Abstract
Background: Single nucleotide polymorphisms (SNPs) near thyroid transcription factor genes (FOXE1 rs965513/NKX2-1 rs944289) have been shown to be associated with differentiated thyroid cancer (DTC) in Caucasoid populations. We investigated the role of those SNPs in German patients with DTC and also extended our analysis to tumor stages and lymphocytic infiltration of the tumors (ITL).
Methods: Patients with DTC (n=243; papillary, PTC; follicular, FTC) and healthy controls (HC; n=270) were analyzed for the rs965513 and rs944289 SNPs.
Results: The case-control analysis for rs965513 SNP showed that the genotypes “AA,” “AG,” and minor allele “A” were more frequent in patients with DTC than in HC (pronounced in PTC pgenotype=0.000084, pallele=0.006 than FTC pgenotype=0.29 and pallele=0.06). Furthermore, subgroup analysis of the DTC patients stratified for primary tumor stage (T1–T2, T3–T4), the absence or presence of regional lymph node metastases (N0, N1), for distant metastases (M0, M1), as well as for ITL, showed an association of rs965513 with stages T1–T2, T1–T3, N1, and absence of ITL. The NKX2-1 SNP rs944289, however, was not associated with DTC.
Conclusion: Our results confirm that the FOXE1 rs965513 SNP confers an increased risk for DTC in the German population, particularly allele “A” and the genotypes “AA” and “AG” for PTC. This increased risk was also observed in advanced tumor stages and absence of ITL, which may reflect the course of a more aggressive disease. The NKX2-1 rs944289 SNP, however, appears to play a secondary role in the development of DTC in the German population.
Introduction
The majority of malignant thyroid tumors derive from follicular epithelial cells. The differentiated thyroid cancers (DTC) are papillary (PTC; ∼85% of cases) and follicular (FTC; ∼10% of cases) subtypes (1), and their etiology is still not well characterized. The transformation of normal into malignant tissue is a multifactorial process involving genetic and environmental factors such as radiation exposure (2). Some environmental factors such as hepatitis C infection or late and multiple pregnancies have been suggested as risk factors, but they have not been confirmed (3,4). Moreover, genetic components—for example proto-oncogenes, which encode receptor tyrosine kinases (RET) and small GTPases (RAS)—may contribute to thyroid cancer formation in familial syndromes (5). However, none of these factors is thyroid specific, and therefore the identification of additional candidate genes is important for a better understanding of the pathogenesis of thyroid cancer. In this context, key genes involved in thyroid organogenesis, for example the thyroid transcription factors (TTFs), have been identified as genetic susceptibility factors of various thyroid conditions. This illustrates the pivotal function of these genes in the development of thyroid diseases (6). Recently, three genome wide association (GWA) studies demonstrated associations of single nucleotide polymorphisms (SNPs) located near the TTF-1 (also known as thyroid-specific enhancer-binding protein: NKX2-1, rs944289) and TTF-2 (also known as forkhead box protein: FOXE1, rs965513) loci with an increased risk of sporadic thyroid carcinoma in Icelandic, British, and Japanese populations (7–9). Radiation-related PTC of Belarusian patients showed the strongest association only with the FOXE1 gene (10). In order to elucidate the role of TTFs as susceptibility genes in DTC of the German population, in the present case-control study, we investigated the SNPs near FOXE1 and NKX2-1 genes (rs965513 and rs944289 respectively). In addition, we explored whether the tumor stage and an immune-cell infiltration in the thyroid gland modified the risk of DTC.
Subjects and Methods
Subjects
In total, 243 patients (160 females, 83 males) with a pathologically confirmed diagnosis of DTC (196 papillary: 130 females and 66 males; 47 follicular: 30 females and 17 males) and known tumor node metastasis (TNM) stage were recruited from the Department of Nuclear Medicine at the University Hospital Frankfurt am Main, Germany. Healthy controls (HC; n=270, 113 females, 157 males) were volunteer blood donors from the staff personnel or medical students from the University Hospital Frankfurt am Main, Germany. All participants were of Caucasian (mostly German) origin and inhabitants of the area surrounding Frankfurt am Main, Germany. The median age of the patients with DTC and HC was 55 years (range 14–92 years) and 40 years (range 23–101 years) respectively. The study protocol was approved by the Ethics Committee of the University Hospital Frankfurt am Main, and informed consent was obtained from all participants.
SNP selection
In total, two SNPs (rs965513 and rs944289) previously associated with thyroid cancer susceptibility in the Icelandic population were investigated (7). While the rs965513 variant (A/G; Chromosome 9) is located ∼59 kb centromeric to the FOXE1 gene, the rs944289 variant (C/T; Chromosome 14) is situated 337 kb telomeric to the NKX2-1 gene. The SNP positions are given according to the National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov).
Genotyping
Genomic DNA from whole blood containing ethylenediaminetetraacetic acid was isolated by salting out procedure (11) and subjected to real time polymerase chain reaction (PCR). For the genotype analysis, the PCR reaction was carried out in 25 μL containing 3 μL (60 ng) DNA template, 6.25 μL ABsoluteTMQPCR mix (Abgene®; Thermo Fischer Scientific, Schwerte, Germany), 15.45 μL H2O, and 0.3 μL Taqman assay (FOXE1 rs965513/C_1593670_20, or NKX2-1 rs944289/ C_1444137_10), and analyzed in an ABI 7300 PCR system (Applied Biosystems, Darmstadt, Germany). The conditions were denaturation at 95°C for 10 min followed by 50 cycles at 92°C for 50 s and 60°C for 1 min. In order to confirm the accuracy of the method, random samples of the FOXE1 rs965513 and the NKX2-1 rs944289 SNPs were genotyped twice with a concordance of 100%.
Lymphocytic infiltration and thyroid antibodies
Fifty-five tissues from patients with DTC were examined for lymphocyte infiltration using the alkaline phosphate anti-alkaline phosphatase (APPAP) method (12). Additionally, thyroglobulin (TG) and thyroid peroxidase (TPO) antibodies (Abs) were measured using an enzyme-linked immunosorbent assay (Phadia, Freiburg, Germany). Concentrations >100 UI/mL for thyroid Abs were defined as positive.
Statistical analysis
Deviation from Hardy–Weinberg equilibrium and differences in genotype and allele distributions between groups were evaluated by chi-square test (BiAS software, package 9.08; Epsilon, Weinheim, Germany). The odds ratio (OR) and its confidence interval [CI] were estimated by unconditional logistic regression as a measure of the associations between genotypes or alleles and thyroid cancer risk. In order to elucidate the role of TTFs as susceptibility genes in DTC of the German population, the comparison of the genotype/allele frequencies between cases and controls were analyzed for each cancer group separately (DTC/PTC/FTC vs. HC respectively, DTCfemale/PTCfemale/FTCfemale vs. HCfemale respectively, and DTCmale/PTCmale/FTCmale vs. HCmale respectively). The observed p-values were corrected by multiplication with the number of genotypes(×3)/alleles(×2) tested. Furthermore, to address whether a certain risk genotype/allele influences the progression of DTC, the patients were classified in four groups with regard to the extent of the primary tumor (T), regional lymph node metastases (N), distant metastatic lesions (M), and thyroid lymphocytic infiltrate (ITL). The comparisons were performed within each of those groups (see Table 2) using the Mantel–Haenszel procedure. With a significance at pglobal≤0.05, a Multiple Mantel–Haenszel comparison was applied and corrected p-values (pc) were calculated using the Bonferroni method (13). The assumed significance level was ≤0.05. A power calculation for each group was performed assuming an allele frequency of 34.1% (rs965513 A) and 57.3% (rs944289 T) (7) and a type 1 error rate of 5%. On the basis of these assumptions, we estimate that we have more than 80% power to detect an allelic odds ratio of 1.44 (DTC), 1.47 (PTC), and 1.9 (FTC) for disease susceptibility in the case/control data set by using the program Power and Sample Size Calculations v3.0 (14).
Table 2.
Distribution of FOXE1 rs965513 Single Nucleotide Polymorphisms in Patients with Differentiated Thyroid Carcinoma According to the Extent of Primary Tumor and the Absence or Presence of Regional Lymph Node Metastases and Distant Metastatic Lesions
| Genotype [n (frequency)] | Allele [n (frequency)] | OR [CI] | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | n | AA | AG | GG | p | pc | A | G | OR [CI] A | OR [CI] G | p | pc |
| FOXE1 rs965513 | ||||||||||||
| HC | 270 | 41 (15.2%) | 110 (40.7%) | 119 (44.1%) | 192 (35.6%) | 348 (64.4%) | ||||||
| T1–T2 | 144 | 23 (16.0%) | 84 (58.3%) | 37 (25.7%) | 0.0075 | 0.02 | 130 (45.1%) | 158 (54.9%) | 1.49 [1.11–2.00] | 0.67 [0.50–0.90] | 0.0071 | 0.02 |
| T3–T4 | 66 | 13 (19.7%) | 36 (54.5%) | 17 (25.8%) | 0.02 | 0.06 | 62 (47.0%) | 70 (53.0%) | 1.61 [1.09–2.36] | 0.62 [0.42–0.92] | 0.01 | 0.03 |
| T1–T2 vs. T3–T4 | 0.70 | 2.10 | 0.72 | 2.16 | ||||||||
| HC | 270 | 41 (15.2%) | 110 (40.7%) | 119 (44.1%) | 192 (35.6%) | 348 (64.4%) | ||||||
| N0 | 71 | 12 (16.9%) | 38 (53.5%) | 21 (29.6%) | 0.09 | 0.27 | 62 (43.7%) | 80 (56.3%) | 1.40 [0.96–2.04] | 0.71 [0.49–1.04] | 0.08 | 0.24 |
| N1 | 78 | 10 (12.8%) | 53 (67.9%) | 15 (19.2%) | 0.01 | 0.03 | 73 (46.8%) | 83 (53.2%) | 1.59 [1.11–2.29] | 0.63 [0.44–0.90] | 0.01 | 0.03 |
| N0 vs. N1 | 0.54 | 1.62 | 0.59 | 1.77 | ||||||||
| HC | 270 | 41 (15.2%) | 110 (40.7%) | 119 (44.1%) | 192 (35.6%) | 348 (64.4%) | ||||||
| M0 | 40 | 8 (20.0%) | 21 (52.5%) | 11 (27.5%) | 0.08 | 0.24 | 37 (46.3%) | 43 (53.8%) | 1.56 [0.97–2.50] | 0.64 [0.40–1.03] | 0.06 | 0.18 |
| M1 | 18 | 4 (22.2%) | 11 (61.1%) | 3 (16.7%) | 0.05 | 0.15 | 19 (52.8%) | 17 (47.2%) | 2.03 [1.03–3.99] | 0.49 [0.25–0.97] | 0.04 | 0.12 |
| M0 vs. M1 | 0.49 | 1.47 | 0.52 | 1.56 | ||||||||
p-Values <0.05 are in bold type. Group T: p(global)genotype=0.0060, p(global)allele=0.0059; group N: p(global)genotype=0.02, p(global)allele=0.02; group M: p(global)genotype=0.04, p(global)allele=0.03.
Results
In our study, patients with DTC (n=243) had 81% papillary and 19% follicular subtypes, and there were more females (66%) than males (34%). All genotypes were in Hardy–Weinberg equilibrium (p>0.05) for the two investigated SNPs.
Genotype and allele analysis and DTC
The case-controls analysis for FOXE1 rs965513 SNP showed that the genotypes “AA” and “AG” (17.3% vs. 15.2% and 57.2% vs. 40.7% respectively) were more frequent in patients with DTC than in HC, while the genotype “GG” was less frequent (25.5% vs. 44.1%; pc=0.00014). Additionally, the allele “A” was observed more often in DTC patients than in HC (45.9% vs. 35.6%; OR 1.54 [CI 1.20–1.97], pc=0.0019). These differences in the genotype and allele distribution between patients and controls was more pronounced in the DTC histological PTC subtype (pc genotype=0.000084 and pc allele=0.006 respectively) than FTC (pc genotype=0.29 and pc allele=0.06 respectively). Subsequently, a sex-stratified analysis revealed differences for the FOXE1 rs965513 genotypes in female as well as in male DTC/PTC patients compared with their corresponding HC group (pc=0.01/pc=0.01 and pc=0.03/pc=0.007 respectively). This result was not confirmed in FTC (pc=0.33 and pc=0.55 respectively). Finally, the comparison of the allele distribution between patients and HC reached significant differences only in the DTC/PTC/FTC females but not in DTC/PTC/FTC males (pc=0.03/pc trend=0.08/pc trend=0.08 and pc=0.32/pc=0.32/pc=1.54 respectively). In contrast, the genotypes and alleles of the NKX2-1 rs944289 SNP showed no associations with DTC (Table 1). In order to detect a possible contribution of the NKX2-1 rs944289 SNP to DTC susceptibility, we analyzed its genotype combinations with FOXE1 rs965513 (presented in Supplementary Table S1; Supplementary Data are available online at www.liebert.com/thy), revealing nine different combinations. DTC patients carry the genotype combination “GG-rs965513/CT-rs944289” less often than HC (DTC, pc=0.0027; PTC, pc=0.02; FTC, pc=0.18). There were also significant differences between patients and controls in the FOXE1 rs965513 carrying the NKX2-1 rs944289 “CT” genotype, but not the NKX2-1 rs944289 “TT” or “CC” genotypes (Supplementary Table S2).
Table 1.
Distribution of FOXE1 rs965513 and NKX2-1 rs944289 Gene Single Nucleotide Polymorphisms in Patients with Differentiated Thyroid Cancer, Papillary Thyroid Cancer, and Follicular Thyroid Cancer Compared with Healthy Controls
| Genotype [n (frequency)] | Allele [n (frequency)] | OR [CI] | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | n | AA | AG | GG | p | pc | A | G | A | G | p | pc | |
| FOXE1 rs965513 | |||||||||||||
| All | HC | 270 | 41 (15.2%) | 110 (40.7%) | 119 (44.1%) | 192 (35.6%) | 348 (64.4%) | ||||||
| DTC | 243 | 42 (17.3%) | 139 (57.2%) | 62 (25.5%) | 0.000046 | 0.00014 | 223 (45.9%) | 263 (54.1%) | 1.54 [1.20–1.97] | 0.65 [0.51–0.84] | 0.00096 | 0.0019 | |
| PTC | 196 | 30 (15.3%) | 118 (60.2%) | 48 (24.5%) | 0.000028 | 0.000084 | 178 (45.4%) | 214 (54.6%) | 1.51 [1.16–1.97] | 0.66 [0.51–0.87] | 0.003 | 0.006 | |
| FTC | 47 | 12 (25.5%) | 21 (44.7%) | 14 (29.8%) | 0.097 | 0.29 | 45 (47.9%) | 49 (52.1%) | 1.66 [1.07–2.59] | 0.60 [0.39–0.93] | 0.03 | 0.06 | |
| Females | HC | 113 | 21 (18.6%) | 43 (38.1%) | 49 (43.4%) | 85 (37.6%) | 141 (62.4%) | ||||||
| DTC | 160 | 34 (21.3%) | 87 (54.4%) | 39 (24.4%) | 0.0034 | 0.01 | 155 (48.4%) | 165 (51.6%) | 1.56 [1.10–2.21] | 0.64 [0.45–0.91] | 0.01 | 0.03 | |
| PTC | 130 | 25 (19.2%) | 73 (56.2%) | 32 (24.6%) | 0.0051 | 0.01 | 123 (47.3%) | 137 (52.7%) | 1.49 [1.04–2.14] | 0.67 [0.47–0.97] | 0.04 | 0.08 | |
| FTC | 30 | 9 (30.0%) | 14 (46.7%) | 7 (23.3%) | 0.11 | 0.33 | 32 (53.3%) | 28 (46.7%) | 1.90 [1.07–3.37] | 0.53 [0.30–0.94] | 0.04 | 0.08 | |
| Males | HC | 157 | 20 (12.7%) | 67 (42.7%) | 70 (44.6%) | 107 (34.1%) | 207 (65.9%) | ||||||
| DTC | 83 | 8 (9.60%) | 52 (62.7%) | 23 (27.7%) | 0.01 | 0.03 | 68 (41.0%) | 98 (59.0%) | 1.34 [0.91–1.98] | 0.74 [0.51–1.10] | 0.16 | 0.32 | |
| PTC | 66 | 5 (7.60%) | 45 (68.2%) | 16 (24.2%) | 0.0023 | 0.007 | 55 (41.7%) | 77 (58.3%) | 1.38 [0.91–2.10] | 0.72 [0.48–1.10] | 0.16 | 0.32 | |
| FTC | 17 | 3 (17.6%) | 7 (41.2%) | 7 (41.2%) | 0.85 | 2.55 | 13 (38.2%) | 21 (61.8%) | 1.20 [0.58–2.49] | 0.84 [0.40–1.73] | 0.77 | 1.54 | |
| Genotype [n (frequency)] | Allele [n (frequency)] | OR [CI] | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | n | CC | CT | TT | p | pc | C | T | C | T | p | pc | |
| NKX2-1 rs944289 | |||||||||||||
| All | HC | 270 | 47 (17.4%) | 128 (47.4%) | 95 (35.2%) | 222 (41.1%) | 318 (58.9%) | ||||||
| DTC | 243 | 42 (17.3%) | 109 (44.9%) | 92 (37.9%) | 0.8 | 2.4 | 193 (39.7%) | 293 (60.3%) | 0.94 [0.73–1.21] | 1.06 [0.83–1.36] | 0.69 | 1.4 | |
| PTC | 196 | 34 (17.3%) | 93 (47.4%) | 69 (35.2%) | 1.0 | 3.0 | 161 (41.1%) | 231 (58.9%) | 1.00 [0.77–1.30] | 1.00 [0.77–1.30] | 0.95 | 1.9 | |
| FTC | 47 | 8 (17.0%) | 16 (34.0%) | 23 (48.9%) | 0.16 | 0.48 | 32 (34.0%) | 62 (66.1%) | 0.74 [0.47–1.17] | 1.35 [0.85–2.14] | 0.24 | 0.48 | |
| Females | HC | 113 | 21 (18.6%) | 56 (49.6%) | 36 (31.9%) | 98 (43.4%) | 128 (56.6%) | ||||||
| DTC | 160 | 30 (18.8%) | 70 (43.8%) | 60 (37.5%) | 0.58 | 1.74 | 130 (40.6%) | 190 (59.4%) | 0.89 [0.63–1.26] | 1.12 [0.79–1.58] | 0.58 | 1.16 | |
| PTC | 130 | 25 (19.2%) | 59 (45.4%) | 46 (35.4%) | 0.79 | 2.37 | 109 (41.9%) | 151 (58.1%) | 0.94 [0.66–1.35] | 1.06 [0.74–1.52] | 0.81 | 1.62 | |
| FTC | 30 | 5 (16.7%) | 11 (36.7%) | 14 (46.7%) | 0.30 | 0.9 | 21 (35.0%) | 39 (65.0%) | 0.70 [0.39–1.27] | 1.42 [0.79–2.57] | 0.31 | 0.62 | |
| Males | HC | 157 | 26 (16.6%) | 72 (45.9%) | 59 (37.6%) | 124 (39.5%) | 190 (60.5%) | ||||||
| DTC | 83 | 12 (14.5%) | 39 (47.0%) | 32 (38.6%) | 0.91 | 2.73 | 63 (38.0%) | 103 (62.0%) | 0.94 [0.64–1.38] | 1.07 [0.72–1.57] | 0.82 | 1.64 | |
| PTC | 66 | 9 (13.6%) | 34 (51.5%) | 23 (34.8%) | 0.72 | 2.16 | 52 (39.4%) | 80 (60.6%) | 1.00 [0.66–1.51] | 1.00 [0.66–1.52] | 0.93 | 1.86 | |
| FTC | 17 | 3 (17.6%) | 5 (29.4%) | 9 (52.9%) | 0.39 | 1.17 | 11 (32.4%) | 23 (67.6%) | 0.73 [0.35–1.56] | 1.37 [0.64–2.90] | 0.53 | 1.06 | |
p-Values <0.05 are in bold type.CI, confidence interval; DTC, differentiated thyroid cancer; FTC, follicular thyroid cancer; HC, healthy controls; OR, odds ratio; pc, corrected p-values; PTC, papillary thyroid cancer.
Genotype and allele analysis of FOXE1 rs965513 SNP in DTC according to tumor stages
The patients were grouped according to the extent of the primary tumor (T1–T2, T3–T4), the absence or presence of regional lymph node metastases (N0, N1), and the absence or presence of distant metastatic lesions (M0, M1). The genotypes “AA,” “AG,” or allele “A” were more frequent both in patients with T1–T2 as well as those with T3–T4 in comparison to controls (both pc genotype and pc allele=0.02; pc genotype=0.06 and pc allele=0.03 respectively). Also, the genotypes “AG” or allele “A” were only found more frequently in the group of patients with N1 compared to HC (both pc genotype and pc allele=0.03). In contrast, no association of the rs965513 SNP with M (M0: pc allele=0.18; M1: pc allele=0.12) was observed. The patient groups with advanced stages T3–T4, N1, or M1 had higher odds ratios in the presence of allele “A” (Table 2). However, the frequency of allele “A” in the advanced stage groups in comparison with the groups with a low/nonadvanced stage did not reach the level of significance (T3–T4: 47% vs. T1–T2: 45.1%; N1: 46.8% vs. N0: 43.7%; and M1: 52.8% vs. M0: 46.3%; pc allele=2.16, pc allele=1.77, pc allele=1.59 respectively).
Thyroid antibodies in DTC patients in relation to the FOXE1 rs965513 genotypes
In 121 DTC patients, thyroid antibodies (TPO-Ab/TG-Ab) levels were available. Among them, nine (7.4%) were positive (pos) and 112 (92.6%) negative (neg) for TPO-Ab and/or TG-Ab titers. A comparison of the rs965513 SNP in patients with TPO-Ab/TG-Abpos and TPO-Ab/TG-Abneg was performed and showed no differences between the groups (pgenotypes=0.79 and pallele=0.92). A subsequent analysis in which the median values of TPO-Ab and TG-Ab in DTC patients were compared revealed no differences between the various genotypes of the rs965513 SNP (TPO-Ab AA: 1 IU/mL, AG: 2 IU/mL, GG 3 IU/mL, pglobal=0.21; TG-Ab AA: 10 IU/mL, AG: 14 IU/mL, GG: 13 IU/mL, pglobal=0.37).
Lymphocytic infiltration in DTC in relation to FOXE1 rs965513 SNP
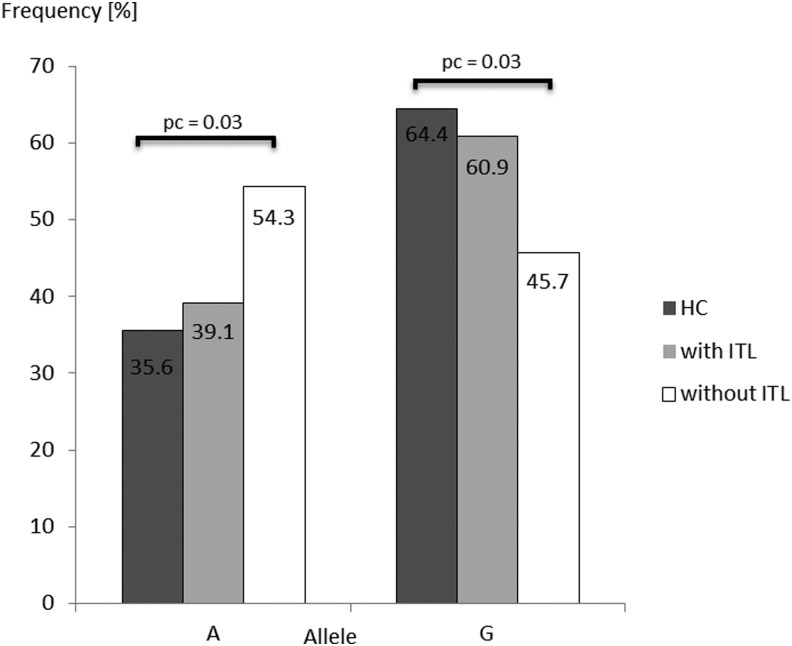
Histopathology reports were available for 55 patients with DTC (33 females, 22 males; median age 47 years). Among them, 58.2% showed evidence for a lymphocytic infiltrate. Among the 32 patients with lymphocytic infiltrates within the thyroid gland, 31% suffered from an autoimmune thyroid disease (nine Hashimoto's thyroiditis and one Graves' disease), and 6% were only positive for thyroid antibodies. Whether the lymphocyte infiltrate interacts with the allele risk of DTC was addressed by dividing into two groups—(i) absence or (ii) presence of lymphocytic infiltration—and comparing both groups with HC and among themselves (p(global)allele=0.04). Only DTC patients without lymphocytic infiltrate had the “A” allele significantly more often and the “G” allele less often compared to HC (54.3% vs. 35.6%, OR 2.16 [CI 1.18–3.96]; 45.7% vs. 64.4%, OR 0.46 [CI 0.25–0.85]; p=0.01, pc=0.03, respectively; Fig. 1). In contrast, patients with lymphocytic infiltration showed no differences in the allele distribution (“A” allele 39.1% vs. 35.6%, OR 1.16 [CI 0.68–1.98]; “G” allele, 60.9% vs. 64.4%, OR 0.86 [CI 0.51–1.41], p=0.58, pc=1.74). Furthermore, no difference in allele distribution was observed between patients with lymphocytic infiltration and those without (p=0.11, pc=0.33).
FIG. 1.
Comparison of allelic (A;G) distribution for FOXE1 rs965513 SNP in DTC patients with or without thyroid lymphocytic infiltration (ITL) and healthy controls (HC). p-Values<0.05 are shown. SNP, single-nucleotide polymorphism; DTC, differentiated thyroid cancer; pc, corrected p-value.
Discussion
The development of the thyroid gland and its normal migration in the embryonic stage, as well as the maintenance of the differentiated state throughout life, depends on the interplay between three TTFs: TTF-1/NKX2-1, TTF-2/FOXE1, and paired box 8 (PAX-8) (15). That set of transcriptional factors is unique in the thyroid follicular cell type and regulates the transcription of genes such as thyrotropin receptor, thyroperoxidase, thyroglobulin, and sodium iodide symporter that are critical for thyroid hormone synthesis (16). Besides their role for normal thyroid physiology, they show an altered expression in thyroid tumors and are probably involved in thyroid oncogenesis (17,18). A GWA study, analyzing Icelandic cases and cohorts of European descent, showed that two SNPs, situated on chromosomes 9q22.33 (near FOXE1: rs965513) and 14q13.3 (near NKX2-1: rs944289), are associated with both PTC and FTC (7). This finding has been confirmed in other populations (British and Japanese) (8,9) but not yet in the German or other Central European population. Also, whether certain genotypes/alleles of the rs965513 and rs944289 SNPs differentiate the risk for tumor progression is unknown so far. Lymphocytic infiltration within the thyroid gland is frequently observed in DTC, and its presence surrounding or within the tumor might be a useful marker predicting a favorable prognosis. It has been described that DTC patients with lymphocytic infiltration tend not to develop recurrent disease (19). Therefore, it is of interest whether certain alleles of the rs965513 and rs944289 SNPs are associated with such intrathyroidal immune reaction in DTC patients.
Our results demonstrate that solely the FOXE1 rs965513 variant affects the risk for DTC, and this finding is more pronounced in PTC than in FTC. Remarkably, the association between FOXE1 rs965513 SNP and DTC was found especially in individuals with extent of the primary tumor (T1–T2 vs. HC, T3–T4 vs. HC), affected lymph nodes (N1 vs. HC), and was detectable in patients without lymphocytic infiltrate (without ITL vs. HC). We therefore assume that patients with this FOXE1 risk allele also carry a risk for more aggressive disease, but the data need to be confirmed with a larger cohort and additional BRAF status screening in thyroid tissues of DTC patients. These examinations have already been performed by Matsuse et al. (9). That work investigated the BRAF mutation status in PTC patients according to the FOXE1 rs965513 in a Japanese cohort and found an association of rs965513 with BRAFV600E-pos but not with BRAFV600E-neg cases. Because there was no difference between the subgroups BRAFV600E-neg and BRAFV600E-pos, the authors concluded that a clear relationship between FOXE1 rs965513 SNP and the BRAFV600E status does not exist. It is important to note that thyroid carcinomas in the Japanese population show a higher incidence of PTC and higher BRAFV600E-pos rate than in the European population (80% and 50% respectively) (9). Also, the role of the intrathyroidal immune cells in DTC needs to be studied with larger numbers and in more detail. In this regard, we found no association between the FOXE1 rs965513 SNP and thyroid antibody status as well as lymphocytic infiltration in DTC patients. However, there was a limited number of patients who were either thyroid antibody positive or who had lymphocytic infiltrations. Future and sufficiently powered studies must address whether the FOXE1 rs965513 SNP is involved if DTC develops in the background of autoimmune thyroid disease.
The association between FOXE1 rs965513 and thyroid cancer in the present work confirms the results of other studies. The association with DTC (only PTC or both cancer subtypes PTC/FTC) is marked by the minor allele “A” of the FOXE1 rs965513 SNP (7–10,20). In contrast, Denny et al. (6) found in a thyroid-related phenotype study only a trend (p=0.09), attributed to the smaller number of patients (n=96). Also, the lack of an association between FTC and the rs965513 SNP observed in the present study may be due to the smaller number of our patients, since the power to detect a suspected OR of 1.75 (7) is too low for this sample size. Here, only an OR of 1.9 can be detected with sufficient power (80%). It is noteworthy that association studies on other SNPs near FOXE reinforce this region on chromosome 9 as a good candidate for thyroid cancer susceptibility: rs965513 is in strong pair-wise linkage disequilibrium with three other SNPs near FOXE (rs925489, rs7850258, and rs10759944) (6,10). These SNPs demonstrated a significant association with Chernobyl radiation-related PTC in Belarusian patients (10). Furthermore, another SNP located in the FOXE1 promoter (rs1867277) and in the same LD region as rs965513 was associated with DTC in Spanish/Italian, Belarusian, and British cohorts (8,21,22). In this context, the risk allele “A” of the SNP rs1867277 is capable of augmenting FOXE1 transcription by creating a binding site for leucine zipper upstream stimulatory factors 1 and 2 (USF1/ USF2) (22). Also, in three recent studies, both the rs1867277 and the FOXE1 polyAla tract variants were associated with DTC cancer in Spanish, Australian, and Portuguese cohorts (20,21,23).
The rs944289 SNP is situated in a 249-kb LD-region. Neighboring genes of that region are Breast Cancer Metastasis Suppressor 1-Like (BRMS1L), MAP3K12 binding inhibitory protein 1 (MBIP), surfactant associated 3 (SFTA3), and NKX2-1. Among these genes, NKX2-1 is a suitable candidate gene for thyroid cancer risk because of its role in thyroid development (24), and its altered expression in thyroid tumors (18). It is interesting to note that whereas the NKX2-1 rs944289 SNP is associated with DTC in Icelandic and Japanese populations, no associations were found in the Belarusian and our present study (7,9,10). However, the case-control comparison for the NKX2-1 rs944289 SNP from Gudmundsson et al. (7) (192 cases and 37,196 controls; p=0.000000002) and Takahashi et al. (10) (507 cases and 2766 controls; p=0.01) reached different p-values based on asymmetric case/control group sizes. We therefore assume that the limited number of our study participants (243 cases and 270 controls) cannot solely explain the lack of association between DTC and the NKX2-1 rs944289 SNP (expected for 80% power OR 1.44, reached OR 1.06). Furthermore, the only differences between cases and controls in our current study concerning this SNP are detected by the combination analysis of the two FOXE1 and NKX2-1 variants. We identified two genotype combinations: risk “AA-rs965513/CT-rs944289” and protective “GG-rs965513/CT-rs944289.” However, this finding has no impact on overall DTC risk. Moreover, analyses of this gene (addressing the A39V germline mutation) in several nonmedullary thyroid carcinoma families did not confirm NKX2-1 as a risk gene (25). Finally, independent groups performed case-control studies of TTFs in thyroid cancer including the NKX2-1 rs944289 SNP and provided contradictory results for an association (7,9,10). Gudmundsson et al. (7) found no association in the non-Icelandic individuals (cohort of Spanish origin) between NKX2-1 and DTC. This may indicate a minor or secondary role of this variant in DTC susceptibility. These observations suggest that differences exist in the genetic profile of the TTFs depending on the population background. It is also conceivable that factors, which are related to rs944289 SNP, have a direct effect on the development of DTC and are more powerful in all population groups. In this direction, Jendrzejewskia et al. (26) investigated the mechanism of how the NKX2-1 rs944289 SNP might predispose to PTC. They propose a dysregulation in the expression of the PTC susceptibility candidate 3 (PTCSC3) located 3.2 kb downstream of rs944289. This long intergenic noncoding RNA gene acts as a tumor suppressor.
In summary, our findings are in agreement with earlier DTC studies that found an association with the FOXE1 rs965513 SNP, indicating that this variant also confers DTC risk in the German population. Furthermore, we show for the first time that the association is also found in patients with advanced stages of DTC disease (T and N) and in the absence of lymphocytic infiltration in the thyroid gland. This observation may reflect more aggressive disease. In contrast, the NKX2-1 rs944289 SNP appears to play a secondary role for DTC risk in Germans, and its effect is probably population specific.
The DTC risk gene FOXE1 may serve to control thyroid growth both in early embryological stages as well as during adult life, and safeguard benign differentiation also under the influence of thyroid cell damage either by carcinogens or radiation effects.
Supplementary Material
Acknowledgment
This study was supported by a grant from the European Union (FP7-NAIMIT: Grant Agreement No. 241447).
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- 1.Aschebrook-Kilfoy B, Ward MH, Sabra MM, Devesa SS.2011Thyroid cancer incidence patterns in the United States by histologic type, 1992–2006. Thyroid 21:125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agate L, Lorusso L, Elisei R.2012New and old knowledge on differentiated thyroid cancer epidemiology and risk factors. J Endocrinol Invest 35:3–9 [PubMed] [Google Scholar]
- 3.Antonelli A, Ferri C, Fallahi P, Pampana A, Ferrari SM, Barani L, Marchi S, Ferrannini E.2007Thyroid cancer in HCV-related chronic hepatitis patients: a case-control study. Thyroid 17:447–451 [DOI] [PubMed] [Google Scholar]
- 4.Rossing MA, Voigt LF, Wicklund KG, Daling JR.2000Reproductive factors and risk of papillary thyroid cancer in women. Am J Epidemiol 151:765–772 [DOI] [PubMed] [Google Scholar]
- 5.Gomez Saez JM.2011Diagnostic and prognostic markers in differentiated thyroid cancer. Curr Genomics 12:597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denny JC, Crawford DC, Ritchie MD, Bielinski SJ, Basford MA, Bradford Y, Chai HS, Bastarache L, Zuvich R, Peissig P, Carrell D, Ramirez AH, Pathak J, Wilke RA, Rasmussen L, Wang X, Pacheco JA, Kho AN, Hayes MG, Weston N, Matsumoto M, Kopp PA, Newton KM, Jarvik GP, Li R, Manolio TA, Kullo IJ, Chute CG, Chisholm RL, Larson EB, McCarty CA, Masys DR, Roden DM, de AM.2011Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet 89:529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Sigurdsson A, Bergthorsson JT, He H, Blondal T, Geller F, Jakobsdottir M, Magnusdottir DN, Matthiasdottir S, Stacey SN, Skarphedinsson OB, Helgadottir H, Li W, Nagy R, Aguillo E, Faure E, Prats E, Saez B, Martinez M, Eyjolfsson GI, Bjornsdottir US, Holm H, Kristjansson K, Frigge ML, Kristvinsson H, Gulcher JR, Jonsson T, Rafnar T, Hjartarsson H, Mayordomo JI, de la Chapelle A, Hrafnkelsson J, Thorsteinsdottir U, Kong A, Stefansson K.2009Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet 41:460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones AM, Howarth KM, Martin L, Gorman M, Mihai R, Moss L, Auton A, Lemon C, Mehanna H, Mohan H, Clarke SE, Wadsley J, Macias E, Coatesworth A, Beasley M, Roques T, Martin C, Ryan P, Gerrard G, Power D, Bremmer C, Tomlinson I, Carvajal-Carmona LG.2012Thyroid cancer susceptibility polymorphisms: confirmation of loci on chromosomes 9q22 and 14q13, validation of a recessive 8q24 locus and failure to replicate a locus on 5q24. J Med Genet 49:158–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuse M, Takahashi M, Mitsutake N, Nishihara E, Hirokawa M, Kawaguchi T, Rogounovitch T, Saenko V, Bychkov A, Suzuki K, Matsuo K, Tajima K, Miyauchi A, Yamada R, Matsuda F, Yamashita S.2011The FOXE1 and NKX2-1 loci are associated with susceptibility to papillary thyroid carcinoma in the Japanese population. J Med Genet 48:645–648 [DOI] [PubMed] [Google Scholar]
- 10.Takahashi M, Saenko VA, Rogounovitch TI, Kawaguchi T, Drozd VM, Takigawa-Imamura H, Akulevich NM, Ratanajaraya C, Mitsutake N, Takamura N, Danilova LI, Lushchik ML, Demidchik YE, Heath S, Yamada R, Lathrop M, Matsuda F, Yamashita S.2010The FOXE1 locus is a major genetic determinant for radiation-related thyroid carcinoma in Chernobyl. Hum Mol Genet 19:2516–2523 [DOI] [PubMed] [Google Scholar]
- 11.Miller SA, Dykes DD, Polesky HF.1988A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cordell JL, Falini B, Erber WN, Ghosh AK, Abdulaziz Z, MacDonald S, Pulford KA, Stein H, Mason DY.1984Immunoenzymatic labeling of monoclonal antibodies using immune complexes of alkaline phosphatase and monoclonal anti-alkaline phosphatase (APAAP complexes). J Histochem Cytochem 32:219–229 [DOI] [PubMed] [Google Scholar]
- 13.Bland JM, Altman DG.1995Multiple significance tests: the Bonferroni method. Br Med J 310:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupont WD, Plummer WD., Jr1998Power and sample size calculations for studies involving linear regression. Control Clin Trials 19:589–601 [DOI] [PubMed] [Google Scholar]
- 15.Trueba SS, Auge J, Mattei G, Etchevers H, Martinovic J, Czernichow P, Vekemans M, Polak M, Attie-Bitach T.2005PAX8, TITF1, and FOXE1 gene expression patterns during human development: new insights into human thyroid development and thyroid dysgenesis-associated malformations. J Clin Endocrinol Metab 90:455–462 [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharyya KK, Coenen MJ, Bahn RS.2005Thyroid transcription factor-1 in orbital adipose tissues: potential role in orbital thyrotropin receptor expression. Thyroid 15:422–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sequeira MJ, Morgan JM, Fuhrer D, Wheeler MH, Jasani B, Ludgate M.2001Thyroid transcription factor-2 gene expression in benign and malignant thyroid lesions. Thyroid 11:995–1001 [DOI] [PubMed] [Google Scholar]
- 18.Zhang P, Zuo H, Nakamura Y, Nakamura M, Wakasa T, Kakudo K.2006Immunohistochemical analysis of thyroid-specific transcription factors in thyroid tumors. Pathol Int 56:240–245 [DOI] [PubMed] [Google Scholar]
- 19.Cunha LL, Morari EC, Guihen AC, Razolli D, Gerhard R, Nonogaki S, Soares FA, Vassallo J, Ward LS.2012Infiltration of a mixture of different immune cells may be related to molecular profile of differentiated thyroid cancer. Endocr Relat Cancer 19:L31–L36 [DOI] [PubMed] [Google Scholar]
- 20.Tomaz RA, Sousa I, Silva JG, Santos C, Teixeira MR, Leite V, Cavaco BM.2012FOXE1 polymorphisms are associated with familial and sporadic nonmedullary thyroid cancer susceptibility. Clin Endocrinol (Oxf) 77:926–933 [DOI] [PubMed] [Google Scholar]
- 21.Kallel R, Belguith-Maalej S, Akdi A, Mnif M, Charfeddine I, Galofre P, Ghorbel A, Abid M, Marcos R, Ayadi H, Velazquez A, Hadj KH.2010Genetic investigation of FOXE1 polyalanine tract in thyroid diseases: new insight on the role of FOXE1 in thyroid carcinoma. Cancer Biomark 8:43–51 [DOI] [PubMed] [Google Scholar]
- 22.Landa I, Ruiz-Llorente S, Montero-Conde C, Inglada-Perez L, Schiavi F, Leskela S, Pita G, Milne R, Maravall J, Ramos I, Andia V, Rodriguez-Poyo P, Jara-Albarran A, Meoro A, del PC, Arribas L, Iglesias P, Caballero J, Serrano J, Pico A, Pomares F, Gimenez G, Lopez-Mondejar P, Castello R, Merante-Boschin I, Pelizzo MR, Mauricio D, Opocher G, Rodriguez-Antona C, Gonzalez-Neira A, Matias-Guiu X, Santisteban P, Robledo M.2009The variant rs1867277 in FOXE1 gene confers thyroid cancer susceptibility through the recruitment of USF1/USF2 transcription factors. PLoS Genet 5:e1000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bullock M, Duncan EL, O'Neill C, Tacon L, Sywak M, Sidhu S, Delbridge L, Learoyd D, Robinson BG, Ludgate M, Clifton-Bligh RJ.2012Association of FOXE1 polyalanine repeat region with papillary thyroid cancer. J Clin Endocrinol Metab 97:E1814–E1819 [DOI] [PubMed] [Google Scholar]
- 24.Parlato R, Rosica A, Rodriguez-Mallon A, Affuso A, Postiglione MP, Arra C, Mansouri A, Kimura S, Di LR, De FM.2004An integrated regulatory network controlling survival and migration in thyroid organogenesis. Dev Biol 276:464–475 [DOI] [PubMed] [Google Scholar]
- 25.Cantara S, Capuano S, Formichi C, Pisu M, Capezzone M, Pacini F.2010Lack of germline A339V mutation in thyroid transcription factor-1 (TITF-1/NKX2.1) gene in familial papillary thyroid cancer. Thyroid Res 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jendrzejewski J, He H, Radomska HS, Li W, Tomsic J, Liyanarachchi S, Davuluri RV, Nagy R, de la Chapelle A.2012The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci USA 109:8646–8651 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.