Abstract
If mathematical modeling is to be used effectively in cancer drug development, future models must take into account both the mechanistic details of cellular signal transduction networks and the pharmacokinetics (PK) of drugs used to inhibit their oncogenic activity. In this perspective, we present an approach to building multiscale models that capture systems-level architectural features of oncogenic signaling networks, and describe how these models can be used to design combination therapies and identify predictive biomarkers in silico.
The Challenge
The last 60 years have witnessed an explosion in scientific data about the molecular underpinnings of health and disease. Unfortunately, this increase in information has not translated into an increase in pharmaceutical research productivity–over the same 60-year period the annual output of new drugs has held roughly steady while R&D expenses have grown exponentially.1 Why is this? Some have speculated that all of the “low-hanging therapeutic fruit” has been harvested, and others have opined that the increasingly competitive market and regulatory environment is demanding more stringent benefit/risk profiles for new therapies.2 While these issues may hold some truth, what has become increasingly clear is that the development of future medicines will require a deeper appreciation of the complexity of disease states, and an understanding of how drug treatments affect not only diseased tissue, but also surrounding healthy tissue and organs across diverse patient populations. One proposed solution to improve drug R&D productivity is to utilize computational modeling and simulation as a foundational platform, as it has been in other high-tech industries. By providing a coherent framework for integrating knowledge and formalizing assumptions, mathematical models can enhance our ability to make informed and quantitative decisions. This is not a new idea – the FDA advocated “model-based drug development” as a pillar of its critical path initiative in 2004.3 The question is: why are notable successes on this front so scarce a decade later?
A simplistic explanation is that biology is too complex–more complex than we comprehend, and still too complex to be formulated mathematically. While we may lack a unified theory of molecular biology, this doesn't necessarily preclude our ability to develop models capable of answering specific questions, within a defined scope.4 Computational modeling has in fact played an important role in biomedical R&D for years. The issue is that the types of models currently employed exist in parallel universes, with different academic journals, university departments, and languages.5 Computational systems biology is largely an academic discipline, focused on discovery-stage research with an emphasis on understanding molecular and cellular mechanisms. Given the complexity of molecular biochemistry in comparison with the sparse data typically available, such models are by nature generally nonidentifiable.6 At the other end of the spectrum, pharmacometrics is focused on utilizing available clinical data to guide pressing drug development decisions. The technical emphasis is thus on robust parameterization and predictive accuracy rather than mechanistic detail, which biases towards more empirical model formulations. Developing multiscale models both with respect to the underlying science (connecting genes and proteins to cellular phenotypes, to tissue, organ, and population-level responses) and the drug development process (linking discovery through clinical development) is a critical challenge.7 While a number of organizational and cultural issues are involved, a key technical difficulty lies in linking together the different time and length scales at play into a single representative format.
Signaling Networks Link Molecular Events to Cellular Responses
Signal transduction networks function at the interface of the cell's external and internal worlds, converting environmental information into biochemical changes, and ultimately discrete cell fate decisions. These molecular circuits thus serve not just as linear messengers, but as computational systems performing highly complex, multivariate signal processing. Many pathologies, in particular cancers, are currently understood to arise from aberrant cellular information processing, a result of genetic and epigenetic perturbations in signaling networks.8 Signal transduction networks thus appear to be an appropriate and perhaps necessary means to mechanistically link PK and pharmacodynamic (PD) models.9 Traditional drug exposure-response models are constructed using empirical transfer functions, though often including semi-mechanistic details based on receptor theory.10 These are generally sufficient for purposes in which they were developed—defining dosing regimens for monotherapies, based on existing clinical data. Such empirical formulations however fall short of the most pressing challenges in modern drug development. Using oncology as an example, where an increasing array of molecular targeted therapies are available for clinical use, meaningful responses to single agents are rare and generally transient, necessitating combination drug regimens.11 In addition, tumors are widely heterogeneous at the molecular level. This molecular diversity mediates differential drug sensitivities between patients, and even between clonal cell populations within individual tumors.12 Predictive molecular biomarkers are thus required to match drug regimens with patient populations expected to respond to therapy. Testing drug combination regimens in an empirical manner is, however, becoming increasingly unfeasible. Given the number of anticancer agents currently available and the genetic diversity of the disease, the combinatorial explosion of possibilities is beyond what can typically be tested through brute force screening. Computational modeling and simulation could provide means to beset this limitation, allowing one to perform ultra-high throughput in silico screening and analysis of synthetic treatment-response data. Developing mathematical models capable of meeting this challenge is, however, a nontrivial task. Below we describe the obstacles and outline some proposed solutions.
Quantitative Logic as a Framework for Representing Signaling Networks
So how does one mathematically formulate network biology-based PK-PD models? Most biochemical network models published to date have been based on mass action kinetic-based ordinary differential equations.13 Motivation for doing so arises from the desire to develop mathematical models based on fundamental physiochemical properties and reaction constants, thus transferable across different cells, tissues, or disease states. Moreover, given the extensive history of mass action kinetic-based modeling in other disciplines, established methods and expertise are widely available to draw from. There are many notable successes, both in fundamental cell biology14,15 as well industry pursuits such as drug target discovery16 and therapeutic antibody design.17,18,19 However, caution should be used when applying the assumptions underlying mass action kinetics to intracellular processes. Most biochemical reactions involved in cellular signal transduction take place as part of multiprotein complexes, often tethered to scaffolds or cell membranes. The kinetics would therefore be expected to deviate from that predicted by the laws of mass action, which assume homogeneous solution-phase reactions. More importantly, as our knowledge of molecular biology is still far from complete, it is very difficult to parse biochemical cascades down to fundamental reaction steps, or to account for all relevant molecular species and reactions.20 This problem is particularly acute for processes downstream of canonical signaling cascades, connecting signaling events to gene expression changes or cellular phenotypes. Even extensively detailed physiochemical-based models thus often contain many lumped parameters, which must be estimated by fitting to experimental data rather than derived from biophysical properties. This may underlie one of the difficulties in extrapolating model parameters across different cell lines.
It is also important to recognize the distinct time scales at play. Dynamic events triggered by cell surface receptor engagement reach (quasi)-steady state within minutes to a few hours, while phenotypic readouts (i.e., measurable changes in bulk tumor size) are typically quantified on the order of days to weeks. As a consequence, many molecular events may be represented algebraically rather than with more arduous differential equations. Another practical consideration is the type of data available for model training. Biochemical measurements are typically semi-quantitative, lacking the precision (molecules/cell) and the coverage (measured vs. inferred species) required to parameterize mass-action kinetic-based models. Quantitative logic provides an alternative and relatively simple formalism to represent the structure and information processing capabilities of signaling networks,21,22 bridging the distinct time scales of biochemical and physiological events. Quantitative logic networks are assembled using Hill-type equations, malleable signal-response curves representing information flow between nodes (i.e., protein species). When a network node contains multiple inputs, quantitative logic gates can be used to represent various types of signal processing.23 These are analogue extensions of Boolean logic truth tables, the most common forms being AND, NAND, OR, and NOR gates, which can be configured to recapitulate biochemical and pharmacological mechanisms. The algebraic equations can be easily extended into differential equation form, so as to capture both fast (steady state) and slower (dynamic) process together, using systems of differential-algebraic equations (Figure 1). The logic gates and hill functions used to describe signal flow in quantitative logic networks are data-driven rather than based upon fundamental biophysical constants. However, they are in fact not as different from mass action kinetic ordinary differential equations as initial appearances suggest, given that such models often contain many data-driven parameters as well. Note there is no single best approach to modeling cell signal transduction. The choice between alternatives, from purely data-driven statistical models to physiochemical ODEs, should be determined by the specific questions at hand, data available, and specifics of the underlying biology.
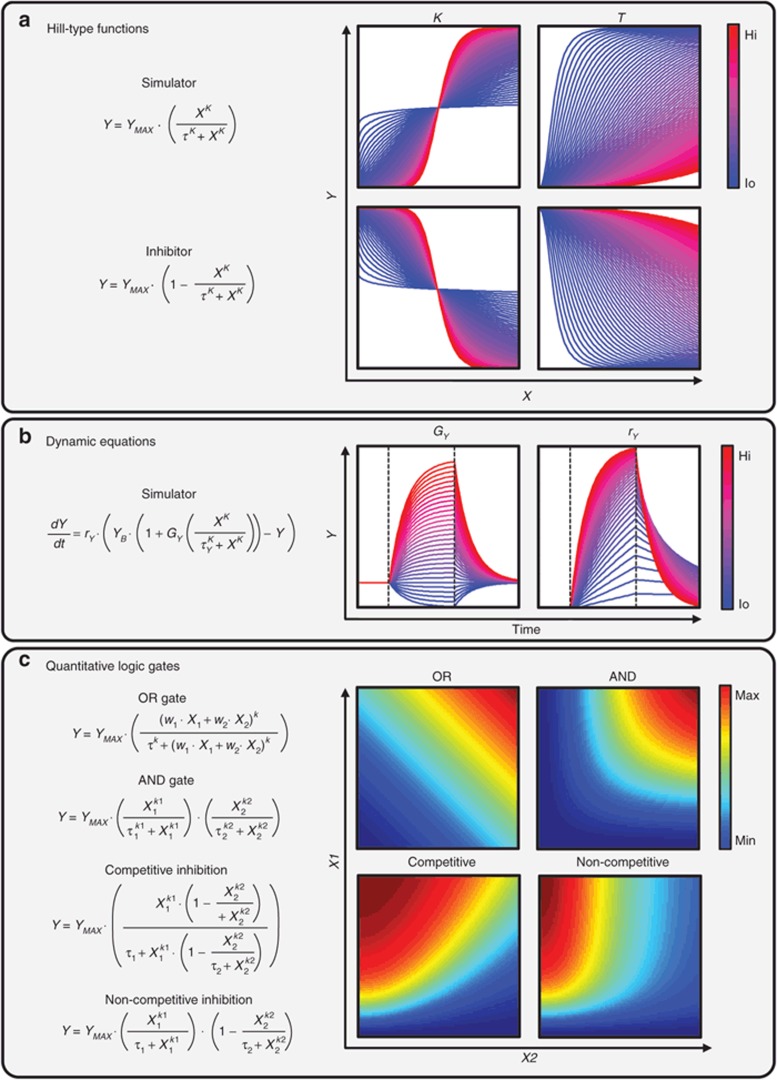
Figure 1.
Mathematical underpinnings of quantitative logic networks. (a) Hill-type equations can be used to describe input–output relationships between individual nodes (i.e., proteins) in a network. Using two parameters, the Hill coefficient (K) and EC50 value (τ), these malleable equations are capable of describing a wide range of stimulatory and inhibitory response curves. (b) Hill-type equations can be modified to capture dynamic as well as algebraic (steady state) relationships. Shown are exemplary time course phase portraits of an output variable (Y) in response to a pulse of stimulator (X), indicated by dashed lines. The system gain (GY) and symmetric rates of production/degradation (rY) can be tuned to capture differing system dynamics, while the basal set-point of Y (YB) quantifies the steady-state level. (c) Various types of quantitative logic gates can be used to capture biologically relevant responses to dual inputs. Shown are four exemplary surface responses: dual stimulatory OR gate (i.e., alternative RTKs activating PI3K, with different relative activation weights (w)), dual stimulatory AND gate (i.e., co-dependent transcription factors activating gene expression), competitive inhibition (i.e., stimulatory ligand vs. ligand-blocking antibody), and non-competitive inhibition (i.e., a stimulatory ligand vs. small molecule RTK inhibitor). PI3K, phosphatidylinositide 3-kinase.
The next challenge is specifying the level of detail required to describe the relevant biology and mechanistically capture exposure-response relationships. Explicitly characterizing all biochemical reactions taking place in a cell (“bottom-up” network reconstruction) is currently impractical given the complexity and remaining knowledge gaps involved in cellular signal transduction.20 Can we then identify archetypal properties of signaling networks necessary to be included in our models, striking a balance between predictive power and mechanistic representation of the biology? Below we describe four systems-level design features as hallmarks of oncogenic signaling networks: modularity, redundancy, adaptation, and heterogeneity. We will outline how these features can be captured mathematically, and the implications for drug development. While our focus is oncology, we believe the principles and approaches described are generalizable to other areas.
Hallmark 1: Modularity
It has been long standing practice in molecular biology to classify proteins and their biochemical interactions into pathways. It is now appreciated that such simple classification schemes, and original depictions of pathways as linear, insulated signaling cascades are vast oversimplifications of more complex integrated networks. However, evidence supports the notion of pathways as functional network modules—clusters of nodes (proteins) which are highly interconnected but less connected to other parts of the network, and mapped to highly overlapping phenotypes.24 This concept is perhaps best supported by data emerging from cancer genome sequencing projects. Even within clinically homogenous diseases, mutations between tumors are found to be incredibly disparate. However, when mutations are mapped onto protein interaction networks, they fall onto a relatively conserved set of oncogenic modules. Most notable are the phosphatidylinositide 3-kinase (PI3K)/AKT and mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) cascades, and the receptor tyrosine kinases (RTKs) which regulate their activities, the TP53 and cell-cycle control modules, the apoptotic caspase machinery, as well as a handful of other more indication and tissue-specific signaling cascades.25,26,27 Moreover, mutations within these network modules are generally found to be mutually exclusive (the prototypic example being PI3KCA-activating mutations vs. PTEN deletions), indicating that what “matters” to the cell is the activation of modules rather than individual molecules.
The implication is that mathematical representations of molecular networks can be condensed to more abstract, modular formats, while maintaining functional relevance. This can be achieved using rigorous bioinformatic28 or graph theoretical approaches,29 or heuristically (as is common practice) by selecting key sentinel nodes representative of modules activation states (i.e., phosphorylated AKT and ERK for the PI3K and MAPK cascades, respectively). This reduction vastly simplifies the computational requirements for parameterizing sparsely sampled and underdetermined systems. However, if the module decomposition scheme is appropriate, functional inferences from such granular descriptions should be equivalent to more detailed molecular forms.
Hallmark 2: Redundancy
Redundancy is observed at multiple levels within biochemical networks – we have already mentioned one example in that mutations within a network modules display functionally equivalent (or at least highly similar) effects. This is a consequence of biochemical information flow – disrupting a signal at different points along the transmission route will have similar effects (“vertical” redundancy). An alternative form of “horizontal” redundancy arises from cell's capacity for alternative biochemical wiring. A consequence of evolutionary recycling of protein-binding domains, network modules and their constitutive proteins are capable of combining in alternate configurations. The family of RTKs for example, though expressed and employed in a functionally diverse manner, all transmit signals across the plasma membrane through binding to a limited set of adaptor proteins. As a result, they inevitably plug into the same PI3K and MAPK cascades, though with differing strengths and dynamics. The functional consequences of RTK activation are thus highly overlapping, as the signals converge on a limited set of core cytoplasmic hub proteins (so called “bow tie” architecture).20 Clinical evidence now supports this theory, as different RTKs have been found to be functionally exchangeable even within individual tumor biopsies.30 The consequences of this architectural feature are becoming increasingly appreciated in oncology through the phenomena of “pathway switching.” Tumors displaying oncogene addiction to a single RTK driver are often capable of switching dependency to an alternate, but functionally equivalent RTK following inhibitor treatment.31
The core cytosolic cascades themselves also display a degree of horizontal redundancy. For example, in a variety of preclinical cancer models it has been demonstrated that complete suppression of either PI3K or MAPK pathways alone results in at best tumor growth suppression (or stasis), while cotargeting both cascades is required to induce apoptosis and tumor regression.32 This phenomenon can be described mathematically using logical OR gates along the signal transmission route from surface receptors to phenotypic readouts (i.e., AKT can be activated by EGFR or ERBB3 receptors, apoptosis is inhibited by active AKT or ERK signals).
Hallmark 3: Adaptation
A distinguishing feature of living vs. nonliving matter is the ability to maintain internal stasis in the face of a continually shifting environment. Homeostatic regulation typically involves feedback control circuits, and many biochemical feedback circuits have been characterized. Feedback controls within signal transduction cascades buffer the effect of targeted inhibitors and other perturbations, an example being phosphorylated ERK inhibiting the activation of its upstream kinases.33 The activities of intracellular signaling cascades are also fine-tuned through regulation of cell surface receptor expression.34,35 These receptor-coupled feedback circuits underlie the so called “Whac-a-Mole®” effect, named after the popular arcade game, wherein suppression of a single RTK or pathway induces compensatory activation of a parallel, functionally redundant pathway. These feedback regulatory circuits can be simulated using methods adapted from engineering control theory, and integrated within logic-based signaling network models (Figure 2).
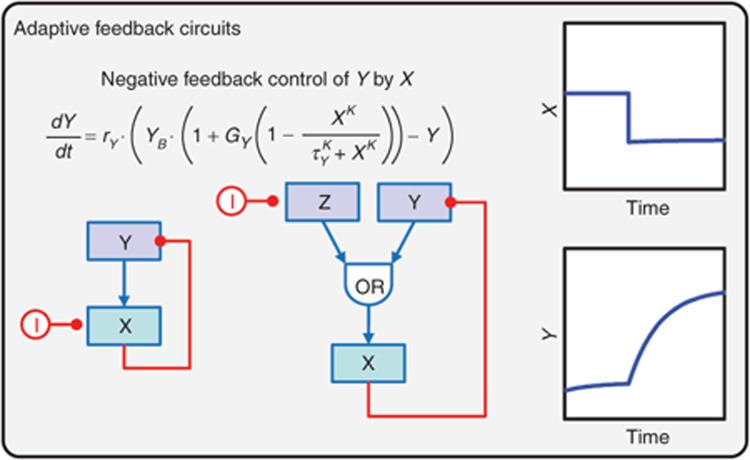
Figure 2.
Adaptive feedback circuits based on control theory. Slight modification of the equation presented in Figure 1b can be used to describe the activation of node (Y) by one of its downstream dependents (X), forming a negative feedback control circuit. Two alternative configurations are depicted, each displaying similar response dynamics to targeted inhibition (I) of X – up-regulation of Y to a new steady-state value. For exemplary purposes, the inhibitor (I) is constant in these simulations, but a pharmacokinetic driving model could be incorporated to account for more complex in vivo pharmacodynamics.
Hallmark 4: Heterogeneity
Whereas tumors are generally believed to be monoclonal in origin, full-blown carcinogenesis results from a branched evolutionary process, resulting in extensive clonal heterogeneity at diagnosis.12,36,37 The genetic and biochemical diversity of clones within a tumor then serves as the fodder of evolutionary adaptation. A result of the diversity in molecular wiring, biochemical responses to drug treatments would be expected to differ between clonal populations, underlying differential drug sensitivities.38,39 Selective pressure then allows for the expansion of rare drug resistant clones and eventual clinical relapse.40 This is perhaps the primary obstacle limiting our ability to achieve sustained remissions with targeted anticancer agents. For translational models to be clinically relevant, it is thus critical to understand molecular mechanisms of drug sensitivity and resistance, both between patients as well as between different clones within an individual tumor, and describe the phenomenon mathematically.
So how do we then attempt this feat? A critical first step is to connect internal signaling states to tumor growth, or other relevant phenotypes. This is a nontrivial task given the multivariate, non-linear nature of cellular signal decoding.41,42 However, by designing appropriate perturbation-response experiments, it is possible to define proliferation and apoptosis rates as functions of a few key intracellular effector molecules (i.e., phospho-AKT and -ERK). Next, given an understanding of the variability in key nodes (protein abundance) and parameters (biochemical rates), Monte Carlo simulations can be employed to simulate heterogeneity in tumor growth rates among molecular diverse tumor “clones.” Performing these simulations in both the control and drug treatment conditions, one can then quantify the sensitivity of each clone to drug treatment using metrics such as percentage tumor growth inhibition. Such synthetic multivariate datasets can then be used to extract statistical relationships between biochemical variability and drug responsiveness (Figure 3). Given the semi-empirical nature of quantitative logic networks, parameter variations reflect relative changes from a prototypic cell type used for model training. Mutations can be represented by their functional effects. For example, gene amplification (or other means of increased protein expression) can be captured by simply increasing the relative basal level of a node, and conversely for gene knockouts. Activating kinase mutations can be captured by decreasing the EC50 value in upstream activating connections and thus simulating hyper-activity, conversely for deactivating mutations. While their parametric changes may be viewed as crude representations of molecular biochemistry, their relative effects on network-level responses to drug treatment may still be informative.
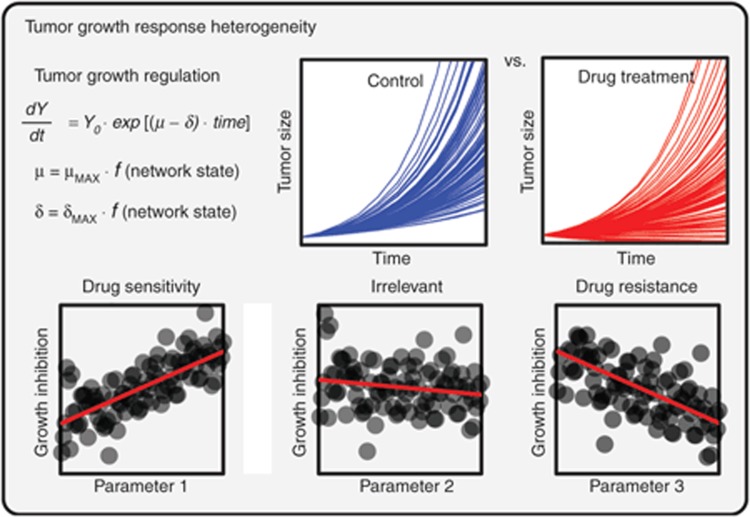
Figure 3.
Heterogeneity in tumor growth and drug responsiveness described using Monte Carlo simulations. Tumor growth models (in this case exponential, but other forms are possible) describe the bulk tumor burden over time as a balance between population-average cell proliferation (µ) vs. death (δ) rates. These phenotypic parameters can in turn be described as functions of cell signaling states. By randomly varying model parameters describing signal flow through the network (i.e., protein levels, biochemical kinetics, and gene mutations) and performing repeated Monte Carlo simulations, it is possible to create synthetically heterogeneous tumor “clones.” Following in silico treatment of tumor populations with drugs or drug combinations, statistical analyses can extract mechanistic biomarkers predictive of drug sensitivity and resistance.
Variations on pieces of this approach have been employed elsewhere. Population pharmacokinetic modeling is widely used to understand how interindividual variability in drug metabolism and physiology affects drug exposure and clinical responses.43 Tumor dynamic models have been used to predict overall survival based on the variability in initial tumor growth responses to drug treatment.44,45 At the cellular level, simulations of molecular variation have been used to recapitulate phenotypic heterogeneity in cell populations.46 Signaling networks provide a means to link these different classes of models, connecting drug exposure, through molecular activities, to clinically relevant responses. This could enable the identification of mechanistic, rather than statistically-based clinical biomarkers for patient stratification.
Strategy and Future Perspectives
Combined, these four characteristic features of oncogenic signaling networks underlie the robustness of tumors to targeted perturbations, limiting the efficacy of drug treatment. Cancer cells addicted to an oncogenic pathway often have multiple built-in escape routes leading to drug resistance. Quantifying the mechanisms by which signaling networks mediate drug sensitivity vs. resistance, however, could enable the design of synergistic drug combinations, and the identification of predictive stratification and PD biomarkers a priori. Note that the mathematical concepts we have described are borrowed from the scientific literature—the use of Boolean networks in molecular cell biology has an extensive history.47 What have been lacking until very recently were the experimental tools necessary to generate sufficiently quantitative and thorough data to parameterize these network models. The availability of highly selective kinase inhibitors against both cell surface receptors and intracellular transducers, coupled with quantitative, multiplex and high-throughput protein measurement technologies48 are finally providing the means to do so. Systematic perturbation-response experiments can be used to produce datasets necessary to specify the network connectivity, logic gates, and adaptive feedback circuits employed by cancer cells. By profiling phenotypic readouts (i.e., proliferation) in tandem with biochemical measurements, it is then possible to specify cell fate decisions as multivariate functions of network activation states, a necessary step in developing translational models.49
We have recently described such an approach to develop targeted combination therapies for the treatment of HER2-amplifed breast cancer,50 the general network schematic of which is summarized in Figure 4. A model connecting ErbB receptor family signaling to cell growth was parameterized using in vitro profiling data, and in silico screening was then performed to identify synergistic drug combinations and predictive biomarkers. Select results were then validated in vivo using xenograft tumor models. ErbB3-mediated signaling was identified as a potent mechanism of resistance to multiple targeted agents, and combining the ErbB3 inhibitor MM-111 with ErbB2 inhibitors was found to induce synergistic tumor regression. Other groups have recently published similar mechanistic PK-PD models (though parameterized entirely in vivo) for other preclinical cancer models and drugs. These include models of MEK inhibitor treatment in BRAF-mutant melanoma,51 rituximab treatment of non-Hodgkin's lymphoma,52 gefitinib treatment of glioblastoma.53
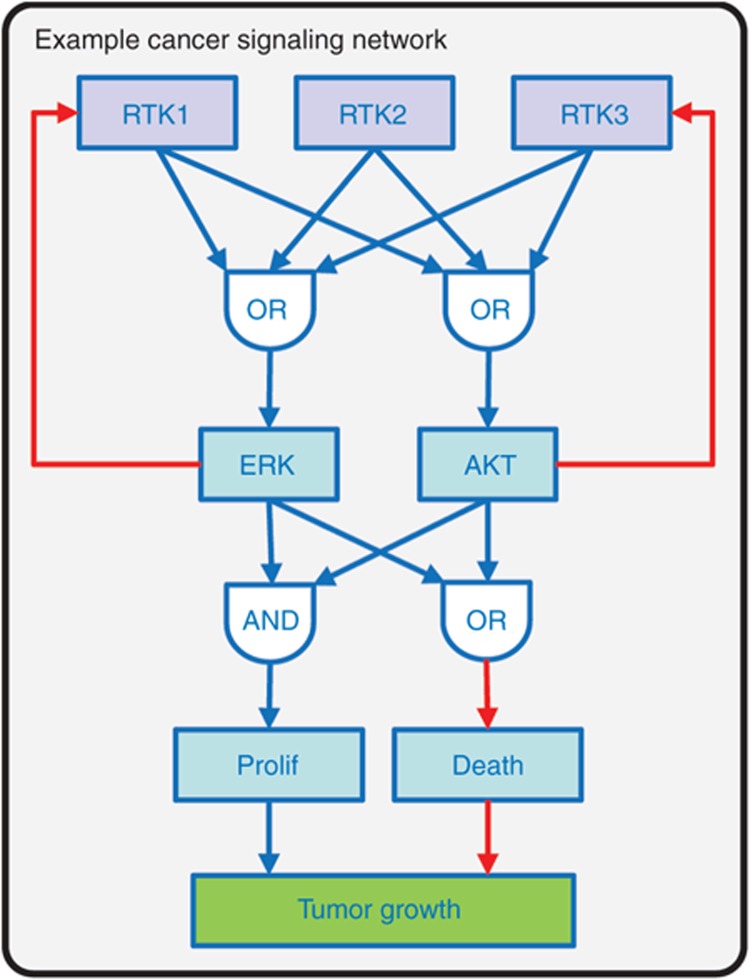
Figure 4.
Example representation of a generalized cancer cell signaling network. We have recently used a variation of this generalized network model for in silico combination drug screening in ERBB2-amplified breast cancer.49 Three receptor tyrosine kinases (RTKs; in our specific case three ERBB-family receptors) are activated in ligand-dependent or independent manner. Active (phosphorylated forms) of the receptors initiate the canonical PI3K/AKT and MAPK/ERK cascades, though with different characteristic biases, captured using multi-input OR gates. Active (phospho-) AKT and ERK in turn inhibit RTK expression via negative feedback regulatory circuits (red lines). Phospho-AKT and ERK were identified as driving cell proliferation and inhibiting cell death via logical OR and AND gates respectively. Measurable tumor cell growth then represents a balance between rates of cell proliferation and death. The signal transduction portion of the model (RTKs to AKT/ERK) was represented algebraically, while the RTK feedback circuits and cell-growth regulation were represented using differential equations. MAPK/ERK, mitogen-activated protein kinase/extracellular signal-regulated kinase; PI3K, phosphatidylinositide 3-kinase.
Whereas this approach has proven fruitful in a few select cases, incorporating cellular networks into PK-PD models is by no means a panacea to drug development challenges. Computational models are only as good as their assumptions, and assumptions which hold in one context may break down in another. First, our knowledge about the topology of intracellular signaling networks remains far from complete. Even extensively studied signal transduction cascades, when reconstructed from unbiased proteomic methods turn have orders of magnitude more interactions than depicted in canonical signaling maps.20 Determining which of these canonical and noncanonical interactions to include in a model thus remains an open problem. More vexing, network connectivity appears to be fluid, such that biochemical circuitry differs between tissues, and even in the same cell under alternate environmental conditions. This poses a particularly acute challenge for translating model predictions from cell culture, to animal models, to clinical strategies. Developing a clearer understanding of the variability in network architectures across different tissues and cancer indications will be essential to broadening the scope of mechanistic PK-PD models. Perhaps tumors can be clustered into distinct groups based on molecular wiring and pathway dependence, rather than histology or molecular profiles? This would provide a more functionally relevant guide to drug development strategies. Another limitation of the above approach is that cancer cells are modeled as autonomous units, ignoring tissue architecture, physiology, and cell–cell interactions.54,55 These processes are known to play important roles in tumor biology and pharmacology, such that multiscale models must eventually describe tumors as complex tissues involving intercellular signaling networks,56 rather than independent collections of cells.
Model development is an iterative process. Successive rounds of model parameterization, verification, and modification require a close collaboration between computational scientists, experimental biologists, and clinicians. Fostering such relationships can be challenging in large organizations with established cultures and clearly defined boundaries between departments and scientific disciplines. Overcoming such cultural barriers is perhaps just as challenging as the technical issues we have discussed. Despite the remaining obstacles, we believe the stepwise introduction of mechanistic, network biology-based mathematical models will be essential for translating knowledge about molecular biology into clinical decisions. The approach described and mathematical tools presented represent our humble attempt to do so. Through the continued development and application of mechanistic model-based drug development, we hope to see a new generation of highly selective and effective drug regimens emerge to better serve the needs of patients.
Conflict of interest
The authors declared no conflict of interest.
Acknowledgments
We thank Bart Hendricks, Charlotte McDonagh, Birgit Schoeberl, Ulrik Nielson, Jinyan Du, Johanna Lahdenranta, Petra Loesch, as well as the reviewers for careful reading of the manuscript and providing helpful comments.
References
- Munos B. Lessons from 60 years of pharmaceutical innovation. Nat. Rev. Drug Discov. 2009;8:959–968. doi: 10.1038/nrd2961. [DOI] [PubMed] [Google Scholar]
- Scannell J.W., Blanckley A., Boldon H., Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 2012;11:191–200. doi: 10.1038/nrd3681. [DOI] [PubMed] [Google Scholar]
- FDA Innovation or Stagnation: Challenge and Opportunity on the Critical Path to New Medical Products. 2004.
- Vicini P. Multiscale modeling in drug discovery and development: future opportunities and present challenges. Clin. Pharmacol. Ther. 2010;88:126–129. doi: 10.1038/clpt.2010.87. [DOI] [PubMed] [Google Scholar]
- Graaf P.H., van der, Benson N. Systems pharmacology: bridging systems biology and pharmacokinetics-pharmacodynamics (PKPD) in drug discovery and development. Pharm. Res. 2011;28:1460–1464. doi: 10.1007/s11095-011-0467-9. [DOI] [PubMed] [Google Scholar]
- Gutenkunst R.N., Waterfall J.J., Casey F.P., Brown K.S., Myers C.R., Sethna J.P. Universally sloppy parameter sensitivities in systems biology models. PLoS Comput. Biol. 2007;3:1871–1878. doi: 10.1371/journal.pcbi.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P.K., et al. Quantitative and Systems Pharmacology in the Post-genomic Era: New Approaches to Discovering Drugs and Understanding Therapeutic Mechanisms An NIH White Paper by the QSP Workshop Group – October, 2011. NIH White paper. 2011.
- Reimand J., Bader G.D. Systematic analysis of somatic mutations in phosphorylation signaling predicts novel cancer drivers. Mol. Syst. Biol. 2013;9:637. doi: 10.1038/msb.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar R., Zhao S., Chung S.W., Mager D.E., Gallo J.M. Merging systems biology with pharmacodynamics. Sci. Transl. Med. 2012;4:126ps7. doi: 10.1126/scitranslmed.3003563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager D.E., Jusko W.J. Development of translational pharmacokinetic-pharmacodynamic models. Clin. Pharmacol. Ther. 2008;83:909–912. doi: 10.1038/clpt.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald J.B., Schoeberl B., Nielsen U.B., Sorger P.K. Systems biology and combination therapy in the quest for clinical efficacy. Nat. Chem. Biol. 2006;2:458–466. doi: 10.1038/nchembio817. [DOI] [PubMed] [Google Scholar]
- Yap T.A., Gerlinger M., Futreal P.A., Pusztai L., Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci. Transl. Med. 2012;4:127ps10. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- Aldridge B.B., Burke J.M., Lauffenburger D.A., Sorger P.K. Physicochemical modelling of cell signalling pathways. Nat. Cell Biol. 2006;8:1195–1203. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- Huang C.Y., Ferrell J.E. Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10078–10083. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla U.S., Iyengar R. Emergent properties of networks of biological signaling pathways. Science. 1999;283:381–387. doi: 10.1126/science.283.5400.381. [DOI] [PubMed] [Google Scholar]
- Schoeberl B., et al. Therapeutically targeting ErbB3: a key node in ligand-induced activation of the ErbB receptor-PI3K axis. Sci. Signal. 2009;2:ra31. doi: 10.1126/scisignal.2000352. [DOI] [PubMed] [Google Scholar]
- McDonagh C.F., et al. Antitumor activity of a novel bispecific antibody that targets the ErbB2/ErbB3 oncogenic unit and inhibits heregulin-induced activation of ErbB3. Mol. Cancer Ther. 2012;11:582–593. doi: 10.1158/1535-7163.MCT-11-0820. [DOI] [PubMed] [Google Scholar]
- Fitzgerald J., Lugovskoy A. Rational engineering of antibody therapeutics targeting multiple oncogene pathways. MAbs. 2011;3:299–309. doi: 10.4161/mabs.3.3.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms B.D., Kearns J.D., Su S.V., Kohli N., Nielsen U.B., Schoeberl B. Optimizing properties of antireceptor antibodies using kinetic computational models and experiments. Meth. Enzymol. 2012;502:67–87. doi: 10.1016/B978-0-12-416039-2.00004-5. [DOI] [PubMed] [Google Scholar]
- Kirouac D.C., Saez-Rodriguez J., Swantek J., Burke J.M., Lauffenburger D.A., Sorger P.K. Creating and analyzing pathway and protein interaction compendia for modelling signal transduction networks. BMC Syst. Biol. 2012;6:29. doi: 10.1186/1752-0509-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M.K., Saez-Rodriguez J., Sorger P.K., Lauffenburger D.A. Logic-based models for the analysis of cell signaling networks. Biochemistry. 2010;49:3216–3224. doi: 10.1021/bi902202q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M.K., Saez-Rodriguez J., Clarke D.C., Sorger P.K., Lauffenburger D.A. Training signaling pathway maps to biochemical data with constrained fuzzy logic: quantitative analysis of liver cell responses to inflammatory stimuli. PLoS Comput. Biol. 2011;7:e1001099. doi: 10.1371/journal.pcbi.1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann D.M., Krumsiek J., Saez-Rodriguez J., Lauffenburger D.A., Klamt S., Theis F.J. Transforming Boolean models to continuous models: methodology and application to T-cell receptor signaling. BMC Syst. Biol. 2009;3:98. doi: 10.1186/1752-0509-3-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H., Hopfield J.J., Leibler S., Murray A.W. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Network et al.Comprehensive genomic characterization of squamous cell lung cancers. Nature 489519–525.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P.J., Oslo Breast Cancer Consortium (OSBREAC) et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E., Friedman N., Koller D., Regev A. A module map showing conditional activity of expression modules in cancer. Nat. Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- Liu Y.Y., Slotine J.J., Barabási A.L. Observability of complex systems. Proc. Natl. Acad. Sci. U.S.A. 2013;110:2460–2465. doi: 10.1073/pnas.1215508110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snuderl M., et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Wilson T.R., et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sos M.L., et al. Identifying genotype-dependent efficacy of single and combined PI3K- and MAPK-pathway inhibition in cancer. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18351–18356. doi: 10.1073/pnas.0907325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirit M., Wang C.C., Haugh J.M. Systematic quantification of negative feedback mechanisms in the extracellular signal-regulated kinase (ERK) signaling network. J. Biol. Chem. 2010;285:36736–36744. doi: 10.1074/jbc.M110.148759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandarlapaty S., et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan J.S., et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S., Breast Cancer Working Group of the International Cancer Genome Consortium et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau D.A., et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish J.M., et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Cohen A.A., et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- Diaz L.A.et al.The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486537–540.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Jensen K., Janes K.A., Brugge J.S., Lauffenburger D.A. Common effector processing mediates cell-specific responses to stimuli. Nature. 2007;448:604–608. doi: 10.1038/nature06001. [DOI] [PubMed] [Google Scholar]
- Lee M.J., et al. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149:780–794. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould D.R., Upton R.N. Basic concepts in population modeling, simulation, and model-based drug development. CPT: Pharma. Syst. Pharmacol. 2012;1:e6. doi: 10.1038/psp.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claret L., et al. Model-based prediction of phase III overall survival in colorectal cancer on the basis of phase II tumor dynamics. J. Clin. Oncol. 2009;27:4103–4108. doi: 10.1200/JCO.2008.21.0807. [DOI] [PubMed] [Google Scholar]
- Wang Y., et al. Elucidation of relationship between tumor size and survival in non-small-cell lung cancer patients can aid early decision making in clinical drug development. Clin. Pharmacol. Ther. 2009;86:167–174. doi: 10.1038/clpt.2009.64. [DOI] [PubMed] [Google Scholar]
- Gaudet S., Spencer S.L., Chen W.W., Sorger P.K. Exploring the contextual sensitivity of factors that determine cell-to-cell variability in receptor-mediated apoptosis. PLoS Comput. Biol. 2012;8:e1002482. doi: 10.1371/journal.pcbi.1002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman S.A. Metabolic stability and epigenesis in randomly constructed genetic nets. J. Theor. Biol. 1969;22:437–467. doi: 10.1016/0022-5193(69)90015-0. [DOI] [PubMed] [Google Scholar]
- Saez-Rodriguez J., Alexopoulos L.G., Stolovitzky G. Setting the standards for signal transduction research. Sci. Signal. 2011;4:pe10. doi: 10.1126/scisignal.2001844. [DOI] [PubMed] [Google Scholar]
- Janes K.A., Yaffe M.B. Data-driven modelling of signal-transduction networks. Nat. Rev. Mol. Cell Biol. 2006;7:820–828. doi: 10.1038/nrm2041. [DOI] [PubMed] [Google Scholar]
- Kirouac D., et al. Computational modeling of ERBB2-amplified breast cancer identifies combined ErbB2/3 blockade as superior to the combination of MEK and AKT inhibitors. Sci. Signal6, ra68 (2013 [DOI] [PubMed]
- Wong H., et al. Bridging the gap between preclinical and clinical studies using pharmacokinetic-pharmacodynamic modeling: an analysis of GDC-0973, a MEK inhibitor. Clin. Cancer Res. 2012;18:3090–3099. doi: 10.1158/1078-0432.CCR-12-0445. [DOI] [PubMed] [Google Scholar]
- Harrold J.M., Straubinger R.M., Mager D.E. Combinatorial chemotherapeutic efficacy in non-Hodgkin lymphoma can be predicted by a signaling model of CD20 pharmacodynamics. Cancer Res. 2012;72:1632–1641. doi: 10.1158/0008-5472.CAN-11-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Guo P., Wang X., Zhou Q., Gallo J.M. Preclinical pharmacokinetic/pharmacodynamic models of gefitinib and the design of equivalent dosing regimens in EGFR wild-type and mutant tumor models. Mol. Cancer Ther. 2008;7:407–417. doi: 10.1158/1535-7163.MCT-07-2070. [DOI] [PubMed] [Google Scholar]
- McMillin D.W., Negri J.M., Mitsiades C.S. The role of tumour-stromal interactions in modifying drug response: challenges and opportunities. Nat. Rev. Drug Discov. 2013;12:217–228. doi: 10.1038/nrd3870. [DOI] [PubMed] [Google Scholar]
- Kirouac D.C., Madlambayan G.J., Yu M., Sykes E.A., Ito C., Zandstra P.W. Cell-cell interaction networks regulate blood stem and progenitor cell fate. Mol. Syst. Biol. 2009;5:293. doi: 10.1038/msb.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirouac D.C., et al. Dynamic interaction networks in a hierarchically organized tissue. Mol. Syst. Biol. 2010;6:417. doi: 10.1038/msb.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]