Abstract
Oncogene MYC is highly expressed in many human cancers and functions as a global regulator of ribosome biogenesis. Previously, we reported that ribosomal protein (RP) L11 binds to c-Myc and inhibits its transcriptional activity in response to ribosomal stress. Here, we show that another ribosomal protein L5, cooperatively with RPL11, guides the RISC complex to c-Myc mRNA and mediates the degradation of the mRNA, consequently leading to inhibition of c-Myc activity. Knocking down of RPL5 induced c-Myc expression at both mRNA and protein levels, while overexpression of RPL5 suppressed c-Myc expression and activity. Immunoprecipitation revealed that RPL5 binds to 3UTR of c-Myc mRNA and two subunits of RISC complex, TRBP and Ago2, mediating the targeting of c-Myc mRNA by miRNAs. Interestingly, RPL5 and RPL11 co-resided on c-Myc mRNA and suppressed c-Myc expression cooperatively. These findings uncover a mechanism by which these two RPs can cooperatively suppress c-Myc expression, allowing a tightly controlled ribosome biogenesis in cells.
Keywords: c-Myc, TRBP2, microRNA, RPL11, RPL5, ribosomal stress
Introduction
The c-Myc oncoprotein is a globally regulatory factor of cell growth and proliferation1, 2. Deregulation of c-Myc is highly associated with a wide range of cancers3–9. In cancer cells, constant activation of c-Myc plays a vital role in cancer development by transcriptionally regulating a number of genes that are involved in cell division, metabolic adaptation, cell survival, and ribosome biogenesis1, 10. A number of studies have shown that amplification of MYC, the c-Myc-encoding gene, through multiple mechanisms, including chromosomal translocation and mutations11–14, is correlated with poor clinical outcome and tumor aggression 15, 16. Proliferation of tumor cells with high levels of c-Myc is no longer dependent on growth-factor stimulation, whereas in normal cells, growth-factor stimulation is required for c-Myc-dependent proliferation, metabolic pathways, cell adhesion, and ribosome biogenesis 13, 14, 17–20. Thus, the role of c-Myc in controlling ribosome biogenesis and translation is crucial for the development and progression of tumors17.
Ribosome biogenesis is essential for fast-growing cancer cells and tightly regulated in normal growing cells. In prokaryotes, free ribosomal proteins resulted from deregulation of ribosome biogenesis can bind to their own mRNAs and inhibit their transcription to avoid unneeded synthesis of ribosomal proteins21. This simple mode of autoregulation of ribosomal protein expression, though not yet found in eukaryotes, has evolved to a more complicated mechanism involving c-Myc in eukaryotic cells. Previously, we discovered that ribosomal protein (RP) L11 binds to c-Myc and inhibits its activity 22. Later on, RPL11 was shown to bind to c-Myc mRNA and promote its degradation via miRNAs23. Because c-Myc stimulates the transcription of ribosomal proteins-encoding mRNAs 17, 22, RPL11 can negate c-Myc activity via a negative-feedback mechanism and consequently lead to a tightly monitored ribosome biogenesis in mammalian cells. A “byproduct” of this negative regulation of c-Myc by free RPL11 is to prevent cell transformation, as ribosome biogenesis is usually elevated in cancer cells17. Indeed, free forms of several ribosomal proteins, generated by unexpected ribosome biogenesis or in response to ribosomal stresses, have been identified to play important roles in preventing tumorigenesis. A number of ribosomal proteins, including RPL11 24–27, RPL5 28, RPL23 29, 30, RPL26 31, RPS332, RPS7 33, 34, RPS1435, RPS2536, RPS27 37 or RPS27a 38, have been shown to suppress tumor cell growth by activating p53 in response to ribosomal stress. Although RPL11 has been shown to negatively regulate c-Myc activity independently of the MDM2-p53 pathway22, and also, RPL11 and RPL5 have been shown to work together to activate p53 39, it remains largely unclear whether these p53-activating ribosomal proteins could also exert their extra-ribosomal function individually or cooperatively toward c-Myc in a sub-ribosomal complex. Thus, this prompted us to determine whether RPL5 also plays a role in regulating c-Myc level or activity, and if true, whether this regulation is executed by cooperating with RPL11.
In this study, we intended to address these two questions. First, we found out that RPL5 binds to c-Myc mRNA and inhibits its expression by mediating the binding of the RISC complex to c-Myc mRNA. Also, we showed that RPL5 co-resides with RPL11 on c-Myc mRNA and inhibits the expression of c-Myc cooperatively with the latter. Hence, our studies as detailed below demonstrate the cooperative action of these two ribosomal proteins on inactivation of c-Myc by guiding the RISC complex to its mRNA and inhibiting its expression.
Results
RPL5 inhibits c-Myc-dependent proliferation by suppressing c-Myc expression
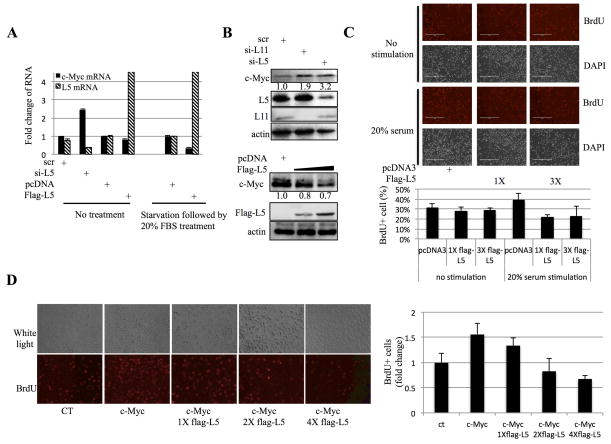
To investigate whether c-Myc expression is affected by RPL5, we first used siRNA to knock down endogenous RPL5 in H1299 cells. As shown in Figure 1A, knockdown of RPL5 led to the increase of c-Myc mRNA level as determined by RT real-time quantitative PCR (RT-qPCR), while overexpression of RPL5 by transfection of a plasmid that expressed a flag-RPL5 fusion protein only caused a mild reduction of c-Myc mRNA (left, Figure 1A). However, interestingly, after stimulating cells by 20% serum to induce c-Myc expression, overexpression of RPL5 led to the reduction of c-Myc mRNA level dramatically (right, Figure 1A). The inhibitory effect of RPL5 on c-Myc level was also confirmed at its protein level by Western blot (WB) analysis. RPL5 knockdown resulted in the elevation of c-Myc protein level, similar to that of RPL11 knockdown (Figure 1B, upper panel); Consistently, RPL5 overexpression led to the reduction of c-Myc protein level (Figure 1B, lower panel). Because of the importance of c-Myc for cell proliferation 18, 40, 41, we next checked if increased RPL5 expression could affect cell proliferation by regulating c-Myc level. As shown in Figure 1C, ectopic RPL5 led to a marked decrease in BrdU (5-bromodeoxyuridine) incorporation in serum-re-stimulated H1299 cells, compared to cells without exogenous RPL5. Interestingly, the inhibitory effect of RPL5 on proliferation could be observed only in serum-re-stimulated cells, suggesting that c-Myc induction might be required for this regulation (Figure 1C). To further determine the effect of RPL5 on c-Myc-enhanced proliferation, we performed a BrdU cell proliferation assay with c-Myc–transfected cells. As expected, c-Myc induced cell proliferation as indicated by the increase in BrdU incorporation. Again, co-expression of Flag-RPL5 in c-Myc-expressing cells resulted in the significant decline of BrdU incorporation in a dose-dependent manor (Figure 1D). Together, these results indicate that RPL5 suppresses c-Myc expression and inhibits its activity, consequently retarding c-Myc promoted cell proliferation.
Figure 1. RPL5 suppresses c-Myc expression and inhibits cell proliferation.
(A) Knocking down RPL5 increases, but overexpression of RPL5 decreases, c-Myc mRNA level in H1299 cells. Left panel: cells were transfected with indicated plasmids or siRNAs and the expression of c-Myc mRNA was measured by q-RT-PCR. Right panel: cells were starved in 0.2% serum for 36 hours and stimulated with 20% serum for 1.5 h before harvesting. Data are presented as mean ± standard error, n = 3. (B) Knocking down RPL5 increases, but overexpression of RPL5 decreases, c-Myc protein level in H1299 cells. Upper panel: cells were transfected with indicated siRNAs and harvested for IB with antibodies as indicated. Lower panel: after transfected with indicated plasmids, cells were starved in 0.2% serum for 36 hours and stimulated with 20% serum for 1.5 h before harvesting for WB. (C) RPL5 inhibits cell proliferation. H1299 cells transfected with an increasing amount of flag-RPL5 plasmid were treated with starvation followed by stimulation as described in panel A. Cell then were labeled with BrdU and subjected to anti-BrdU staining (red) and counter-stained with DAPI. Percentage of BrdU-positive cells is shown in right panel. (D) Serum stimulation of c-Myc is dispensable for the inhibitory effect of RPL5 on proliferation. The assay as that in panel C was conducted except the indicated cells were not treated with serum-stimulation.
RPL5 inhibits induction of c-Myc by serum stimulation
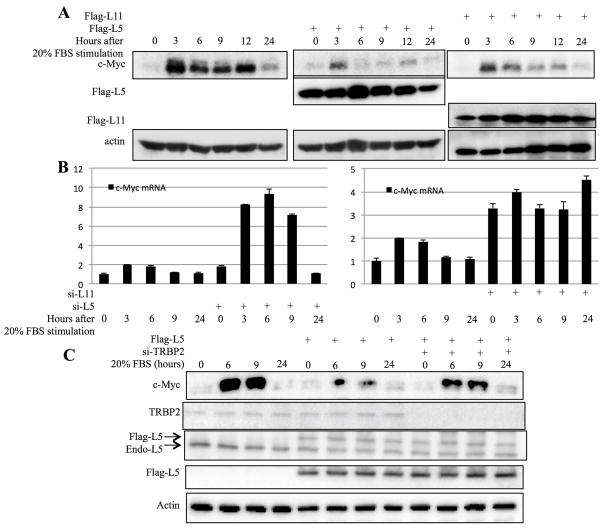
c-Myc is rapidly induced in response to serum stimulation. To determine if this serum responsive induction could be affected by RPL5, we compared c-Myc levels in H1299 cells transfected with RPL5, RPL11 or pcDNA vector upon serum stimulation. H1299 cells were serum-starved for 24h and then cultured in medium containing 20% bovine serum. Cells were then harvested at different time points after serum stimulation. As expected23, the induction of c-Myc expression was drastically reduced in cells that overexpressed RPL11 at each time point after serum stimulation (Figure 2A). Similarly and interestingly, overexpression of RPL5 also markedly decreased the level of endogenous c-Myc, in comparison with control cells (Figure 2A). Consistent with this result, knockdown of either of these two endogenous RPs in H1299 cells led to the induction of c-Myc mRNA levels, compared to control cells (Figure 2B). These results indicate that RPL5 also regulates c-Myc expression after serum stimulation.
Figure 2. RPL5 and RPL11 suppress c-Myc expression in response to serum stimulation.
(A) and (B) RPL5 and RPL11 suppress c-Myc expression in response to serum stimulation. H1299 cells transfected with indicated plasmids or siRNAs were starved in 0.2% serum for 36 hours and stimulated with 20% serum for indicated time. c-Myc protein and mRNA were measured by WB (A) and q-RT-PCR (B) respectively. (C) Knocking down TRBP2 impairs the inhibitory effect of RPL5 on c-Myc expression. H1299 cells transfected with indicated plasmids were treated with serum starvation and stimulation and harvested at indicated time points. Cell lysates were subjected to WB with indicated antibodies.
Because RPL5 suppresses c-Myc mRNA expression (Figures 1A and 2B) similarly to what RPL11 does23, and RPL11 was previously shown to target c-Myc mRNA via miRNAs23, we suspected that RPL5 might employ the same mechanism to inhibit c-Myc expression. First, we investigated whether disruption of the miRNA machinery would affect the regulation of c-Myc by RPL5 by knocking down TRBP (HIV-1 TAR RNA-binding protein), an important subunit of RISC42, 43, in H1299 cells, as TRBP knockdown was previously shown to impair the miRNA processing and silencing steps44, 45. Indeed, disruption of miRNA functions by TRBP knockdown rescued the inhibitory effect of RPL5 on c-Myc induction by serum-stimulation, at least partially (Figure 2C), suggesting that miRNAs might be involved in the regulation of c-Myc by this ribosomal protein. Because H1299 cells are p53-deficient, this effect is p53-independent.
RPL5 targets c-Myc 3′UTR
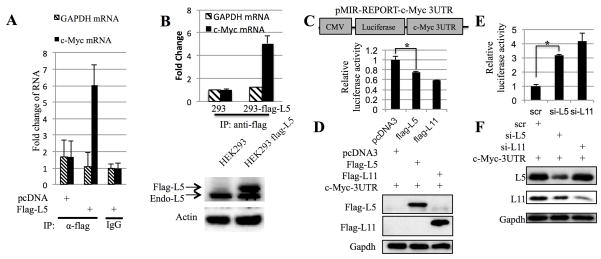
We assumed that RPL5 might bind to c-Myc 3′UTR and thus suppress c-Myc expression by utilizing a mechanism similar to that for RPL1123. To test this idea, we first checked the binding of RPL5 with c-Myc mRNA in HEK 293 cells by ribonucleoprotein immunoprecipitation (RNP IP) assay23, 46. As shown in Figure 3A, c-Myc mRNA, but not gapdh mRNA, was enriched more than six-fold in Flag-L5 IP samples than that in IgG IP samples. Also, this enrichment of c-Myc mRNA depended on RPL5 expression, for c-Myc mRNA was not enriched by the anti-flag antibody in cells without overexpression of flag-RPL5. This association between RPL5 and c-Myc mRNA was further confirmed using an HEK293 cell line that stably expresses flag-RPL5, as c-Myc mRNA was only pulled down by the flag antibody in flag-RPL5-HEK293 stable cells, but not in parental HEK293 cells (Figure 3B). Thus, it is likely that RPL5 can form a complex with c-Myc mRNA in cells.
Figure 3. RPL5 binds to c-Myc mRNA and targets its 3UTR.
(A) RPL5 binds to endogenous c-Myc mRNA. HEK 293 cells transfected with indicated plasmids were treated with serum starvation and stimulation and harvested for immunoprecipitation assay with anti-flag or mouse IgG. c-Myc and GAPDH mRNAs pulled down by indicated antibody were determined by q-RT-PCR. (B) RPL5 binds to endogenous c-Myc mRNA. Flag-RPL5-293 stable cells or HEK293 cells were treated with serum starvation and stimulation, and harvested for immunoprecipitation (IP) assays with anti-flag. c-Myc and GAPDH mRNAs pulled down by indicated antibodies were determined by q-RT-PCR. (C) and (E) RPL5 and RPL11 target c-Myc 3UTR. H1299 cells were cotransfected with pMIR-c-Myc 3UTR and indicated plasmids. Forty-eight hours after transfection, luciferase activities were measured. Luciferase activity was normalized to beta-gal expression. RPL5 and RPL11 protein expressions were measured by WB as shown in panel (D) and (F). Schematic representation of the pMIR-REPORT-c-Myc 3UTR expression vector is shown in the top part of panel C.
Next, we tested if RPL5 could directly target c-Myc 3′UTR by conducting a luciferase assay with the pMIR-REPORT vector that contains c-Myc 3′UTR at the 3′ end of the luciferase ORF. As shown in Figures 3C–3D, when HEK293 cells were cotransfected with flag-RPL5 and pMIR-REPORT containing the intact c-Myc 3′UTR, the luciferase reporter expression was significantly reduced, similar to the reduction of luciferase activity by RPL11, compared to cells transfected with pMIR-REPORT-c-Myc 3′UTR and pcDNA3. Conversely, knockdown of either RPL5 or RPL11 in HEK293 cells led to a marked induction of the luciferase activity (Figures 3E–F). These results suggest that RPL5, similarly to RPL11, may regulate c-Myc expression by interacting with the 3′UTR region of its mRNA.
RPL5 binds to the TRBP2 and Ago2 subunits of the RISC complex
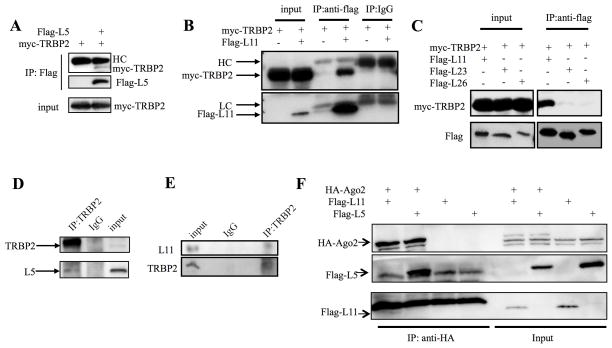
Our finding that RPL5 associates with c-Myc mRNA and targets its 3′UTR suggests that this RP might, like RPL1123, regulate c-Myc mRNA level via miRNA as well. To begin to test this possibility, we first determined if RPL5 could bind to the RNA-induced silencing complex (RISC). As one of the RISC subunits, TRBP has been shown to be an essential component of this complex and play a vital role in facilitating the binding of the other two subunits of RISC, Dicer and Ago2 42, 43. Therefore, we tested the association of RPL5 with TRBP2 by transfecting HEK293 cells with myc-TRBP2 and flag-RPL5, followed by carrying out immunoprecipitation-WB (IP-WB) analysis. Interestingly, myc-TRBP2 was pulled down by anti-flag antibodies in cells expressing both flag-RPL5 and myc-TRBP2, but not in cells without flag-RPL5, indicating that RPL5 binds to TRBP2 in cells (Figure 4A). We also tested whether other ribosomal proteins might bind to TRBP2 by performing a set of co-IP-WB experiments. As shown in Figures 4B and 4C, myc-TRBP2 was co-immunoprecipitated with flag-RPL11 as well, but not flag-RPL23 and flag-RPL26, suggesting that the interaction with TRBP2 is specific to RPL5 and RPL11, but not other RPs as tested here. Also, this specific interaction was not due to the different expression levels of these proteins, as WB analysis of HEK293 cell extracts confirmed that their steady state levels were equivalent in all of the transfected cells (Figure 4B and C). The interaction of TRBP2 with RPL5 and RPL11 was also verified using endogenous proteins, as anti-TRBP2, but not non-specific IgG, antibodies could pull down endogenous RPL5 and RPL11 in HEK293 cells as well (Figures 4D and 4E). These results indicate that RPL5 and RPL11 can bind to TRBP2 in cells. As RPL11 was previously reported to bind to another RISC subunit Ago223, we also tested if this is true to RPL5. Indeed, RPL5 was co-immunoprecipitated with Ago2 as shown in Figure 4F. Taken together, these results demonstrate that RPL5, similar to RPL11, can bind to the RISC complex in cells.
Figure 4. RPL5 and RPL11 interact with TRBP2 in cells.
(A) RPL11 binds to TRBP2. HEK 293 Cells were transfected with myc-TRBP2 and Flag-RPL11 as indicated in the figure and harvested for IP (0.5 mg total proteins) using anti-Flag or mouse IgG, followed by IB with the antibodies as indicated. (B) RPL5 binds to TRBP2. HEK 293 Cells were transfected with myc-TRBP2 and Flag-RPL5 as indicated in the figure and harvested for IP (0.5 mg total proteins) using anti-Flag, followed by IB with the antibodies as indicated. (C) RPL23 and RPL26 cannot bind to TRBP2. HEK 293 Cells were transfected with indicated plasmids and harvested for IP (0.5 mg total proteins) using anti-Flag, followed by IB with the antibodies as indicated. (D) Endogenous RPL11 and RPL5 interact with endogenous TRBP2 in HEK 293 cells. Cells lysates (0.6 mg) were prepared from HEK 293 cells after treatment with Act D for 8 h. IP was conducted with anti-TRBP2 or rabbit IgG followed by IB with anti-TRBP2 or anti-ribosomal proteins. All the experiments were repeated more than 2 times and true to the rest of data.
RPL5 mediates the binding between RISC and c-Myc 3′UTR
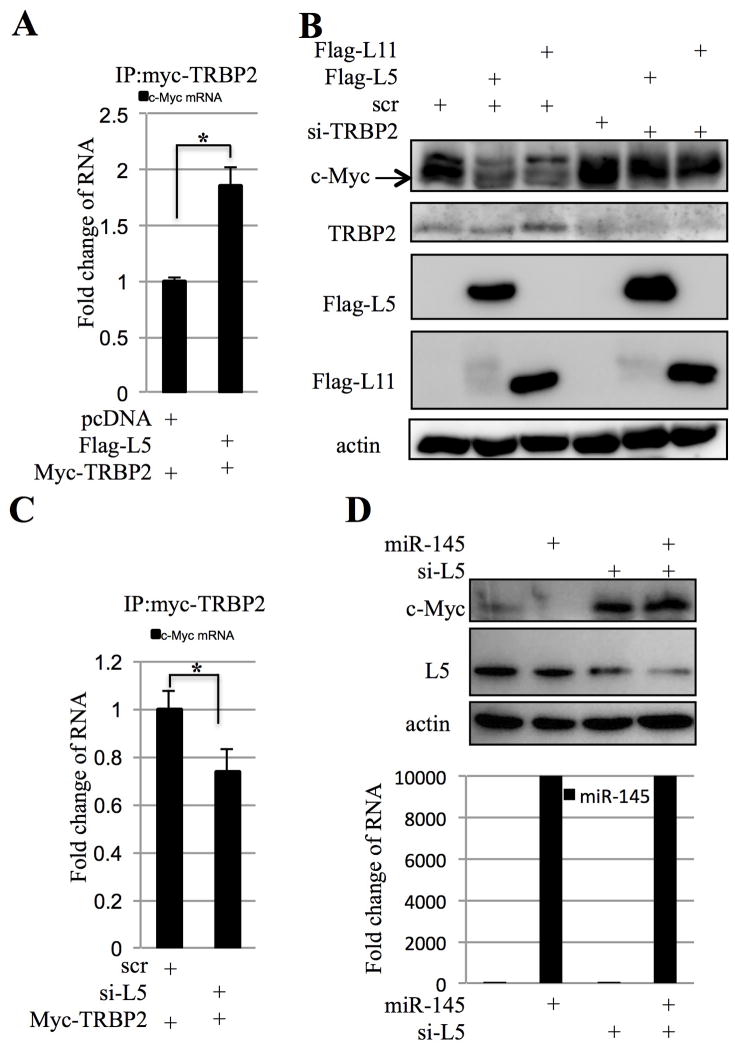
Our findings that RPL5 binds to RISC subunits and c-Myc mRNA via its 3′UTR suggest that RPL5 might mediate the association of RISC with c-Myc 3′UTR. To test this idea, we performed an RNP-IP assay by introducing myc-TRBP2 with flag-L5 or pcDNA3 control plasmid into HEK293 cells. Then, ribonucleoprotein complexes were immunoprecipitated from cell lysates using an antibody against myc-TRBP2; Subsequently, RNA was extracted, reverse-transcribed, and subjected to qRT-PCR with primers for c-Myc mRNA. As shown in Figure 5A, the c-Myc transcript level as immunoprecipitated in the complex was significantly increased in the presence of flag-RPL5 compared to the pcDNA3 control, suggesting that RPL5 may promote the binding of TRBP2 to c-Myc mRNA.
Figure 5. RPL5 mediates the binding of RISC to c-Myc mRNA.
(A) Overexpression of RPL5 promotes TRBP2 and c-Myc mRNA binding. HEK 293 cells transfected with indicated plasmids were treated with serum starvation and stimulation and harvested for immunoprecipitation assay with anti-myc. c-Myc mRNAs pulled down by myc-TRBP2 were determined by q-RT-PCR. (B) Knocking down TRBP2 abrogates the inhibitory effect of RPL5 on c-Myc expression. H1299 cells transfected with indicated plasmids were treated with serum starvation and stimulation and harvested for WB with indicated antibodies. (C) Knocking down RPL5 suppresses TRBP2 and c-Myc mRNA binding. HEK 293 cells transfected with indicated plasmids were harvested for immunoprecipitation assay with anti-myc. c-Myc mRNAs pulled down by myc-TRBP2 were determined by q-RT-PCR. (D) RPL5 is required for the targeting of c-Myc by miR-145. H1299 cells transfected with indicated plasmids were harvested for WB with indicated antibodies. The expression of miR-145 was determined by q-RT-PCR as shown in the lower part of this panel.
Since TRBP2 was previously reported to be essential for miRNA functions 44, 45, we examined whether TRBP2 is required for RPL5 regulation of c-Myc expression in cells. Indeed, this was the case as siRNA-mediated silencing of TRBP2 expression in H1299 cells abrogated the inhibitory effect of RPL5 on c-Myc expression (Figure 5B). To check if RPL5 is required for the binding of TRBP2 to c-Myc mRNA, we carried an RNP-IP assay after knocking down endogenous RPL5 by siRNA. As expected, RPL5 knockdown impaired the binding of TRBP2 to c-Myc mRNA (Figure 5C). Consistent with this result, knockdown of RPL5 by siRNA abrogated the inhibitory effect of overexpressed miR-145 on c-Myc expression, as in the absence of RPL5 siRNA, ectopically expressed miR-145 led to the marked decrease of endogenous c-Myc in H1299 cells as determined by WB (Figure 5D). Taken together, these results suggest that RPL5 can act as a mediator for the targeting of c-Myc mRNA by miR-145-quided RISC.
RPL5 and RPL11 inactivate c-Myc cooperatively
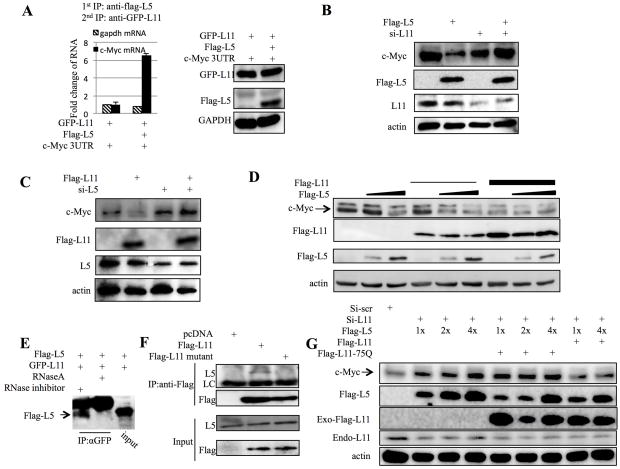
Since both RPL5 and RPL11 had been shown to protect each other from degradation47 and to stimulate p53 cooperatively39, we would like to test if RPL5 might also cooperate with RPL11 to inhibit c-Myc mRNA expression via miRNAs. To this end, we first carried sequential RNP-IP to examine whether these two RPs could simultaneously interact with c-Myc mRNA. Flag-RPL5, GFP-RPL11, and c-Myc 3′UTR were co-expressed in HEK293 cells and their expression was confirmed by WB (Right, Figure 6A). Sequential RNP-IP analysis was then performed with anti-flag antibodies for the first IP to pull down flag-RPL5 followed by secondary IP with anti-GFP antibodies to pull down GFP-RPL11 after eluting the RPL5-associated complexes with flag peptides. Then the captured GFP-RPL11- and Flag-RPL5-containing complexes were used for RNA extraction, which was used for Q-RT-PCR assays. As shown in Figure 6A, a ~7-fold enrichment in c-Myc 3′UTR, but not gapdh mRNA, was clearly evident in the complexes, demonstrating that RPL5 and RPL11 do co-interact with c-Myc mRNA.
Figure 6. RPL5 and RPL11 cooperatively inactivate c-Myc.
(A) RPL5 and RPL11 co-bind to c-Myc mRNA. HEK 293 cells transfected with indicated plasmids were treated with serum starvation and stimulation and harvested for immunoprecipitation assay with anti-flag antibody. The IPed complex were then washed off by flag peptide and subjected to a second immunoprecipitation assay with anti-GFP antibody. c-Myc and GAPDH mRNAs were determined by q-RT-PCR. The expressions of RPL11 and RPL5 were shown in the right part of this panel. (B) Knocking down RPL11 abrogates the inhibitory effect of RPL5 on c-Myc expression. H1299 cells transfected with indicated plasmids were treated with serum starvation and stimulation and harvested for WB with indicated antibodies. (C) Knocking down RPL5 abrogates the inhibitory effect of RPL11 on c-Myc expression. H1299 cells transfected with indicated plasmids were treated with serum starvation and stimulation and harvested for WB with indicated antibodies. (D) RPL5 and RPL11 inactivate c-Myc cooperatively. H1299 cells transfected with indicated plasmids were treated with serum starvation and stimulation and harvested for WB with indicated antibodies. (E) RPL5 binds to RPL11 in an RNase dependent fashion. HEK 293 cells were transfected with flag-RPL5 and GFP-RPL11 plasmids. Cells were harvested for IP-WB with the anti-GFP antibodies as indicated. RNaseA or RNase inhibitor was added into indicated samples. (F) RPL11 R75Q mutant fails to pull down endogenous RPL5. HEK 293 cells were transfected with pcDNA3, flag-RPL11, or flag-RPL11 R75Q. Cells were harvested for immunoprecipitation with anti-flag antibody. LC: lighter chain of IgG. (G) RPL11 R75Q fails to reduce c-Myc level cooperatively with RPL5. H1299 cells transfected with indicated plasmids were harvested for WB with indicated antibodies.
Next, we tested whether RPL5 and RPL11 act cooperatively to inhibit c-Myc expression via miRNAs. To determine if RPL11 is required for the inhibitory effect of RPL5 on c-Myc expression, RPL11 gene expression in H1299 cells was silenced using siRNA methodology. As expected, RPL11 siRNA transfection decreased the level of endogenous RPL11; interestingly, this decrease of RPL11 level impaired the inhibitory effect of RPL5 on c-Myc expression, as determined by WB (Figure 6B). Similarly, RPL5 knockdown also impaired the declining of c-Myc level by RPL11 (Figure 6C). These findings suggest RPL5 and RPL11 need each other to regulate c-Myc level in cells. To further determine if they could cooperatively help each other, we introduced different amounts of flag-RPL5 or flag-RPL11 expression vectors in H1299 cells and monitored the c-Myc level by WB. Indeed, although c-Myc protein level was not obviously affected by each of these RPs alone at lower concentrations (lane 2 and 4, Figure 6D), co-expression of RPL5 and RPL11 at the same levels significantly reduced the expression of c-Myc (lane 5, Figure 6D), indicating the two RPs could cooperate with each other in inhibiting the expression of c-Myc. Because RPL5 and RPL11 can form a complex via 5S rRNA, we tested whether this association is essential for their cooperative effect on c-Myc expression by employing a mutant RPL11, R75Q, as this mutant was defective in binding to 5S rRNA and RPL539. As expected, GFP-RPL11 bound to flag-RPL5 in an RNase-dependent fashion (Figure 6E); Also, flag-RPL11 bound to endogenous RPL5, whereas flag-RPL11 R75Q failed to do so (Figure 6F). To determine whether the binding between RPL5 and RPL11 is essential for the regulation of c-Myc expression, we knocked down RPL11 and investigated whether RPL5 or RPL11 R75Q mutant can prevent the c-Myc induction by knocking down endogenous RPL11. As shown in Figure 6G, overexpression of ectopic RPL5 failed to do so (lane 1 to 4); This result, consistent with the above results, again indicates that RPL11 is required for the targeting of c-Myc by RPL5. Interestingly, co-overexpressing ectopic wild-type RPL11, but not its mutant R75Q, with RPL5 reversed the RPL11 depletion-induced c-Myc expression (Figure 6G). These results confirm that the binding between RPL5 and RPL11 is essential for this regulation. In summary, these results indicate that RPL5 and RPL11 cooperatively suppress the expression of c-Myc via RISC.
Discussion
c-Myc overexpression is highly associated with tumorigenesis, which is at least partially due to the global deregulation of ribosomal biogenesis17. Previously we showed that RPL11 inhibits c-Myc activity through direct binding to c-Myc protein22. Later on, this protein was found to also suppress c-Myc activity by recruiting miRNAs to c-Myc mRNAs23. These findings indicate the existence of the autoregulation of c-Myc by some of its downstream transcriptional target genes that encode ribosomal proteins. Our study, as presented here identified RPL5 as another regulator of c-Myc. Moreover, RPL5 can cooperate with RPL11 in suppressing the expression of c-Myc through a RISC-mediated miRNA targeting mechanism. First, we showed that RPL5 suppresses the expression of c-Myc and consequently inhibits proliferation induced by this oncoprotein (Figure 1). Interestingly, we found that TRBP, one of the RISC components, is critical for this inhibition by RPL5, as knocking down TRBP partially rescued the inhibitory effect of RPL5 on c-Myc induction in response to serum-stimulation (Figure 2). Also, RPL5 facilitated the targeting of c-Myc mRNA by miRNAs, such as miR145, by binding to c-Myc mRNA and two RISC subunits, TRBP and Ago2 (Figure 3, 4, and 5). This binding to RISC appears specific to these two RPs, as neither RPL23 nor RPL26 could bind to this complex (Figure 4C). Finally, we uncovered that RPL5 and RPL11 form a complex with c-Myc mRNA, acting together to repress c-Myc expression in a cooperative fashion (Figure 6). These results demonstrate that RPL5 and RPL11 are the two partners that act as critical feedback regulators of c-Myc during ribosomal biogenesis. These two ribosomal proteins have also been previously shown to work together in activating p53 by cooperatively inhibiting the activity of the MDM2 oncoprotein39. Our study that recapitulated their cooperative effect on another oncoprotein c-Myc further validates the notion that RPL5 and RPL11 can indeed work as one complex in cells to control cell proliferation and growth (Figures 1–6), although it remains unclear why eukaryotic cells utilize two, instead of one, RPs to control c-Myc activity.
Interestingly, our finding also presents an evolutionarily conserved molecular mechanism underlying the feedback regulation of ribosomal biogenesis. In prokaryotes, in response to ribosomal stress or shortage of complementary rRNA, ribosomal proteins could bind to their own mRNA and repress ribosomal protein synthesis21. This negative feedback loop of ribosomal proteins and their mRNAs plays a vital role in fine-tuning of ribosome biogenesis in prokaryotic cells. Although this simple autoregulatory feedback circuit of ribosomal biogenesis has not yet been reported thus far in eukaryotes, this mechanism is resembled by the feedback regulation of c-Myc level and activity by RPL11 and RPL5, as the genes encoding both of the RPs are the transcriptional targets of c-Myc. Different from the prokaryotic mechanism, these eukaryotic RPs target c-Myc mRNA, instead of their own mRNAs, to execute their inhibitory action of c-Myc through a more complicated and RISC-mediated mechanism. Through this unique and sophisticated mechanism, ribosomal biogenesis is finely tuned so that no excess amounts of ribosomal proteins would be produced under normal growth conditions, as extra amounts of these proteins would also be toxic to cells by activating the p53 pathway48–51.
A number of ribosomal proteins have been shown to activate p53 by inhibiting MDM2-induced polyubiquitination of p53 in response to ribosomal stresses24–31, 33–35, 37, 38. However, how free ribosomal proteins act in p53-null cells is still largely unknown. Here, we show that RPL5 can suppress c-Myc expression by targeting its mRNA via RISC and miRNA independently of the p53 pathway (Figures 1–5). Together with RPL11, RPL5 among all of the MDM2-interacting RPs 22, 23 might play a more important role in preventing neoplasia by acting on both of the p53 and c-Myc pathways important in tumor development and progression. Indeed, both of RPL11 and RPL5 have been shown to negate tumor growth in genetic studies48–51. The negative action of RPL5 and RPL11 on c-Myc and MDM2 could be utilized as a strategy for the development of anti-cancer therapy in the future.
Materials and Methods
Cell lines, plasmids, and antibodies
Human HEK293 and H1299 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 U/ml penicillin and 0.1 mg/ml streptomycin at 37°C in a 5% CO2 humidified atmosphere. RPL5, RPL11, RPL23, RPL26, TRBP and c-Myc expression plasmids were described previously 28, 29, 43, 52. RPL11 mutant R75Q plasmids was a gift from Dr. Vousden39. c-Myc 3′UTR was amplified from HCT116 cells and inserted into pMIR Reporter vector. The primers used were: P1: 5′-GGACTAGTTCTCAGAGGCTTGGCGGGAAAAAGA-3′ and P2: 5′-CCCAAGCTTGGCTCAATGATATATTTGCCAGTTA-3′. Anti-L5 28 and anti-L11 22 have been previously described. Anti-Flag (Sigma), anti-c-Myc (N262, Santa Cruz Biotechnology), anti-actin (Sigma), anti-BrdU (IIB5, Santa Cruz Biotechnology), anti-TRBP2 (1D9, LifeSpan BioSciences, Inc), anti-HA (F-7, Santa Cruz Biotechnology), anti-myc (9E10, Santa Cruz Biotechnology) were commercially purchased.
Transient transfection, Western blot and co-immunoprecipitation analyses
As described previously 53, briefly, cells were transfected with indicated plasmids as shown in each figure by using TransFectin (Bio-Rad), following the company’s instruction. Unless specifically mentioned, forty eight hrs post transfection, cells were harvested and lysed in lysis buffer. The total protein concentration for each sample was determined and equal amount of total proteins were then subjected to SDS-PAGE, followed by Western blot (WB). Immunoprecipitation (IP) was conducted by using antibodies as indicated in the figure legends and described previously 54. Beads were washed three times with lysis buffer. Bound proteins were detected by IB with antibodies as indicated in the figure legends.
Reverse transcriptase-polymerase chain reaction and quantitative real-time PCR analysis
RT and Q-PCR for mRNAs were done by using the methods described previously28, 55. Briefly, quantitative real-time PCR was performed on an ABI 7300 real-time PCR system (Applied Biosystems) using SYBR Green Mix (Applied Biosystems). Relative gene expression was calculated using the ΔCT method, following the manufacturer’s instruction. All reactions were carried out in triplicate.
Luciferase reporter assays
Cells were transfected with pMIR-c-Myc-3′UTR and indicated plasmids (total plasmid DNA 1 μg/well) as indicated in figures. Luciferase activity was determined and normalized by a factor of β-gal activity in the same assay as described previously 56.
Ribonucleoprotein immunoprecipitation (RNP IP) assay
The RNP-IP assay was performed as described previously23 using anti-flag, anti-myc, or anti-GFP for ectopic proteins. Immunoprecipitated RNAs were extracted and analyzed by real-time PCR amplification using primers for c-Myc.
BrdU incorporation assays
BrdU incorporation assays were carried out by basically following the previously described protocol55. Cells were incubated in the presence of 10 μM of BrdU for 5 h. Cells were then fixed with 95% of ethanol and 5% of acetic acid, treated with 2 M HCl containing 1% Triton X-100, and stained with the monoclonal anti-BrdU antibody, followed by staining with Alexa Fluor 546 (red) goat anti-mouse antibodies and DAPI. Stained cells were analyzed under a Zeiss Axiovert 25 fluorescent microscope.
Knockdown of the endogenous mRNAs
siRNAs for RPL5 and RPL11 were described previously22, 28. siRNA for TRBP was purchased from Santa Cruz Biotechnology. Transfection of siRNAs were performed the same as that of normal siRNA as described previously 57 by using siLentFect™ Lipid (Bio-Rad), following the manufacturer’s protocol.
Acknowledgments
We thank Karen Vousden for providing us with the RPL11 mutant plasmid, Mu-shui Dai for offering some reagents, and Shelya X. Zeng as well as other members of the Lu laboratory for advices and active discussion. This work was supported in part by NIH-NCI grants CA095441, CA079721, CA129828, and CA172468 to H.L.
Footnotes
Conflict of interest:
The authors declare no conflict of interest.
References
- 1.Dang C, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verbeek S, van Lohuizen M, van der Valk M, Domen J, Kraal G, Berns A. Mice bearing the E mu-myc and E mu-pim-1 transgenes develop pre-B-cell leukemia prenatally. Molecular and cellular biology. 1991;11:1176–1179. doi: 10.1128/mcb.11.2.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soucek L, Whitfield J, Martins C, Finch A, Murphy D, Sodir N, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sodir N, Swigart L, Karnezis A, Hanahan D, Evan G, Soucek L. Endogenous Myc maintains the tumor microenvironment. Genes & development. 2011;25:907–916. doi: 10.1101/gad.2038411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Molecular cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 7.Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, et al. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science (New York, NY) 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- 8.Flores I, Murphy D, Swigart L, Knies U, Evan G. Defining the temporal requirements for Myc in the progression and maintenance of skin neoplasia. Oncogene. 2004;23:5923–5930. doi: 10.1038/sj.onc.1207796. [DOI] [PubMed] [Google Scholar]
- 9.Felsher D, Bishop J. Reversible tumorigenesis by MYC in hematopoietic lineages. Molecular cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Chu J, Shen X, Wang J, Orkin S. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright J, Brown S, Cole M. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Molecular and cellular biology. 2010;30:1411–1420. doi: 10.1128/MCB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pomerantz M, Ahmadiyeh N, Jia L, Herman P, Verzi M, Doddapaneni H, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nature genetics. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer N, Penn L. Reflecting on 25 years with MYC. Nature reviews Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 14.Eilers M, Eisenman R. Myc’s broad reach. Genes & development. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nesbit C, Tersak J, Prochownik E. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 16.Beroukhim R, Mermel C, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Riggelen J, Yetil A, Felsher D. MYC as a regulator of ribosome biogenesis and protein synthesis. Nature reviews Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 18.Gallant P. Myc, cell competition, and compensatory proliferation. Cancer research. 2005;65:6485–6487. doi: 10.1158/0008-5472.CAN-05-1101. [DOI] [PubMed] [Google Scholar]
- 19.Dang C. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai M-S, Lu H. Crosstalk between c-Myc and ribosome in ribosomal biogenesis and cancer. Journal of cellular biochemistry. 2008;105:670–677. doi: 10.1002/jcb.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gourse RL, Thurlow DL, Gerbi SA, Zimmermann RA. Specific binding of a prokaryotic ribosomal protein to a eukaryotic ribosomal RNA: implications for evolution and autoregulation. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:2722–2726. doi: 10.1073/pnas.78.5.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai MS, Arnold H, Sun XX, Sears R, Lu H. Inhibition of c-Myc activity by ribosomal protein L11. The EMBO journal. 2007;26:3332–3345. doi: 10.1038/sj.emboj.7601776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Challagundla KB, Sun XX, Zhang X, DeVine T, Zhang Q, Sears RC, et al. Ribosomal protein L11 recruits miR-24/miRISC to repress c-Myc expression in response to ribosomal stress. Molecular and cellular biology. 2011;31:4007–4021. doi: 10.1128/MCB.05810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 25.Sasaki M, Kawahara K, Nishio M, Mimori K, Kogo R, Hamada K, et al. Regulation of the MDM2-P53 pathway and tumor growth by PICT1 via nucleolar RPL11. Nature medicine. 17:944–951. doi: 10.1038/nm.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. The EMBO journal. 2004;23:2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Molecular and cellular biology. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. The Journal of biological chemistry. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 29.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Molecular and cellular biology. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin A, Itahana K, O’Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Molecular and cellular biology. 2004;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Wang J, Yuan Y, Zhang W, Guan W, Wu Z, et al. Negative regulation of HDM2 to attenuate p53 degradation by ribosomal protein L26. Nucleic acids research. 38:6544–6554. doi: 10.1093/nar/gkq536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yadavilli S, Mayo LD, Higgins M, Lain S, Hegde V, Deutsch WA. Ribosomal protein S3: A multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA repair. 2009;8:1215–1224. doi: 10.1016/j.dnarep.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, et al. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26:5029–5037. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 34.Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, et al. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Molecular cell. 2009;35:316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X, Hao Q, Liao J, Zhang Q, Lu H. Ribosomal protein S14 unties the MDM2-p53 loop upon ribosomal stress. Oncogene. 2013;32:388–396. doi: 10.1038/onc.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Wang W, Wang H, Wang MH, Xu W, Zhang R. Identification of ribosomal protein S25 (RPS25)-MDM2-p53 regulatory feedback loop. Oncogene. 2013;32:2782–2791. doi: 10.1038/onc.2012.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong X, Zhao Y, He H, Sun Y. Ribosomal protein S27-like and S27 interplay with p53-MDM2 axis as a target, a substrate and a regulator. Oncogene. 2011;30:1798–1811. doi: 10.1038/onc.2010.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun XX, DeVine T, Challagundla KB, Dai MS. Interplay between ribosomal protein S27a and MDM2 protein in p53 activation in response to ribosomal stress. The Journal of biological chemistry. 2011;286:22730–22741. doi: 10.1074/jbc.M111.223651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horn HF, Vousden KH. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene. 2008;27:5774–5784. doi: 10.1038/onc.2008.189. [DOI] [PubMed] [Google Scholar]
- 40.Miyazaki T, Liu Z, Kawahara A, Minami Y, Yamada K, Tsujimoto Y, et al. Three distinct IL-2 signaling pathways mediated by bcl-2, c-myc, and lck cooperate in hematopoietic cell proliferation. Cell. 1995;81:223–231. doi: 10.1016/0092-8674(95)90332-1. [DOI] [PubMed] [Google Scholar]
- 41.Pfeifer-Ohlsson S, Goustin A, Rydnert J, Wahlström T, Bjersing L, Stehelin D, et al. Spatial and temporal pattern of cellular myc oncogene expression in developing human placenta: implications for embryonic cell proliferation. Cell. 1984;38:585–596. doi: 10.1016/0092-8674(84)90513-0. [DOI] [PubMed] [Google Scholar]
- 42.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, et al. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO reports. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koscianska E, Starega-Roslan J, Krzyzosiak WJ. The role of Dicer protein partners in the processing of microRNA precursors. PloS one. 2011;6:e28548. doi: 10.1371/journal.pone.0028548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ota H, Sakurai M, Gupta R, Valente L, Wulff BE, Ariyoshi K, et al. ADAR1 Forms a Complex with Dicer to Promote MicroRNA Processing and RNA-Induced Gene Silencing. Cell. 2013;153:575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zarnack K, Konig J, Tajnik M, Martincorena I, Eustermann S, Stevant I, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bursac S, Brdovcak MC, Pfannkuchen M, Orsolic I, Golomb L, Zhu Y, et al. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20467–20472. doi: 10.1073/pnas.1218535109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macias E, Jin A, Deisenroth C, Bhat K, Mao H, Lindstrom MS, et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer cell. 2010;18:231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Golomb L, Bublik DR, Wilder S, Nevo R, Kiss V, Grabusic K, et al. Importin 7 and exportin 1 link c-Myc and p53 to regulation of ribosomal biogenesis. Mol Cell. 2012;45:222–232. doi: 10.1016/j.molcel.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu H. A ribosomal tactic to halt cancer. Nature medicine. 2011;17:930–931. doi: 10.1038/nm.2434. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Q, Xiao H, Chai SC, Hoang QQ, Lu H. Hydrophilic residues are crucial for ribosomal protein L11 (RPL11) interaction with zinc finger domain of MDM2 and p53 protein activation. The Journal of biological chemistry. 2011;286:38264–38274. doi: 10.1074/jbc.M111.277012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin Y, Zeng SX, Sun XX, Lee H, Blattner C, Xiao Z, et al. MDMX promotes proteasomal turnover of p21 at G1 and early S phases independently of, but in cooperation with, MDM2. Molecular and cellular biology. 2008;28:1218–1229. doi: 10.1128/MCB.01198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeng SX, Dai MS, Keller DM, Lu H. SSRP1 functions as a co-activator of the transcriptional activator p63. The EMBO journal. 2002;21:5487–5497. doi: 10.1093/emboj/cdf540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao JM, Lu H. Autoregulatory suppression of c-Myc by miR-185-3p. The Journal of biological chemistry. 2011;286:33901–33909. doi: 10.1074/jbc.M111.262030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin Y, Dai MS, Lu SZ, Xu Y, Luo Z, Zhao Y, et al. 14-3-3gamma binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. The EMBO journal. 2006;25:1207–1218. doi: 10.1038/sj.emboj.7601010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun XX, Dai MS, Lu H. Mycophenolic acid activation of p53 requires ribosomal proteins L5 and L11. The Journal of biological chemistry. 2008;283:12387–12392. doi: 10.1074/jbc.M801387200. [DOI] [PMC free article] [PubMed] [Google Scholar]