Abstract
Curcumin from Curcuma longa may exert putative neuroprotective properties in the brain. Impaired mitochondrial function is implicated in Alzheimer’s disease and the presence of the apolipoprotein (APO) E4 genotype, which is a risk factor for late-onset Alzheimer’s disease, may aggravate mitochondrial malfunction. Here, we report that in the brain of 16-month-old APOE4-targeted replacement mice, adenosine triphosphate (ATP) concentrations were significantly lower than in APOE3 mice. A 3-month dietary supplementation of 0.2 % curcumin numerically increased ATP concentrations in APOE3 and significantly in APOE4 mice compared to the respective controls. Curcumin significantly induced the transcription of peroxisome proliferator-activated receptor (PPAR) γ and mitochondrial transcription factor A (TFAM) in APOE3, but not in APOE4 mice. Moreover, PPARγ coactivator (PGC)-1α and guanine–adenine repeat binding protein α (GABPa) mRNA was only increased in APOE3 mice. Consistent with these observations, protein expression of mitochondrial respiratory complexes, especially of complex IV, also appeared to be increased in APOE3 mice. In conclusion, we provide evidence that curcumin affects mitochondrial function and gene and protein expression in the murine brain despite its low bioavailability and carriers of the Alzheimer’s disease-risk genotype APOE4 may be less responsive to dietary curcumin than APOE3 carriers.
Keywords: Curcumin, APOE, Mitochondrial function, ATP synthesis, Mice
Introduction
Curcumin or diferuloylmethane makes up 50–60 % of the total curcuminoids isolated from Curcuma longa (family Zingiberaceae) and is chemically a bis-α,β-unsaturated β-diketone of two ferulic acid units (Esatbeyoglu et al. 2012; Chin et al. 2013). Curcumin has been attributed with neuroprotective potential and the Indo-US Cross National Dementia Study reported that India had a lower incidence and a lower prevalence of Alzheimer’s disease (AD) compared to a reference American population (Chandra et al. 1998, 2001). To date, there have been two completed clinical trials investigating the neuroprotective efficacy of curcumin with regard to AD, but both studies have not reported significant neuroprotective effects (Baum et al. 2008; Ringman et al. 2012). One possible explanation for the lack of neuroprotective efficacy may be the low bioavailability of curcumin. Curcumin, being a fat soluble molecule, exhibits poor aqueous solubility, poor stability in solution, and rapid intestinal and hepatic metabolism (Anand et al. 2007). Due to its low toxicity, many studies are administering high doses of curcumin in the hope of increasing its concentration in the brain. We have previously reported that curcumin cannot be detected in the brain of NMRI mice after oral administration of 50 mg/kg BW curcumin (Schiborr et al. 2010). However, a recent report from Japan observed that behavioural symptoms of patients with AD could be improved by applying a low dose of approximately 100 mg/day curcumin (Hishikawa et al. 2012). This suggests that even very small concentrations of curcumin or curcumin metabolites below the analytical detection limit may induce effects on the molecular level (neuro- or mitohormesis) and account for the observed neuroprotective effects (Mattson and Cheng 2006; Calabrese et al. 2010).
The apolipoprotein (APO) E4 allele is a major genetic risk factor for sporadic or late-onset AD (Strittmatter and Roses 1996; Jofre-Monseny et al. 2008). APOE is involved in the distribution and cellular uptake of cholesterol and is the major apolipoprotein in the brain (Huebbe et al. 2007). Although the exact mechanism through which APOE4 contributes to an increased risk of AD is as yet unclear, it is likely that APOE is involved in many different mechanisms that contribute to AD. One of these mechanisms might involve mitochondrial function. Mitochondrial dysfunction has been observed repeatedly in AD and favours inflammation and cell loss, which are central events in the pathogenesis of AD (Hauptmann et al. 2009). Decreased expression of mitochondrial respiratory complexes and lowered mitochondrial respiratory capacity, as observed in the cortex of APOE4 mice, could further promote mitochondrial dysfunction and therefore contribute to AD (Chen et al. 2011). Interestingly, it has recently been shown that mitochondrial dysfunction associated with APOE4 could be improved by small molecule structure correctors which inhibit the domain interaction of APOE4 involved in mediating its detrimental effects (Chen et al. 2012).
We have previously reported that dietary curcumin can modulate markers of mitochondrial dysfunction in the brain of senescence-accelerated mouse prone 8 (Eckert et al. 2013). Despite widespread interest in curcumin as a potential therapeutic or preventive agent for neurodegenerative diseases (Eckert et al. 2012), to the best of our knowledge, the neuroprotective efficacy of curcumin has never been investigated in the context of APOE. We hypothesised that dietary curcumin might increase adenosine triphosphate (ATP) concentrations and modulate genes involved in the regulation of mitochondrial biogenesis and function in the cortex of APOE3- and APOE4-targeted gene replacement mice.
Materials and methods
Mice and diet
Female APOE3- and APOE4-targeted gene replacement mice aged 6–8 weeks old were purchased from Taconic Europe (Ry, Denmark). APOE-targeted gene replacement mice were originally developed at the laboratory of Nobuya Maeda and are well characterized (Knouff et al. 1999; Sullivan et al. 1997, 1998). A detailed description of how these mice were created can be found in our previous publication (Huebbe et al. 2011). Briefly, the murine APOE gene was targeted in E14TG2a embryonic stem cells for replacement with the human APOE alleles. The targeted cells were injected into blastocytes, and the resultant chimeras were backcrossed to C57BL/6 mice for 9 generations. Colonies (N9) are maintained at Taconic through in-crossing of homozygotes.
The animal study was performed according to German animal welfare laws and regulations, and with permission from the responsible authorities. The mice were housed in macrolon cages under controlled environmental conditions (55 % relative humidity, 22–24 °C, and 12-h light–dark cycle). The mice were maintained on a semi-synthetic Western-type diet (C1000 mod., Altromin, Lage, Germany) containing 40 % carbohydrate, 18 % protein, 20 % milk fat, 1.25 % cholesterol, as well as a vitamin and mineral premix. The mice had free access to tap water. For the first 11 months, all mice were fed the Western-type diet ad libitum. Afterwards, the mice were randomly assigned to two diet groups (control and 0.2 % curcumin-supplemented). The supplementation period was 3 months, during which both groups were given ad libitum access to the respective diets. At the end of the trial, the mice were fasted for 12 h and killed by cervical dislocation. For the ATP measurements, dissociated brain cells were freshly prepared (Eckert et al. 2013), and for the other measurements, parts of the cortex were stored in RNAlater™ (Qiagen, Hilden, Germany) at −20 °C or snap-frozen in liquid nitrogen and stored at −80 °C.
Preparation of dissociated brain cells
One brain hemisphere was used for the preparation of dissociated brain cells and quickly dissected on ice after removal. The cerebellum was excluded and the remaining tissue was first washed, and then minced in 2 mL of medium I (138 mM NaCl, 5.4 mM KCl, 0.17 mM Na2HPO4, 0.22 mM KH2PO4, 5.5 mM glucose, 58.4 mM sucrose, pH 7.35) with a scalpel. The brain cells were further dissociated first by filtration through a crude filter using a Pasteur pipette and later through a fine filter. The cell suspension was then washed twice in medium II (110 mM NaCl, 5.3 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 25 mM glucose, 70 mM sucrose, 20 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), pH 7.4). After a final centrifugation step (1,000g, 5 min, 4 °C), the cell pellet was resuspended in 4.5 mL Dulbecco’s modified Eagle’s medium (Gibco®, Invitrogen, Germany) and distributed 50 μL/well in white 96-well plates for ATP measurements. Plated dissociated brain cells were incubated at 37 °C in 5 % CO2 for 3 h prior to measurement. ATP concentrations are expressed as fluorescence per milligram protein, and the protein content was determined using the Bio-Rad DC Protein Assay (Bio-Rad, Munich, Germany).
Measurement of ATP concentrations
Basal ATP concentrations were measured using the Via Light™ Plus Bioluminescence Kit (Lonza, Walkersville, USA) according to the manufacturer’s protocol. The Via Light™ Plus Kit is based on the production of light from ATP and luciferin in the presence of luciferase. The emitted light (bioluminescence) correlates linearly with ATP concentration and can be determined using an external standard curve. Briefly, dissociated brain cells were removed from the incubator and allowed to cool to room temperature for 10 min. After that, cells were lysed with the provided lysis reagent for 10 min and then incubated with the ATP monitoring reagent for 5 min. Measurements were carried out in a Tecan Infinite F200 microplate reader (Tecan Group Ltd., Crailsheim, Germany). At least five animals per group were included for the measurement.
RNA isolation and quantitative real-time polymerase chain reaction (RT-PCR)
Total RNA of the murine cortex was isolated using the NucleoSpin® miRNA Kit according to the manufacturer’s protocol (Macherey & Nagel, Düren, Germany). RNA concentration was determined by measuring the absorbance at 260 nm on a spectrophotometer (Beckmann Instruments, Munich, Germany), and RNA purity was determined by calculating 260/280 and 260/230 nm ratios. RNA aliquots were then stored at −80 °C until PCR analysis. The RT-PCR primers were designed using Primer3 Input software (version 0.4.0) (Table 1) and purchased from Eurofins MWG (Ebersberg, Germany). One-step quantitative reverse transcriptase PCR was carried out with the SensiMix™ SYBR No-ROX One-step Kit (Bioline, Luckenwalde, Germany) and with SYBR Green detection using the Rotorgene 6000 cycler (Corbett Life Science, Sydney, Australia). At least three animals per group were included for the measurement. An external standard curve was applied. Relative mRNA concentrations of target genes were related to housekeeping gene expression. The mean value of expression in the control APOE3 group was set to an arbitrary unit of 1.
Table 1.
Nucleotide sequences (5′–3′) of murine primers used for the quantitative real-time PCR
| Gene | Gene ID | Primer sequence forward | Primer sequence reverse |
|---|---|---|---|
| β-Actin | 11461 | GACAGGATGCAGAAGAGATTACT | TGATCCACATCTGCTGGAAGGT |
| GAPBa | 14390 | GTCATCTGGAGCTTCTTCG | TAATCTGCACAGGCTGGTC |
| PGC1α | 19017 | AGAAGCGGGAGTCTGAAAGG | CAGTTCTGTCCGCGTTGTG |
| PPARγ | 19016 | AAGAGCTGACCCAATGGTTG | ACCCTTGCATCCTTCACAAG |
| TFAM | 21780 | TAGGAAAATTGCAGCCCTGT | GCTGAACGAGGTCTTTTTGG |
Western blotting
Whole cell lysates of murine cortex were used to investigate the protein concentrations of mitochondrial complexes. Protein concentrations were first determined using the Pierce BCA protein assay kit (Thermo Scientific, Rockford, USA). Samples were mixed with Tris/glycine reducing buffer and denaturing loading buffer (both from Invitrogen™, Germany). Subsequently, the samples were loaded and electrophoresed on NuPAGE 4–12 % Bis–Tris Gels (Invitrogen™, Germany). Proteins were then transferred to PVDF membranes (Amersham Biosciences) and incubated with the respective primary antibodies: MitoProfile Total OXPHOS (ab110411), complex IV (ab110259) (both from Abcam, Cambridge, UK) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (MAB374) (Millipore, Billerica, USA), and secondary antibodies (Calbiochem, Germany) conjugated with horseradish peroxidase. The bands were then visualized by using ECL Plus reagent (Amersham Biosciences). GAPDH served as a loading control. At least three animals per group were included for the measurement, and two replicates of each measurement were carried out. Band analysis was performed using BioRad’s Quantity One software.
Measurement of proteasomal activity
Proteasomal activity was assayed as described previously by Stolzing and Grune (Stolzing and Grune 2003) with minor modifications. At least three animals per group were included for the measurement. Cortex tissue was homogenized in 20 μL lysis buffer (250 mM sucrose, 25 mM HEPES, 10 mM MgCl2, 1 mM EDTA, and 1.7 mM dithiothreitol (DTT)) per milligram tissue. Following homogenization, cells were centrifuged at 4 °C and 15,000g for 25 min and the supernatants were used for protein determination using Bradford assay and for proteasomal activity measurement. For proteasomal activity, the supernatants were incubated in 225 mM Tris buffer (pH 7.8), 45 mM KCl, 7.5 mM magnesium acetate, 7.5 mM MgCl2, and 1 mM DTT. To measure chymotrypsin-like activity of the proteasome, the fluorogenic peptide Suc-LLVY-MCA was used as a substrate at a concentration of 200 μM. Samples were incubated for 30 min at 37 °C, followed by MCA liberation measurements with a fluorescence reader (Reader EL 340, BioTek Instruments, Bad Friendrichshall, Germany) at 360-nm excitation and 460-nm emission. Free MCA was used as the standard for quantification.
Statistical analysis
Statistical analysis was performed using SPSS version 19.0 (SPSS GmbH Software, Munich, Germany). Normal distribution of data was tested by Kolmogorov–Smirnov and Shapiro–Wilk tests. Normally distributed data were analysed using Student’s t test. In cases where data were not normally distributed, Mann–Whitney U test was performed. Results are presented as mean ± SEM. p values <0.05 were considered statistically significant.
Results
Curcumin supplementation increased ATP concentrations in the brain
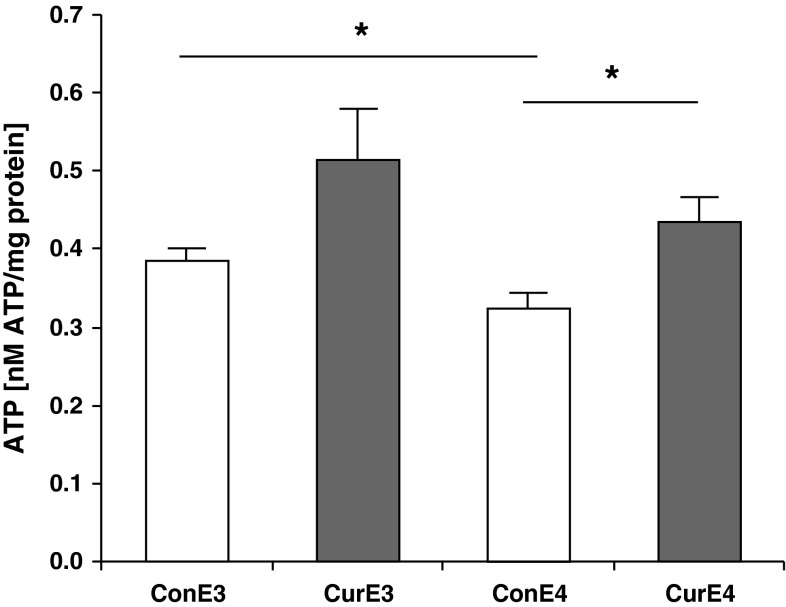
ATP concentrations were measured in dissociated brain cells of APOE3 and APOE4 mice fed with a control and curcumin-supplemented diet. ATP concentrations were significantly lower in APOE4 than in APOE3 control mice (Fig. 1). Curcumin supplementation significantly increased ATP concentrations in APOE4 mice (p < 0.05) and numerically in APOE3 mice (p = 0.087) relative to the respective controls.
Fig. 1.
Adenosine triphosphate (ATP) concentrations in dissociated brain cells of control and curcumin-supplemented APOE3 and APOE4 mice per milligram protein. ATP concentrations in dissociated brain cells are significantly lower in control APOE4 mice compared to control APOE3 mice. A similar trend can be observed comparing curcumin-supplemented APOE4 and APOE3 mice. Curcumin supplementation increased ATP concentrations significantly in APOE4 mice and numerically in APOE3 mice (p = 0.087). ConE3, APOE3 control group; CurE3, APOE3 curcumin group; ConE4, APOE4 control group; CurE4, APOE4 curcumin group. All values are means + SEM. *p < 0.05
Curcumin supplementation differentially regulated transcription factors involved in mitochondrial biogenesis in APOE3 and APOE4 mice
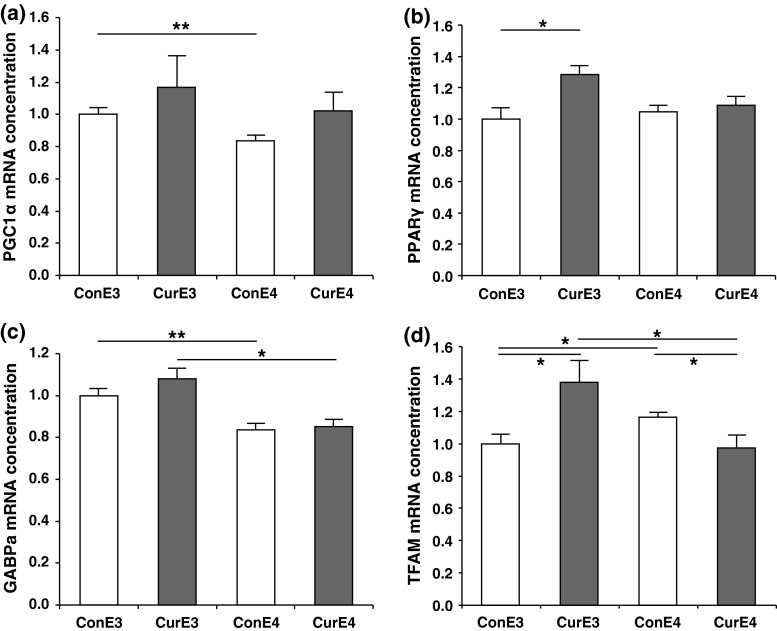
Consistent with reduced ATP concentrations (Fig. 1), the mRNA concentrations of the PGC1α, a key factor of mitochondrial biogenesis, were significantly lower in the cortex of APOE4 compared to APOE3 control mice. However, curcumin supplementation had no significant effect on PGC1α mRNA concentrations, probably due to high standard errors of the mean within the curcumin-supplemented groups (Fig. 2a). We then investigated the expression of PPARγ, as curcumin is a known PPARγ agonist (Rinwa et al. 2010; Wang et al. 2010), and PPARγ has been reported to interact with PGC1α (Puigserver et al. 1998). Curcumin supplementation induced a significant increase in PPARγ mRNA in the cortex of APOE3 (p < 0.05), but not in APOE4 mice (Fig. 2b). Furthermore, curcumin differentially modulated the mRNA concentrations of GABPa (Fig. 2c), another transcription factor that was also reported to interact with PGC1α and regulate mitochondrial biogenesis (Wu et al. 1999). GABPa mRNA concentrations in APOE3 mice were significantly higher than those in APOE4 mice, in both the curcumin-supplemented and control groups (p < 0.05 and p < 0.01, respectively). In accordance with these observations, mRNA concentrations of TFAM, which is a downstream effector of GABPa, were significantly increased following curcumin supplementation in APOE3 mice (p < 0.05) but were significantly reduced in APOE4 mice (p < 0.05; Fig. 2d). Taken together, a general trend of differential regulation of PPARγ, GABPa, and TFAM, where curcumin was effective in APOE3, but not in APOE4 mice, could be observed. Nuclear respiratory factor (NRF) 1, another transcription factor which regulates TFAM, was not significantly regulated (data not shown).
Fig. 2.
Expression of a peroxisome proliferator-activated receptor (PPAR) γ coactivator-1α (PGC1α), b PPARγ, c GA repeat binding protein alpha subunit (GABPa), and d mitochondrial transcription factor A (TFAM) mRNA in the cortex of control and curcumin-supplemented APOE3 and APOE4 mice. Relative mRNA concentration was assessed using RT–PCR and related to the housekeeping gene β–actin. A general trend of differential regulation where curcumin was effective in APOE3, but not in APOE4 mice could be observed for PPARγ, GABPa, and TFAM. For PGC1α, the effects of curcumin supplementation were not significant. The mRNA expression of PPARγ was significantly higher in curcumin-supplemented APOE3 mice compared to control APOE3 mice but was not significantly regulated in APOE4 mice. GABPa was significantly reduced comparing control and curcumin-supplemented APOE3 mice to APOE4 mice. In the case of TFAM, curcumin significantly upregulated TFAM mRNA concentrations in APOE3 mice, but downregulated TFAM in APOE4 mice. ConE3, APOE3 control group; CurE3, APOE3 curcumin group; ConE4, APOE4 control group; CurE4, APOE4 curcumin group. Values are expressed as means + SEM. *p < 0.05, **p < 0.01
Curcumin supplementation increased protein concentrations of mitochondrial respiratory complexes in the cortex
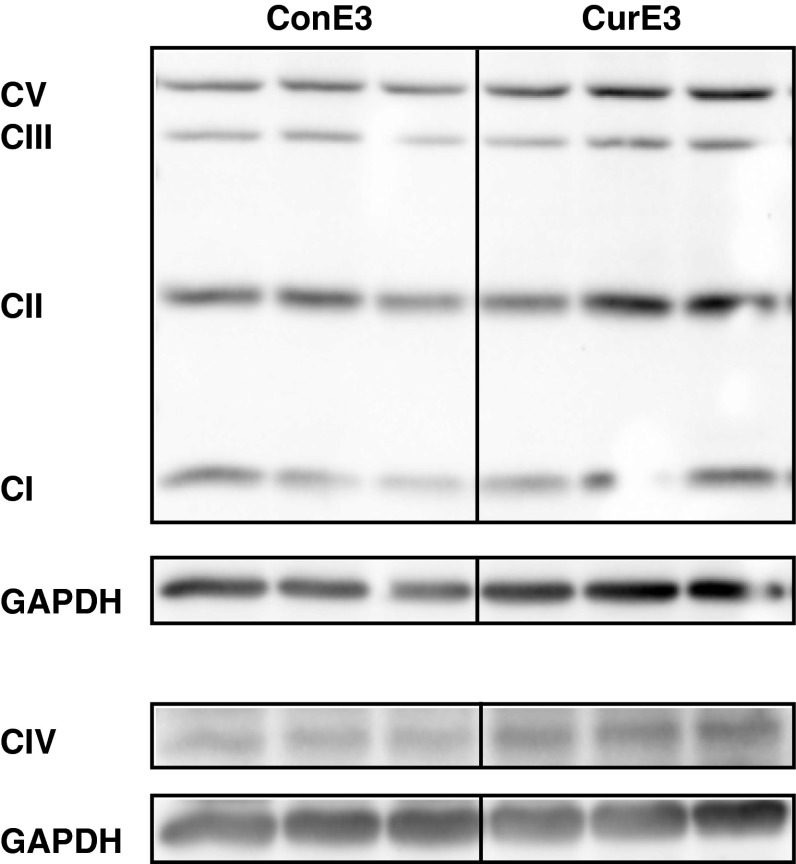
Consistent with the increased mRNA concentrations of transcription factors involved in the regulation of mitochondrial biogenesis and function, the protein concentrations of mitochondrial respiratory complexes I–V seemed slightly increased following curcumin supplementation in APOE3 mice (Fig. 3). In particular, complex IV showed a trend towards increased protein expression (densitometry normalized to ConE3 ± SEM: ConE3 = 1.0 ± 0.1, CurE3 = 1.5 ± 0.2; p = 0.059). There were no differences observed in APOE4 mice in response to curcumin supplementation (data not shown).
Fig. 3.
Western blotting of the mitochondrial respiratory complexes I–V in the cortex of control and curcumin-supplemented APOE3 mice. Relative intensities of bands were quantified by densitometry and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a loading control. There is a trend towards increased protein concentrations of mitochondrial respiratory complexes in curcumin-supplemented APOE3 mice, especially for complex IV. ConE3, APOE3 control group; CurE3, APOE3 curcumin group; CI, complex I; CII, complex II; CIII, complex III; CIV, complex IV; CV, complex V
Proteasomal activity in the cortex did not differ between curcumin-supplemented and control APOE3 and APOE4 mice
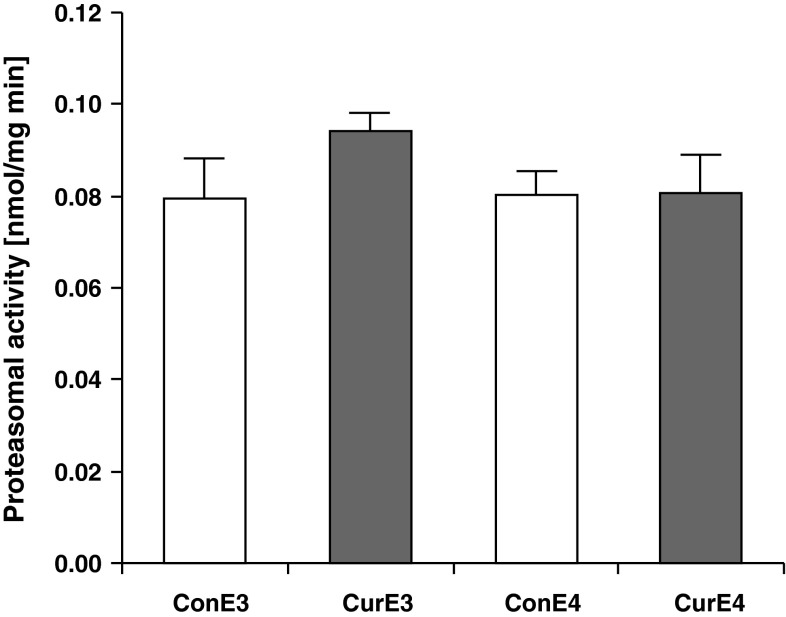
Since the proteasomal system degrades transcription factors and thereby affects gene transcription, we decided to measure whether the proteasomal activity differed between APOE3 and APOE4 mice or after curcumin supplementation. However, when measuring the proteasomal activity in the tissue lysates of the cortex, no significant differences between the groups were observed (Fig. 4).
Fig. 4.
Proteasomal activity in the cortex of curcumin-supplemented and control APOE3 and APOE4 mice. There were no significant differences observed between curcumin and control mice or between APOE3 and APOE4 mice. ConE3, APOE3 control group; CurE3, APOE3 curcumin group; ConE4, APOE4 control group; CurE4, APOE4 curcumin group. Values are expressed as means + SEM
Discussion
Despite widespread interest in curcumin as a potential therapeutic or preventive agent for neurodegenerative diseases, the neuroprotective efficacy of curcumin has not been investigated in the context of APOE even though APOE4 is a major genetic risk factor for late-onset AD. The principal finding of our study is that dietary curcumin supplementation may increase ATP concentrations in the brain of APOE3- and APOE4-targeted gene replacement mice, which is in agreement with our previous observations in SAMP8 mice (Eckert et al. 2013). Moreover, our APOE4 mice showed significantly lower ATP concentrations compared to APOE3 mice, which is consistent with the previous observation of lowered mitochondrial respiratory capacity in the cortex of APOE4 mice (Chen et al. 2011). We also observed that APOE4 mice had increased concentrations of mitochondrial superoxide dismutase 2 (data not shown), which might be indicative of increased oxidative damage as previously observed in patients with AD carrying the APOE4 allele (Tayler et al. 2010). Contrarily, we did not observe any changes in the proteasomal activity in the cortex although curcumin has been reported to modulate proteasomal degradation (Dikshit et al. 2006; Hasima and Aggarwal 2013).
Interestingly, our attempt to identify potential underlying mechanisms revealed that curcumin induced the expression of transcription factors (PPARγ, GABPa, and TFAM) and coactivators (PGC1α) of mitochondrial biogenesis in APOE3 mice. However, this was not the case for APOE4 mice since the expression of three out of four of these genes did not increase in response to curcumin. With the exception of PGC1α, the expression of PPARγ, GABPa, and TFAM remained unchanged or was even decreased in curcumin-fed APOE4 mice. The induction of PGC1α by curcumin in APOE3 and APOE4 mice confirms our previous observations in SAMP8 mice fed curcumin for 5 months (Eckert et al. 2013). PGC1α is considered the master regulator of mitochondrial biogenesis and function (Puigserver and Spiegelman 2003), modulating the expression of other transcription factors such as PPARγ, NRF1, and GABPa (Wu et al. 1999). Therefore, the curcumin-induced, albeit minor, increase in PGC1α mRNA may at least partly account for the higher ATP concentrations in curcumin-fed mice of both genotypes, APOE3 and APOE4.
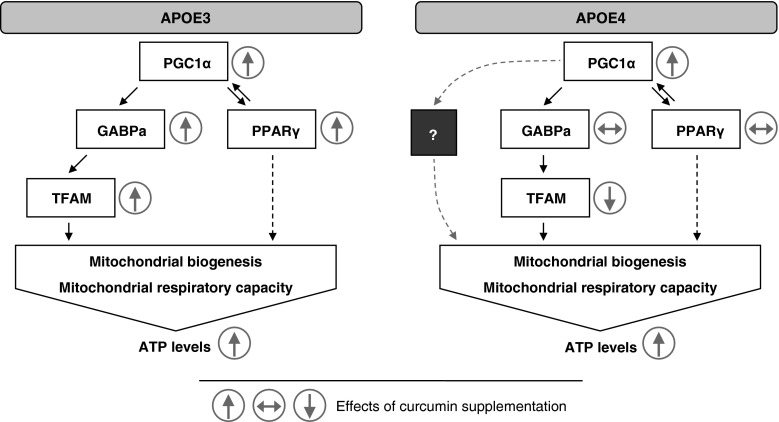
In contrast, the curcumin-mediated induction of PPARγ, GABPa, and TFAM mRNA was only observed in APOE3 mice. Similar results were obtained from protein concentrations of mitochondrial respiratory complexes, which showed a moderate induction upon curcumin supplementation in APOE3, but not APOE4 mice. The proposed interaction between the transcription factors that were regulated in response to curcumin supplementation in APOE3 mice is illustrated in Fig. 5. In APOE3 mice, curcumin induced PGC1α which interacted with GABPa and PPARγ to increase mitochondrial biogenesis and mitochondrial respiratory capacity. These effects could be mediated by the activation of TFAM, the putative downstream effector of GABPa. In APOE3 mice, our observation that increased GABPa and TFAM mRNA concentrations correlated with an increase in mitochondrial respiratory chain proteins (in particular, complex IV) is consistent with previous findings showing that silencing of GABPa reduced TFAM and cytochrome c oxidase (complex IV) expression in rat primary cortical neurons (Ongwijitwat et al. 2006). Curcumin, a known PPARγ agonist (Rinwa et al. 2010; Wang et al. 2010), also induced PPARγ in our APOE3 mice. Although PPARγ has not been associated with the regulation of mitochondrial respiratory complexes, PPARγ may indirectly enhance mitochondrial biogenesis by activating the PPARγ responsive element found in the gene promoter region of PGC1α, thus increasing the transcription of PGC1α (Hondares et al. 2006).
Fig. 5.
Possible interaction between various transcription factors and transcriptional coactivators contributing to the observed increase in adenosine triphosphate (ATP) concentrations in response to curcumin supplementation in APOE3 and APOE4 mice. Peroxisome proliferator-activated receptor (PPAR) γ coactivator (PGC)-1α is a transcriptional coactivator that functions as a key regulator for energy homeostasis and mitochondrial biogenesis. PGC1α was previously shown to interact with the guanine–adenine repeat binding protein alpha subunit (GABPa) to induce increased mitochondrial biogenesis and mitochondrial respiratory capacity. This mechanism could explain the increased mitochondrial respiratory capacity after curcumin supplementation in APOE3 mice since the activation of mitochondrial transcription factor A (TFAM) through GABPa possibly led to the increased protein concentrations of mitochondrial respiratory complexes and increased ATP concentrations. Curcumin effects in APOE3 mice might also be mediated by PPARγ. Although PPARγ has not been associated with the regulation of mitochondrial respiratory complexes, PPARγ may indirectly enhance mitochondrial biogenesis by activating the PPARγ responsive element found in the gene promoter region of PGC1α, thereby increasing the transcription of PGC1α. In APOE4 mice, curcumin only induced a slight increase in PGC1α but did not seem to induce downstream PPARγ or GABPa. TFAM was even decreased in APOE4 mice. An alternative pathway might be responsible for the increased ATP concentrations in APOE4 mice
Our data suggest that at least some of the effects of curcumin are less pronounced in APOE4 compared to APOE3 mice and thus that APOE3 is more responsive to dietary curcumin intervention. This is in accordance with our previous findings that APOE4 mice are less sensitive to dietary intervention with vitamin E and quercetin than APOE3 mice (Huebbe et al. 2007; Boesch-Saadatmandi et al. 2009).
We have previously shown that oral gavage of 50 mg/kg BW curcumin did not lead to detectable amounts of curcumin in the brain (Schiborr et al. 2010). Nevertheless, these data suggest that curcumin could affect mitochondrial function in the cortex despite its low bioavailability. This is consistent with previous observations that curcumin prevented mitochondrial dysfunction in the brain of senescence-accelerated mice (Eckert et al. 2013). The finding that curcumin induced a positive response in the cortex although it could not be detected in these tissues suggests that curcumin may exert secondary effects without being physically present in the brain. Previously, curcumin metabolites such as ferulic and vanillic acid have been shown to be enriched in serum upon application of curcumin and could therefore contribute to its metabolic effects (Vitaglione et al. 2012). Furthermore, it has been shown that curcumin is an iron chelator and could interact with iron metabolism (Chin et al. 2014; Jiao et al. 2009), thus modulating mitochondrial function. The possibility that local antimicrobial effects of curcumin (Bhawana et al. 2011) might modify the oral or gut microflora which could then trigger systemic alterations and exert effects on the brain is also particularly interesting. In fact, there are indications that the oral microflora may influence the pathology of AD. Certain strains of periodontal pathogens have been found more frequently in AD brains, and serum levels of antibodies against common periodontal pathogens have been associated with an increased risk of developing AD (Riviere et al. 2002; Kamer et al. 2009; Sparks Stein et al. 2012). Alternatively, the concept of hormesis which describes the adaptive process where exposure to a (very) low dose of a stimulus can activate cellular stress response pathways and thereby increase the ability to cope with more severe stress (Mattson and Cheng 2006; Calabrese et al. 2010) may also explain the observed effects of curcumin. According to this concept, even very low curcumin brain concentration could affect gene and protein expression and improve the organism’s stress response (Mattson and Cheng 2006; Calabrese et al. 2010).
Conclusion
In conclusion, we provide evidence that dietary curcumin affects gene and protein expression in the murine brain despite its low bioavailability. Furthermore, with regard to mitochondrial function and ATP synthesis, our data support the notion that the APOE3 genotype may be more responsive to dietary curcumin intervention compared to the APOE4 genotype.
Acknowledgments
We are grateful to the German Federal Ministry of Education and Research (BMBF) for financial support (Grant # 0315679).
Conflict of interest
Dawn Chin, Stephanie Hagl, Annika Hoehn, Patricia Huebbe, Kathrin Pallauf, Tilman Grune, Jan Frank, Gunter P. Eckert, and Gerald Rimbach declare that they have no conflict of interest.
Ethical standard
All institutional and national guidelines for the care and use of laboratory animals were followed.
References
- Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. Bioavailability of curcumin: problems and promises. Mol Pharm. 2007;4(6):807–818. doi: 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- Baum L, Lam CW, Cheung SK, Kwok T, Lui V, Tsoh J, Lam L, Leung V, Hui E, Ng C, Woo J, Chiu HF, Goggins WB, Zee BC, Cheng KF, Fong CY, Wong A, Mok H, Chow MS, Ho PC, Ip SP, Ho CS, Yu XW, Lai CY, Chan MH, Szeto S, Chan IH, Mok V. Six-month randomized, placebo-controlled, double-blind, pilot clinical trial of curcumin in patients with Alzheimer disease. J Clin Psychopharmacol. 2008;28(1):110–113. doi: 10.1097/jcp.0b013e318160862c. [DOI] [PubMed] [Google Scholar]
- Bhawana Basniwal RK, Buttar HS, Jain VK, Jain N. Curcumin nanoparticles: preparation, characterization, and antimicrobial study. J Agric Food Chem. 2011;59(5):2056–2061. doi: 10.1021/jf104402t. [DOI] [PubMed] [Google Scholar]
- Boesch-Saadatmandi C, Wolffram S, Minihane AM, Rimbach G. Effect of apoE genotype and dietary quercetin on blood lipids and TNF-alpha levels in apoE3 and apoE4 targeted gene replacement mice. Br J Nutr. 2009;101(10):1440–1443. doi: 10.1017/S0007114508102434. [DOI] [PubMed] [Google Scholar]
- Calabrese V, Cornelius C, Dinkova-Kostova AT, Calabrese EJ, Mattson MP. Cellular stress responses, the hormesis paradigm, and vitagenes: novel targets for therapeutic intervention in neurodegenerative disorders. Antioxid Redox Signal. 2010;13(11):1763–1811. doi: 10.1089/ars.2009.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V, Ganguli M, Pandav R, Johnston J, Belle S, DeKosky ST. Prevalence of Alzheimer’s disease and other dementias in rural India: the Indo-US study. Neurology. 1998;51(4):1000–1008. doi: 10.1212/WNL.51.4.1000. [DOI] [PubMed] [Google Scholar]
- Chandra V, Pandav R, Dodge HH, Johnston JM, Belle SH, DeKosky ST, Ganguli M. Incidence of Alzheimer’s disease in a rural community in India: the Indo-US study. Neurology. 2001;57(6):985–989. doi: 10.1212/WNL.57.6.985. [DOI] [PubMed] [Google Scholar]
- Chen HK, Ji ZS, Dodson SE, Miranda RD, Rosenblum CI, Reynolds IJ, Freedman SB, Weisgraber KH, Huang Y, Mahley RW. Apolipoprotein E4 domain interaction mediates detrimental effects on mitochondria and is a potential therapeutic target for Alzheimer disease. J Biol Chem. 2011;286(7):5215–5221. doi: 10.1074/jbc.M110.151084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HK, Liu Z, Meyer-Franke A, Brodbeck J, Miranda RD, McGuire JG, Pleiss MA, Ji ZS, Balestra ME, Walker DW, Xu Q, Jeong DE, Budamagunta MS, Voss JC, Freedman SB, Weisgraber KH, Huang Y, Mahley RW. Small molecule structure correctors abolish detrimental effects of apolipoprotein E4 in cultured neurons. J Biol Chem. 2012;287(8):5253–5266. doi: 10.1074/jbc.M111.276162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin D, Huebbe P, Pallauf K, Rimbach G. Neuroprotective properties of curcumin in Alzheimer’s disease–merits and limitations. Curr Med Chem. 2013;20(32):3955–3985. doi: 10.2174/09298673113209990210. [DOI] [PubMed] [Google Scholar]
- Chin D, Huebbe P, Frank J, Rimbach G, Pallauf K (2014) Curcumin may impair iron status when fed to mice for six months. Redox Biology. doi:http://dx.doi.org/10.1016/j.redox.2014.01.018 [DOI] [PMC free article] [PubMed]
- Dikshit P, Goswami A, Mishra A, Chatterjee M, Jana NR. Curcumin induces stress response, neurite outgrowth and prevent NF-kappaB activation by inhibiting the proteasome function. Neurotox Res. 2006;9(1):29–37. doi: 10.1007/BF03033305. [DOI] [PubMed] [Google Scholar]
- Eckert GP, Renner K, Eckert SH, Eckmann J, Hagl S, Abdel-Kader RM, Kurz C, Leuner K, Muller WE. Mitochondrial dysfunction—a pharmacological target in Alzheimer’s disease. Mol Neurobiol. 2012;46(1):136–150. doi: 10.1007/s12035-012-8271-z. [DOI] [PubMed] [Google Scholar]
- Eckert GP, Schiborr C, Hagl S, Abdel-Kader R, Muller WE, Rimbach G, Frank J. Curcumin prevents mitochondrial dysfunction in the brain of the senescence-accelerated mouse-prone 8. Neurochem Int. 2013;62(5):595–602. doi: 10.1016/j.neuint.2013.02.014. [DOI] [PubMed] [Google Scholar]
- Esatbeyoglu T, Huebbe P, Ernst IM, Chin D, Wagner AE, Rimbach G. Curcumin—from molecule to biological function. Angew Chem Int Ed Engl. 2012;51(22):5308–5332. doi: 10.1002/anie.201107724. [DOI] [PubMed] [Google Scholar]
- Hasima N, Aggarwal BB (2013) Targeting proteasomal pathways by dietary curcumin for cancer prevention and treatment. Curr Med Chem. doi:10.2174/09298673113206660135 [DOI] [PubMed]
- Hauptmann S, Scherping I, Drose S, Brandt U, Schulz KL, Jendrach M, Leuner K, Eckert A, Muller WE. Mitochondrial dysfunction: an early event in Alzheimer pathology accumulates with age in AD transgenic mice. Neurobiol Aging. 2009;30(10):1574–1586. doi: 10.1016/j.neurobiolaging.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Hishikawa N, Takahashi Y, Amakusa Y, Tanno Y, Tuji Y, Niwa H, Murakami N, Krishna UK. Effects of turmeric on Alzheimer’s disease with behavioral and psychological symptoms of dementia. Ayu. 2012;33(4):499–504. doi: 10.4103/0974-8520.110524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondares E, Mora O, Yubero P, Rodriguez de la Concepcion M, Iglesias R, Giralt M, Villarroya F. Thiazolidinediones and rexinoids induce peroxisome proliferator-activated receptor-coactivator (PGC)-1alpha gene transcription: an autoregulatory loop controls PGC-1alpha expression in adipocytes via peroxisome proliferator-activated receptor-gamma coactivation. Endocrinology. 2006;147(6):2829–2838. doi: 10.1210/en.2006-0070. [DOI] [PubMed] [Google Scholar]
- Huebbe P, Jofre-Monseny L, Boesch-Saadatmandi C, Minihane AM, Rimbach G. Effect of apoE genotype and vitamin E on biomarkers of oxidative stress in cultured neuronal cells and the brain of targeted replacement mice. J Physiol Pharmacol. 2007;58(4):683–698. [PubMed] [Google Scholar]
- Huebbe P, Nebel A, Siegert S, Moehring J, Boesch-Saadatmandi C, Most E, Pallauf J, Egert S, Muller MJ, Schreiber S, Nothlings U, Rimbach G. APOE epsilon4 is associated with higher vitamin D levels in targeted replacement mice and humans. FASEB J. 2011;25(9):3262–3270. doi: 10.1096/fj.11-180935. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wilkinson JT, Di X, Wang W, Hatcher H, Kock ND, D’Agostino R, Jr, Knovich MA, Torti FM, Torti SV. Curcumin, a cancer chemopreventive and chemotherapeutic agent, is a biologically active iron chelator. Blood. 2009;113(2):462–469. doi: 10.1182/blood-2008-05-155952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofre-Monseny L, Minihane AM, Rimbach G. Impact of apoE genotype on oxidative stress, inflammation and disease risk. Mol Nutr Food Res. 2008;52(1):131–145. doi: 10.1002/mnfr.200700322. [DOI] [PubMed] [Google Scholar]
- Kamer AR, Craig RG, Pirraglia E, Dasanayake AP, Norman RG, Boylan RJ, Nehorayoff A, Glodzik L, Brys M, de Leon MJ. TNF-alpha and antibodies to periodontal bacteria discriminate between Alzheimer’s disease patients and normal subjects. J Neuroimmunol. 2009;216(1–2):92–97. doi: 10.1016/j.jneuroim.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouff C, Hinsdale ME, Mezdour H, Altenburg MK, Watanabe M, Quarfordt SH, Sullivan PM, Maeda N. Apo E structure determines VLDL clearance and atherosclerosis risk in mice. J Clin Invest. 1999;103(11):1579–1586. doi: 10.1172/JCI6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Cheng A. Neurohormetic phytochemicals: low-dose toxins that induce adaptive neuronal stress responses. Trends Neurosci. 2006;29(11):632–639. doi: 10.1016/j.tins.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Ongwijitwat S, Liang HL, Graboyes EM, Wong-Riley MT. Nuclear respiratory factor 2 senses changing cellular energy demands and its silencing down-regulates cytochrome oxidase and other target gene mRNAs. Gene. 2006;374:39–49. doi: 10.1016/j.gene.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24(1):78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–839. doi: 10.1016/S0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Ringman JM, Frautschy SA, Teng E, Begum AN, Bardens J, Beigi M, Gylys KH, Badmaev V, Heath DD, Apostolova LG, Porter V, Vanek Z, Marshall GA, Hellemann G, Sugar C, Masterman DL, Montine TJ, Cummings JL, Cole GM. Oral curcumin for Alzheimer’s disease: tolerability and efficacy in a 24-week randomized, double blind, placebo-controlled study. Alzheimers Res Ther. 2012;4(5):43. doi: 10.1186/alzrt146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinwa P, Kaur B, Jaggi AS, Singh N. Involvement of PPAR-gamma in curcumin-mediated beneficial effects in experimental dementia. Naunyn Schmiedebergs Arch Pharmacol. 2010;381(6):529–539. doi: 10.1007/s00210-010-0511-z. [DOI] [PubMed] [Google Scholar]
- Riviere GR, Riviere KH, Smith KS. Molecular and immunological evidence of oral treponema in the human brain and their association with Alzheimer’s disease. Oral Microbiol Immunol. 2002;17(2):113–118. doi: 10.1046/j.0902-0055.2001.00100.x. [DOI] [PubMed] [Google Scholar]
- Schiborr C, Eckert GP, Rimbach G, Frank J. A validated method for the quantification of curcumin in plasma and brain tissue by fast narrow-bore high-performance liquid chromatography with fluorescence detection. Anal Bioanal Chem. 2010;397(5):1917–1925. doi: 10.1007/s00216-010-3719-3. [DOI] [PubMed] [Google Scholar]
- Sparks Stein P, Steffen MJ, Smith C, Jicha G, Ebersole JL, Abner E, Dawson D., 3rd Serum antibodies to periodontal pathogens are a risk factor for Alzheimer’s disease. Alzheimers Dement. 2012;8(3):196–203. doi: 10.1016/j.jalz.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolzing A, Grune T. Impairment of protein homeostasis and decline of proteasome activity in microglial cells from adult Wistar rats. J Neurosci Res. 2003;71(2):264–271. doi: 10.1002/jnr.10478. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Roses AD. Apolipoprotein E and Alzheimer’s disease. Annu Rev Neurosci. 1996;19:53–77. doi: 10.1146/annurev.ne.19.030196.000413. [DOI] [PubMed] [Google Scholar]
- Sullivan PM, Mezdour H, Aratani Y, Knouff C, Najib J, Reddick RL, Quarfordt SH, Maeda N. Targeted replacement of the mouse apolipoprotein E gene with the common human APOE3 allele enhances diet-induced hypercholesterolemia and atherosclerosis. J Biol Chem. 1997;272(29):17972–17980. doi: 10.1074/jbc.272.29.17972. [DOI] [PubMed] [Google Scholar]
- Sullivan PM, Mezdour H, Quarfordt SH, Maeda N. Type III hyperlipoproteinemia and spontaneous atherosclerosis in mice resulting from gene replacement of mouse Apoe with human Apoe*2. J Clin Invest. 1998;102(1):130–135. doi: 10.1172/JCI2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayler H, Fraser T, Miners JS, Kehoe PG, Love S. Oxidative balance in Alzheimer’s disease: relationship to APOE, Braak tangle stage, and the concentrations of soluble and insoluble amyloid-beta. J Alzheimers Dis. 2010;22(4):1363–1373. doi: 10.3233/JAD-2010-101368. [DOI] [PubMed] [Google Scholar]
- Vitaglione P, Barone Lumaga R, Ferracane R, Radetsky I, Mennella I, Schettino R, Koder S, Shimoni E, Fogliano V. Curcumin bioavailability from enriched bread: the effect of microencapsulated ingredients. J Agric Food Chem. 2012;60(13):3357–3366. doi: 10.1021/jf204517k. [DOI] [PubMed] [Google Scholar]
- Wang HM, Zhao YX, Zhang S, Liu GD, Kang WY, Tang HD, Ding JQ, Chen SD. PPARgamma agonist curcumin reduces the amyloid-beta-stimulated inflammatory responses in primary astrocytes. J Alzheimers Dis. 2010;20(4):1189–1199. doi: 10.3233/JAD-2010-091336. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]