Abstract
From an epistemological point of view, nutritional physiology has been developed, like other factual sciences such as physics, from a purely descriptive to a mechanismic-explanatory scientific discipline. Nowadays, nutritional physiology has entered the molecular stage. Based on this micro-reductionism, molecular targets (e.g., transcription factors) of energy intake, certain nutrients (e.g., zinc) and selected plant bioactives (e.g., flavonoids) have been identified. Although these results are impressive, molecular approaches in nutritional physiology are limited by nature since the molecular targets of nutrients seem to have no ontic priority to understand the nutritional phenotype of an organism. Here we define, to the best of our knowledge, for the first time Nutri-informatics as a new bioinformatics discipline integrating large-scale data sets from nutritional studies into a stringent nutritional systems biology context. We suggest that Nutri-informatics, as an emerging field, may bridge the gap between nutritional biochemistry, nutritional physiology and metabolism to understand the interactions between an organism and its environment.
Keywords: Micro-reductionism, Nutrigenomics, Nutritional systems biology, Nutri-informatics
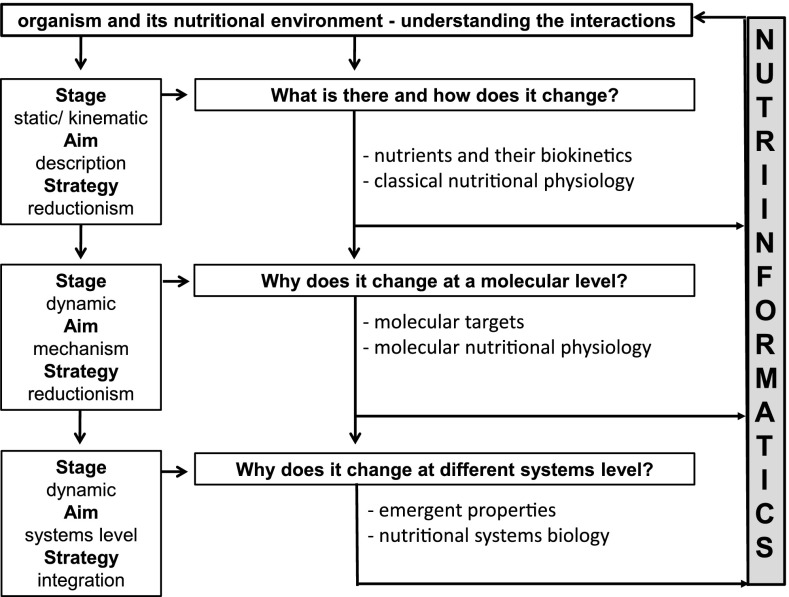
Based on a view from history of science, it is interesting to note that factual sciences such as physics, biology and psychology have developed through the stages of statics, kinematics and dynamics (Bunge 2004). For nutritional physiology (Fig. 1), a similar trend from a purely descriptive to mechanismic1-explanatory scientific discipline can be recognized (Strohle and Doring 2010). For example, in the stages of statics (“What is there?”) and kinematics (“How does it change?”), nutritional physiologists identified the essentials components of our diet and answered the question, how the status of nutrients may change as function of intake. On this basis, robust dietary recommendations were made for many nutrients. In the stage of dynamics (“Why does it change?”), the physiological and biochemical functions of the nutrients were unraveled. During the last two decades, nutritional physiology has entered a molecular stage (“What is the molecular mechanism?”). This kind of micro-reductionism has contributed to a deeper understanding of nutritional processes. For example, pioneering work on vitamin A as transcription factor ligands has been published for more than 20 years (Chambon 1996). More recently, by applying gene chip technology, we and others have identified molecular targets differentially regulated by dietary restriction (Giller et al. 2013), vitamins (Fischer et al. 2001), trace elements (Fischer et al. 2001; tom Dieck et al. 2003), and plant bioactives (Boomgaarden et al. 2010). Notably, dietary factors do not only regulate gene expression on the mRNA, but also on the micro-RNA level (Boesch-Saadatmandi et al. 2011; Gaedicke et al. 2008). Moreover, genome-wide association studies have been successfully used to pinpoint allelic variants explaining individual differences in macronutrient intake (Tanaka et al. 2013). More recently, nutritional epigenomics has been established to get insights, for instance, into persistent effects of prenatal exposure to famine (Heijmans et al. 2008). Even more sophisticated, the groups of M. Ristow and S. Schuster have shown that silico analyses of genome-scale human metabolic networks per se may help to discover novel pathways that might explain body weight reduction of low-carb diets (Kaleta et al. 2011). A comprehensive overview regarding the advancement of molecular nutrition research has been given by the groups of H. Daniel and B. van Ommen (Daniel et al. 2008).
Fig. 1.
Nutri-informatics as a specialized bioinformatics discipline to integrate the different stages, aims, strategies and questions of basic nutritional science
No doubt, the success of the molecular approach in nutritional science is impressive. Nevertheless, the initial euphoria about the potential of molecular nutritional physiology has given way to a greater realism (see e.g., van Ommen 2007). In fact, molecular targets (i.e., transcription factors) of nutrients seem to have no ontic2 priority to understand the nutritional phenotype of an organism: the function of a molecular target depends simply on its epigenomic and cellular environment. For example, the identification of genes differentially expressed under caloric restriction conditions in nearly 600 experiments is not meaningful to predict the resulting phenotype (i.e., longevity) of a restricted organism (Swindell 2008). We have recently shown that dedicated so-called Nutri-informatics tools and algorithms (i.e., Ortho2ExpressMatrix) enable functional classification, pathway analysis and phylogenetic allocations are useful to relate molecular data with nutritional phenotypes (Ludewig et al. 2014). In this context, the nutritional phenotype initiative (i.e., dbNP) seems to be a very useful enterprise to understand the emergent3 properties of an organism in response to nutritional cues (Norheim et al. 2012; van Ommen et al. 2010). To the best of our knowledge, the coinage Nutri-informatics was mentioned for the first time more than 10 years ago (www.molnut.uni-kiel.de/pdfs/Popl_Vortrag/kurzfassung_nutriomic.pdf). Recently, the group of J. Zempleni has provided a comprehensive guidance regarding bioinformatics resources that are useful in nutrition sciences (Malkaram et al. 2012). Thus, the need of bioinformatics in nutritional research emerged with the “omic” era. Overall, Nutri-informatics can be defined as a specialized bioinformatics discipline to integrate large-scale data sets from nutritional studies into a stringent nutritional systems biology context (Fig. 1). In particular, Nutri-informatics should focus, for instance, on analyses of metabolic networks, mathematical simulation of metabolism [i.e., fluxomics (Winter and Kromer 2013)], genome-based identification of essentials nutrients, food genomics, integration of large-scale data sets derived from transcriptomics, proteomics and metabolomics, and promoter-framework analysis to define nutrient-dependent regulons. Thus, we suggest that Nutri-informatics, as an emerging field, may bridge the gap between nutritional biochemistry, nutritional physiology and metabolism. We emphasize that Nutri-informatics should be developed into a basic scientific discipline to understand the interactions between an organism and its nutritional environment, one of the most noble objectives of nutritional science. In addition, Nutri-informatics has a heuristic potential to foster rather applied disciplines. However, the scientific success of Nutri-informatics depends primarily on the formulation of unsolved fundamental and interesting questions, an inherent problem in nutrition science (Strohle and Doring 2010).
Conflict of interest
Frank Döring declares that he has no conflict of interest. Gerald Rimbach declares that he has no conflict of interest.
Footnotes
We used explicitly the term “mechanismic” (and not mechanistic) because not all mechanisms are physical—dependent on the systems level cellular and physiological mechanisms are relevant (see also footnote 3).
Philosophical term: the reality, irrespective of how or whether it is known.
Philosophical term: emergence, a gain of a property of a system which cannot be derived from the characteristics of the individual elements (i. e. molecular targets such as transcription factors) of the system.
References
- Boesch-Saadatmandi C, Loboda A, Wagner AE, Stachurska A, Jozkowicz A, Dulak J, Doring F, Wolffram S, Rimbach G. Effect of quercetin and its metabolites isorhamnetin and quercetin-3-glucuronide on inflammatory gene expression: role of miR-155. J Nutr Biochem. 2011;22(3):293–299. doi: 10.1016/j.jnutbio.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Boomgaarden I, Egert S, Rimbach G, Wolffram S, Muller MJ, Doring F. Quercetin supplementation and its effect on human monocyte gene expression profiles in vivo. Br J Nutr. 2010;104(3):336–345. doi: 10.1017/S0007114510000711. [DOI] [PubMed] [Google Scholar]
- Bunge M. How does it work? The search for explanatory mechanisms. Philos Soc Sci. 2004;34(2):182–210. doi: 10.1177/0048393103262550. [DOI] [Google Scholar]
- Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J Off Publ Fed Am Soc Exp Biol. 1996;10(9):940–954. [PubMed] [Google Scholar]
- Daniel H, Drevon CA, Klein UI, Kleemann R, van Ommen B. The challenges for molecular nutrition research 3: comparative nutrigenomics research as a basis for entering the systems level. Genes Nutr. 2008;3(3–4):101–106. doi: 10.1007/s12263-008-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Pallauf J, Gohil K, Weber SU, Packer L, Rimbach G. Effect of selenium and vitamin E deficiency on differential gene expression in rat liver. Biochem Biophys Res Commun. 2001;285(2):470–475. doi: 10.1006/bbrc.2001.5171. [DOI] [PubMed] [Google Scholar]
- Gaedicke S, Zhang X, Schmelzer C, Lou Y, Doering F, Frank J, Rimbach G. Vitamin E dependent microRNA regulation in rat liver. FEBS Lett. 2008;582(23–24):3542–3546. doi: 10.1016/j.febslet.2008.09.032. [DOI] [PubMed] [Google Scholar]
- Giller K, Huebbe P, Hennig S, Dose J, Pallauf K, Doering F, Rimbach G. Beneficial effects of a 6-month dietary restriction are time-dependently abolished within 2 weeks or 6 months of refeeding–genome-wide transcriptome analysis in mouse liver. Free Radic Biol Med. 2013;61C:170–178. doi: 10.1016/j.freeradbiomed.2013.03.023. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA. 2008;105(44):17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaleta C, de Figueiredo LF, Werner S, Guthke R, Ristow M, Schuster S. In silico evidence for gluconeogenesis from fatty acids in humans. PLoS Comput Biol. 2011;7(7):e1002116. doi: 10.1371/journal.pcbi.1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig AH, Klapper M, Doring F. Identifying evolutionarily conserved genes in the dietary restriction response using bioinformatics and subsequent testing in Caenorhabditis elegans. Genes Nutr. 2014;9(1):363. doi: 10.1007/s12263-013-0363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkaram SA, Hassan YI, Zempleni J. Online tools for bioinformatics analyses in nutrition sciences. Adv Nutr. 2012;3(5):654–665. doi: 10.3945/an.112.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norheim F, Gjelstad IM, Hjorth M, Vinknes KJ, Langleite TM, Holen T, Jensen J, Dalen KT, Karlsen AS, Kielland A, Rustan AC, Drevon CA. Molecular nutrition research: the modern way of performing nutritional science. Nutrients. 2012;4(12):1898–1944. doi: 10.3390/nu4121898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohle A, Doring F. Molecularization in nutritional science: a view from philosophy of science. Mol Nutr Food Res. 2010;54(10):1385–1404. doi: 10.1002/mnfr.201000078. [DOI] [PubMed] [Google Scholar]
- Swindell WR. Genes regulated by caloric restriction have unique roles within transcriptional networks. Mech Ageing Dev. 2008;129(10):580–592. doi: 10.1016/j.mad.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Ngwa JS, van Rooij FJ, Zillikens MC, Wojczynski MK, Frazier-Wood AC, Houston DK, Kanoni S, Lemaitre RN, Luan J, Mikkila V, Renstrom F, Sonestedt E, Zhao JH, Chu AY, Qi L, Chasman DI, de Oliveira Otto MC, Dhurandhar EJ, Feitosa MF, Johansson I, Khaw KT, Lohman KK, Manichaikul A, McKeown NM, Mozaffarian D, Singleton A, Stirrups K, Viikari J, Ye Z, Bandinelli S, Barroso I, Deloukas P, Forouhi NG, Hofman A, Liu Y, Lyytikainen LP, North KE, Dimitriou M, Hallmans G, Kahonen M, Langenberg C, Ordovas JM, Uitterlinden AG, Hu FB, Kalafati IP, Raitakari O, Franco OH, Johnson A, Emilsson V, Schrack JA, Semba RD, Siscovick DS, Arnett DK, Borecki IB, Franks PW, Kritchevsky SB, Lehtimaki T, Loos RJ, Orho-Melander M, Rotter JI, Wareham NJ, Witteman JC, Ferrucci L, Dedoussis G, Cupples LA, Nettleton JA. Genome-wide meta-analysis of observational studies shows common genetic variants associated with macronutrient intake. Am J Clin Nutr. 2013;97(6):1395–1402. doi: 10.3945/ajcn.112.052183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tom Dieck H, Doring F, Roth HP, Daniel H. Changes in rat hepatic gene expression in response to zinc deficiency as assessed by DNA arrays. J Nutr. 2003;133(4):1004–1010. doi: 10.1093/jn/133.4.1004. [DOI] [PubMed] [Google Scholar]
- van Ommen B. Personalized nutrition from a health perspective: luxury or necessity? Genes Nutr. 2007;2(1):3–4. doi: 10.1007/s12263-007-0018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ommen B, Bouwman J, Dragsted LO, Drevon CA, Elliott R, de Groot P, Kaput J, Mathers JC, Muller M, Pepping F, Saito J, Scalbert A, Radonjic M, Rocca-Serra P, Travis A, Wopereis S, Evelo CT. Challenges of molecular nutrition research 6: the nutritional phenotype database to store, share and evaluate nutritional systems biology studies. Genes Nutr. 2010;5(3):189–203. doi: 10.1007/s12263-010-0167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G, Kromer JO. Fluxomics—connecting ‘omics analysis and phenotypes. Environ Microbiol. 2013;15(7):1901–1916. doi: 10.1111/1462-2920.12064. [DOI] [PubMed] [Google Scholar]