Abstract
The US National Institutes of Health (NIH) has developed the Biomedical Translational Research Information System (BTRIS) to support researchers’ access to translational and clinical data. BTRIS includes a data repository, a set of programs for loading data from NIH electronic health records and research data management systems, an ontology for coding the disparate data with a single terminology, and a set of user interface tools that provide access to identified data from individual research studies and data across all studies from which individually identifiable data have been removed. This paper reports on unique design elements of the system, progress to date and user experience after five years of development and operation.
Keywords: Health Information Exchange, Patient Privacy, Patient Advocacy, Electronic Health Records, Clinical research, translational research, research data repository, research data warehouse, ontology
1. Introduction
Institutions that engage in both patient care and clinical research are increasingly turning to the development of clinical data repositories (or “warehouses”) as resources for gathering data collected during routine patient care with the intent of re-using them to answer secondary questions.[1] The National Institutes of Health (NIH) is no exception. The Clinical Center, a 240-bed hospital on the NIH's Bethesda, Maryland campus is devoted exclusively to research. Since 1976, the Clinical Center has maintained two contiguous electronic health record systems (EHRs). The data from those systems, together with data from research systems at many of the NIH's 26 other institutes and centers, comprise an important collection of longitudinal information about common and rare conditions, diagnosed and treated in conventional and unconventional ways. In order to maximize the value of these data, the NIH launched the development of the Biomedical Translational Research Information System (BTRIS; pronounced BEE-triss), charged with providing a single, central resource that could support the data requirements of primary researchers, as well as those who seek to re-use old data to answer new questions related to research or hospital operations.
Some aspects of BTRIS have been previously described briefly, including its general design,[2] and a query tool for retrieval of data with personal identifiers removed.[3] The purpose of this paper is to provide further details about BTRIS's database design, terminology management, and query tools, and to provide a comparison with the architectures of other major current clinical data repository efforts.
2. Background
2.1 NIH Clinical Research Data Environment
From 1976 to 2004, the Clinical Center's electronic health record was a version of the Medical Information Management System from Technicon Data Systems (Tarrytown, NY). Locally referred to as MIS (for “Medical Information System”), it supported entry and review of vital signs and orders, and provided access to dictated reports and clinical and pathology laboratory test results. While most of the data were presented in text form, a dedicated graphics computer was integrated with MIS to support graphical review of clinical laboratory values, vital signs and medication administration events.[4]
In 2004, the Clinical Center installed Sunrise Clinical Manager from the Eclipsys Corporation (Boca Raton, FL; now Allscripts, Chicago, IL). Known as CRIS (for “Clinical Research Information System”) , the new system includes the same types of data collected in MIS, as well as clinical documents entered manually by clinical personnel, messages from CRIS's automated alerting function, reports from additional ancillary systems (such as the pulmonary function and cardiology systems), and document images in Portable Document Format (PDF).
Despite a number of innovative customizations to support clinical research, MIS and CRIS are essentially patient care systems. Although all the patients in the Clinical Center are research subjects in one or more studies at any given time, many of the study-specific data are collected separately, for example as responses to surveys, laboratory results obtained from research laboratories, or data entered into case report forms. Much of this information is placed in various clinical trials data management systems (CTDMSs). Table 1 lists the major systems used by research groups in NIH institutes.
Table 1.
NIH Clinical Research Systems
| System Name | Institute | System Type | Developer | Data Domains |
|---|---|---|---|---|
| Medical Information System (MIS) * | Clinical Center | EHR | Commercial | Demographics, Laboratory Tests, Blood Bank, Medications, Microbiology, Radiology, Vital Signs, Clinical Notes, Anatomic Pathology |
| Clinical Research Information System (CRIS) | Clinical Center | EHR | Commercial | Demographics, Subject-Study Attribution, Vital Signs, Clinical Documentation, Alerts, Allergies, Observations, Document Images, Medication Administration, Medication Orders, Other Orders, Admission/Discharge/Transfer |
| Softlab | Clinical Center | Ancillary Department System | Commercial | Clinical Laboratory Tests, Microbiology Tests, Anatomic Pathology, Blood Bank Tests, Blood Bank Products |
| Softmed | Clinical Center | Medical Records Department System | Commercial | Admission Notes, Discharge Notes, Diagnoses, Other Dictated Notes |
| Vmax * | Clinical Center, NHLBI | Ancillary Department System | Commercial | Pulmonary Function Reports |
| Jaeger | Clinical Center, NHLBI | Ancillary Department System | Commercial | Pulmonary Function Reports |
| Pain and Palliative Care System | Clinical Center | Ancillary Department System | Clinical Center | Pain and Palliative Care Notes |
| LinkTools | Clinical Center, NHLBI | Ancillary Department System | Commercial | Electrocardiograms |
| ProSolv | Clinical Center, NHLBI | Ancillary Department System | Commercial | Echocardiology Reports |
| RadNet | Clinical Center | Ancillary Department System | Commercial | Radiology Reports |
| Carestream | Clinical Center | Picture Archiving and Communication System | Commercial | Radiographic Images |
| Protrak | Clinical Center | Protocol Services Department System | Clinical Center | Studies, Investigators |
| Clinical Research Information Management System of the NIAID (CRIMSON) | NIAID | CTDMS | NIAID | Study-Subject Attribution, Laboratory Tests, Medications, Patient Problems |
| Clinical Research Database (CRDB) | NIAAA | CTDMS | NIAAA | Assessments (Surveys) |
| Labmatrix | NCI | CTDMS | Commercial | Biospecimens |
| Cancer Central Clinical Database (C3D) | NCI | CTDMS | NCI | Study Attribution, Laboratory Tests, Case Report Forms |
| Clinical Trials Database (CTDB) | NICHD, NIAAA, NIDDK, Clinical Center | CTDMS | NICHD | Encounter Forms |
| Labmatrix | NHGRI | CTDMS | Commercial | Biospecimens, Case Report Forms |
| Varsifter | NHGRI, NIMH | Laboratory Database | NHGRI | Whole Exome Sequences |
indicates archived data, domains in italics are not yet loaded into BTRIS
CTDMS - clinical trials data management system, EHR - electronic health record, NCI - National Cancer Institute, NHGRI - National Human Genome Research Institute, NHLBI - National Heart, Lung and Blood Institute, NIAAA - National Institute on Alcohol Abuse and Alcoholism, NIAID - National Institute of Allergy and Infectious Diseases, NICHD - National Institute of Child Health and Human Development, NIDDK - National Institute of Diabetes and Digestive and Kidney Diseases, NIMH - National Institute of Mental Health
While it would be inappropriate to intermingle many of these data with the patient care record (for example, test results from non-certified laboratories), merging data from the EHR with those of the CTDMs is necessary to support research processes. However, some institute system managers have found that extracting data from CRIS is difficult, while many users of these systems have found that the high quality of the systems’ data capture functions are not always matched by their data reporting functions, nor by their exporting functions, to support analysis by other programs. Furthermore, none of these systems supports secondary studies across previous studies, institute sources, and data types. Such studies have generally been accomplished with manual chart reviews in the Medical Records Department. The needs for improved access to data prompted the NIH to pursue development of a trans-institute clinical data repository.
2.2 Initial Planning
The concept of an NIH-wide clinical research data repository has been a prominent feature of strategic planning since 1999. In 2005, a group of intramural investigators developes a high level list of requirements, entitled the “Clinical Research Data Warehouse Blueprint Report”, that identified core system requirements, use cases, data standards and recommendations for the governance of this trans-NIH initiative. An outside consulting group then led the development of the business case, including interviews of key stakeholders within the NIH intramural community, and an analysis of the clinical research process. Interviews conducted at other healthcare institutions that had developed successful clinical data repositories (Partners Healthcare, Kaiser Permanente, Intermountain Healthcare, Medstar Health and Regenstrief Institute) were used to benchmark system capabilities and implementation timelines. The final business case and initial request for funding was presented to NIH governance in January of 2007. In March of 2007, Dr. Elias Zerhouni, then Director of NIH, approved the clinical research data repository project. Recruitment for a program director was begun in April 2007 by one of us (EJA) and one of us (JJC) was selected to begin in December 2007. The repository was officially named BTRIS and the plan was presented to the NIH community on February 28, 2008.[5]
2.3 Data Access Policy Development
As with any system that contains sensitive personally identifiable information (PII), clear data access policies needed to be established from the outset. The NIH's original vision for BTRIS included support for active clinical research using identifiable research subject data. NIH has clear policies on which researchers can see which data on which subjects, with oversight by institutional review boards (IRBs). Since permission for data access is obtained in advance of actual data collection (through IRB approval and subject consent), appropriate data access controls that BTRIS needed to implement were well-defined.
A second purpose envisioned for BTRIS was to allow access to previously collected data that excluded personal individual identifiers. Again, the NIH has clear policy for such use, regulated by the Office of Human Subjects Research and Protection (OHSRP). However, requiring a BTRIS user to submit paperwork every time a data query was planned would stifle creative data exploration. We therefore sought to establish a policy that would provide immediate, automatic approval for data retrieval if the user provided required information and agreed to adhere to relevant data use policies.
Further complicating matters was the need to protect not only the research subjects, but the researchers themselves. There was general agreement in the research community that data should be shared, but concerns were raised about the possibility of a BTRIS user obtaining EHR data that might also be important for an ongoing study. Publication of findings based on an analysis of BTRIS data could inadvertently “scoop” the research being carried out by the researchers who originally commissioned the capturing of the data. While this risk had always been inherent in the use of data obtained from the Medical Records Department or from MIS and CRIS, researchers anticipated an increased risk with the ready availability of data that BTRIS would provide.
Neither OHSRP nor the IRBs have, in their mission, the protection of the intellectual interests of the investigators. While NIH clearly needs to balance its duty to the investigators (by helping them get credit for their research) with its duty to the American people (by obtaining the maximum benefit from data collected with public funds), no formal written policy existed that clearly laid out when and how investigators could control the reuse of their data. The establishment of BTRIS operating procedures, then, became an opportunity to clarify these policies and also to enforce them, thereby offsetting the increased exposure of data with increased transparency of access to and ownership of the data in a way that was not possible with paper or electronic health records.
2.4 Influential Architectures
2.4.1. The Columbia University Clinical Data Repository
The initial NIH study that led to the BTRIS project identified an environment with many current and historic data sources in a constant state of evolution, with a set of data needs that varied greatly across the various research and administrative constituencies. In many ways, the NIH situation was analogous to that of the Columbia Presbyterian Medical Center in New York City, which found itself in the late 1980s with a growing set of ancillary data systems and no central mechanisms for data storage or access. The Columbia University Center for Medical Informatics (now the Department of Biomedical Informatics) responded to that need with an innovative design, based on lessons from HELP system, developed decades earlier at LDS Hospital and the University of Utah in Salt Lake City.[6]
One key aspect of the Columbia system is a relatively simple, entity-attribute-value (EAV) database,[7] in which all patient data are represented using a single, linked data model that minimizes the number of data tables needed to accommodate information from an ever-changing set of ancillary departmental systems. By 2008, the Columbia model had been in place for almost 20 years, having successfully supported a succession of clinical applications, including one of the first Web-based electronic health record systems that remains in use today.[8-10] While other designs for clinical data repositories were beginning to emerge, the tried-and-true Columbia design appeared to be a good fit for the environment and requirements at NIH, with similar approaches being adopted elsewhere.[11]
2.4.2 The Medical Entities Dictionary (MED)
A key component of the Columbia design is a single, unified controlled terminology, called the Medical Entities Dictionary (MED),[12] that is used to code all data from the various sources. Applications that interface with the repository (including both data display systems and data entry systems) make use of the terminology to adapt to changing data and functions in a dynamic manner, based on knowledge in the terminology, rather than reprogramming.[13]
The MED follows a well-established set of “desiderata”[14,15] for biomedical concept representation and includes a number of key design features for representing local and generic universal terms:
Every local term from a data source terminology is considered to correspond to a unique concept unless the source was referencing some outside concept set,
Terms are organized into hierarchies, with no inherent limit on the number of parents or children a term can have, nor how deep it can be placed in the hierarchy,
Terms can be related to each other with semantic relationships that convey information about the meaning of the terms, and
Terms are never deleted once they have been used in programs or the database.
Each term was added to the MED as a distinct concept and then knowledge about the term was added; e.g., the relationships among panels, specimens and tests. This knowledge was useful for organizing the terms into a natural hierarchy,[12] for automating a variety of decision support tools,[13] and for ongoing maintenance as systems renamed existing terms and added new terms, and as the hospital introduced new systems with new terminologies.[16] This experience informed a a similar approach to terminology management in BTRIS (see section 4.4).
2.4.3 Other Contemporary Technologies
By 2007, development of single-institution clinical data repositories was becoming commonplace in the US and elsewhere. As is often the case throughout the history of clinical informatics, individual efforts began to give rise to interest in developing and applying standards to support interoperability and reusability. For example, the Starbrite[17] and BRIDG[18] projects had just recently published their experience with exchanging clinical trials data among several institutions using data models from the Clinical Data Interchange Standards Consortium (CDISC) and Health Level 7 (HL7). These efforts appeared relevant to the NIH goals for consolidating data from multiple sources into a single repository. However, because the NIH source systems adhered to HL7, a deliberate attempt to convert data to CDISC prior to integration did not seem expedient, especially given that the standards themselves were evolving.
Terminology standards were also emerging during this time. However, with the exception of the International Classification of Diseases, 9th Edition with Clinical Modification (ICD-9-CM), source systems had largely ignored standard terminologies for capturing or coding their data. The Common Terminology Services 2 (CTS2) standard was fairly mature at that time, but offered few advantages over a terminology service modeled on the MED, given that our internal systems needed terminology queries not supported by the standard (such as text-based searches restricted to terminology classes) and that no external systems required the ability to query our terminology.
Finally, we examined the data and ontology models of Informatics for Integrating Biology and the Bedside (i2b2), a project with the goal of providing investigators with a suite of tools to support clinical and translational research.[19] A careful comparison of the i2b2 repository and ontology models was made with those of Columbia. The i2b2 repository model did not seem to offer any particular advantage over Columbia's with the Columbia model having a tighter connection to its ontology model. The i2b2 ontology model had many similarities to the MED, but failed to support the multiple hierarchies and other inter-concept relationships that are a critical aspect of the MED's functionality.
In each case, the emerging standards were not rejected, but rather considered carefully and had some influence on the directions in which BTRIS evolved. Indeed, throughout the project, we made sure that our designs would be compatible with these standards if a need arose to exchange data in CDISC, respond to queries to our terminology server, or act as an i2b2 data repository “cell”.
2.4.4 Development of the BTRIS “Demo” Version
The core of the BTRIS development team (described in the Results section, below) was in place by March of 2008 and initial data sets were obtained from MIS, CRIS and one CTDMS (CRIMSON; see Table 1) in May. In order to better understand the requirements for data representation, storage and retrieval, as well as to elicit use requirements, the BTRIS team developed a prototype that provided limited, but diverse, functionality.
For the initial data set, we selected a convenience sample of 29 studies with approximately 4,000 subjects, and created a simple database containing demographic data (CRIS), vital signs (CRIS), laboratory test results (CRIS, MIS, CRIMSON), medication administration data (CRIS, CRIMSON), radiology reports (CRIS), allergies (CRIS) and patient diagnoses (CRIS). We then constructed an initial controlled terminology to represent and code the terms found in these data sets.
We chose a “business intelligence” tool (Business Objects, SAP America, Inc., Newtown Square, PA) to serve as the initial user query tool. Data queries were carried out using query templates that were created for each data type. Templates required the user to specify one or more research studies of interest and then allowed optional specifications for subsets of research subjects, date ranges, specific data based on controlled terms, and value ranges for those data. Principal investigators were able to view identified data associated with their own studies. For demonstration purposes, we created a copy of the database in which the individual identifiers and data values were scrambled to prevent actual reuse, but could be used as a proof of concept for other users.
The demonstration instance was made available to the NIH research community for two months starting in early August, 2008.[21] Although the development of the prototype delayed initiation of the first version of the full BTRIS system by four months, the information gathered about both data and user requirements was invaluable for gaining an understanding beyond the initial consultant report of what BTRIS would need to become. Demonstration of the prototype was also invaluable for eliciting feedback from future potential users and obtaining support from various stakeholders, such as researchers, clinical directors, administrators, and funding committees.
3. Methods
In general, BTRIS development has followed good software development practices, with a focus on four basic requirements: the ability to accommodate any type of data that might be encountered, a database design optimized for the kinds of queries likely to be performed, use of a controlled terminology that would include detailed terms encountered in data as well as the high-level concepts that users were likely to include in their queries, and a user interface that would empower NIH researchers to carry out their own queries.
3.1 Data Sources
As noted in Table 1, the NIH information environment includes a wide variety of data sources, including hospital departmental systems, central EHRs and institute CTDMSs. Methods required for accessing the data from these systems are equally varied. In some cases, data are exported as standard HL7 messages, of the sort typically seen in hospitals.[22] In other cases, archived data sets are available, each with its own unique structure. In still other cases, data are extracted using software that can directly query the source system databases using Open Database Connectivity (ODBC).[23]
In each case, a specific extraction-transformation-load (ETL) process was developed, tested and then set to run at appropriate intervals (once for archived data sets; daily or weekly for active systems). The design of ETL processes for health data repositories, including the use of translation tables to convert data coded with terminologies from source systems into the repository's terminology, is a mature process.[24]
3.2 Database Design Goals
As with the ETL processes, we designed the BTRIS database to accommodate the wide variety of data from our sources. As is common in repository design, we distinguished between data elements that represent relatively stable statements about real-world objects (such as a subject's race, gender and date of birth) and facts about those objects that would be added to the database in a monotonic manner (such as body weights, diagnoses and laboratory results). As with most repositories at the time, we chose to implement BTRIS using a relational database.
While the flexibility of pure EAV data models held some appeal for dealing with new types of data as they appeared, we also sought to organize data elements to maximize efficiency for retrieval. For example, while we expected that users would want to query for laboratory test results grouped in many different ways, we also expected that they would always want those data related to a research subject identifier as well as particular times and dates, and that some attributes such as units of measure and normal range would be required as well (hard coded columns).[25] We did not, on the other hand, expect users to retrieve only normal ranges or only units of measure. It therefore seemed practical to include them in the same table rows as the laboratory test results, rather than require additional complexity in the query (that is, an additional relational joins to other rows in the same table or some subordinate tables).
3.3 Controlled Terminology Philosophy
The use of controlled terminologies in local NIH systems is completely analogous to the situation at the Columbia University Medical Center: each source system has one or more of its own local terminologies.[12] Standard terminologies or mappings from local to standard terms are rarely used, terminologies are constantly changing, and good terminology practices are not fully embraced. We therefore established from the outset the Research Entities Dictionary (RED) to be a terminological knowledge resource that, like the MED for Columbia, would support as many aspects of BTRIS as possible, including data modeling, data coding, ETL processes, user interface functions, and data retrieval.
Although the MED was originally developed using a commercial knowledge engineering tool, available software was found to be inadequate in the late 1980s and early 1990s, which led to the development of “home-grown” terminology management tools. Twenty years later, the state of the art has advanced to the point where a number of tools are available commercially or as open-source products. We therefore sought an ontology development environment that would support both our desire to create a principled knowledge base of clinical research data concepts, and meet the practical needs for data modeling and storage in the real world.
3.4 User Interface Requirements
The principle requirement for the BTRIS user interface was that users can access data (whether from their own studies or from across all studies) in an independent, “self-service” manner so as to allow users to be creative in their queries and be able to carry them out in a timely manner. This requirement was also necessary due to the limited customer support resources available in the project budget. To meet this requirement, the development process included gathering user requirements, assembling a user group to provide feedback on design features and user interface look-and-feel, and conducting user acceptance testing prior to release of any new function. In general, the users input translated to requests for particular types of reports, features for customizing the data selection criteria of the reports, and formatting of the report output. As with any complex system, this was an iterative process, with users able to provide more precise feedback after preliminary versions of new features were available for use.
A second key requirement was for the system to enforce data access policies. Besides user authentication, authorization to view specific identifiable data is limited based on the subject's participation in a study, the period of involvement in the study, the study status (studies must be active for users to see identifiable data), the user's role on the study (obtained from the NIH protocol tracking system; see Protrak in Table 1 and Section 4.3.1), and whether the study's principal investigator has chosen to give the user access to the study's data. Different policies related to the re-use of data from other studies must also be enforced, including removal of individual personal identifiers, obtaining a supervisor's approval to conduct queries, and notification of original investigators when their data are being accessed. While all users have training in these policies, many will be unfamiliar with their enforcement. For example, CRIS provides users access to patient data based on their clinical roles, not research roles, which are regulated by very different policies.
4. Results
The development and deployment of the BTRIS prototype in August, 2008[5] provided the means by which to refine user requirements and NIH policy. Subsequent acquisition of data, development of the user interface, and specification of data queries and reports proceeded in parallel, with deployment of the full version of BTRIS in July, 2009.[26] Since that time, requirements have shifted, and budgets have tightened. The current status of BTRIS is reported here; lessons learned are described in the Discussion.
4.1 Development Team
Staffing of the BTRIS project has shifted over time, with make-up varying as the focus shifted from requirements gathering through development, to deployment (with its attendant maintenance and user support tasks). The current composition of the staff is shown in Table 2.
Table 2.
BTRIS Development and Maintenance Staff
| Role | Number | Responsibilities |
|---|---|---|
| Project Director, Deputy Director | 2 | Project leadership; strategy; presentation; overall management |
| Program Manager | 1 | Project scheduling; budget; staffing; coordinating team communication |
| Data Architects, Administrators | 3 | Data warehouse management; design ETL processes; tune SQL queries; backups |
| ETL Developers | 3 | Develop and maintain all ETL processes |
| Business and Data Analyst, Data Mapping, Customer Support | 3 | Manage the requirements gathering and documenting process throughout the software development lifecycle; manage system configuration, map incoming data to BTRIS data model; provide customer support, including training |
| Subject Matter Expert | 1 | Evaluation of all requirements documentation; mediate queries; data quality control |
| Application Development | 3 | Architect applications; design and develop non-ETL applications; business intelligence tool administrator; design and development reports |
| Quality Assurance and Testing | 2 | Test all ETL data loads, applications, and reports prior to release to production; report back to team on pass/fail status of requirements; document traceability of requirements, test plans, test scripts. |
| System Administrators, Technical Support | 2 | Manage all servers with updates and backups. |
| Medical Ontologists | 2 | Annotate and classify all terminology from disparate data sources |
4.2 Extraction, Transformation and Loading (ETL) Processes
For the most part, the ETL processes developed for BTRIS have faced challenges, and developed solutions, similar to those of other repository projects. Due to the heterogeneous, dynamic nature of the NIH data environment, some unique aspects of those solutions are worth noting.
From the start of the project, and continuing into the foreseeable future, data sources become available to BTRIS in a sporadic manner. The framework for carrying out loading processes has therefore demanded flexibility to accommodate new sources on an ad hoc basis. Despite this variability some commonalities have appeared, such as error handling, auditing, logging and reporting, which have allowed us to reuse extraction techniques and add them to our daily routine in a modular way. Furthermore, we find that some new sources are similar to existing sources, such as a new HL7 interface, a different local installation of LabMatrix (a CTDMS from Biofortis, Columbia, MD), or new table from CRIS. This allows us to adapt software routines for reuse and incorporate them into the daily ETL work flow.
A second unusual aspect of the BTRIS ETL process is that it makes use of the RED to create the translation tables used to transform the controlled terms used by local systems into the coding system used in the BTRIS tables. This approach is similar to the one used at Columbia.[10] When data source system managers notify the BTRIS team of terminology updates, the updates are applied to the RED (typically as new terms and term codes). New ETL tables are generated nightly, prior to initiation of the daily ETL processes, so that new data from the sources are immediately recognized. In the case where the data source system manager does not notify the BTRIS team (which happens more often than not), new data will not match the ETL translation table, and will be stored with a place-holder RED code. This allows the data to be stored and retrieved in limited ways; information about the non-matching codes is placed in a “RED Pending” table. RED managers use this table to apply updates to the RED, so that the subsequent ETL table will have the new codes. The nightly ETL process includes the replacement of the placeholder codes with more specific codes as they become available in the RED.
Archived data have been loaded from the MIS database. On-going data are loaded daily from CRIS and eight other institute systems. Table 1 shows the data sources and data domains. Table 3 shows the daily records counts from each source.
Table 3.
Record counts for a typical daily load from BTRIS data sources (see Table 1 for system names)
| System | Data Domain | Row Count |
|---|---|---|
| CRIS | Subjects | 524 |
| CRIS | Study-Subject Attribution | 257 |
| CRIS | Vital Signs | 31,930 |
| CRIS | Medication Orders | 2,998 |
| CRIS | Medication Administration | 9,724 |
| CRIS | Client/Clinical Documents | 16,942 |
| CRIS | Alerts | 7,724 |
| CRIS | Allergies | 183 |
| CRIS | Observations | 210,548 |
| CRIS | Document Images | 1401 |
| CRIMSON | Medications | 1,961 |
| CRIMSON | Laboratory Tests | 7,923 |
| CRIMSON | Study-Subject Attribution | 28 |
| CRIMSON | Problem Package | 32,626 |
| Protrak | Studies | 112 |
| Protrak | Investigator Notifications | 51 |
| SoftLab | Microbiology | 1,969 |
| SoftLab | General Laboratory | 34,300 |
| SoftLab | Anatomic Pathology | 152 |
| SoftLab | Blood Bank Products | 124 |
| SoftLab | Blood Bank Tests | 574 |
| TDE | Red Code Updates | 100 |
| CD3 | Study Attribution | 14 |
| CD3 | Laboratory Tests | 1,756 |
| CD3 | Non-Laboratory Data | 17,645 |
| NIAAA | Assessments | 12,646 |
| LinkTools | EKG | 1,203 |
| RadNet | Radiology | 948 |
| SoftMed | Diagnoses | 2,264 |
| ProSolv | Echocardiology | 753 |
| Labmatrix | Biomaterials (Specimens) | 32 |
| CTDB | Encounter Forms | 3,757 |
4.3 Data Model
The BTRIS data model comprises four broad sets of information: relatively stable data about various entities (subjects, studies, investigators and users), subject facts that are acquired in a monotonic fashion (that is, new data that add to, rather than over-write, existing data), knowledge from the RED (discussed below), and various tables of data related to the ETL processes (such as data importation error logs, staging tables, etc).
4.3.1. Subject-Study-Investigator-User Data
The data that relate studies to the responsible investigators, the subjects enrolled in the studies, and the users with permission to access the studies are stored in a traditional entity-relation model. In general, we use CRIS as the system of reference for information about the subjects, such as demographic information. The Protrak system (managed by the Office of Protocol Services) is the system of reference for study and investigator information. CRIS is the main source for information relating subjects to studies, but can be over-ridden by information from other institutional systems or provided by the investigator using BTRIS Preferences (described below). All subjects are assigned a global unique identifier (GUID) in a master person index that allows mapping of their data from sources that may use different subject identifiers. A more complete entity-relation diagram is available on line.*
4.3.2. Subject Facts
All facts must have a subject identifier (typically the Clinical Center medical record number, although others are permitted; all are mapped to subject GUIDs), a date/time stamp, and at least one codable concept from the RED. We generally classify facts into those that provide some additional data obtained from the subject (which we refer to as “observations”) and those that do not (which we refer to as “events”). In general, we find that events and observations are related. For example a medication order (event) may relate to one or more medication administration records (observations that note such things as drug amounts given and reactions noted), while laboratory orders (events) may relate to one or more specific test results (observations).
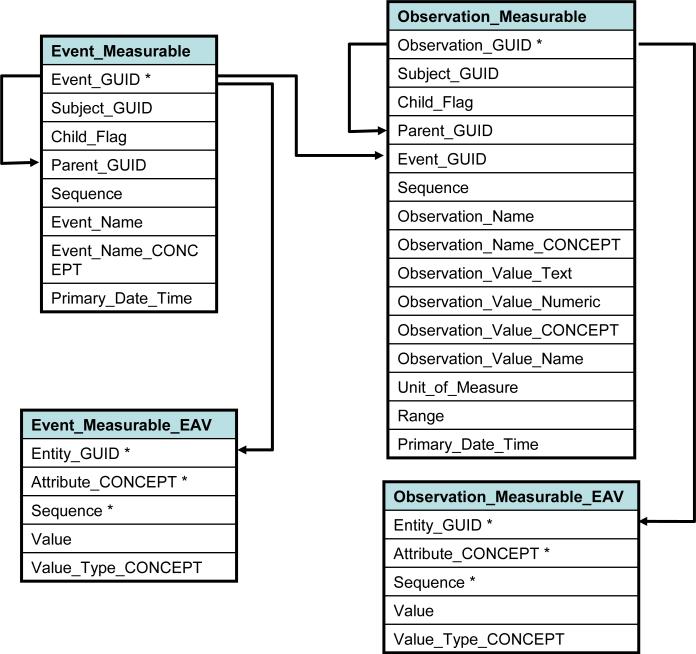
We further characterize observations as those that report some specific measurement (typically including units of measure and normal values or ranges), those that report information about a substance administered (such as a medication), and those that do not fall into either of these two categories (such as diagnoses and text reports). We refer to these three types of observations as “measurable”, “substance” and “general”, respectively. Although the three types of events and three types of observations have many common elements, for performance reasons we have divided data into three pairs of event-observation tables corresponding to the three observation types. Figure 1 shows a simplified data model relating events and observations. The full data model diagram is available on line, as noted above.
Figure 1.
Simplified entity-relation diagram showing table relationships in BTRIS. Table names are in bold, column names in regular font; column names marked with (*) indicate unique key for table; solid arrows show one-to-many relationships between table rows. Tables shown are for measureable observations and their corresponding events. “GUID” stands for :”globally unique identifier”, with Subject_GUID being the identifier for human subjects. Parent, Child and Sequence rows convey structure among entities and observations. The full specifications are available at http://people.dbmi.columbia.edu/~ciminoj/BTRIS-Data-Model.xhtml.
As described in the Methods section, we sought to include in each row of the fact tables those data elements that would either be the subject of a user's search (such as a test name) or be requested frequently with search results (such as the actual test result). Current row counts for each of the six event and observation tables are shown in Table 4. Additional attributes that are provided by source systems that do not meet the search-and-retrieval criteria described above, but are nonetheless deemed to be important for specific purposes, are stored in EAV tables. For performance reasons, one EAV table was created for each of the six “parent” event/observation tables, yielding a fact database of twelve tables in total. The EAV tables store additional rows of information in a one-to-many manner, linked to their master tables by unique event identifiers (commonly referred to as “nesting”[25]). The relationships between EAV and parent tables, along with the attributes found in each EAV table are shown in Figure 1. Table 4 lists the current row counts for the EAV tables.
Table 4.
Current Sizes of Main BTRIS Tables
| Table Name | Row Count | EAV Row Count |
|---|---|---|
| Event_General | 23,857,814 | 252,435,445 |
| Event_Measurable | 87,275,050 | 333,692,094 |
| Event_Substance | 23,530,200 | 344,287,179 |
| Total Events | 134,663,064 | 930,414,718 |
| Observation_General | 217,996,211 | 1,836,001,442 |
| Observation_Measurable | 223,138,934 | 790,428,492 |
| Observation_Substance | 17,085,460 | 74,955,346 |
| Total Observations | 458,220,605 | 2,701,385,280 |
| Total Events and Observations | 592,883,669 | 3,631,799,998 |
| Subject Table (one row per subject) | 485,234 | Not Applicable |
4.3.3. Primary Date and Time
In addition to conversion of subject identifiers to GUIDs and local terms to RED codes, we also take special care in modeling the dates and times associated with data. While some data come with a single date/time stamp, some data come with multiple values. We store each of these values in the appropriate EAV table with an appropriate RED code; however, we choose the one value that we determine to be the most clinically relevant to serve as the “primary” date/time stamp for the data and store that in the appropriate main event or observation table. For example, when laboratory data are tagged with multiple dates and times, we selected from the following precedence (with increasing clinical relevance): date reported, date/time reported, date/time specimen received in laboratory, and date/time specimen obtained from subject.
4.4 The Research Entities Dictionary (RED)
We chose the Terminology Development Environment (TDE, Apelon, Ridgefield, CT) as the platform for development and maintenance of the RED. TDE provides a suite of ontology management tools that support our terminology requirements, including support for multiple hierarchies, non-hierarchical semantic relationships, attribute inheritance, and additional terminology attributes that can be individually annotated with “qualifier” values. We used a modified version of TDE, obtained from the National Cancer Institute's Center for Biomedical Informatics and Information Technology (CBIIT), that provided additional practical functionality for ontology management.[27]
With each source system using one or more controlled terminologies, integrating their terms into the RED has proved challenging. This is particularly true for terms used to represent older data, where the precise meanings of the terms have not been clearly documented. The intent of the RED is to include formal, explicit representation of the meanings of each term. However, the first priority is to assign a unique identifier for each term, in order to facilitate storage and retrieval of data encountered in the ETL processes (Figure 2). Terminology inclusion follows a sequence of: reduction of terms to concepts, bulk loading of concepts, addition of local terminology information, initial classification of concepts, addition of semantic information, and further refinement of classification.
Figure 2.
Representative example of a RED Concept. Attributes (properties, roles and associations) are shown in bold, qualifiers are shown in italic, and the values of attributes and qualifiers are shown in plain text. The concept corresponds to a particular NICHD research study from the CTDB system. Of particular note, it has two parents (“superconcepts”) and is related to several other RED concepts that correspond to specific encounter forms for the study. Parts of this concept description have been edited out for clarity.
The reduction of terms to concepts is sometimes straightforward: if each coded term in a local source terminology is recognized as representing a unique concept that is specific to the source system, then there is a one-to-one mapping from terms to concepts. This is not always the case, however. For example, we found 87,161 unique medication terms used in the MIS pharmacy data set, of which many were variant spellings and abbreviations of each other. Automated lexical analysis, together with a great deal of manual review, allowed us to reduce the terminology to 69,543 unique concepts.
Once the final set of concepts is established, loading them into TDE, along with local term names and codes, is straightforward. Classification of terms into hierarchical relationships is more time-consuming. Available standards for some terminology domains, such as diseases, are available, but are lacking for our largest term sets. We examined, for example, LOINC (for laboratory tests) and RxNorm (for medications) and found that low-level terms are abundant but that useful higher level classes are inadequate for our purposes. Instead, we relied on a combination of automated lexical processes with review by human domain experts to organize the concepts into a hierarchy that was evaluated during user testing to determine its ability to meet users’ needs for browsing and data retrieval. This organization process is on-going and perhaps never-ending. Table 5 presents some statistics on the major source terminologies included in the RED. Table 6 presents some statistics on RED content. Figure 2 shows the representation of a typical RED concept, as it appears in TDE.
Table 5.
Source Terminology Statistics with Reduction of Unique Terms to Concepts (see Table 1 for source system names)
| Data Source | Number of Original Terms | Number of RED Concepts |
|---|---|---|
| CRIS | 324859 | 119635 |
| MIS | 107258 | 83424 |
| Blood Bank | 3163 | 2720 |
| NICHD Forms, Sections, and Protocols | 11355 | 11145 |
| NICHD Questions | 17690 | 15080 |
| NICHD All | 29045 | 26225 |
| NIAAA | 4758 | 4693 |
| NCI LabMatrix | 343 | 342 |
| NCI C3D | 2142 | 2084 |
| NCI all | 2485 | 2426 |
| NIGRI | 428 | 388 |
Table 6.
Selected Statistics for the Research Entities Dictionary
| Entity | Number of Entity Types | Total Instances |
|---|---|---|
| Concepts | 1 | 276,600 |
| Diagnosis, Procedure, Problem and Rare Disease | 40,976 | |
| Laboratory | 31,522 | |
| Radiology and Imaging | 10,711 | |
| Specimens | 1,856 | |
| Medications | 86,477 | |
| Survey Forms and Questions | 26,106 | |
| is-a Relationships | 1 | 477,845 |
| Roles | 91 | 25,160 |
| Associations | 17 | 71,365 |
| Synonyms | 3 | 873,915 |
| Non-Synonym Properties | 106 | 1,554,876 |
| Qualifiers | 28 | 1,813,821 |
Concept: the main components of the RED, which correspond to a term from a source system; is-a Relationship: the hierarchical relationship in the RED; Role: a concept attribute that expresses inheritable relationships between concepts; Association: a concept attribute that expresses non-inherited relationships between concepts; Property: a concept attribute that holds a literal value (text, number, date, etc.) that is associated with a concept, often derived from a source terminology or related to ETL requirements; Synonym: a text-valued property that holds a searchable name for a concept; Qualifier: an attribute that related to a specific instance of a concept's role, association or property value. Figure 2 shows examples of attributes, qualifiers, and their values.
The RED also stores and manages system operational metadata to support a variety of administrative procedures and BTRIS functions (such as the ETL processes, described above). All BTRIS database tables, columns, attributes, domains, and data sources are coded with RED codes. Relationships between table columns and the classes of RED codes they maintain provide a form of computable documentation of the BTRIS data model. In addition to TDE, RED content can be accessed using a Web-based user interface (Distributed Terminology Service from Apelon), a BTRIS report that queries RED tables, and Web-based search tool described below.
The RED content is exported from the TDE environment to a set of tables in the BTRIS database. These tables are used for creating ETL translation tables (described above) and for term look-up by users creating data queries (described below). One of the most important RED tables is the Ancestor_Descendant_Identity (ADI) table, which includes one row for every RED ancestor-descendant relationship, including parent-child, grandparent-grandchild, etc. “Identity” refers to the fact that the table contains one row in which each RED concept is in both the ancestor and descendant columns. This arrangement supports class-based queries to the database in a reliable way that does not depend on knowing the particular level of the RED hierarchy in which data may be coded. So, for example, if a user or application seeks to query for all of a subject's cancer diagnoses, the query begins by finding all the values from the Children column of the ADI table where the Ancestor column contains the value C4194479 (Malignant Neoplasm). This will retrieve all instances of the RED Codes for Malignant Neoplasm of Lung, Small Cell (C2171008), Small Cell Lung Carcinoma (C2168040), and even Malignant Neoplasm itself. This set of RED Codes can then be used to retrieve instances of data from the relevant BTRIS tables. In practice, this is accomplished with a query that is a simple join between the ADI table and the BTRIS table.
4.5 User Interfaces
4.5.1 Identified Data Access
Although the BTRIS demo version was created with Business Objects, our development requirements evolved in ways that made a different commercial product, Cognos (IBM, Armonk, NY), more appropriate for rapid development and terminology browsing related to study-related reporting of identified data. After logging on, users can select from a variety of “canned” reports, each of which is associated with a “prompt page” that allows the user to specify report parameters. Most reports are available to all users, with institute-specific reports available to users from the relevant institute. BTRIS does not allow users to construct their own queries or reports.
Each prompt page displays a list of the studies for which the user is authorized, and a variety of optional fields that allow for selection of specific subjects from selected studies, data ranges, values, etc. (Figure 3). Once the report is executed, results are displayed in a spreadsheet format, which may include links to associated documents or images (Figure 4), and may be downloaded to the user's computer for further processing, including loading into visualization software (Figure 5).[28]
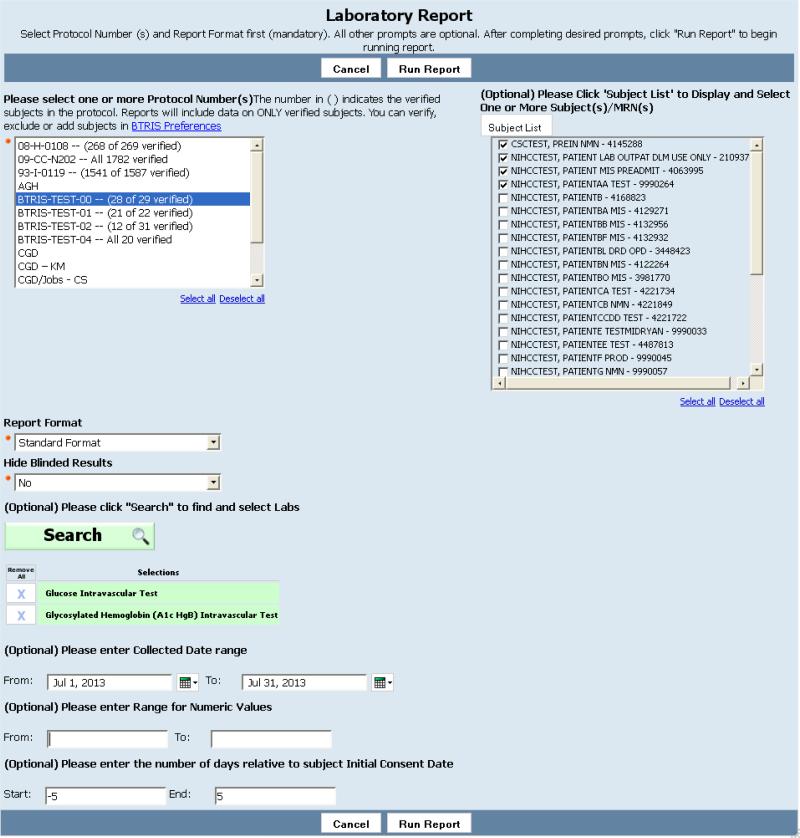
Figure 3.
Example of “prompt page” for capturing a user's specifications for a study-specific BTRIS identified data report; in this case, the report will retrieve laboratory test results. Selection of a study (“Protocol Number” at upper left) is required. All other parameters are optional; clicking on “Run Report” with no other parameter values would retrieve all results for all subjects on the selected study. In this case, the user has selected four subjects, a date range, and specified that the results should be within five days of the date that the subjects signed their respective consent forms (the dates they started the study). The user has also used the RED browser (not shown here but similar to Figure 8) to specify that results should be in the test classes “Glucose Intravascular Test” and “Glycosylated Hemoglobin (A1c Hgb) Intravascular Test”.
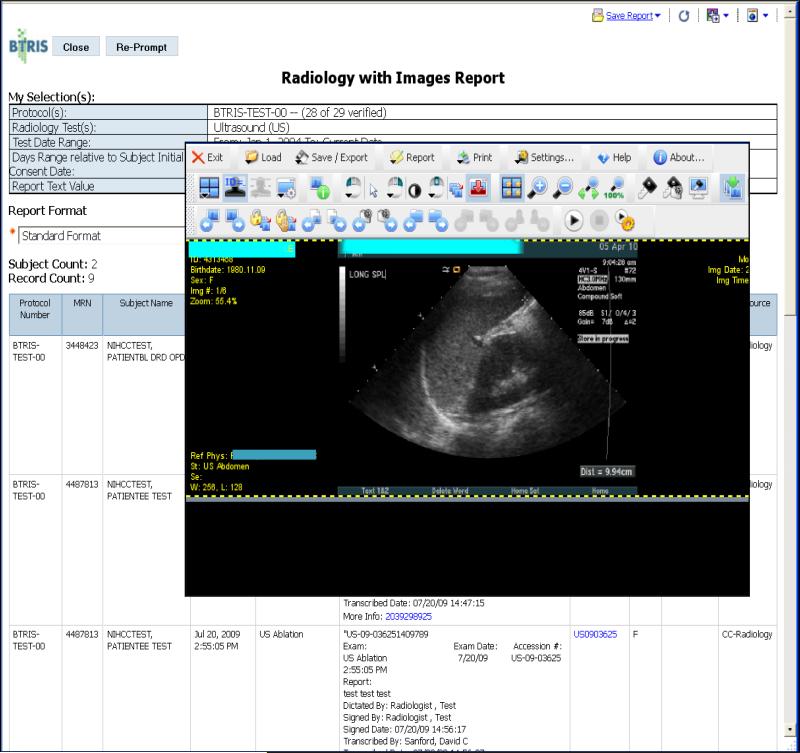
Figure 4.
Results of running a BTRIS identified data report; in this case, the Radiology Report. The user has completed the appropriate prompt page (not shown, but similar to Figure 3) and selected a study and the RED class “Ultrasound”. The report is in spreadsheet form with one row per result. The 7th column contains identifiers such as “US0903625” that are hyperlinks. Clicking on one of these links evokes a viewer application that retrieves the relevant images from the Radiology Departments picture archiving and communication system (PACS). This figure shows the result of clicking on one such link from an ultrasound report.
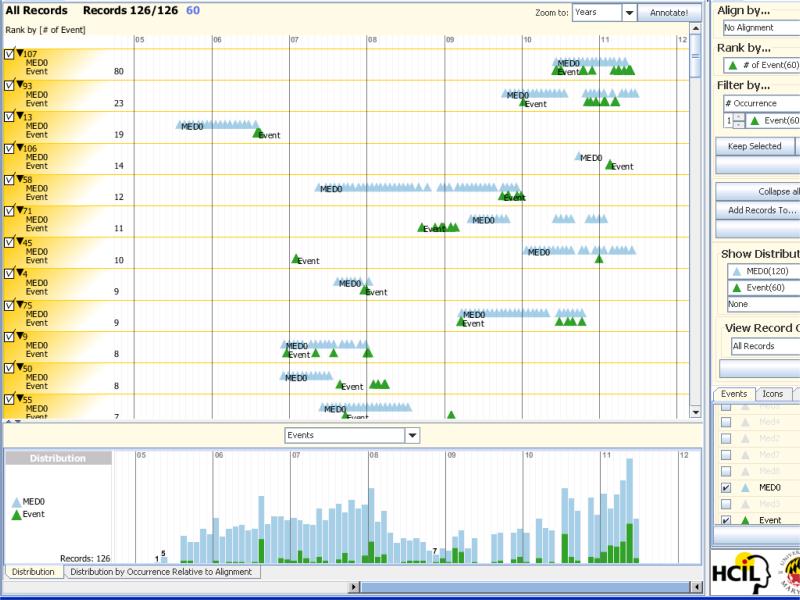
Figure 5.
Display of BTRIS data using the Lifelines2 visualization tool from the University of Maryland's Human Computer Interface Laboratory.[28] Data consist of timing of administration of a study medication (blue triangles, labeled “MED0”) and occurrences of a particular adverse event (green triangles, labeled “Event”), with each horizontal row corresponding to a particular study subject; 12 subjects are shown in the top panel. The panel at the bottom shows the monthly counts for medications and events for all 126 subjects in the study. Together, they show that the event often appears to follow the administration of the medication and that increased events noted in 2011 (right side of both panels) correlate with increased use of the medication.
A complementary application, called “BTRIS Preferences,” was developed to allow users to provide their own information related to their studies, including corrections to the list of enrolled subjects, setting authorizations for access to the data, and thresholds for notifications when data are being reused by others with personal identifiers removed (see below). BTRIS Preferences also allows users to enter clinical trials data, such as treatment arm assignments, outcomes and adverse events, to allow BTRIS to automatically submit results to ClincialTrials.gov.[29,30]
4.5.2 Retrievals of Data with Personal Identifiers Removed
The initial release of BTRIS in 2009 included a simple query tool that allowed users to obtain summary results for individual data types, such as the number of subjects that met certain demographic characteristics or the number of laboratory results of a certain type, date range and value range. The tool also permitted retrieval of detailed coded data sets* associated with studies that had been terminated. Because NIH policy requires permission from original investigators prior to re-use of their data, this initial version of BTRIS did not retrieve more recent data, pending development of a mechanism to notify investigators when access to their data occurred. This initial version of the system allowed us the opportunity to study the kinds of studies users wished to conduct.
Although some users found the initial version met their needs, most non-study-related user queries over the ensuing years required human-mediated searching by BTRIS staff using ad hoc SQL queries.[31] The most frequent requirement was the ability to combine data from multiple domains with Boolean (“and” and “or”) relationships in an ad hoc manner. We found that the methods for dynamic queries offered by Cognos, in which the output of one query was passed to a second query in a “drill through” approach, would be too awkward for our users. Queries across the entire BTRIS database pose an additional challenge: re-use of research data requires permission, in many cases, from the original investigators. This entails identifying those investigators for each datum retrieved, providing this information to the BTRIS user, and also notifying the investigators when a data download occurs. Attempting to insert this process into the Cognos workflow proved to be difficult in the first version of BTRIS; so much so, that we avoided the issue entirely by only retrieving data that required no permission (that is, data from studies terminated more than five years prior to the query).
We therefore chose to develop a new tool, modeled on the i2b2 user interface[31] that supports drag-and-drop query specifications, including Boolean relationships among data types found in individual subject records. The new application (shown in Figures 6 and 7) was released in February of 2013.[33] Written in .NET, HTML5 and JavaScript/jQuery, it uses XML and JSON to provide a modular approach to specifying the application features and communication between the Web services and the BTRIS database server. Based on user feedback from the first version of BTRIS, we initially limited the second version to four data domains (demographics, laboratory test results, medication administration records and patient diagnoses), with query features that include the ability to combine data across multiple domains, the use of Boolean relationships (including “not”), date and value ranges, and a sophisticated term look-up application called RED Web Search (described below).
Figure 6.
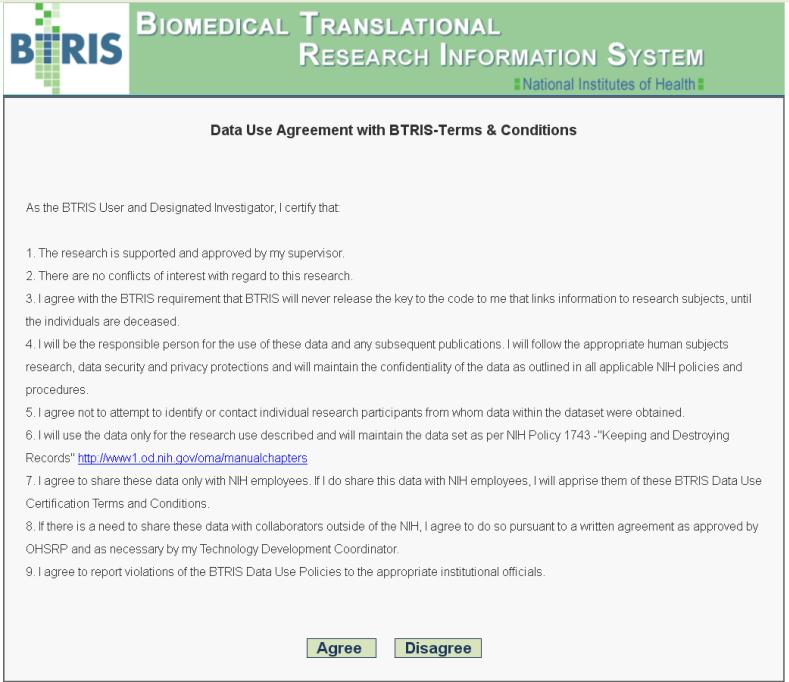
Data use agreement for retrieval of BTRIS data across all studies (with personal identifiers removed). The use must click on “Agree” prior to any use of the query tool (shown in Figure 7).
Figure 7.
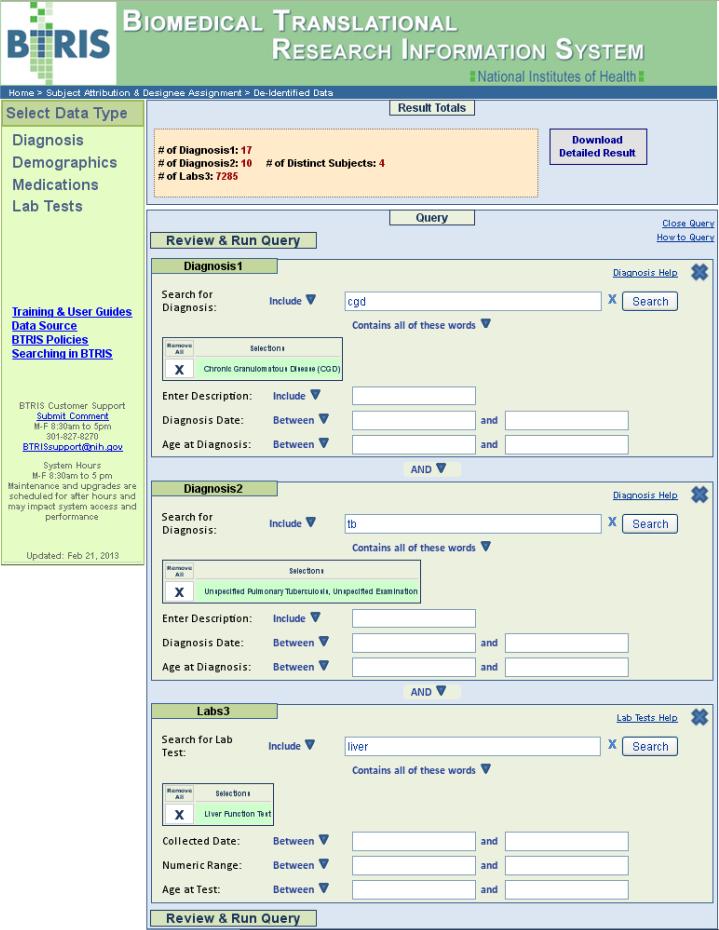
User interface for retrieval of BTRIS data across all studies (with personal identifiers removed). In this example, the user has constructed a query to find subjects that have had a diagnosis of chronic granulomatous disease (“Diagnosis1”) and tuberculosis (“Diagnosis2”) and have had liver function tests performed (“Labs3). The user has selected disease and test terms from the RED (see Figure 8). Execution of the query shows that there are 7,285 test results on four unique subjects. Clicking on the “Download Detailed Result” button initiates the data retrieval and notification processes, as described in the text.
Users may perform unlimited searches to obtain summary statistics; queries with interesting summary results can be re-executed to obtain detailed data. When the user downloads data from these queries, original investigators are notified (according to NIH policy) about access to data from their active studies or studies terminated within the past two years. The policy for investigator notifications is described in Section 4.6, Data Access Policies, below.
4.5.3 Retrievals for Non-Research Purposes
The availability of data from multiple systems (especially MIS and CRIS) in one database, together with the ability to query across large populations using RED classes, has proven to be valuable for a variety of non-research purposes. These have included calculations of total body radiation exposure from routine radiographic studies, reporting on safety of an anti-leprosy drug to the FDA, identifying fungal infection outbreaks related to steroid injections, characterizing changing patterns in lengths of hospital stays, and detecting anomalous patterns in routine laboratory analyses, to name a few. Neither the Cognos-based study-specific query interface nor the new cross-study tool are appropriate for these queries because they are study-independent, allow inclusion of personally identifiable information and do not require investigator notification. For the time being, these queries will be mediated manually by BTRIS staff.
4.5.4 Terminology look-up
Both BTRIS query tools use the RED Web Search to allow users to select concepts that correspond to specific terms from source systems, as well as superclasses of those terms. Built using .NET, HTML5, JavaScript/jQuery, XML and JSON, the tool can be evoked to allow users to conduct searches against specified concept attributes (such as names, synonyms, and local codes) within a specified part of the RED hierarchy.
Terms that match the user's search term are assembled into their corresponding hierarchy and displayed with two levels of the hierarchy expanded. Users can further expand the hierarchy to reveal more specific terms. For example, the query shown Figure 7 required the user to identify terms from different parts of the RED hierarchy (diseases, laboratory tests and medications). Figure 8 shows the RED query needed to identify one of the disease terms used in the query and shows the more specific terms in the RED. Only terms that actually appear in the data, and the ancestors of those terms, are retrieved. Hovering the cursor over a particular term evokes a popup display showing the number of records in the database corresponding to that term and its descendants.
Figure 8.
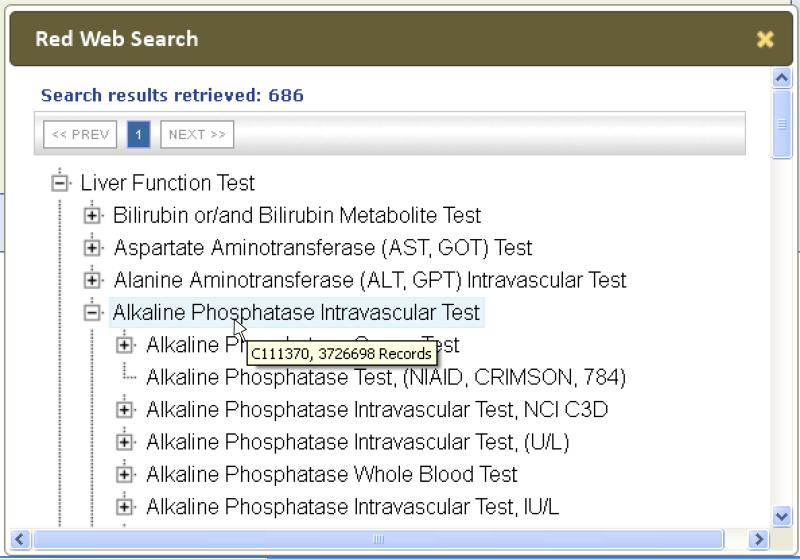
Web-based tool for retrieving concepts from the RED. This screen shows the result of the search for tests and test classes with the word “Liver” (see the “Lab3” panel in Figure 7). In this example, the user has expanded the hierarchy under “Alkaline Phosphatase Intravascular Test” so show more specific test terms, including some from the CRIMSON and C3D systems. The mouse pointer is hovering over the intravascular test class term, causing the hover box to be displayed showing the RED Code for the concept (C111370) and the total number of results (3,727,698) in BTRIS for all tests in this class.
4.6 Data Access Policies
BTRIS provides unprecedented access to NIH subject data. Naturally, there is a great deal of policy relevant to such access to protect the rights of research subjects and researchers. A Code of Conduct and a Data Use Disclaimer were developed to detail the policy requirements as well as to ensure users follow rules related to subject privacy, data security and research community citizenship. In many cases, these policies have had to be refined to make them relevant to the kinds of access that BTRIS affords. For example, policies about who may view which data are related to the role of the investigator in a study and the relevance of particular data to the study. BTRIS provides the opportunity for principal investigators to extend explicit permission to specific individuals involved in their studies (using BTRIS Preferences).
BTRIS Data Sharing and Use Policies were vetted by the intramural research community and approved by the NIH Institute Directors in May of 2009. Key policy elements include: a) access to identified data for active studies by the principal investigator or a designee also named as an investigator on the study, and b) access to coded data for hypothesis testing for data without study attribution or for data associated with studies terminated over five years. Policies have continued to evolve, for example the recognition of additional non-research purposes for data access, the reduction of the requirement for permission on access to terminated studies from five years post-termination to two years, and reduction in the requirements for automatic notification of original investigators.
By default, notifications are issued if the BTRIS data set contains data on at least 1% of the subjects from the original study and those subjects constitute at least 5% of the subjects in the data set. Investigators may change the notification thresholds to “never notify”, “always notify” or any numeric criteria in between. To date, some investigators have selected the “never notify” option for a small number of studies, but otherwise they have accepted the default thresholds. Some investigators have responded to the notifications with surprise or concern but have generally been satisfied after receiving a complete explanation of the BTRIS process, the research question being asked, and the data obtained. In a small number of cases, investigators have asked users not to use the data due to conflicts with their own active intellectual interests.
4.7 User Experience to Date
At the initial release of BTRIS in July of 2009, all of the approximately 400 active principle investigators (PIs) were automatically granted access to BTRIS Data Access and BTRIS Preferences. Many PIs have since indicated additional personnel who should have access, including IRB-approved associate investigators and those in non-research roles such as data extractors and managers. Access is also provided to those who perform nightly data queries to obtain data for institute clinical trial management systems and administrative users who perform queries for quality assurance purposes. By September 2009, 682 licenses to BTRIS had been granted; this number has remained stable as new users are added and dormant users are removed.
BTRIS usage is tracked daily. In an average month, 95 unique users will access the system between one and 100 times, generate about 1200 study-specific reports, carry out about 150 cross-study summary queries (such as the one shown in Figure 7), and a smaller number (zero to ten) downloads of detailed cross-study query results. In addition, two clinical trials data management systems perform nightly downloads of data from BTRIS, serving many users of those systems. Reports are run on all types of studies, including natural history, interventional and data re-use studies.
BTRIS conducts an annual survey to gauge user satisfaction and solicit input on developmental priorities. Overall BTRIS users are satisfied with the system and have noted that BTRIS improves their research productivity. Although the full impact of BTRIS on research is difficult to measure, since 2008, authors of 34 journal papers, 23 conference papers and 26 abstracts have credited BTRIS (although not always officially so in the Acknowledgements section) with providing their data.
Users have expressed less satisfaction with some of the user interface features related to report prompt pages and with system performance when large data sets are being retrieved. Users routinely ask for additional sources of structured clinical data to be added to BTRIS, most frequently data that are not available in the EHR, or that might only be found in unstructured text such as medications or herbal supplements taken at home. Users have also noted the need to include study milestones, organized by visit, to help with data analysis. Finally, some users have expressed the need for support with data analysis. To date, no clear common requirements have emerged; BTRIS therefore continues to focus on providing results in forms that support analysis with external analytic tools.
5. Discussion
5.1 BTRIS in 2013
BTRIS is a complex, multi-faceted system that combines data from clinical care and clinical research in a way that supports several perspectives, including specific, prospective research as well as retrospective re-use of data. Due to space constraints, many additional features of BTRIS, such as access to document images and genomic data, support for a variety of data formats, integration of visualization tools, automatic data reporting to ClinicalTrials.gov,[34] and management of a diverse set of data access policy requirements are not discussed here.
The BTRIS database has been designed to be flexible enough to accommodate a variety of data types in anticipation of receiving new data types that may yet emerge. The separation of high-use data into main tables and low-use data into EAV tables has proven generally successful and stable. Since the original design in 2009, only one field has been “promoted” from storage as EAV rows to become a column in the main table. We are continuing to add new data sources to BTRIS, based on the priorities our users convey to us through surveys, committees and other feedback. With the exception of genomic data (see Limitations, below), the original model appears to be appropriate moving forward.
The RED imparts a functionality that is found in only a small number of clinical data repositories. As a knowledge resource about data source terminologies, it is invaluable for BTRIS developers and users alike. The ability to drive both ETL and reporting processes to handle new data as they appear in data sources allows BTRIS to function seamlessly, without programming changes or service interruptions. The complexity of the RED content reflects the complexity of the underlying terminology sources. However, when a laboratory system adds, for example, a new glucose test term, its incorporation into the RED means that ETL systems will be able to code new data without missing a beat and that users searching for glucose tests results can continue to search for “Glucose Intravascular Test”. They do not need to know that the new test has been added, let alone whether it joined one of the 11 tests in the class “Glucose, Whole Blood Tests”, one of the 54 tests in the class “Glucose Plasma Tests, one of the 97 tests in the class “Glucose Serum Tests, or one of the 11 tests in the class “Glucose Intravascular Test, Unspecified”.
The complexity of the RED hierarchy is also one of its strengths. By allowing multiple classification hierarchies to co-exist, with no artificial limits on depth or breadth, data can be stored and retrieved based on real-world attributes that do not have to be crammed into some predetermined structure. Thus, the addition of a class like “Glucose Intravascular Test, NCI” can be added under “Glucose Intravascular Test” to support NCI-specific ETL and reporting requirements without disrupting storage or reporting.
The ability of the RED to incorporate non-hierarchical inter-concept semantic relationships provides other advantages for BTRIS. For example, the ability to relate a report (e.g., Glucose Serial Intravascular Test Spreadsheet Report) to multiple classes of tests (e.g., Blood Glucose Serial Test 01, Blood Glucose Serial Test 02, etc.) allows users to obtain a pre-specified range of data without having to know which classes exist in the RED, while the report query itself does not need to change if additional tests are added to the classes or additional classes desired in the report.
Finally, the user interfaces in BTRIS provide NIH intramural researchers with unprecedented access to the data from their own studies and those of others. Before BTRIS, researchers would typically need to transcribe data from the EHR into their own analytic tools, one result at a time, for each subject, for each day (usually with intervening paper notes). With BTRIS, they can obtain an electronic data set that includes all data for all subjects at literally the touch a few buttons. BTRIS's is already having a strong impact on access to identified data for active studies. Use of the new query tool, is starting to increase as well, as Medical Records Department personnel are now referring investigators with data abstraction requests the BTRIS.
5.2 Comparison of BTRIS to i2b2
A number of institutions have developed clinical data repositories based on their local EHRs, including Columbia University,[35] Intermountain Healthcare,[36] Mayo Clinic,[37] Stanford University,[38] and Duke University[39]. Because BTRIS draws data from multiple NIH institutes, it is in some ways more analogous to multi-institutional repositories, such as i2b2,[18,40] the Observational Medical Outcomes Partnership (OMOP),[41] the HMO Research Network's Virtual Data Warehouse,[42,43] and the Distributed Ambulatory Research in Therapeutics Network (DARTNet).[44] A complete review of these systems and an in-depth comparison to BTRIS is beyond the scope of this paper. However, due to the wide-spread success of i2b2, some contrast may help provide understanding of the significant contributions of BTRIS. i2b2 comprises a suite of technologies and a standard for integrating them into coherent applications, of which most relevant to the current discussion are the Ontology, the Data Repository and the i2b2 Workbench.
As noted in the Background section, the i2b2 Ontology serves as the controlled terminology for i2b2 applications. The Ontology is analogous to the RED in BTRIS, in that it is composed of concepts drawn from other terminologies and includes hierarchical (but not non-hierarchical) relationships between concepts.[45] While the content and function of both ontologies are still evolving, a key design difference between the two ontologies relates to the hierarchy. As also noted, the i2b2 ontology is a strict hierarchy, limiting concepts to having a single parent. This arrangement has a number of advantages by keeping the structure simple and adhering to good philosophical principles.[45] However, it has proven to be an awkward, artificial restraint when representing data in clinical domains.[15] The need for multiple hierarchies was the main reason we did not choose the i2b2 architecture when designing the RED. Our experience to date confirms that this capability is invaluable for all aspects of data representation and use.
Other distinctions between the i2b2 ontology and the RED exist but are relatively minor. For example, the RED uses a unique, meaningless identifier as a concept code and represents its hierarchy through explicit inter-concept relationships while i2b2 uses unique names as concept identifiers and represents the hierarchy with “full names” for each term that are sequential, delimited character strings containing the names of all ancestor terms. Another minor difference is that the RED specifies concept modifier sets through relationships to other concepts, either as an explicit list of relationships or as a relationship to a class of concepts that can serve as reusable modifiers, while i2b2 provides explicit modifiers for each concept where desired and catalogues modifiers in a separate terminology table.[46] Each of these approaches has relative advantages and disadvantages that are beyond the scope of this paper; in any case, with the exception of the multiple-versus-strict hierarchy and the non-hierarchical inter-concept relationships, translation between the two (that is, representing the RED concepts in the i2b2 ontology structure and vice versa) is probably feasible.
At a high level, the BTRIS database and the i2b2 Data Repository have a similar design for relating facts and dimensions. They are also similar in the use of EAV modeling to capture specific data attributes, but differ in the degree to which they do so. BTRIS models events and observations separately, with only observations having “results”, while i2b2 treats all data facts as observations. Both BTRIS and i2b2 include columns for a number of universal attributes (such as a subject identifier, date-time stamp, and status codes). BTRIS handles additional attributes (such as modifiers) through the use of dependent EAV tables; in 2011, i2b2 began using a modifier_cd column to capture additional attributes, creating new rows in its observation table that repeat the universal attributes for each row if more than one modifier is stored.[47]
Another distinction between database models is the handling of time and date; i2b2 tags observations with a “start date/time”, while BTRIS selects a “best date/time” as described above. While the former method provides a consistent rule for modeling temporal information, the latter provides a simple way to get the temporal information most relevant to the majority of BTRIS users.
BTRIS and i2b2 also have some interesting differences in their approach to user interfaces. Unlike i2b2, BTRIS uses a “canned report” design to provide access to study-specific data in identified form. This restricts flexibility (for example, users have only limited control over the data columns returned) but makes common queries extremely easy to conduct (for example, retrieving all laboratory test data only requires selecting the report type, selecting the research study of interest, and selecting “Run Report”). This user interface has proven to be extremely popular with investigators with active studies.
The new BTRIS query interface takes a different approach, inspired by the i2b2 Workbench “drag and drop” paradigm for constructing complex, cross-domain queries with a few differences. Users of the Workbench select terms from the ontology as their search parameters, while BTRIS users select domains (demographics, medications, etc.) and then select terms from the RED if desired. In both applications, modifiers can include date ranges and Boolean relationships between selection criteria. Some minor differences include specification of subject age (BTRIS) and number of occurrences (i2b2).
5.3 Limitations
Clinical data at NIH differ from those found in other clinical data repositories. Users of BTRIS need to understand these differences in order to use the system effectively. Most of the data in BTRIS are derived from patient care systems, yet they do not represent complete patient records, since NIH subjects generally get their routine health care elsewhere, from their own providers. On the other hand, some of the data collected have no equivalent anywhere else, due to the focus on rare conditions, innovative diagnostic studies, and experimental interventions.
Although the data in BTRIS are all derived from clinical studies of one form or another, they are not readily reducible to study-specific data sets, such as might be found in the Database of Genotypes and Phenotypes (dbGAP).[48] Metadata about the relevance of a particular datum to a particular data collection point in a study is generally not available in a computable form. Moreover, because patients may be subjects on multiple studies simultaneously, a particular datum cannot be reliably attributed to one or another (or even multiple) studies. On the other hand, the data available in BTRIS go beyond what is often included in a study data set. For example, in addition to a single, representative laboratory outcome measure, BTRIS includes all previous and subsequent values of that measure, as well as all other laboratory tests performed, and the co-occurring vital signs, radiology reports, clinical notes, etc.
Although user survey responses are generally positive, we know that there is much more to be done to the BTRIS user interface. The Cognos-based data reports are inflexible in terms of options for inclusion of data attributes, while the new query interface covers only a subset (albeit the most voluminous and important subset) of the data. There are also issues with system response time, particularly with large, complex queries that can take 10 minutes or more. We will attempt to address these issues in future versions of the software (see below).
Perhaps the next greatest challenge for BTRIS will be to develop a mechanism for access to its data by outside researchers in a way that protects the privacy of our research subjects and respects the intellectual contributions of our investigators. We have shown that even simple laboratory data, stripped of identifiers date/time information can still be used to re-identify subject records.[49] Access to BTRIS data will therefore require a significant authorization process, perhaps a data access committee review process similar to the one used by dbGAP.
5.4 Next Directions
Although the current version of BTRIS became operational in 2009, it has continued to grow in data content and reporting capabilities since that time. The new query tool shown in Figure 7 is the latest major advancement; however, other changes are occurring almost monthly. For example, in July of this year, BTRIS added the ability to accept CDISC-compliant spreadsheets, offering great potential for users to preserve, manipulate and share their clinical research data.
The BTRIS work plan includes a long list of new data to be imported, including additional data from existing data sources (such as pulmonary function test parameters that have not been exported to CRIS but that are of interest to researchers) and others are new sources (such as new clinical trials data management systems being installed at various institutes). While most of these new data appear to fit our event-observation-EAV model, we have found that we need to extend our model to accommodate information about samples, analytic techniques and reference information for storing genetic variant data derived from whole exome sequences. A preliminary model for these data is in place now and holds 951 genomes. Initial exploration of user requirements for composite phenome-genome reporting are under way.
New retrieval capabilities are also on the current work plan, including the expansion of the new query tool to handle new data domains and allow the users to specify temporal and cardinality parameters. We are also beginning to use natural language processing technology from the National Library of Medicine to index text documents and to remove personal identifiers.[50] The priorities in which we take on these new projects are set by our user community; the rate at which they are achieved are based on the availability of funding resources.
6. Conclusions
The Biomedical Translational Research Information System fills an important gap in the information infrastructure at NIH. In addition to improving current methods for data access and use, the formal modeling of research entities and terminologies are supporting new methods by bringing together data from patient care, clinical research and genomic research into a unified conceptual search environment. Although the NIH's primary mission is research, the data in BTRIS, and the reasons for their re-use, are similar in many ways to those found in institutions whose primary mission is patient care. BTRIS's design, in particular, the data model, Research Entities Dictionary, user query interface, and data sharing policies will be relevant to others who seek to represent disparate clinical and genomic data to support translational research.
Highlights.
- The NIH has created a repository for clinical and translational research data
- The repository includes data from over 19 systems
- Data are indexed with a single controlled unified terminology
- Self-service user interfaces support retrieval of identified and de-identified data
Acknowledgments
The authors thank NIH Clinical Center Director John Gallin and former NIH Director Elias Zerhouni for the vision to launch the BTRIS project. We have had additional invaluable guidance from Dr. Richard Cannon and Dr. Richard Davey on many design and policy decisions. We also thank all the BTRIS staff, past and present, for their contributions to the project, especially Mark Budd, James Rohrbaugh and Donald Griffin. We are grateful to the National Cancer Institute's Center for Biomedical Informatics and Information Technology (CBIIT) for sharing their ontology development tools and experience. Drs. Cimino and Huser are supported by research funding from the NIH Clinical Center and the National Library of Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Although often called “de-identified data”, data sets in which personal identifiers are removed but which retain a data-set-specific identifier that can be used to link back to other data on the same subjects is technically and legally referred to as a “coded data set”.
References
- 1.Bernstam EV, Hersh WR, Johnson SB, et al. CTSA Biomedical Informatics Key Function Committee. Synergies and distinctions between computational disciplines in biomedical research: perspective from the Clinical and Translational Science Award programs. Acad Med. 2009 Jul;84(7):964–70. doi: 10.1097/ACM.0b013e3181a8144d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cimino JJ, Ayres EJ. The clinical research data repository of the US National Institutes of Health. Stud Health Technol Inform. 2010;160(Pt 2):1299–303. [PMC free article] [PubMed] [Google Scholar]
- 3.Cimino JJ, Ayres EJ, Beri A, Freedman R, Oberholtzer E, Rath S. Leong TY, Aronsky D, editors. Developing a Self-Service Query Interface for Re-Using De-Identified Electronic Health Record Data. Proceedings of the 2013 Medinfo. (in press) [PMC free article] [PubMed]
- 4.Cimino JJ, Munson PJ, Lewis T, Rodbard D. Graphical Display of Patient Data Using a Desk-Top Computer. In: Heffernan HG, editor. Proceedings of the Fifth Annual Symposium on Computer Applications in Medical Care. Washington, D.C.: Oct, 1981. pp. 1085–1088. [Google Scholar]
- 5.NIH Videocast BTRIS Town Hall Meeting; February 28, 2008; [July 29, 2013]. http://videocast.nih.gov/summary.asp?Live=6575. [Google Scholar]
- 6.Warner HR, Olmsted CM, Rutherford BD. HELP--a program for medical decision-making. Comput Biomed Res. 1972 Feb;5(1):65–74. doi: 10.1016/0010-4809(72)90007-9. [DOI] [PubMed] [Google Scholar]
- 7.Johnson SB. Generic data modeling for clinical repositories. J Am Med Inform Assoc. 1996;3:328–39. doi: 10.1136/jamia.1996.97035024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cimino JJ, Socratous SA, Clayton PD. Internet as clinical information system: application development using the World Wide Web. J Am Med Inform Assoc. 1995 Sep-Oct;2(5):273–84. doi: 10.1136/jamia.1995.96073829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hripcsak G, Cimino JJ, Sengupta S. WebCIS: large scale deployment of a Web-based clinical information system. Proc AMIA Symp. 1999:804–8. [PMC free article] [PubMed] [Google Scholar]
- 10.Wilcox AB, Vawdrey DK, Chen YH, Forman B, Hripcsak G. The evolving use of a clinical data repository: facilitating data access within an electronic medical record. AMIA Annu Symp Proc. 2009;2009:701–5. [PMC free article] [PubMed] [Google Scholar]
- 11.Nadkarni PM. QAV: querying entity-attribute-value metadata in a biomedical database. Comput Methods Programs Biomed. 1997 Jun;53(2):93–103. doi: 10.1016/s0169-2607(97)01815-4. [DOI] [PubMed] [Google Scholar]
- 12.Cimino JJ, Clayton PD, Hripcsak G, Johnson SB. Knowledge-based approaches to the maintenance of a large controlled medical terminology. J Am Med Inform Assoc. 1994 Jan-Feb;1(1):35–50. doi: 10.1136/jamia.1994.95236135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cimino JJ. From data to knowledge through concept-oriented terminologies: experience with the Medical Entities Dictionary. J Am Med Inform Assoc. 2000 May-Jun;7(3):288–97. doi: 10.1136/jamia.2000.0070288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cimino JJ. Desiderata for controlled medical vocabularies in the Twenty-First Century. Methods of Information in Medicine. 1998;37(4-5):394–403. [PMC free article] [PubMed] [Google Scholar]
- 15.Cimino JJ. In Defense of the Desiderata. Journal of Biomedical Informatics. 2006;39(3):299–306. doi: 10.1016/j.jbi.2005.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cimino JJ, Johnson SB, Hripcsak G, Hill CL, Clayton PD. Managing Vocabulary for a Centralized Clinical System. In: Kaihara S, Greenes RA, editors. Proceedings of the World Congress on Medical Informatics - Medinfo ‘95. Healthcare Computing and Communications Canada, Edmonton, Alberta; Vancouver, Canada: 1995. pp. 117–120. [Google Scholar]
- 17.Kush R, Alschuler L, Ruggeri R, Cassells S, Gupta N, Bain L, Claise K, Shah M, Nahm M. Implementing Single Source: the STARBRITE proof-of-concept study. J Am Med Inform Assoc. 2007 Sep-Oct;14(5):662–73. doi: 10.1197/jamia.M2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fridsma DB, Evans J, Hastak S, Mead CN. The BRIDG project: a technical report. J Am Med Inform Assoc. 2008 Mar-Apr;15(2):130–7. doi: 10.1197/jamia.M2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy SN, Mendis M, Hackett K, Kuttan R, Pan W, Phillips LC, Gainer V, Berkowicz D, Glaser JP, Kohane I, Chueh HC. Architecture of the open-source clinical research chart from Informatics for Integrating Biology and the Bedside. AMIA Annu Symp Proc. 2007 Oct;11:548–52. [PMC free article] [PubMed] [Google Scholar]
- 20.Common Terminology Services 2 [July 29, 2013]; https://wiki.nci.nih.gov/display/VKC/Common+Terminology+Services+2.
- 21.NIH Videocast BTRIS Town Hall Meeting; September 16, 2008; [July 29, 2013]. http://videocast.nih.gov/summary.asp?Live=6943. [Google Scholar]
- 22.Health Level 7 (HL7) [July 29, 2013]; http://en.wikipedia.org/wiki/HL7.
- 23.Open Database Connectivity (ODBC) [July 29, 2013]; http://en.wikipedia.org/wiki/ODBC.
- 24.Extract, Transform, and Load (ETL) [July 29, 2013]; http://en.wikipedia.org/wiki/Extract,_transform,_load.
- 25.Huser V, Cimino JJ. Desiderata for Healthcare Integrated Data Repositories Based on Architectural Comparison of Three Public Repositories. In: Holmes JH, editor. Proc of 2013 AMIA Fall Symposium. Washington, DC.: 2013. (in press) [PMC free article] [PubMed] [Google Scholar]
- 26.NIH Videocast BTRIS Town Hall Meeting; September 15, 2009; [July 29, 2013]. http://videocast.nih.gov/summary.asp?Live=7588. [Google Scholar]
- 27.Noy NF, de Coronado S, Solbrig H, Fragoso G, Hartel FW, Musen MA. Representing the NCI Thesaurus in OWL DL: Modeling tools help modeling languages. Appl Ontol. 2008 Jan 1;3(3):173–190. doi: 10.3233/AO-2008-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang TD, Wongsuphasawat K, Plaisant C, Shneiderman B. Extracting insights from electronic health records: case studies, a visual analytics process model, and design recommendations. J Med Syst. 2011 Oct;35(5):1135–52. doi: 10.1007/s10916-011-9718-x. [DOI] [PubMed] [Google Scholar]
- 29.Zarin DA, Tse T, Williams RJ, Califf RM, Ide N. The ClinicalTrials.gov results database – update and key issues. N Engl J Med. 2011 Mar 3;364(9):852–860. doi: 10.1056/NEJMsa1012065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cimino JJ, Ayres E, Rath S, Freedman R. Automated Submission of Clinical Trials Results to ClinicalTrials.gov (Poster). In: Bernstam E, editor. 2013 AMIA Clinical Research Informatics Summit. San Francisco, CA: p. 41. [Google Scholar]
- 31.Cimino JJ, Ayres EJ, Beri A, Freedman R, Oberholtzer E, Rath S. Aronsky D, Leong TY, editors. Developing a Self-Service Query Interface for Re-Using De-Identified Electronic Health Record Data. Proceedings of Medinfo 2013. (in press) [PMC free article] [PubMed]
- 32.Weber GM, Murphy SN, McMurry AJ, Macfadden D, Nigrin DJ, Churchill S, Kohane IS. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. J Am Med Inform Assoc. 2009 Sep-Oct;16(5):624–30. doi: 10.1197/jamia.M3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.NIH Videocast BTRIS Town Hall Meeting; February 19, 2009; [July 29, 2013]. http://videocast.nih.gov/summary.asp?Live=12520. [Google Scholar]
- 34.Cimino JJ, Ayres E, Rath S, Freedman R. Automated Submission of Clinical Trials Results to ClinicalTrials.gov (Poster). In: Bernstam E, editor. 2013 AMIA Clinical Research Informatics Summit. San Francisco, CA: p. 41. [Google Scholar]
- 35.Wilcox AB, Vawdrey DK, Chen YH, Forman B, Hripcsak G. The evolving use of a clinical data repository: facilitating data access within an electronic medical record. AMIA Annu Symp Proc. 2009;2009:701–5. [PMC free article] [PubMed] [Google Scholar]
- 36.Huff SM, Rocha RA, Bray BE, Warner HR, Haug PJ. An event model of medical information representation. J Am Med Inform Assoc. 1995;2:116–34. doi: 10.1136/jamia.1995.95261905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chute CG, Beck SA, Fisk TB, Mohr DN. The Enterprise Data Trust at Mayo Clinic: a semantically integrated warehouse of biomedical data. J Am Med Inform Assoc. 2010;17:131–5. doi: 10.1136/jamia.2009.002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowe HJ, Ferris TA, Hernandez PM, Weber SC. STRIDE--An integrated standards-based translational research informatics platform. AMIA Annu Symp Proc. 2009;2009:391–5. [PMC free article] [PubMed] [Google Scholar]
- 39.Horvath MM, Winfield S, Evans S, Slopek S, Shang H, Ferranti J. The DEDUCE Guided Query tool: providing simplified access to clinical data for research and quality improvement. Journal of biomedical informatics. 2011;44:266–76. doi: 10.1016/j.jbi.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kohane IS, Churchill SE, Murphy SN. A translational engine at the national scale: informatics for integrating biology and the bedside. J Am Med Inform Assoc. 2012 Mar-Apr;19(2):181–5. doi: 10.1136/amiajnl-2011-000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hornbrook MC, Hart G, Ellis JL, Bachman DJ, Ansell G, Greene SM, et al. Building a virtual cancer research organization. J Natl Cancer Inst Monogr. 2005:12–25. doi: 10.1093/jncimonographs/lgi033. [DOI] [PubMed] [Google Scholar]
- 42.Chapter 4: Virtual Data Warehouse, Collaboration toolkit. 2012 http://www.hmoresearchnetwork.org/resources/toolkit/HMORN_CollaborationToolkit.pdf#4.
- 43.Pace WD, Cifuentes M, Valuck RJ, Staton EW, Brandt EC, West DR. An electronic practice-based network for observational comparative effectiveness research. Annals of internal medicine. 2009;151:338–40. doi: 10.7326/0003-4819-151-5-200909010-00140. [DOI] [PubMed] [Google Scholar]
- 44.Stang PE, Ryan PB, Racoosin JA, Overhage JM, Hartzema AG, Reich C, et al. Advancing the science for active surveillance: rationale and design for the Observational Medical Outcomes Partnership. Ann Int Med. 2010;153:600–6. doi: 10.7326/0003-4819-153-9-201011020-00010. [DOI] [PubMed] [Google Scholar]
- 45. [July 29, 2013];i2b2 Ontology. https://www.i2b2.org/software/projects/ontologymgmt/Ontology_Design_15.pdf.
- 46.Smith B. From Concepts to Clinical Reality: An Essay on the Benchmarking of Biomedical Terminologies. J Biomed Inform. 2006;39(3):288–298. doi: 10.1016/j.jbi.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 47. [July 29, 2013];Use of modifiers in i2b2. https://community.i2b2.org/wiki/display/DevForum/Modifiers+in+i2b2+Data+Model.
- 48.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Lee M, Shao Y, Wang ZY, Sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, Sherry ST. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007 Oct;39(10):1181–6. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cimino JJ. The False Security of Blind Dates: Chrononymization's Lack of Impact on Data Privacy of Laboratory Data. Applied Clin Informatics. 2012;3(4):392–403. doi: 10.4338/ACI-2012-07-RA-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker L, Starks H, West KM, Fullerton SM. dbGaP data access requests: a call for greater transparency. Sci Transl Med. 2011 Dec 14;3(113):113cm34. doi: 10.1126/scitranslmed.3002788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fung KW, Jao CS, Demner-Fushman D. Extracting drug indication information from structured product labels using natural language processing. J Am Med Inform Assoc. 2013 May 1;20(3):482–8. doi: 10.1136/amiajnl-2012-001291. [DOI] [PMC free article] [PubMed] [Google Scholar]