 28 dpi compared with naïve spinal cords. B: The specificity of the anti-PPAR-δ antibody is demonstrated by the presence of a single specific band for purified recombinant PPAR-δ protein but not for recombinant PPAR-α or γ proteins. The antibody also produced a single band when probed against spinal cord-injured homogenates, with a slight decrease at 1 dpi and subsequent return to naïve levels by 28 dpi. N, naïve; d, day post injury. #, P < 0.05 vs. 14 dpi; ˆˆP < 0.01 vs. 7 dpi; **, P < 0.01 vs. Naive.
NG2
The specificity of the antibody was confirmed via Western blot of rat brain tissue and recombinant protein by the manufacturer, giving a ~280-kDa band. This antibody shows a similar pattern in CNS tissue to the NG2 antibody generated by William Stallcup, whose specificity was confirmed by its absence in NG2 knockout mice (Grako et al., 1999; McTigue et al., 2006)
CC1 clone of the APC antibody
The APC antibody was described to recognize oligodendrocytes and was shown to recognize a single band of 300 kDa from rat brains (Bhat et al., 1996). It is raised against recombinant human APC amino acid residues 1–226. The CC1 antibody co-localizes with Olig2, Nogo-A, and GST-pi (Linares et al., 2006; Tripathi and McTigue, 2007; Kuhlmann et al., 2007). Additionally, mice expressing proteo-lipid protein (PLP; expressed only in mature oligodendrocytes) under a green fluorescent protein (GFP) promoter showed co-localization of GFP with this antibody (Fuss et al., 2000).
Bromodeoxyuridine
This antibody was derived from a mouse myeloma cell line raised against (BrdU)16-BSA. As expected, immunohistochemistry reveals nuclear localization and an absence of labeling in tissue from animals that did not receive BrdU injections.
NeuN
This antibody clone, called A60, is raised against purified cell nuclei from mouse brain by repeated immunization to recognize neurons in the CNS and peripheral nervous system (PNS). Immunoblotting reveals three bands and it binds to DNA in vitro (Mullen et al., 1992). This antibody primarily is nuclear but has some cytoplasmic reactivity and has been well characterized in developing and adult nervous systems in work by Mullen et al. (1992).
α-Tubulin
This antibody was used as a loading control for Western blot and produced a single band at 53 kDa as expected. It complies with previous reports for Western blot of spinal cord tissue (Mutti et al., 2007).
Data analysis
PPAR-δ counts and PPAR-δ+ CC1 cell counts
High-power images were collected from sections double-labeled for PPAR-δ and CC1 (and counterstained with Draq5) located at the epicenter and ± 1.35 and 2.25 mm by using a Zeiss 510 Meta Laser Scanning Confocal microscope (40× objective, Zoom × 2). The images were used to place sample boxes (0.05 mm2) in different regions of interest, which included spared white matter (along the pial border), spared gray matter, white matter lesion border, and gray matter lesion border, as shown in Figure 2A. Lesion borders, first mapped on adjacent sections immunolabeled for neurofilament and myelin, were defined as the border between frank cavitation or tissue necrosis and the edge of spared tissue containing intact myelin and axons (white matter borders) or intact gray matter (gray matter borders). These maps were then used to locate lesion borders on the adjacent sections labeled for PPAR-δ and CC1. This technique has been used previously to identify lesion borders in cross sections from contused rat spinal cords (Tripathi and McTigue, 2007). Bias in sample box placement along lesion borders was avoided by placing the first sample box in the dorsomedialmost aspect of the lesion border, typically located within the dorsal funicular region. Sequential boxes were then placed around the lesion border two box widths (0.45 mm) away from the previous sample box.
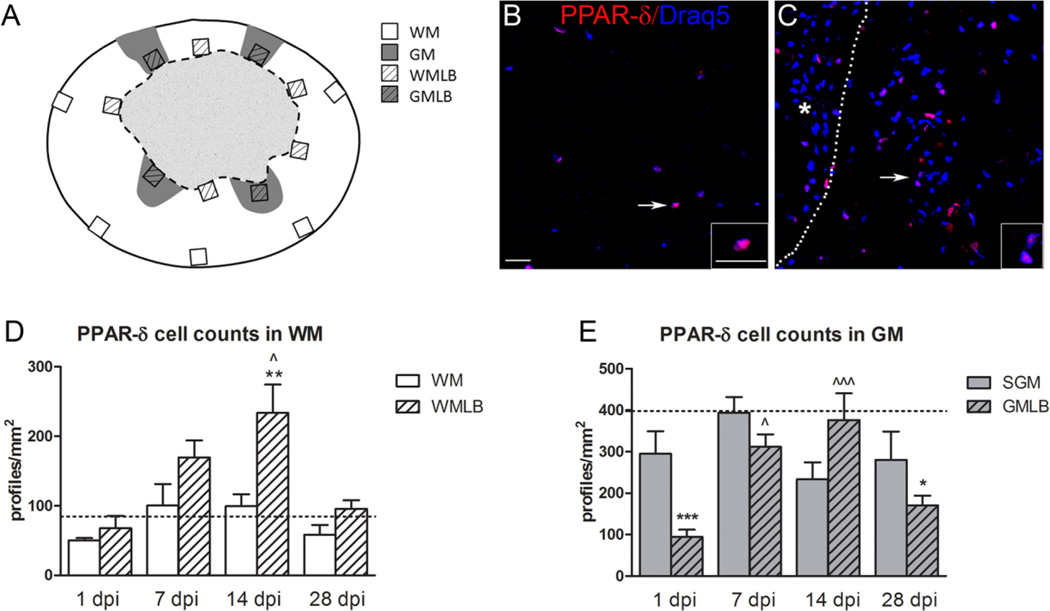
Figure 2.
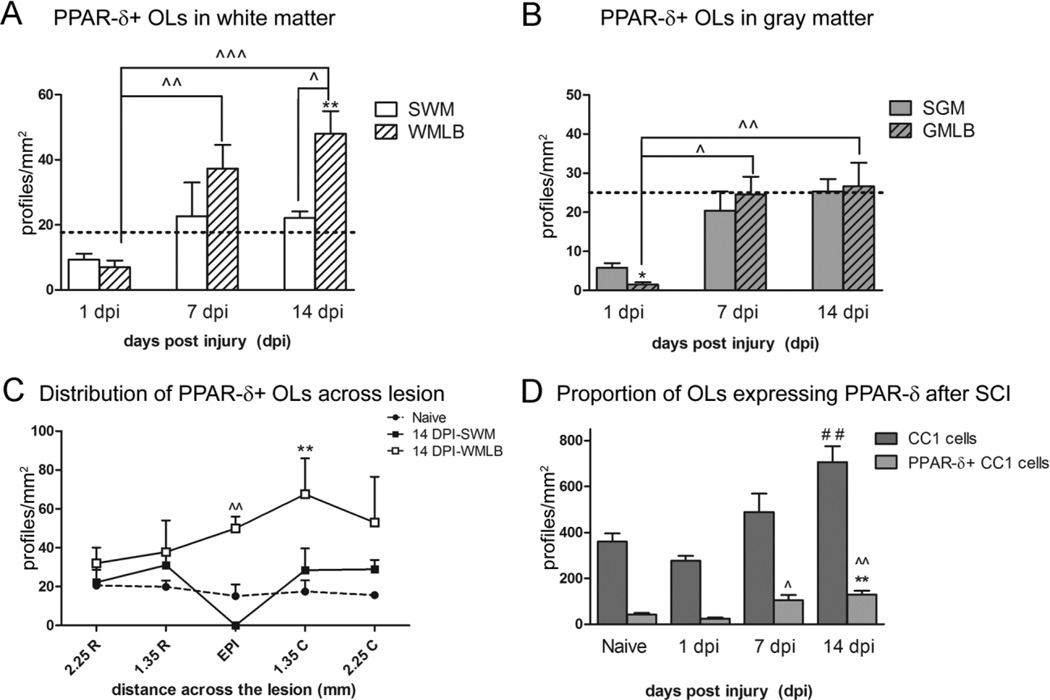
Peroxisome proliferator-activated receptor-δ (PPAR-δ)+ cells increase after spinal cord injury (SCI). A: Diagram of an injured spinal cord section demonstrating the sample box placement used to obtain cell counts here and in Figures 3 and 5. WM, white matter; GM, gray matter; WMLB, white matter lesion border; GMLB, gray matter lesion border. B: PPAR-δ+ cell nuclei (red) were present in white matter of naïve spinal cords; sections were counterstained with Draq5 (blue). C: Elevated PPAR-δ+ cell numbers were present along the WMLB at 14 dpi. The dotted line delineates the lesion border (lesion denoted by asterisk). Arrows in B and C point to cells shown at higher magnification in the insets. D: PPAR-δ+ cells in WMLB increased by 7 dpi and were significant higher at 14 dpi compared with naïve WM and outlying spared WM in the same sections, P < 0.05 vs. 14 dpi WM; **, P < 0.01 vs. Naive. E: PPAR-δ cell counts in spared GM and GMLB revealed a decline at 1 dpi, especially along the lesion borders. Cells increased significantly in GMLB at 7 and 14 dpi and then declined again at 28 dpi compared with naïve. SGM, spared gray matter, P < 0.05 vs. 1 dpi GMLB; ˆˆˆ, P < 0.001 vs. 1 dpi GMLB; *, P < 0.05 vs. Naive; ***, P < 0.001 vs. Naive. Dashed lines in D and E represent the number of PPAR-δ+ cells in naïve spinal cord WM and GM. Scale bar = 20 µm in B (applies to B,C) and inset to B (applies to both insets). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
It was noted whether box location fell in white matter or gray matter bordering the lesion. If a box spanned gray and white matter, it was moved laterally until it was located entirely within gray or white matter. In spared white matter, sample boxes were consistently placed along the pial border in the dorsolateral white matter, the lateral funiculus, and the ventral funiculus. For spared gray matter, sample boxes were centered in dorsal or ventral horns, when present. Within each sample box, the total number of cells (Draq5+) and PPAR-δ cells were counted, and then the number of PPAR-δ+ nuclei belonging to CC1+ cells was determined. PPAR-δ+ profiles were identified as immuno-reactivity co-localized with a Draq5+ nucleus. Double-labeled cells were identified by merging the CC1 image with the PPAR-δ/Draq5 image and counting the number of cells in which CC1 completely surrounded the PPAR-δ/Draq5+ nucleus; single-labeled CC1+ cells with Draq5+ nuclei were also counted.
To correct for size of the profiles being counted, an Abercrombie correction was performed (Coggeshall et al., 1996; Guillery et al., 1997). For this, the size of PPAR-δ+ nuclei in the immunolabeled sections was determined by using the confocal software to measure the distance from the top to bottom of PPAR-δ+ nuclei sampled from injured and naïve tissue. The correction factors obtained by measuring average nuclear size for PPAR-δ+ cells and PPAR-δ+ oligodendrocytes were 0.62 and 0.64, respectively, which were multiplied to all the cell counts. Final data are presented as profiles/mm2.
PPAR-δ+ NG2 cell counts
Sections double-labeled for PPAR-δ and NG2 and counterstained with methyl green were used to quantify the total number of NG2 cells and the number of NG2 cells expressing PPAR-δ+ nuclei. Cells single-labeled for NG2 or double-labeled for NG2 and PPAR-δ were manually counted by using a 63× oil objective on a Zeiss Axioplan 2 Imaging light microscope. A standardized sample box (0.025 mm2) was placed in the regions of interest as above; sequential sample boxes were placed along lesion borders 0.32 mm (two box widths) distal to the previous box. Boxes within spared tissue were placed as described above. Positively labeled cells were identified as PPAR-δ+ or methyl green+ nuclei surrounded by NG2 immunoreactivity. The Abercrombie correction was again applied to these data. For this, Axiovision software (Zeiss) controlling a light microscope with an automated stage was used to measure the distance between the top and bottom of positively labeled nuclei within these sections. The average nuclear size was multiplied to obtain a correction factor of 0.625, which was used to multiply all cell counts; final counts are reported as profiles/mm2.
Statistics
GraphPad Prism 5.01 (GraphPad Software, La Jolla, CA) was used for statistical analyses, which included one-way ANOVA followed by Bonferroni post hoc tests for total profiles/animal. For counts across different distances along the spinal cord, a two-way repeated measures ANOVA followed by Bonferroni post hoc test was used. P < 0.05 was considered significant. All data are graphed as mean ± SEM. All microscopic images were arranged into plates by using Adobe Photoshop; contrast and brightness were enhanced when needed to reproduce images as viewed through the microscope.
RESULTS
PPAR-δ mRNA and protein are expressed after SCI
PPAR-δ mRNA expression after SCI was determined by quantitative real-time PCR on injured spinal cord tissue at 7, 14, and 28 dpi with naïve tissue serving as control. The ratio of PPAR-δ versus the internal control 18s ribosomal RNA shows that PPAR-δ mRNA is elevated threefold by 28 dpi, which is significantly greater than that in naïve and 7 and 14 dpi (Fig. 1A). The presence of PPAR-δ protein after SCI was confirmed by Western blot (Fig. 1B). The PPAR-δ antibody used for the study recognizes a specific single band at the appropriate molecular weight in spinal cord homogenates; this antibody did not bind to other PPAR isoforms (left; Fig. 1B) when probed by using recombinant PPAR-α, δ and γ proteins. The PPAR-δ levels at 1 and 3 dpi appear lower than those in naïve animals, which probably reflect the loss of neurons at the injury site. This early loss is confirmed below (Fig. 2E).
PPAR-δ+ cells increase after SCI
The spatial and temporal distribution of PPAR-δ+ cells after SCI was compared in cross sections of naïve and injured spinal cords. Regions of interest include spared white matter (WM) and gray matter (GM) along the pial border, and white matter and gray matter along the lesion border (WMLB and GMLB, respectively; Fig. 2A). Uninjured spinal cords contained PPAR-δ+ cells scattered throughout the gray and white matter, as expected (Fig. 2B). After SCI, the number of PPAR-δ+ cells progressively increased in WMLB during the first 2 weeks (Fig. 2C). At 14 dpi, the number of cells was significantly greater in WMLB compared with spared and naïve WM (Fig. 2D). PPAR-δ+ cell numbers returned to naïve levels by 28 dpi.
In GMLB, PPAR-δ+ cells were significantly reduced by 1 dpi (likely due to neuron loss) and then increased threefold through 7 dpi and 14 dpi (Fig. 2E). Intriguingly, the number dropped again by 28 dpi in GMLB such that it was significantly lower than controls. In spared GM, PPAR-δ+ cell numbers were not significantly altered after SCI (Fig. 2E).
NG2 cells expressing PPAR-δ increase after SCI
NG2 cells are thought to be comprised at least partially of oligodendrocyte progenitors in the adult CNS and after SCI (Yoo and Wrathall, 2007). To determine whether potential oligodendrocyte progenitors expressed PPAR-δ after SCI, we performed double-label immunohistochemistry for PPAR-δ and NG2 and quantified the number of single-and double-labeled NG2 cells as above. PPAR-δ+ NG2 cells were detected in uninjured spinal tissue and markedly increased after injury, especially along lesion borders (Fig. 3A–C).
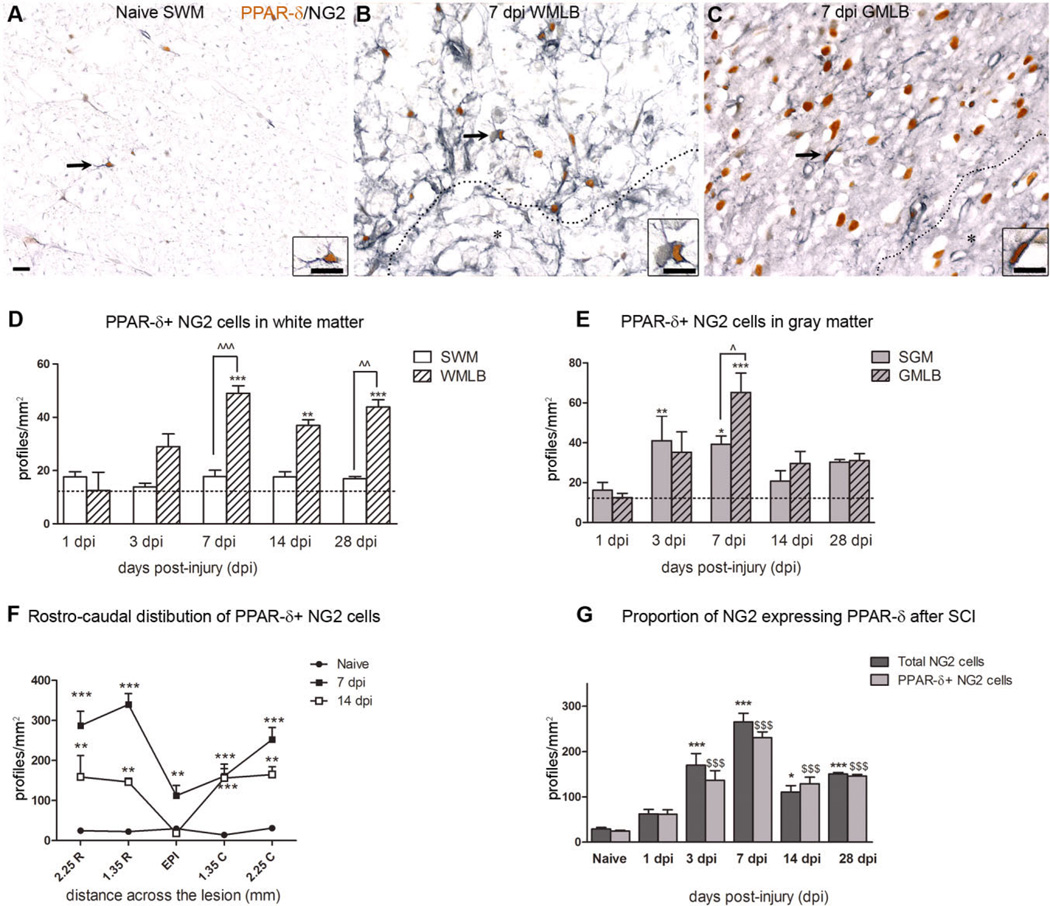
Figure 3.
PPAR-δ+ NG2 cells are elevated chronically after spinal cord injury (SCI). A: PPAR-δ (orange) and NG2 (black) double-labeled cells were present in white matter of naïve animals. B,C: At 7 dpi, PPAR-δ+ NG2 cells along white matter lesion border (WMLB; B) and gray matter lesion border (GMLB; C) were prominent (lesion border delineated by dashed line; asterisk denotes lesion). Arrows (A–C) indicate cells shown at higher magnification in insets. The 7-dpi images were taken 1.35 mm rostral to epicenter. SWM, spared white matter. D: PPAR-δ+ NG2 cells increased in WMLB at 7, 14, and 28 dpi compared with naïve white matter. Lesion borders also contained more PPAR-δ+ NG2 cells than outlying spared white matter (SWM) along the pial border at 7 and 28 dpi. **, P < 0.01 vs. Naive; ***P < 0.001 vs. Naive; ˆˆ, P <0.01 vs. 28 dpi SWM; ˆˆˆ, P < 0.001 vs. 7 dpi SWM. E: PPAR-δ+ NG2 cells increased compared with naïve in spared gray matter (SGM) at 3 dpi and in SGM and GM lesion borders (GMLB) 7 dpi. GMLB also contained more cells than SGM in the same sections. Dashed lines in D and E represent the number of PPAR-δ+ cells in uninjured tissue. *, P < 0.05 vs. Naive; **, P < 0.01 vs. Naive; ***, P < 0.001 vs. Naive; ˆ, P < 0.05 vs. 7 dpi SGM. F: PPAR-δ+ NG2 cells were elevated compared with naïve tissue in the lesion epicenter at 7 dpi and rostral and caudal to the epicenter at 7 and 14 dpi. **, P < 0.01 vs. Naive; ***, P < 0.001 vs. Naive. G: The total numbers of NG2 cells and PPAR-δ+NG2 cells followed a similar pattern post SCI, with significant increases at 3–28 dpi compared with naïve tissue. *, P < 0.05 vs. Naive Total NG2 cells; ***, P < 0.001 vs. Naive Total NG2 cells; $$$, P < 0.001 vs. Naive PPAR-δ NG2 cells. Scale bar = 10 µm in A (applies to A–C and insets before A–C). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Quantification of PPAR-δ+ NG2 cells in WMLB revealed that cell numbers doubled between 1 and 3 dpi, and then increased threefold by 7 dpi, which was significantly greater than that in naïve WM (Fig. 3D). PPAR-δ+ NG2 cells remained significantly elevated in WMLB through at least 28 dpi. Furthermore, there were significantly more PPAR-δ+ NG2 cells along lesion borders compared with outlying white matter at 7 and 28 dpi within the same sections (Fig. 3D).
Gray matter changes in PPAR-δ+ NG2 cell numbers differed from white matter in terms of timing and distribution. An increase in GMLB was detected at 3 dpi, which was significantly higher at 7 dpi (P < 0.01; Fig. 3E). At 14 and 28 dpi, however, PPAR-δ+ NG2 cell counts had declined to nonsignificant levels. In contrast to spared white matter, spared gray matter contained significantly more PPAR-δ+ NG2 cell numbers than naïve at 3 and 7 dpi.
Examination of the rostral-caudal distribution of PPAR-δ+ NG2 cells at 7 dpi revealed a significant elevation throughout the entire extent of the injury site (Fig. 3F). This distribution was similar at 14 dpi (Fig. 3F) and 28 dpi (data not shown), with the exception of a reduction to naïve levels in epicenter sections. Thus, most PPAR-δ+ NG2 cells present chronically after SCI are distributed rostral and caudal to epicenter.
We next determined the ratio of NG2 cells that expressed PPAR-δ by comparing the number of PPAR-δ+ NG2 cells with the total number of NG2 cells present (Fig. 3G). In naïve tissue, the majority of NG2 cells (~86%) expressed PPAR-δ. By 3 dpi, the number of NG2 cells and NG2/PPAR-δ double-labeled cells increased significantly but the ratio of cells expressing PPAR-δ remained similar to naïve. By 7 dpi, the total number of NG2 cells had again increased, but only 65% of NG2 cells expressed PPAR-δ at this time, which suggests that NG2 cells may upregulate PPAR-δ subsequent to dividing. At later times post injury, NG2 cell numbers had declined and the ratio of cells expressing PPAR-δ returned to ~ 86%. Overall, NG2 cell numbers were significantly greater than in naïve tissue at 3–28 dpi, which indicates that the majority of newly derived NG2 cells present after SCI express PPAR-δ.
PPAR-δ+ oligodendrocyte numbers increase after SCI
Because PPAR-δ may be involved in oligodendrocyte differentiation and is present in the majority of NG2 cells, we next examined whether PPAR-δ was expressed by oligo-dendrocytes after SCI. Immunolabeling for CC1, a marker for oligodendrocyte cell bodies, confirmed our previous results showing that oligodendrocyte numbers increase along lesion borders and revealed that many more oligo-dendrocytes in this region compared with outer spared white matter expressed PPAR-δ (Fig. 4).
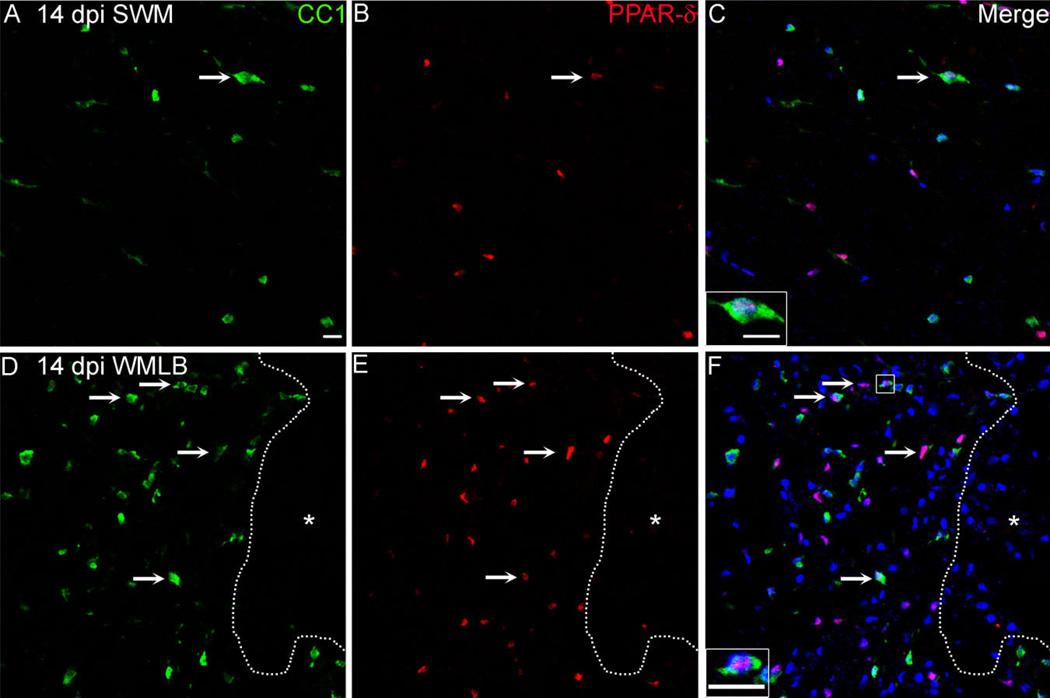
Figure 4.
Peroxisome proliferator-activated receptor-δ (PPAR)-δ+ oligodendrocytes (OLs) increase at 14 dpi in white matter lesion borders. A–F: Single channel and merged confocal images of CC1+ OLs (A,D; green) and PPAR-δ (B,E; red) taken from outlying spared white matter (SWM) and WM lesion border (WMLB) in the same cross section at 14 dpi. The number of double-labeled PPAR-δ+ CC1 cells (arrows) increased robustly along lesion borders compared with SWM. Sections were counterstained with Draq5 (blue) and images taken 2.25 mm caudal to epicenter. The dotted line delineates the lesion border and insets show higher magnification PPAR-δ+ CC1 cells. Scale bar= 20 µm in F (applies to A–F) and insets to C,F. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Quantification of double-labeled PPAR-δ+ CC1 cells in white matter revealed ~50% reduction in spared white matter and WMLB at 1 dpi (Fig. 5A). PPAR-δ+ oligodendrocyte numbers rose between 1 and 7 dpi such that numbers in spared white matter returned to naïve levels and numbers in WMLB exceeded naïve levels and were significantly greater than WMLB levels at 1 dpi (Fig. 5A). PPAR-δ+ CC1 cells continued to increase through 14 dpi, at which time the numbers became significantly greater the naïve and adjacent outlying spared white matter along the pial border (Fig. 5A). Thus, PPAR-δ+ oligodendrocyte numbers continually rise in WMLB between 1 and 14 dpi.
Figure 5.
Peroxisome proliferator-activated receptor-δ (PPAR)-δ+ oligodendrocytes (OLs) increase after spinal cord injury (SCI). A: PPAR-δ+ CC1 cells decreased slightly at 1 dpi in white matter (WM). At 7 and 14 dpi, PPAR-δ+ OLs had significantly increased in WM lesion border (WMLB) compared with 1 dpi WMLB. Furthermore, PPAR-δ+ CC1 cells along the WMLB at 14 dpi were significantly greater than naïve WM and spared white matter (SWM) within the same sections. **, P < 0.01 vs. Naive WM; ˆ, P < 0.05 vs. 14 dpi SWM; ˆˆ, P < 0.01 vs. 1 dpi WMLB; ˆˆˆ, P < 0.001 vs. 1 dpi SWM. B: PPAR-δ+ OLs were significantly reduced at 1 dpi in gray matter lesion borders (GMLB) followed by a subsequent rise at 7 and 14 dpi. *, P < 0.05 vs. Naive GM; ˆ, P <, 0.05 vs. 1 dpi GMLB; ˆˆ, P < 0.01 vs. 1 dpi GMLB. C: PPAR-δ+ OL distribution in WM at 14 dpi revealed the highest numbers were present in the WMLB at the epicenter and caudal sections. **, P < 0.01 vs. Naive WM; ˆˆ, P < 0.01 vs. 14 dpi SWM. D: The number of PPAR-δ+ OLs had doubled by 7 dpi compared with naïve. At 14 dpi, the total number of OLs and the number of OLs with PPAR-δ+ nuclei were both significantly increased. **, P < 0.01 vs. Naive PPAR-δ+ CC1 cells; ˆ, P < 0.05 vs. 1 dpi PPAR-δ+ CC1 cells; ˆˆ, P < vs. 1 dpi PPAR-δd+ CC1 cells; ##, P < 0.01 vs. Naive CC1 cells. SGM, spared gray matter.
Along gray matter lesion borders, the numbers of PPAR-δ+ oligodendrocytes declined significantly to near zero by 1 dpi. The numbers rose over the first week post injury such that at 7 and 14 dpi, GMLB contained significantly more PPAR-δ+ oligodendrocytes than comparable regions at 1 dpi. Thus, similar to WMLB, oligodendrocytes expressing PPAR-δ increased over time in gray matter bordering the lesion.
The rostral-caudal distribution of PPAR-δ+ oligodendrocytes in white matter was examined at 14 dpi. The largest accumulation of double-labeled cells was detected in lesion borders at the epicenter and caudal to the lesion site (Fig. 5C). Indeed, caudal to epicenter, the number of PPAR-δ+ oligodendrocytes was significantly greater than naïve levels.
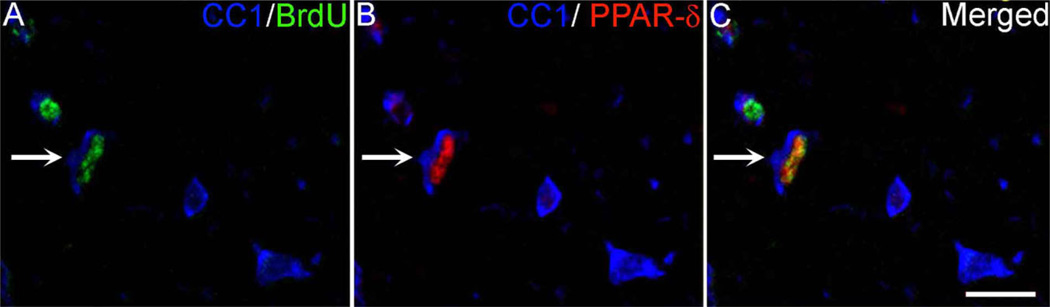
As previously shown, there was a robust increase in total oligodendrocyte number compared with naïve by 14 dpi (P < 0.01; Fig. 5D). Within the population of oligodendrocytes present after SCI, the number that expressed PPAR-δ rose significantly over time compared with 1 dpi and naïve spinal cords (P < 0.01; Fig. 5D). The fact that this number increased over time suggests that new oligodendrocytes generated after SCI expressed PPAR-δ. To verify this, confocal analysis was used to examine sections triple-labeled for oligodendrocytes (CC1), PPAR-δ, and BrdU (Fig. 6). Indeed, PPAR-δ+ oligodendrocytes with BrdU+ nuclei were detected, thereby confirming that PPAR-δ is expressed in new oligodendrocytes that differentiated from proliferating progenitors after SCI.
Figure 6.
PPAR-δ is expressed in new oligodendrocytes (OLs) after SCI. A–C: Single-channel and merged confocal images of triple-label immunofluorescence for OLs (CC1; blue), bromodeoxyuridine (BrdU; green), and peroxisome proliferator-activated receptor-δ (PPAR)-δ (red) from a 7-dpi section. The arrow indicates a triple-labeled OL. Scale bar = 20 µm in C (applies to A–C). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
PPAR-δ is present in neurons in spared tissue
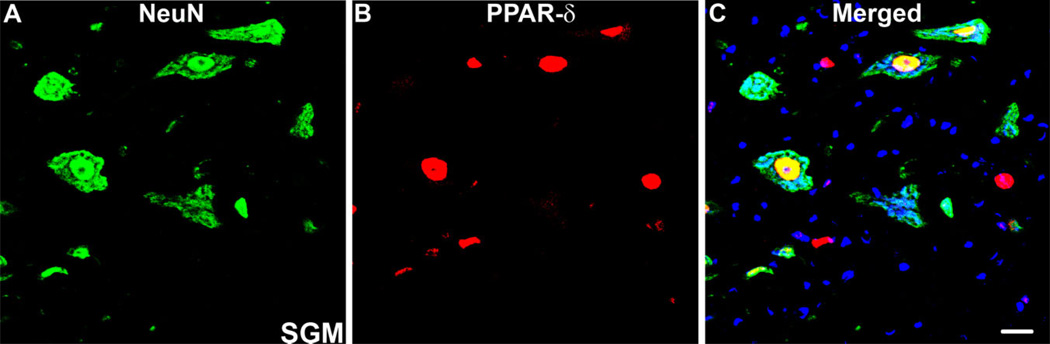
Because PPAR-δ has also been detected in neurons in the normal CNS, we investigated whether neurons present after SCI expressed PPAR-δ. Indeed, PPAR-δ co-localized with NeuN, a neuronal marker, in spared gray matter present at the lesion poles (Fig. 7A–C). Thus, neurons near lesioned tissue maintain PPAR-δ+ expression after SCI.
Figure 7.
PPAR-δ is present in neurons after SCI. A–C: Confocal images of double-labeling for peroxisome proliferator-activated receptor-δ (PPAR)-δ (red) and NeuN (green), a neuronal marker, revealed co-localization in SCI tissue in almost all neurons in spared gray matter (SGM). Image is from 14-dpi tissue, 2.25 mm caudal to epicenter. Scale bar = 20 µm in C (applies to A–C). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
DISCUSSION
The main finding of this work is that NG2 cells and oligodendrocytes expressing the transcription factor PPAR-δ increase after SCI and correlate in time and location to tissue regions known to exhibit robust oligodendrogenesis. Studies from our lab and others examining the dynamic nature of cell death and replacement in spinal cord injured tissue show that oligodendrocytes are lost early and then replenished in the first 2 weeks following injury (Zai and Wrathall, 2005; Horky et al., 2006; Tripathi and McTigue, 2007). Formation of new oligodendrocytes is especially pronounced in peri-lesion tissue, in which they vastly outnumber oligodendrocytes present in outlying spared tissue and in naïve white matter (Tripathi and Mc-Tigue, 2007). These new oligodendrocytes are likely derived from NG2+ progenitor cells that undergo robust proliferation during the first week post injury and accumulate in tissue adjacent to the lesion (McTigue et al., 1998, 2001; Tripathi and McTigue, 2007; Yoo and Wrathall, 2007).
Oligodendrocyte genesis and survival are regulated and influenced by multiple external and internal signaling molecules. Several studies have detected upregulation of growth factors after SCI that may positively influence oligodendrocyte survival and/or generation. For example, neurotrophin-3, brain-derived neurotrophic factor, glial growth factor, fibroblast growth factor-2 (FGF-2), and ciliary neurotrophic factor (CNTF) are all elevated in the vicinity of SCI lesions (Koshinaga et al., 1993; Mocchetti et al., 1996; Widenfalk et al., 2001; Ikeda et al., 2001; Zai et al., 2005; Talbott et al., 2007; Tripathi and McTigue, 2008). In addition to extracellular growth factors, intracellular signals play a key role in regulating oligodendrocyte lineage cell proliferation and differentiation. For example, the transcription factors Olig-1 and Olig-2 are integral for normal oligodendrocyte development (Lu et al., 2001; Talbott et al., 2005).
Here we examined expression of a transcription factor from a different molecular family, the peroxisome proliferator-activated receptors (PPARs). Although the range of functions of these molecules is still under investigation, one known role for PPARs is regulating expression of genes involved in fatty acid metabolism and cholesterol/lipid uptake and efflux (Oliver et al., 2001;Schmuth et al., 2004). Because oligodendrocytes have a clear need for lipid production and metabolism during myelin synthesis, a prominent role for PPARs in oligodendrocyte function is conceivable. In fact, previous work has shown that PPAR-δ is present in oligodendrocytes (and neurons) in the adult CNS and that PPAR-δ activation promotes oligodendrocyte differentiation and myelin membrane formation in vitro (Granneman and others, 1998; Saluja et al., 2001). As a first step to understanding the function of PPAR-δ in the injured CNS, we characterized the spatial and temporal expression of PPAR-δ after SCI to determine whether it correlates with known regions of robust oligodendrogenesis.
Our data reveal that despite the total loss of PPAR-δ+ neurons at the injury site, PPAR-δ mRNA and protein are maintained early after SCI and then increase over time along the rostral-caudal extent of the lesion. In WM lesion borders, PPAR-δ+ cell counts were elevated by 7 dpi and peaked at 14 dpi, at which time they were significantly greater than naïve WM. Intriguingly, the numbers dropped back to control levels by 28 dpi. A similar pattern was observed in the GM lesion border, where the PPAR-δ+ cell numbers dropped at 1 dpi, returned to naïve levels at 7–14 dpi, and then dropped again to less than control values at 28 dpi. Whereas overall PPAR-δ cell numbers appear to decline in lesion borders between 2 and 4 weeks post injury, PPAR-δ mRNA rises significantly at 28 dpi, suggesting that a subsequent increase in PPAR-δ expression may occur.
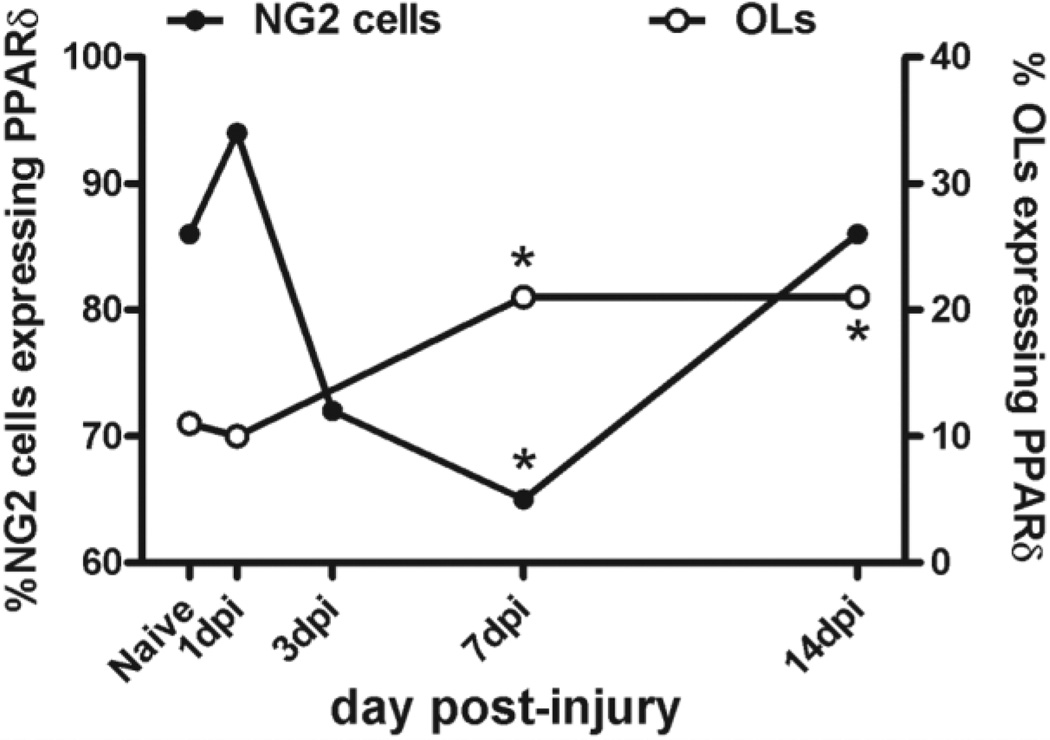
We next determined whether the PPAR-δ+ cell population present after SCI included NG2+ progenitor cells or oligodendrocytes. In uninjured tissue, ~10% of oligodendrocytes and 86% of NG2 cells express PPAR-δ, implicating a constitutive role of PPAR-δ in these cells. By 1 day after injury, the total number of NG2 cells had doubled, and 94% of these cells expressed PPAR-δ. This increase in expression concomitant with increased cell numbers indicates that PPAR-δ is expressed by newly formed and/or infiltrating NG2 cells. Overall NG2 cell counts continued to climb between 1 and 7 dpi, whereas the percentage of double-labeled cells dropped to ~65%. The number of oligodendrocytes also rose during this time; in contrast to NG2 cells, however, the percentage of oligodendrocytes expressing PPAR-δ more than doubled between 1 and 7 dpi to levels significantly greater than 1 dpi or naïve levels. Because new oligodendrocytes are thought to arise from proliferating NG2+ progenitors, a plausible explanation for the timing displayed in these results is that some PPAR-δ+ NG2 cells arising early after SCI differentiated into new PPAR-δ+ oligodendrocytes, which would account for the drop in the ratio of PPAR-δ+ NG2 cells at 7 dpi (see Fig. 8 for comparison of timing of PPAR-δ+ expression in NG2 cells and oligodendrocytes). This idea is supported by our data showing that some PPAR-δ+ oligodendrocytes were also labeled with BrdU, indicating that these are new cells that arose after injury.
Figure 8.
Comparison of the percentage of NG2 cells and oligodendrocytes (OLs) expressing peroxisome proliferator-activated receptor-δ (PPAR)-δ over time after SCI. A high percentage of NG2 cells express PPAR-δ in the normal spinal cord (86%). This proportion dropped significantly at 7 dpi compared with naïve and then returned to near normal levels by 14 dpi. In contrast, the ratio of OLs with PPAR-δ+ nuclei was ~11% in uninjured tissue and approximately double at 7 and 14 dpi. *, P < 0.05 vs. naïve.
By 14 dpi, the total number of NG2 cells was lower than at 7 dpi and the percentage of the remaining cells expressing PPAR-δ returned to baseline levels. In contrast, the number of oligodendrocytes continued to rise between 7 and 14 dpi, which is in accordance with our previous data showing ongoing oligogenesis during this time (Tripathi and McTigue, 2007). The percentage of PPAR-δ+ oligodendrocytes at 14 dpi remained at 21%, suggesting that newly formed oligodendrocytes maintained PPAR-δ expression at this time.
Although it is possible that the detected changes in PPAR-δ+ cell numbers reflect general changes in cell density after SCI due to tissue swelling and shrinkage over time, we feel this is unlikely to be the only or even major reason PPAR-δ+ cell numbers rose/fell after SCI. Indeed, counts of the total cell number (counting all nuclei within each sampled region) revealed a different overall pattern of cell distribution changes compared with that for PPAR-δ+ cells (see Suppl. Fig. 1). In addition, changes within PPAR-δ+ cells within a particular area were often different depending on the cellular phenotype examined. For instance, at 7 dpi, PPAR-δ+ NG2 cells were significantly increased in gray matter, whereas PPAR-δ+ oligodendrocyte numbers in the same region were unchanged. Therefore, changes in PPAR-δ+ cell numbers likely reflect changes within specific subpopulations of cells and not an overall increase or decrease in all cells due to tissue shrinkage or swelling.
When the spatial distribution of PPAR-δ+ cells was examined in detail, it was clear that most PPAR-δ+ NG2 cells and oligodendrocytes were located along lesion borders, particularly in white matter adjacent to the lesion. Interestingly, PPAR-δ+ NG2 cells peaked in this region at 7 dpi whereas PPAR-δ+ oligodendrocytes were highest 1 week later. This again supports the notion that PPAR-δ+ progenitors gave rise to oligodendrocytes expressing PPAR-δ in this region. We have previously shown that the lesion border area, typically referred to as the glial scar, is a zone of active proliferation and gliogenesis after SCI (Tripathi and McTigue, 2007). We have further illustrated that the oligogenic growth factors CNTF and FGF-2 are prominently upregulated along SCI lesion borders, suggesting a potential role for these factors in post-SCI gliogenesis (Tripathi and McTigue, 2007). Notably, CNTF and FGF-2 can upregulate PPAR-α and PPAR-γ expression, respectively, and PPAR-γ can potentiate the effect of FGF-2 on osteoblasts (Neubauer et al., 2004; Yasuda et al., 2005; Ligon et al., 2006; Liu et al., 2007; Kakudo et al., 2007). Therefore, it is feasible that interactions between these growth factors and PPAR-δ may also exist, especially considering they show a similar distribution and time course of expression after SCI.
The role PPAR-δ plays in postinjury oligodendrocyte function is not yet known. However, logical directions for future studies may be gleaned from its role in other studies. For instance, PPAR-δ agonist treatment in an animal model of multiple sclerosis led to reduced proinflammatory signals and lesion load revealing anti-inflammatory and neuroprotective characteristics for PPAR-δ (Polak et al., 2005). The same study also noted increased expression of the myelin gene proteolipid protein (PLP) (Polak et al., 2005), which is in accordance with previous work showing that the PLP gene contains a peroxisome-proliferator response element (PPRE) to which PPAR-δ can bind and promote expression (Saluja et al., 2001). As stated above, PPAR-δ activation in cultured oligodendrocytes promotes process outgrowth and myelin sheet formation (Granneman et al., 1998). Collectively, these data suggest that activation of PPAR-δ in the injured and/or demyelinated CNS may promote oligodendrocyte differentiation and remyelination. Considering that endogenous ligands for PPAR-δ are thought to include fatty acids such as linoleic acid (Moya-Camarena et al., 1999), PPAR-δ may be a prime candidate for therapeutic approaches targeting demyelination and oligodendrocyte loss.
A role for PPAR-δ in cell differentiation after SCI is also supported by results from other models. For instance, PPAR-δ expression during rat embryonic development is high in the CNS and peaks at E11.5, correlating to the time of greatest differentiation, especially in the CNS (Braissant and Wahli, 1998). Furthermore, a role of PPAR-δ in cellular differentiation has been demonstrated in skin keratinocytes, in which PPAR-δ null mice display unrestricted skin proliferation resulting in enhanced skin carcinogenesis (Kim et al., 2006). Accordingly, administration of the PPAR-δ agonist GW0742 upregulated the expression of skin differentiation markers, making PPAR-δ an attractive target for cancer studies (Kim et al., 2006). Similar reports have corroborated a role for PPAR-δ in differentiation of adipocytes (Matsusue et al., 2004), trophoblasts (Nadra et al., 2006), and endothelial cells (Muller-Brusselbach et al., 2007). PPAR-δ also has been reported to constrain endothelial tumor genesis via regulation of the cell cycle inhibitor p57KIP2. Notably, recent work demonstrated a role for p57KIP2 in oligodendrocyte differentiation (Dugas et al., 2007), suggesting a possible common mechanism.
In the adult rat, the presence of PPAR-δ in the CNS is localized to oligodendrocytes and neurons in the spinal cord and other CNS structures such as the cerebellum, hippocampus, and thalamus (Woods et al., 2003; Benani et al., 2004). The continued expression of PPAR-δ in adult neurons in the intact CNS may provide ongoing baseline neuroprotection. For instance, PPAR-δ agonists protect neurons treated with amyloid β in vitro by promoting glutathione synthesis and curbing oxidative damage (Madrigal et al., 2007). PPAR-δ activation also reduces cell death of cerebellar granule neurons induced by low-KCl media (Smith et al., 2004). Reports from in vivo studies using models of ischemia reperfusion and Parkinson’s disease also have shown neuroprotection conferred by PPAR-δ (Iwashita et al., 2007). Thus, the persistent expression of PPAR-δ in adult neurons may serve to promote baseline neuronal health as well as protect neurons surviving a nearby CNS insult such as SCI.
In summary, this study reports the novel finding that PPAR-δ is significantly upregulated for a protracted time after SCI. It is expressed by oligodendrocyte lineage cells and its presence in newly formed oligodendrocytes suggests it may play a role in oligodendrocyte differentiation as it does in other cell populations. Thus, this work sets the stage for ongoing and future studies that will focus on determining the specific role of PPAR-δ after SCI using therapeutic approaches as well as mice deficient in PPAR-δ expression.
Supplementary Material
TABLE 1.
Primary Antibodies Used in This Study
| Antibody | Manufacturer | Cat. no. | Immunogen | Species |
|---|---|---|---|---|
| PPAR-δ | Cayman Chemicals, Ann Arbor, MI | 101720 | The antibody was raised against human PPAR-δ amino acids 39–54 (SSSYTDLSRSSSPPSL) | Rabbit |
| NG2 | US Biological, Swamscott, MA | C5067-70F | Cell line expressing truncated rat NG2 | Mouse |
| CC1/APC | Abcam, Cambridge, MA | Ab16794 | Recombinant human APC, amino acid residues 1–226 | Mouse |
| NeuN | Chemicon, Billerica, MA | MAB377 | Purified cell nuclei from mouse brain | Mouse |
| BrdU | Developmental Studies Hybridoma Bank, Iowa City, Iowa | G3G4 | Mouse BrdU was derived from mouse myeloma cell line generated by the immunogen (BrdUrd)16-BSA | Mouse |
| α-Tubulin | Abcam, Cambridge, MA | Ab7291 | Full-length native protein of chicken α-tubulin (from brain) | Mouse |
Abbreviations: PPAR-δ, peroxisome proliferator-activated receptor-δ; BrdU, bromodeoxyuridine; BSA, bovine serum albumin.
ACKNOWLEDGMENTS
The authors gratefully acknowledge Dr. Jeffery Peters for the generous gift of the PPAR-δ antibody and the COS cell lysate (positive control). We thank Ping Wei and A. Todd Lash for excellent technical assistance and Dr. Daniel Ankeny and David Schonberg for critical review of the manuscript. We also thank Dr. John Gensel for help in automated data analysis of some confocal images. The BrdU antibody developed by S.J. Kaufman was obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 552242.
Grant sponsor: the National Institute of Neurological Disorders and Stroke; Grant numbers: NS059776 and P30-NS045758; Grant sponsor: The Christopher and Dana Reeve Foundation; Grant number: MA1-0702-2.
Footnotes
Additional Supporting Information may be found in the online version of this article.
LITERATURE CITED
- Benani A, Heurtaux T, Netter P, Minn A. Activation of peroxisome proliferator-activated receptor alpha in rat spinal cord after peripheral noxious stimulation. Neurosci Lett. 2004;369:59–63. doi: 10.1016/j.neulet.2004.07.056. [DOI] [PubMed] [Google Scholar]
- Bhat RV, Axt KJ, Fosnaugh JS, Smith KJ, Johnson KA, Hill DE, Kinzler KW, Baraban JM. Expression of the APC tumor supressor protein in oligodendroglia. Glia. 1996;17:169–174. doi: 10.1002/(SICI)1098-1136(199606)17:2<169::AID-GLIA8>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Braissant O, Wahli W. Differential expression of peroxisome proliferator-activated receptor-alpha,-beta, and -gamma during rat embryonic development. Endocrinology. 1998;139:2748–2754. doi: 10.1210/endo.139.6.6049. [DOI] [PubMed] [Google Scholar]
- Coggeshall RE, Lekan HA. Methods for determining numbers of cells and synapses: a cse for more uniform standards of review. J Comp Neurol. 1996;364:6–15. doi: 10.1002/(SICI)1096-9861(19960101)364:1<6::AID-CNE2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Crowe MJ, Bresnahan JC, Shuman SL, Masters JN, Beattie MS. Apoptosis and delayed degeneration after spinal cord injury in rats and monkeys [published erratum appears in Nat Med 1997;3:240] Nat Med. 1997;3:73–76. doi: 10.1038/nm0197-73. [DOI] [PubMed] [Google Scholar]
- Dugas JC, Ibrahim A, Barres BA. A crucial role for p57(Kip2) in the intracellular timer that controls oligodendrocyte differentiation. J Neurosci. 2007;27:6185–6196. doi: 10.1523/JNEUROSCI.0628-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss B, Mallon B, Phan T, Ohlemeyer C, Kirchoff F, Nishiyama A, Macklin WB. Purification and analysis of in vivo-differentiated oligodendrocytes expressing the green fluorescent protein. Dev Biol. 2000;218:259–274. doi: 10.1006/dbio.1999.9574. [DOI] [PubMed] [Google Scholar]
- Grako KA, Ochiya T, Barritt D, Nishiyama A, Stallcup WB. PDGF (alpha)-receptor is unresponsive to PDGF-AA in aortic smooth muscle cells from the NG2 knockout mouse. J Cell Sci. 1999;112:905–915. doi: 10.1242/jcs.112.6.905. [DOI] [PubMed] [Google Scholar]
- Granneman J, Skoff R, Yang X. Member of the peroxisome proliferator-activated receptor family of transcription factors is differentially expressed by oligodendrocytes. J Neurosci Res. 1998;51:563–573. doi: 10.1002/(SICI)1097-4547(19980301)51:5<563::AID-JNR3>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Grossman SD, Rosenberg LJ, Wrathall JR. Temporal-spatial pattern of acute neuronal and glial loss after spinal cord contusion. Exp Neurol. 2001;168:273–282. doi: 10.1006/exnr.2001.7628. [DOI] [PubMed] [Google Scholar]
- Guillery RW, Herrup K. Quantification without pontification: choosing a method for counting objects in sectioned tissues. J Comp Neurol. 1997;386:2–7. doi: 10.1002/(sici)1096-9861(19970915)386:1<2::aid-cne2>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Hall MG, Quignodon L, Desvergne B. Peroxisome proliferator-activated receptor beta/delta in the brain: facts and hypothesis. PPAR Res. 2008;2008:780452. doi: 10.1155/2008/780452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SW, Ritzenthaler JD, Zheng Y, Roman J. PPAR-β/δ agonist stimulates human lung carcinoma cell growth through inhibition of PTEN expression: the involvement of P13K and NF-κB signals. Am J Physiol Lung Cell Mol Physiol. 2008;294:L1238–L1249. doi: 10.1152/ajplung.00017.2008. [DOI] [PubMed] [Google Scholar]
- Hihi AK, Michalik L, Wahli W. PPARs: transcriptional effectors of fatty acids and their derivatives. Cell Mol Life Sci. 2002;59:790–798. doi: 10.1007/s00018-002-8467-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horky LL, Galimi F, Gage FH, Horner PJ. Fate of endogenous stem/progenitor cells following spinal cord injury. J Comp Neurol. 2006;498:525–538. doi: 10.1002/cne.21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda O, Murakami M, Ino H, Yamazaki M, Nemoto T, Koda M, Nakayama C, Moriya H. Acute up-regulation of brain-derived neurotrophic factor expression resulting from experimentally induced injury in the rat spinal cord. Acta Neuropathol. 2001;102:239–245. doi: 10.1007/s004010000357. [DOI] [PubMed] [Google Scholar]
- Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- Iwashita A, Muramatsu Y, Yamazaki T, Muramoto M, Kita Y, Yamazaki S, Mihara K, Moriguchi A, Matsuoka N. Neuroprotective efficacy of the peroxisome proliferator-activated receptor delta-selective agonists in vitro and in vivo. J Pharmacol Exp Ther. 2007;320:1087–1096. doi: 10.1124/jpet.106.115758. [DOI] [PubMed] [Google Scholar]
- Kakudo N, Shimotsuma A, Kusumoto K. Fibroblast growth factor-2 stimulates adipogenic differentiation of human adipose-derived stem cells. Biochem Biophys Res Commun. 2007;359:239–244. doi: 10.1016/j.bbrc.2007.05.070. [DOI] [PubMed] [Google Scholar]
- Kim DJ, Bility MT, Billin AN, Willson TM, Gonzalez FJ, Peters JM. PPARbeta/delta selectively induces differentiation and inhibits cell proliferation. Cell Death Differ. 2006;13:53–60. doi: 10.1038/sj.cdd.4401713. [DOI] [PubMed] [Google Scholar]
- Koshinaga M, Sanon HR, Whittemore SR. Altered acidic and basic fibroblast growth factor expression following spinal cord injury. Exp Neurol. 1993;120:32–48. doi: 10.1006/exnr.1993.1038. [DOI] [PubMed] [Google Scholar]
- Kuhlmann T, Remington L, Maruschak B, Owens T, Bruck W. Nogo-A is a reliable oligodendroglial marker in adult human and mouse CNS and in demyelinated lesions. J Neuropathol Exp Neurol. 2007;66:238–246. doi: 10.1097/01.jnen.0000248559.83573.71. [DOI] [PubMed] [Google Scholar]
- Ligon KL, Kesari S, Kitada M, Sun T, Arnett HA, Alberta JA, Anderson DJ, Stiles CD, Rowitch DH. Development of NG2 neural progenitor cells requires Olig gene function. Proc Natl Acad Sci U S A. 2006;103:7853–7858. doi: 10.1073/pnas.0511001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares D, Taconis M, Mana P, Correcha M, Fordham S, Staykova M, Willenberg DO. Neuronal nitric oxide synthase plays a key role in CNS demyelination. J Neurosci. 2006;26:12672–12681. doi: 10.1523/JNEUROSCI.0294-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QS, Gao M, Zhu SY, Li SJ, Zhang L, Wang QJ, Du GH. The novel mechanism of recombinant human ciliary neurotrophic factor on the anti-diabetes activity. Basic Clin Pharmacol Toxicol. 2007;101:78–84. doi: 10.1111/j.1742-7843.2007.00092.x. [DOI] [PubMed] [Google Scholar]
- Lu QR, Cai L, Rowitch D, Cepko CL, Stiles CD. Ectopic expression of Olig1 promotes oligodendrocyte formation and reduces neuronal survival in developing mouse cortex. Nat Neurosci. 2001;4:973–974. doi: 10.1038/nn718. [DOI] [PubMed] [Google Scholar]
- Madrigal JL, Kalinin S, Richardson JC, Feinstein DL. Neuroprotective actions of noradrenaline: effects on glutathione synthesis and activation of peroxisome proliferator activated receptor delta. J Neurochem. 2007;103:2092–2101. doi: 10.1111/j.1471-4159.2007.04888.x. [DOI] [PubMed] [Google Scholar]
- Matsusue K, Peters JM, Gonzalez FJ. PPARbeta/delta potentiates PPARgamma-stimulated adipocyte differentiation. FASEB J. 2004;18:1477–1479. doi: 10.1096/fj.04-1944fje. [DOI] [PubMed] [Google Scholar]
- McTigue DM, Horner PJ, Stokes BT, Gage FH. Neurotrophin-3 and brain-derived neurotrophic factor induce oligodendrocyte proliferation and myelination of regenerating axons in the contused adult rat spinal cord. J Neurosci. 1998;18:5354–5365. doi: 10.1523/JNEUROSCI.18-14-05354.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTigue DM, Wei P, Stokes BT. Proliferation of NG2-positive cells and altered oligodendrocyte numbers in the contused rat spinal cord. J Neurosci. 2001;21:3392–3400. doi: 10.1523/JNEUROSCI.21-10-03392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTigue DM, Tripathi R, Wei P. NG2 colocalizes with axons and is expressed by a mixed cell population in spinal cord lesions. J Neuropathol Exp Neurol. 2006;65:406–420. doi: 10.1097/01.jnen.0000218447.32320.52. [DOI] [PubMed] [Google Scholar]
- Mocchetti I, Rabin SJ, Colangelo AM, Whittemore SR, Wrathall JR. Increased basic fibroblast growth factor expression following contusive spinal cord injury. Exp Neurol. 1996;141:154–164. doi: 10.1006/exnr.1996.0149. [DOI] [PubMed] [Google Scholar]
- Moya-Camarena SY, Van den Heuvel JP, Belury MA. Conjugated linoleic acid activates peroxisome proliferator-activated receptor alpha and beta subtypes but does not induce hepatic peroxisome proliferation in Sprague-Dawley rats. Biochim Biophys Acta. 1999;1436:331–342. doi: 10.1016/s0005-2760(98)00121-0. [DOI] [PubMed] [Google Scholar]
- Muller-Brusselbach S, Komhoff M, Rieck M, Meissner W, Kaddatz K, Adamkiewicz J, Keil B, Klose KJ, Moll R, Burdick AD, Peters JM, Muller R. Deregulation of tumor angiogenesis and blockade of tumor growth in PPARbeta-deficient mice. EMBO J. 2007;26:3686–3698. doi: 10.1038/sj.emboj.7601803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutti E, Veber D, Stampachiacchiere B, Triaca V, Gammella E, Tacchini L, Aloe L, Scalabrino G. Cobalamin deficiency-induced downregulation of p75-immunoreactive cell levels in rat central nervous system. Brain Res. 2007;1157:92–99. doi: 10.1016/j.brainres.2007.04.055. [DOI] [PubMed] [Google Scholar]
- Nadra K, Anghel SI, Joye E, Tan NS, Basu-Modak S, Trono D, Wahli W, Desvergne B. Differentiation of trophoblast giant cells and their metabolic functions are dependent on peroxisome proliferator-activated receptor beta/delta. Mol Cell Biol. 2006;26:3266–3281. doi: 10.1128/MCB.26.8.3266-3281.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer M, Fischbach C, Bauer-Kreisel P, Lieb E, Hacker M, Tessmar J, Schulz MB, Goepferich A, Blunk T. Basic fibroblast growth factor enhances PPARgamma ligand-induced adipogenesis of mesenchymal stem cells. FEBS Lett. 2004;577:277–283. doi: 10.1016/j.febslet.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Oliver WR, Jr, Shenk JL, Snaith MR, Russell CS, Plunket KD, Bodkin NL, Lewis MC, Winegar DA, Sznaidman ML, Lambert MH, Xu HE, Sternbach DD, Kliewer SA, Hansen BC, Willson TM. A selective peroxisome proliferator-activated receptor delta agonist promotes reverse cholesterol transport. Proc Natl Acad Sci U S A. 2001;98:5306–5311. doi: 10.1073/pnas.091021198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Lee SS, Li W, Ward JM, Gavrilova O, Everett C, Reitman ML, Hudson LD, Gonzalez FJ. Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor beta-(delta) Mol Cell Biol. 2000;20:5119–5128. doi: 10.1128/mcb.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak PE, Kalinin S, Dello RC, Gavrilyuk V, Sharp A, Peters JM, Richardson J, Willson TM, Weinberg G, Feinstein DL. Protective effects of a peroxisome proliferator-activated receptor-beta/delta agonist in experimental autoimmune encephalomyelitis. J Neuroimmunol. 2005;168:65–75. doi: 10.1016/j.jneuroim.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Qi C, Zhu Y, Reddy JK. Peroxisome proliferator-activated receptors, coactivators, and downstream targets. Cell Biochem Biophys. 2000;32:187–204. doi: 10.1385/cbb:32:1-3:187. [DOI] [PubMed] [Google Scholar]
- Rabchevsky AG, Sullivan PG, Scheff SW. Temporal-spatial dynamics in oligodendrocyte and glial progenitor cell numbers throughout ventrolateral white matter following contusion spinal cord injury. Glia. 2007;55:831–843. doi: 10.1002/glia.20508. [DOI] [PubMed] [Google Scholar]
- Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- Saluja I, Granneman JG, Skoff RP. PPAR delta agonists stimulate oligodendrocyte differentiation in tissue culture. Glia. 2001;33:191–204. [PubMed] [Google Scholar]
- Schmittgen TD, Lival KJ. Analyzing real-time PCR data by the comparative CT method. Nature protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Schmuth M, Haqq CM, Cairns WJ, Holder JC, Dorsam S, Chang S, Lau P, Fowler AJ, Chuang G, Moser AH, Brown BE, Mao-Qiang M, Uchida Y, Schoonjans K, Auwerx J, Chambon P, Willson TM, Elias PM, Feingold KR. Peroxisome proliferator-activated receptor (PPAR)-beta/delta stimulates differentiation and lipid accumulation in keratinocytes. J Invest Dermatol. 2004;122:971–983. doi: 10.1111/j.0022-202X.2004.22412.x. [DOI] [PubMed] [Google Scholar]
- Smith SA, Monteith GR, Robinson JA, Venkata NG, May FJ, Roberts-Thomson SJ. Effect of the peroxisome proliferator-activated receptor beta activator GW0742 in rat cultured cerebellar granule neurons. J Neurosci Res. 2004;77:240–249. doi: 10.1002/jnr.20153. [DOI] [PubMed] [Google Scholar]
- Talbott JF, Loy DN, Liu Y, Qiu MS, Bunge MB, Rao MS, Whittemore SR. Endogenous Nkx2.2+/Olig2+ oligodendrocyte precursor cells fail to remyelinate the demyelinated adult rat spinal cord in the absence of astrocytes. Exp Neurol. 2005;192:11–24. doi: 10.1016/j.expneurol.2004.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbott JF, Cao Q, Bertram J, Nkansah M, Benton RL, Lavik E, Whittemore SR. CNTF promotes the survival and differentiation of adult spinal cord-derived oligodendrocyte precursor cells in vitro but fails to promote remyelination in vivo. Exp Neurol. 2007;204:485–489. doi: 10.1016/j.expneurol.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan NS, Icre G, Montagner A, Bordier-ten-Heggeler B, Wahli W, Michalik L. The nuclear hormone receptor peroxisome proliferator-activated receptor beta/delta potentiates cell chemotactism, polarization, and migration. Mol Cell Biol. 2007;27:7161–7175. doi: 10.1128/MCB.00436-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi R, McTigue DM. Prominent oligodendrocyte genesis along the border of spinal contusion lesions. Glia. 2007;55:698–711. doi: 10.1002/glia.20491. [DOI] [PubMed] [Google Scholar]
- Tripathi RB, McTigue DM. Chronically increased ciliary neurotrophic factor and fibroblast growth factor-2 expression after spinal contusion in rats. J Comp Neurol. 2008;510:129–144. doi: 10.1002/cne.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widenfalk J, Lundstromer K, Jubran M, Brene S, Olson L. Neurotrophic factors and receptors in the immature and adult spinal cord after mechanical injury or kainic acid. J Neurosci. 2001;21:3457–3475. doi: 10.1523/JNEUROSCI.21-10-03457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods JW, Tanen M, Figueroa DJ, Biswas C, Zycband E, Moller DE, Austin CP, Berger JP. Localization of PPARdelta in murine central nervous system: expression in oligodendrocytes and neurons. Brain Res. 2003;975:10–21. doi: 10.1016/s0006-8993(03)02515-0. [DOI] [PubMed] [Google Scholar]
- Yasuda E, Tokuda H, Ishisaki A, Hirade K, Kanno Y, Hanai Y, Nakamura N, Noda T, Katagiri Y, Kozawa O. PPARgamma ligands up-regulate basic fibroblast growth factor-induced VEGF release through amplifying SAPK/JNK activation in osteoblasts. Biochem Biophys Res Commun. 2005;328:137–143. doi: 10.1016/j.bbrc.2004.12.163. [DOI] [PubMed] [Google Scholar]
- Yoo S, Wrathall JR. Mixed primary culture and clonal analysis provide evidence that NG2 proteoglycan-expressing cells after spinal cord injury are glial progenitors. Dev Neurobiol. 2007;67:860–874. doi: 10.1002/dneu.20369. [DOI] [PubMed] [Google Scholar]
- Zai LJ, Wrathall JR. Cell proliferation and replacement following contusive spinal cord injury. Glia. 2005;50:247–257. doi: 10.1002/glia.20176. [DOI] [PubMed] [Google Scholar]
- Zai LJ, Yoo S, Wrathall JR. Increased growth factor expression and cell proliferation after contusive spinal cord injury. Brain Res. 2005;1052:147–155. doi: 10.1016/j.brainres.2005.05.071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.