SUMMARY
Innate lymphoid cells (ILCs) are critical in innate immune responses to pathogens and lymphoid organ development. IL-7Rα + ILC subsets, similar to CD4+T helper (Th) cell subsets, produce distinct sets of effector cytokines. However, the molecular control of IL-7Rα + ILC development and maintenance is unclear. Here we report that GATA3 was indispensable for the development of all IL-7Rα + ILC subsets in addition to T cells, but not required for the development of classical NK cells. Gata3 conditional deficient mice had no lymph nodes and were susceptible to Citrobactor rodentium infection. After the ILCs have fully developed, GATA3 remained important for the maintenance and functions of ILC2s. Genome-wide gene expression analyses indicated that GATA3 regulated a similar set of cytokines and receptors in Th2 cells and ILC2s, but not in ILC3s. Thus, GATA3 plays parallel roles in regulating the development and functions of CD4+ T cells and IL-7Rα + ILCs.
INTRODUCTION
CD4+ T helper (Th) cells are central in orchestrating adaptive immune responses; distinct Th subsets are involved in protective immune responses to a variety of pathogens (Kanno et al., 2012; Zhu et al., 2010). For example, type 1 T helper (Th1) cells are critical for eradicating intracellular bacteria and viruses, whereas type 2 T helper (Th2) cells are indispensable for the expulsion of helminths. Interleukin-17 (IL-17)-producing Th (also known as Th17) cells are critical for defending against extracellular bacterial and fungal infections.
It usually takes several (5–10) days for antigen-specific CD4+ T cells to expand from rare precursors in the naïve population and reach a meaningful number to execute host defense functions. Therefore, many innate effector cells including natural killer (NK) cells are responsible for early control of invading pathogens. Recently, a new class of innate effector cells, whose development relies on signaling through the IL-2 receptor (IL-2R) common γ chain and IL-7Rα, has drawn much attention. These cells, together with classical NK cells, are often referred to as innate lymphoid cells (ILCs) (Sonnenberg and Artis, 2012; Spits and Cupedo, 2012; Spits and Di Santo, 2011). Because distinct subsets of ILCs are capable of making the same characteristic effector cytokines as produced by different T helper cell subsets, they are similarly classified into type 1 innate lymphoid cells (ILC1s) including classical NK cells that produce interferon-γ (IFN-γ), type 2 innate lymphoid cells (ILC2s) that produce IL-5 and IL-13, and type 3 ILCs including lymphoid tissue inducer (LTi) cells that produce IL-17 and IL-22(Spits et al., 2013; Walker et al., 2013).
By producing Th2 cell effector cytokines such as IL-13, ILC2s play an important role during early immune responses to helminth infection (Fallon et al., 2006; Moro et al., 2010; Neill et al., 2010; Price et al., 2010; Saenz et al., 2010). Mice with dysfunctional ILC2s have a significant delay in worm expulsion in Nippostrongylus brasiliensis infection whereas expanding the number of ILC2s by IL-25 injection can eliminate the need for Th2 cells in effective resistance to helminth infection. ILC2s are also important for allergen-induced airway inflammation and lung tissue repair in animal models (Chang et al., 2011; Halim et al., 2012a; Monticelli et al., 2011) and human cells corresponding to the ILC2s found in mice have been identified (Mjosberg et al., 2011). The ILCs that produce IL-17 and IL-22 also participate in the early phase of responses to infections and in inflammatory disorders (Buonocore et al., 2010; Lee et al., 2012; Powell et al., 2012; Satoh-Takayama et al., 2008). Thus, understanding the molecular mechanisms controlling the development and functions of ILCs is essential to develop strategies to control responses to pathogens and autoimmunity.
GATA3 is the key transcription factor for Th2 cell differentiation (Yagi et al., 2011). GATA3 expression is indispensable for proper induction of Th2 cytokines including IL-4, IL-5 and IL-13 both in vitro and in vivo (Zhu et al., 2004). Interestingly, GATA3 is critical not only for regulating Th2 cell differentiation, but also for CD4+T cell development in the thymus at multiple stages (Ho et al., 2009; Pai et al., 2003; Ting et al., 1996).
It has been reported that GATA3 is highly expressed by ILC2 cells (Moro et al., 2010; Price et al., 2010). Conditional in activation of the Gata3 gene with a transgenic Cre whose expression is driven by the Il13 locus completely eliminated IL-13-producing ILC2 cells (Liang et al., 2012). GATA3 has been shown to be critical for the maintenance of ILC2 cell number and IL-13 production by these cells both in mice and in humans (Furusawa et al., 2013; Hoyler et al., 2012; Klein Wolterink et al., 2013; Mjosberg et al., 2012; Yang et al., 2013). However, because GATA3 affects ILC2 cell number, IL-13 regulation by GATA3 in the previous studies requires more careful assessment, i.e. at a single cell level with proper controls. Furthermore, other important target genes that are regulated by GATA3 in ILC2s are largely unknown. Finally, whether other ILC subsets require GATA3 to develop remains unclear.
Here we report that GATA3 is not only critical for T cell development, but also indispensable for the development of all the IL-7Rα+ ILC lineages including ILC2s, lymphoid tissue inducer (LTi) cells, IL-7Rα-expressing NK cells, Nkp46+RORγt+, and Nkp46+RORγt− ILCs. Both ILC2 progenitors and LTi progenitors were diminished in the absence of GATA3. Genome-wide analysis of GATA3-regulated genes in ILC2s and ILC3s suggests that GATA3 function during ILC development seems to be independent of many known key transcription factors including Id2, RORα, RORγt and Tcf7. GATA3 regulates many critical genes including Il5, Il13, Il1rl1, Il2ra, Il9r and Ccr8, but not Klrg1in ILC2s. Comparing to GATA3-regulated genes in Th2 cells, we found that while many genes, such as Pth, Cysltr1, Htr1b and Tph1, are regulated by GATA3 in a cell type specific manner, most of the key type 2 immune response related genes are regulated by GATA3 in both ILC2s and Th2 cells. These results demonstrate that GATA3 plays parallel roles in regulating the development and functions of CD4+ T cells and IL-7Rα+ ILCs, that is, the development of both CD4+T cells and all IL-7Rα+ ILCs requires GATA3, and GATA3 is especially critical for the maintenance and functions of ILC2s, similar to its role in Th2 cells.
All ILCs express GATA3 at different amounts
To determine the tissue distribution of ILCs, and ILC2s in particular, we assessed lineage negative (Lin−) CD127 (IL-7Rα) positive cells by flow cytometry analysis of many lymphoid tissues and organs. Lin−CD127+ cells represented <1% of the total cells in spleen, mesenteric lymph nodes and in the lung, but somewhat enriched in the small intestine lamina propria (siLP, FigureS1A). Among these ILCs, some are ILC2s, identified by expression of T1/ST2 (IL-33R)(Moro et al., 2010; Neill et al., 2010). ILC2s (T1/ST2+Lin−CD127+) constituted the largest fraction of recovered ILCs in the lung-derived cells (> 80% of total Lin−CD127+ cells, Figure S1B). In contrast, only ~16% of Lin−CD127+ cells in the spleen were T1/ST2+. In the mesenteric lymph nodes, ~40% of the Lin−CD127+ cells were T1/ST2+.
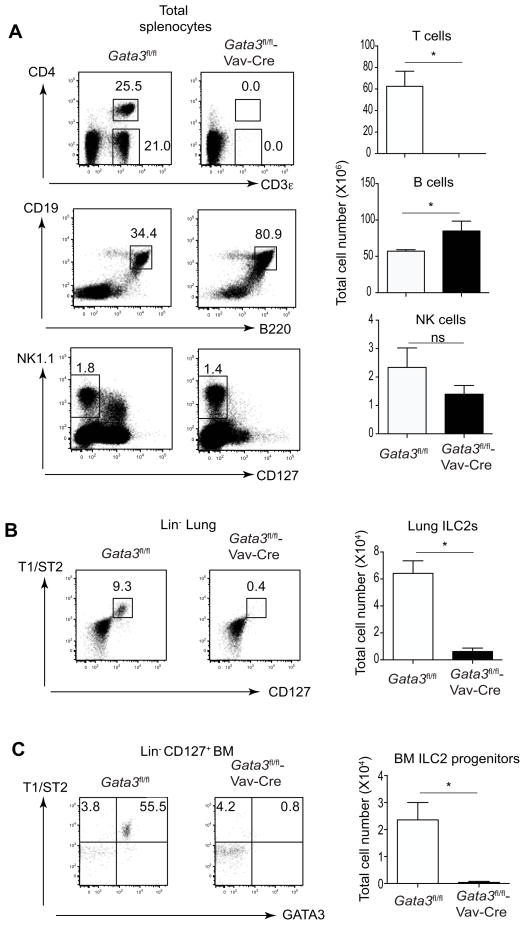
Figure 1. GATA3 is critical for the development of T cells and ILC2s, but not B cells and classical NK cells.
(A) Total splenocytes from Gata3fl/fl or Gata3fl/fl-Vav-Cre mice (3 mice per group) were stained for CD3ε, CD4, CD19, B220, NK1.1 and CD127. Dot plots (left panel) and total cell numbers of T, B and NK cells were shown (right panel).
(B) Cells prepared from the lungs of Gata3fl/fl or Gata3fl/fl-Vav-Cre mice (3 mice per group) were stained with a cocktail of antibodies to various lineage markers, CD127 and T1/ST2. Dot plots gated on Lin− cells (left panel) and total cell numbers of lung ILC2s were shown (right panel).
(C) Cells prepared from bone marrows of Gata3fl/fl or Gata3fl/fl-Vav-Cre mice (3 mice per group) were stained with a cocktail of antibodies to various lineage markers, CD127, T1/ST2 and GATA3. Dot plots gated on Lin− (left panel) and total cell numbers of ILC2 progenitors in the bone marrow were shown (right panel). Numbers indicate the percentages in each box or quadrant. Error bars represent Mean ± SD. “ns” indicates non-significant. “*” indicates p<0.05. Data are representative of three independent experiments.
See also Figure S1.
To assess GATA3 expression in the various subsets of ILCs, we performed ex vivo GATA3 intracellular staining of Lin−CD127+ cells harvested from different tissues. Most ILC2s expressed high levels of GATA3 whether they were from spleen, mesenteric lymph node, lung or siLP (Figure S1B). Interestingly, other ILCs (T1/ST2−Lin−CD127+) expressed intermediate amounts of GATA3. In the siLP, most of the Lin−CD127+GATA3int cells in the siLP are RORγt+ ILCs and all Lin−CD127+GATA3hi cells express another ILC2 marker, KLRG-1, but not RORγt (Figure S1C). Thus, all IL-7Rα ILCs express GATA3 albeit at different amounts.
GATA3 is indispensable for the development of all IL-7Rα+ ILCs
To study the role of GATA3 in ILC development in vivo, we generated Gata3fl/fl-Vav-Cre mice in which Gata3 exon 4 is deleted in hematopoietic stem cells. As expected, no T cells were detected in Gata3fl/fl-Vav-Cre mice (Figure 1A), confirming a previous report on the critical role of GATA3 in T cell development using germline Gata3 deletion (Ting et al., 1996). In contrast, there was a modest but significant increase of B cells in Gata3fl/fl-Vav-Cre mice (Figure 1A), arguing against a loss of stem cell function. In line with a previous report showing that GATA3 is critical for the development of IL-7Rα+ NK cells (Vosshenrich et al., 2006), IL-7Rα+NK1.1+ cells including CD3ε−IL-7Rα+NK1.1+ were absent in the spleen of Gata3fl/fl-Vav-Cre mice, whereas classical IL-7Rα −NK1.1+ cells were not significantly affected (Figure 1A). Staining of cells from the lung of Gata3fl/fl-Vav-Cre mice showed complete absence of ILC2s (Figure 1B). In the bone marrow of wild-type mice, T1/ST2+ cells were detectable among the Lin−CD127+ population; these cells have been regarded as the progenitors of ILC2s and they were all GATA3+ (Figure 1C). However, this population was absent in the bone marrow of Gata3fl/fl-Vav-Cre mice (Figure 1C). Overall, our data indicate that GATA3 is indispensable for the development of ILC2s and T cells but not B cells or IL-7Rα-negative NK cells.
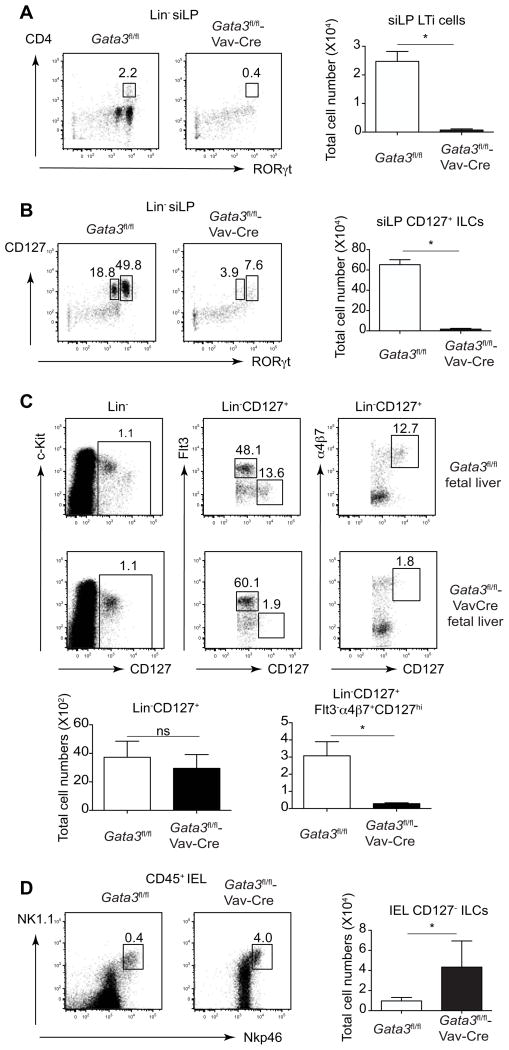
We also noticed a defect in lymphoid organ development in the Gata3fl/fl-Vav-Cre mice; these mice had no lymph nodes including the brachial and axillary lymph nodes similar to the phenotype of RORγt deficient mice. This finding suggested that GATA3 regulates the development of LTi cells, which are Lin−CD4+RORγt+ and critical for the development of lymph node structure (Eberl et al., 2004a; Sun et al., 2000). Indeed, Lin−CD4+RORγt+ LTi cells, which are found with a high frequency in the siLP of Gata3fl/fl mice, were markedly reduced in the Gata3fl/fl-Vav-Cre mice (Figure 2A). Furthermore, total Lin−RORγt+ cells were reduced in the siLP of Gata3fl/fl-Vav-Cre mice (Figure 2B). In fact, IL-7Rα+ cells, whether they express RORγt or not, were abolished in Gata3fl/fl-Vav-Cre mice suggesting GATA3 is critical for development of all IL-7Rα-expressing ILCs. The loss of RORγt+ ILCs is not due to a possible Cre-mediated genotoxicity since all the Gata3fl/+-Vav-Cre mice had normal lymph node structure and these mice developed RORγt+ ILCs normally (Figure S2).
Figure 2. GATA3 is critical for the development of all IL-7Rα + ILCs but not IL-7Rα− ILCs.
(A) Cells prepared fromsmall intestine lamina propria (siLP) of Gata3fl/fl or Gata3fl/fl-Vav-Cre mice (3 mice per group) were stained with a cocktail of antibodies to various lineage markers, CD4 and RORγt. Plots gated on Lin− cells (left panel) and total cell numbers of LTi cells (right panel) were shown.
(B) Cells prepared from siLP of Gata3fl/fl or Gata3fl/fl-Vav-Cre mice (3 mice per group) were stained with a cocktail of antibodies to various lineage markers, CD127 and RORγt. Plots gated on Lin- cells (left panel) and total cell numbers of IL-7Rα+ ILCs (right panel) were shown.
(C) Fetal liver CLPs (Lin−CD127+) from Gata3fl/fl (n=6) or Gata3fl/fl-Vav-Cre (n=2) embryos at embryonic day 15.5 (E15.5) were fractionated by the expression of Flt3 and α4β7 expression. Plots were gated on Lin− (upper left panels) or Lin−CD127+ cells (upper middle and right panels). Total cell numbers of CLPs (Lin−CD127+) cells and LTi progenitors (Lin−CD127+Flt3−α4β7+CD127hi) were calculated (lower panels). (D) Intraepithelial lymphocytes (IELs) prepared from Gata3fl/fl or Gata3fl/fl-Vav-Cre mice (3 mice per group) were stained with a cocktail of antibodies to various lineage markers, CD45, NK1.1 and Nkp46. Plots gated on CD45+ cells (left panel) and total cell numbers of IEL ILCs (right panel) were shown.
Numbers indicate the percentages in each box. Error bars represent Mean ±SD. “ns” indicates non-significant. “*” indicates p<0.05. Data are representative of three independent experiments (A and B) and two independent experiments (C and D). See also Figure S2.
LTi progenitors have been identified in the mouse fetal liver (Cherrier et al., 2012; Possot et al., 2011). To assess the function of GATA3 in the development of LTi progenitors, we stained fetal liver cells from Gata3fl/fl-Vav-Cre embryos. Lin−CD127+ cells were present at a similar frequency and in total cell number between the Gata3fl/fl and Gata3fl/fl-Vav-Cre fetal livers (Figure 2C). However, LTi progenitors that are Flt3−α4β7+CD127hi were greatly diminished in Gata3fl/fl-Vav-Cre fetal livers suggesting GATA3 is critical for the development of such cells. Together with the finding that GATA3 is required for the generation of ILC2 progenitors in the bone marrow (Figure 1C), our data suggest that GATA3 may act in a common innate lymphoid progenitor (CILP) that gives rise to all IL-7Ra-expressing ILCs.
IL-7Rα negative ILCs found in the compartment of intraepithelial lymphocytes (IELs) have been reported, and these cells express both NK1.1 and Nkp46 (Fuchs et al., 2013). Interestingly, these ILCs were significant increased in Gata3fl/fl-Vav-Cre mice compared to those in Gata3fl/fl mice (Figure 2D), indicating GATA3 is critical for the development of IL-7Rα+ ILCs, but not IL-7Rα-negative ILCs.
Gata3fl/fl-Vav-Cre mice are susceptible to Citrobacter rodentium infection
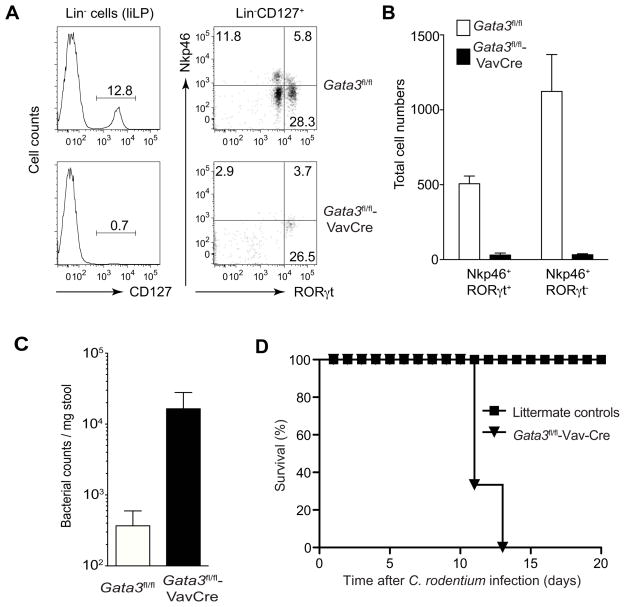
IL-7Rα+ ILCs also include IL-22-producing ILCs, some of which express Nkp46. These cells play a critical role in host defense against Citrobacter rodentium infection. Thus, we tested whether Gata3fl/fl-Vav-Cre mice are susceptible to Citrobacter rodentium infection. On day 4 after infection, a substantial number of Lin−IL-7Rα+ cells, many of which expressed Nkp46, were detected in the large intestine lamina propria (liLP) of littermate mice (Figures 3A and 3B); these cells were absent in the liLP of the Gata3fl/fl-Vav-Cre mice. Accordingly, Gata3fl/fl-Vav-Cre mice had much higher bacterial load than their littermates (Figure 3C). All the Gata3fl/fl-Vav-Cre mice died after C. rodentium infection whereas all the control mice survived (Figure 3D). These data further establish the role of GATA3 in the development of IL-22-producing ILCs.
Figure 3. GATA3 deficiency results in susceptibility to Citrobacter rondentium infection.
Gata3fl/fl or Gata3fl/fl-Vav-Cre mice (3–4 mice per group) were orally infected with Citrobacter rondentium. Four days after infection (A–C), cells were harvested from the large intestine lamina propria (liLP) and stained with a cocktail of antibodies to various lineage markers and CD127, RORγt and Nkp46.
(A) Plots were gated on Lin− cells (left panel) or Lin− CD127+cells (right panel).
(B) Absolute number of Nkp46+ populations from liLP of each mouse was plotted.
(C) Feces were collected to assess bacterial loads. Numbers indicate the percentages in each quadrant or gate. Error bars represent Mean ± SD.
(D) Survival curves for the Gata3fl/fl or Gata3fl/fl-Vav-Cre mice after infection. A–C represents one experiment and D represents another experiment.
GATA3 effect on the development of IL-7Rα + ILCs is cell intrinsic
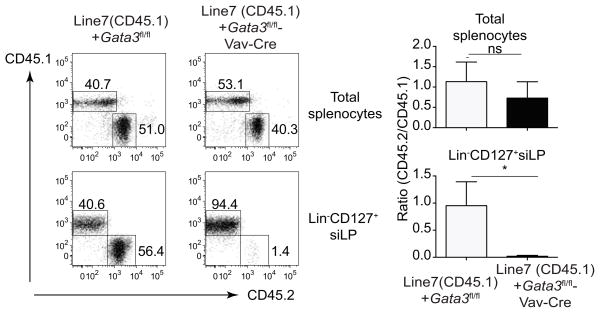
Vav-Cre deletes floxed genes specifically in hematopoietic stem cells; therefore, the failure of ILC development in Gata3fl/fl-Vav-Cre mice is not due to a GATA3 function in non-hematopoietic cells. To further confirm this, we performed mixed bone marrow chimera experiments to directly examine whether the influence of GATA3 on ILC development is cell intrinsic. As expected, GATA3 sufficient and deficient bone marrow cells efficiently repopulated CD45+ cells in the spleen of Rag2−/−Il2rg−/− mice (Figure 4). However, GATA3-deficient bone marrow cells failed to give rise to IL-7Rα+ ILCs in the mixed bone marrow chimeras. In contrast, B cells developed normally, where as T cells failed to develop from GATA3-deficient bone marrow progenitors. These results indicate that the effect of GATA3 on the development of IL-7Rα+ ILCs is cell intrinsic.
Figure 4. GATA3 effect on the development of IL-7Rα+ ILCs is cell intrinsic.
Bone marrow cells from Gata3fl/fl or Gata3fl/fl-Vav-Cre were mixed with bone marrow cells from CD45.1 congenic (Line7) mice and co-transferred into irradiated Rag2−/ −Il2rg−/ − mice. Eight weeks after mixed bone marrow transplant, cells were harvested from spleen or siLP of chimeric mice (3 mice per group) and stained with CD45.1 and CD45.2 together with a cocktail of antibodies to various lineage markers and CD127. Plots were gated on total splenocytes (upper panel), or Lin− CD127+ cells from siLP (lower panel). Ratio of CD45.2/CD45.1 was calculated and shown on the right. Numbers indicate the percentages in each box. Error bars represent Mean ± SD. “ns” indicates non-significant.“*” indicates p<0.05.
GATA3 is required for maintaining ILC2 cell number both in vivo and in vitro
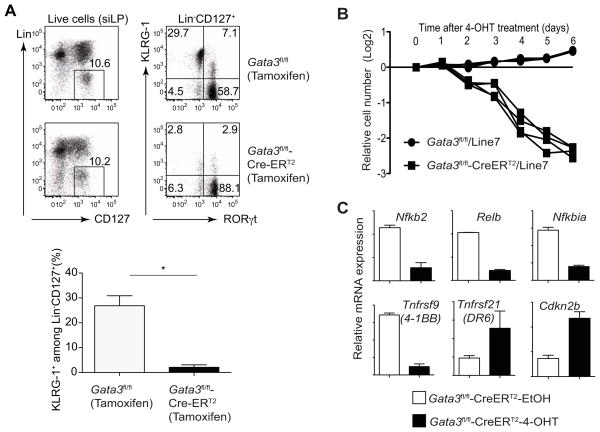
At first consideration, our results contradict an earlier report showing that GATA3 is required for the development of ILC2 but not other ILCs (Hoyler et al., 2012). However, in that study, Gata3 gene was deleted by inducible Cre driven by Id2 locus after all the ILCs had fully developed. Therefore, it is likely that the survival of ILC2s is sensitive to Gata3 deletion. To test this possibility, we generated Gata3fl/fl-CreERT2 mice and treated these mice with tamoxifen. Three weeks after treatment, we observed that only KLRG1+ ILC2s but not RORγt+ ILCs in the siLP were diminished (Figure 5A), confirming that GATA3 is critical only for the maintenance of ILC2s in vivo.
Figure 5. GATA3 is critical for the maintenance of ILC2 cell numbers both in vivo and in vitro.
(A) Gata3fl/fl or Gata3fl/fl-CreERT2 mice (2–3 mice per group) were injected 5 times with tamoxifen i.p., every other day. Three weeks after first injection, siLP cells were prepared and stained with a cocktail of antibodies to various lineage markers, CD127 (IL-7Rα), KLRG-1 and RORγt. Plots were gated on live cells (left panel) or Lin−CD127+ cells (right panel). Numbers indicate the percentages in each quadrant.
(B) Lineage negative cells were purified from mesenteric lymph node of IL-25-treated Gata3fl/fl-CreERT2, Gata3fl/fl and CD45.1 congenic (Line7) mice (2–3 mice per group) by cell sorting. After cultured with IL-7, IL-25 and IL-33 for 5 days, they were mixed as indicated at a 1:1 ratio and then treated with 4-hydroxytamoxifen (4-OHT). The relative cell numbers were calculated based on the change of ratio over a period of 6 days. Four independently mixed cultures were tested in both groups. Data are representative of two independent experiments.
(C) Lineage negative cells were purified from mesenteric lymph node of IL-25-treated Gata3fl/fl-CreERT2 mice (2–3 mice per group in duplicates) by cell sorting. After cultured with IL-7, IL-25 and IL-33 for 5 days, they were then treated with either 4-hydroxytamoxifen (4-OHT) or ethanol (EtOH) for 2 days. RNA-seq analysis was carried out. Relative mRNA expression of several genes was calculated based on the RNA-seq results with duplicates.
Error bars represent Mean ± SD.
See also Figure S3.
Since GATA3 is required for the development and maintenance of ILC2s in vivo, it is difficult to assess the function of GATA3 in ILC2s without obtaining a pure ILC2 population. ILC2s, although present at a low frequency, expand during type 2 immune responses, presumably as a result of stimulation by IL-25 and/or IL-33, which are produced by epithelial cells. Indeed, injection of IL-25 induced substantial expansion of ILC2s (Neill et al., 2010; Saenz et al., 2013). We also observed that IL-25 injection induced the expansion of ILC2s. Culturing sorted Lin− cells harvested from mesenteric lymph nodes of IL-25-injected mice with IL-33, IL-25 and IL-7 in vitro further expanded these cells. After 5–7 days of culture of purified Lin− cells, virtually all cells were GATA3hi (Figure S3A). These cells also expressed two cell-surface molecules, Sca-1 and KLRG1, which are highly expressed by ILC2 (Figures S3B and S3C). Therefore, we chose this approach in an effort to obtain large numbers of ILC2s at high purity; staining and/or sorting for T1/ST2 and IL-7Rα-expressing cells is not required until the end of the culture, alleviating the concern that antibody staining would affect IL-33 or IL-7 signaling.
To determine the function of GATA3 in ILC2s, we prepared ILC2s, as described in Fig. S3, from Gata3fl/fl-CreERT2 or Gata3fl/fl mice. After 5 days of culture, samples were split into two groups and treated either with 4-hydroxytamoxifen (4-OHT), the active metabolite of tamoxifen that binds to CreERT2 to induce Cre activity, or with a vehicle control, ethanol (EtOH). To test whether GATA3 affects ILC2 maintenance in culture in a cell intrinsic manner, we mixed Gata3fl/fl-CreERT2 or Gata3fl/fl ILC2s with ILC2s generated from CD45.1 congenic mice. After 4-OHT treatment, there was a progressive decrease in the percentage of Gata3-deficient ILC2s over a 6-day period (Figure 5B). To comprehensively profile the genes that are regulated by GATA3 in ILC2s at a genome-wide level, we sorted live ILC2s that had or had not undergone Gata3 inactivation on day 2, a time point when no dramatic ILC2 loss was observed, and performed RNA-seq analyses. Interestingly, we found that many TNF and TNFR superfamily genes, such as Tnfrsf9 (encoding 4–1BB which provides co-stimulation) and Tnfsf21 (encoding DR6, which induces cell apoptosis), and NFκb family members, including Nfkb2 and Relb, showed altered expression patterns (Figure 5C). In addition, the cell cycle inhibitor, Cdkn2b, was upregulated upon Gata3 inactivation (Figure 5C). These changes are consistent with and may contribute to the loss of Gata3-deficient ILC2s.
GATA3 regulates many critical genes in ILC2s and in Th2 cells
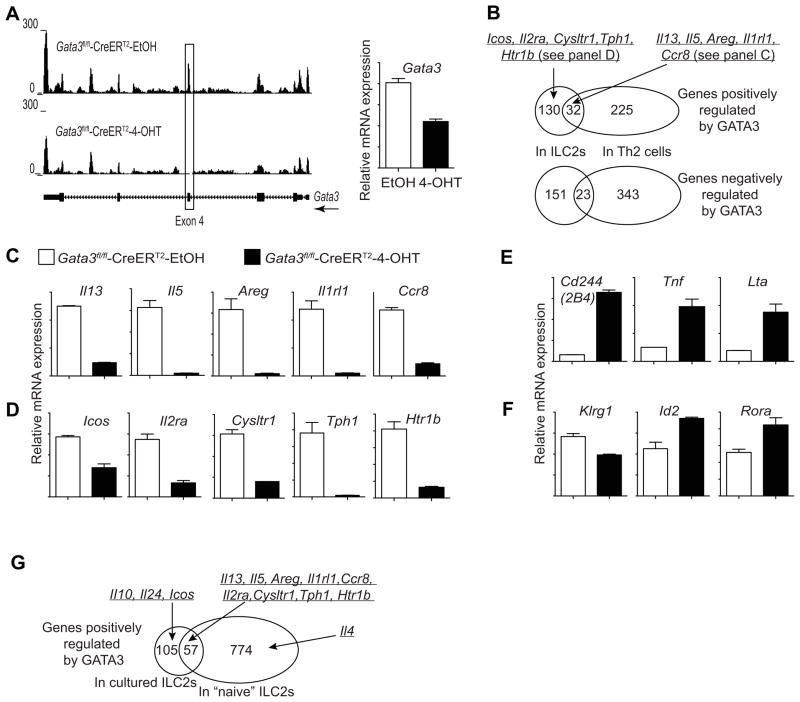
RNA-seq result confirmed efficient Gata3 inactivation by Cre-ERT2 after 4-OHT treatment since a nearly 100% reduction of the reads at the Gata3 exon 4 was observed (Figure 6A). Interestingly, reads at the other exons of the Gata3 gene were only modestly affected implying that GATA3 is not a major player in maintaining its own transcription in ILC2s (Figure 6A).
Figure 6. GATA3 positively and negatively regulates the expression of many critical genes in ILC2s.
(A–F) Lineage negative cells were purified from mesenteric lymph node of IL-25-treated Gata3fl/fl-CreERT2 mice (2–3 mice per group in duplicates) by cell sorting. After cultured with IL-7, IL-25 and IL-33 for 5 days, they were then treated with either 4-hydroxytamoxifen (4-OHT) or ethanol (EtOH) for 2 days.
(A) UCSC genome browser view of the RNA-seq data at the Gata3 locus (A, left panel). Boxed area indicates the deletion of exon 4 by Cre-ERT2. Relative Gata3 mRNA expression was calculated based on the RPKM values obtained from RNA-seq results with duplicates (A, right panel).
(B) Venn diagrams showing overlap of total genes that are positively (upper) or negatively (lower) regulated by GATA3 in ILC2s and Th2 cells.
(C–F) Representative genes regulated by GATA3 in different categories were shown. Relative mRNA expression was calculated based on the RPKM values obtained from RNA-seq results with duplicates.
(C) Examples represent 32 genes that were positively regulated by GATA3 in both ILC2s and Th2 cells.
(D) Examples represent 130 genes that were positively regulated by GATA3 in ILC2s but not in Th2 cells.
(E) Examples represent 151 genes that were negatively regulated by GATA3 in ILC2s but not in Th2 cells.
(F) Expression of Klrg1, Id2 and Rora was not affected by Gata3 inactivation in ILC2s.
(G) Gata3fl/fl or Gata3fl/fl-CreERT2 mice (4 mice per group) were treated with tamoxifen in vivo for 2–3 days and then ILC2s were sorted based on KLRG1 expression. RNA-seq analyses were performed with Gata3-sufficient and Gata3-deficient cells. The Venn diagram showing overlap of total genes that are positively regulated by GATA3 in cultured ILC2s and ILC2s at the steady state.
Error bars represent Mean ± SD.
We then compared genes that are either positively or negatively regulated by GATA3 in ILC2s and in Th2 cells (Wei et al., 2011). A total of 32 genes that are positively regulated by GATA3 in Th2 cells were also affected in ILC2s upon Gata3 inactivation (Figures 6B and 6C, and Table S1). Il13 transcripts were dramatically reduced when GATA3 was absent in ILC2s (Figure 6C and Table S1). Other genes that are positively regulated by GATA3 in both ILC2s and Th2 cells include Areg (encoding Amphiregulin), Il5, Ccr8 and Lif (Figure6Cand Table S1). Interestingly, Il1rl1 (encoding IL-33R) and Il9r were expressed at much higher levels in ILC2s than in Th2 cells; their expression was dramatically reduced when Gata3 gene was disrupted in ILC2s (Figure6Cand Table S1).
We also identified 225 genes including Il4, Maf, Pth and Ikzf3 (encoding Aiolos) that are positively regulated by GATA3 in Th2 cells but not affected by GATA3 in ILC2s (Figure6B). On the other hand, 130 genes are positively regulated by GATA3 in ILC2s but not in Th2 cells; they include Icos, Il2ra, Kit, Il1r2, Cysltr1, Htr1band Tph1, many of which are cell surface markers of ILC2s (Figures 6B and 6D, and Table S1). These data indicate that GATA3 has unique functions in ILC2s and Th2 cells in addition to its common regulation of many type 2 effector cytokines.
Gata3 inactivation in Th2 cells results in de-repression of several Th1 cell-associated genes, such as Fasl, Il12rb2 and Stat4 (Table S1). However, Gata3 inactivation in ILC2s did not alter the expression of these genes; instead, it de-repressed several other genes such as Cd244 (encoding 2B4), Lta and Tnf (Figure 6E and Table S1) that are characteristic of type I ILCs and/or NK cells.
Gata3 inactivation did not reduce Klrg1 transcription (Figure6F), suggesting that Klrg1 is not a GATA3 target gene. Other cell surface molecules expressed by ILC2s include CD127, Sca-1 and Thy1. The expression of Il7r (encoding CD127) and Ly6a/e (encoding Sca-1) were reduced less than 2 fold, whereas Thy1 expression was increased upon Gata3 inactivation (Table S1). The expression of Rora and Id2, encoding two transcription factors critical for the development of ILC2s, were not reduced upon Gata3 inactivation (Figure6F).
To determine whether GATA3 has a similar function in ILC2s at the steady state, we performed RNA-seq using ex vivo purified KLRG1+ ILC2s from tamoxifen-treated Gata3fl/fl or Gata3fl/fl-Cre-ERT2 mice. The results indicate that among the 162 genes positively regulated by GATA3 in activated ILC2s, 57 genes were also positively regulated by GATA3 in “naïve” ILC2s (Figure 6G and Table S1). These include Il5, Il13, Areg, Il1r1, Ccr8, Il2ra, Cysltr1, Tph1 and Htr1b. Interestingly, Il10 and Il24 were regulated by GATA3 in activated ILC2s but not in “naïve” ILC2s (Figure 6G) suggesting that GATA3-mediated regulation of Il10 and Il24 expression requires cofactors that are only present in activated ILC2s. Il4 expression was affected by Gata3 deletion in “naïve” ILC2s but not in activated ILC2s (Figure 6G), consistent with our previous finding that GATA3 is required for the expression of IL-4 in developing Th2 cells but not essential in fully differentiated Th2 cells.
To verify the RNA-seq data at the protein level, we performed flow cytometry analysis. Compared to control samples, GATA3 protein was lost in 4-OHT-treated Gata3fl/fl-CreERT2 ILC2 culture as judged by intracellular staining (Figure S4A), implying that the deletion of the Gata3 gene was efficient and the pre-existing GATA3 protein was degraded in ILC2s over a two-day period. By contrast, consistent with the RNA-seq data, two cell surface molecules highly expressed by ILC2s, KLRG1 and Sca-1, were only modestly, if at all affected, by inactivation of Gata3 (Figures S4B and S4C), demonstrating that ILC2s maintain some features of their phenotype in the absence of GATA3. Consistent with the role of GATA3 in Th2 cells, IL-13 production by ILC2s in response to IL-33 and IL-7 stimulation was dramatically reduced upon Gata3 inactivation (Figure S4D). In agreement with RNA-seq data, T1/ST2 surface expression was also abolished when Gata3 was inactivated in ILC2s (Figure S4E). This result raised the question whether defective expression of IL-13 was due to a failure of the Gata3-deficient cells to respond to IL-33 or to a direct effect of GATA3 on Il13. Therefore, we stimulated these cells with PMA and ionomycin. Even with this potent stimulus, IL-13 production was dramatically decreased in Gata3-deficient cells (Figure S4F) suggesting that GATA3 directly regulates IL-13 production in ILC2s, independent of its role in maintaining IL-33R expression. This is consistent with the established capacity of GATA3 to directly bind to the promoter of the Il13 gene in many T cell subsets (Wei et al., 2011).
GATA3 regulates common and unique sets of genes in ILC2s and ILC3s
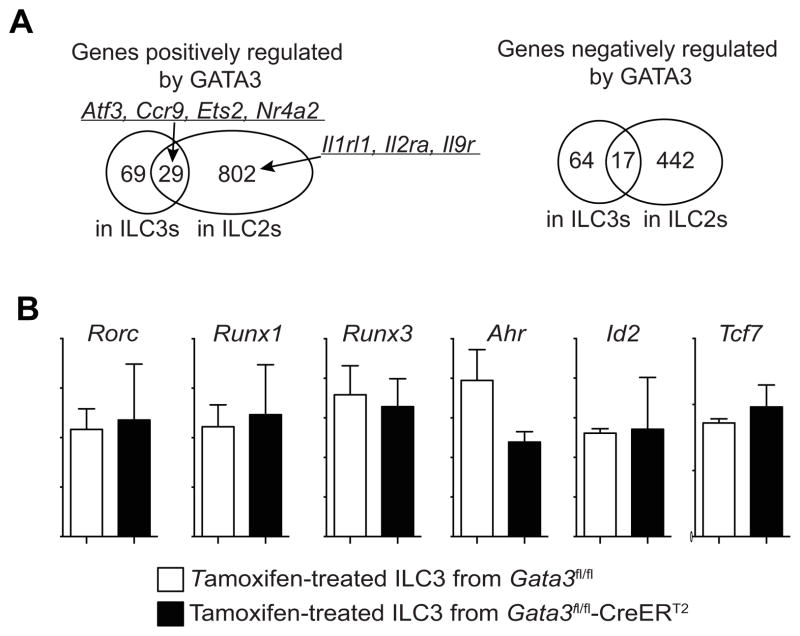
To determine the GATA3 target genes in ILC3s, we performed RNA-seq on ILC3s with or without Gata3 deletion. Ninety-eight and 81 genes were either positively or negatively regulated by GATA3 in ILC3s. Among the 98 genes that were positively regulated by GATA3 in ILC3s, 29 of them, including Atf3, Ccr9, Ets2 and Nr4a2, were also positively regulated by GATA3 in ILC2s (Figure 7A and Table S1). Similarly, among the 81 genes that are negatively regulated byGATA3 in ILC3s, 17 of them are negatively regulated by GATA3 in ILC2s. The functions of these 46 genes that were commonly regulated by GATA3 in ILC2s and ILC3s require further investigation. There are many more genes regulated by GATA3 in ILC2s than in ILC3s possibly because GATA3 is expressed at much higher amounts in ILC2s than that in ILC3s. Some genes encoding cytokine receptors that were regulated by GATA3 in ILC2s, such as Il1rl1, Il2ra and Il9r, were not regulated by GATA3 in ILC3s. The expression of key transcription factors that were reported to be critical for the development of ILC3s, including Rorc, Runx1, Runx3, Ahr, Id2 and Tcf7, was not affected by Gata3 deletion in ILC3s (Figure 7B). Together with the finding that GATA3 does not affect the expression of Id2 and Rora in ILC2s (Figure 6F), our data indicate that the mechanism of GATA3 in regulating the development of IL-7Rα-expressing ILCs is independent of, or in parallel with, all the known key transcription factors.
Figure 7. GATA3 does not regulate known key transcription factors in ILC3s.
(A) Gata3fl/fl or Gata3fl/fl-CreERT2 mice (4 mice per group) were treated with tamoxifen in vivo for 2–3 days and then KLRG1 negative ILCs (mostly ILC3s) were sorted. RNA-seq analyses were performed with Gata3-sufficent and Gata3-deficient ILC3s. RNA-seq analyses were also performed with Gata3-sufficent and Gata3-deficient ILC2s as described in Fig. 6G. The Venn diagram showing overlap of total genes that are positively or negatively regulated by GATA3 in ILC2s and ILC3s.
(B) Expression of Rorc, Runx1, Runx3, Id2, Ahr and Tcf7 was not affected by Gata3 inactivation in ILC3s. Error bars represent Mean ± SD.
DISCUSSION
Research on the ILCs has exploded since the description and characterization of the ILC2s in 2010 by four different groups (Moro et al., 2010; Neill et al., 2010; Price et al., 2010; Saenz et al., 2010) and it is now well accepted that ILCs play important roles in innate immunity by producing effector cytokines(Spits et al., 2013). Since these effector cytokines are virtually identical to those that are produced by T helper cells, ILCs can be divided into group 1, 2, and 3 ILCs representing innate versions of Th1, Th2, and Th17-like cells, respectively (Spits et al., 2013). There are also similarities in the development of ILCs and T helper cells because the key transcription factor that determines a particular Th cell fate is also critically involved in the development of its ILC counterpart (Bernink et al., 2013; Eberl et al., 2004b; Hoyler et al., 2012; Klose et al., 2013; Mjosberg et al., 2012; Sciume et al., 2012; Spits et al., 2013; Vonarbourg et al., 2010). However, much less is known about the early development of ILCs. Our observation, that the development of all IL-7Rα+ ILCs but not classical NK cells requires GATA3, suggests that all IL-7Rα+ ILC subsets may have a common innate lymphoid progenitor (CILP). Furthermore, by comparing gene expression in mature ILC2s with or without GATA3 expression at a genome-wide level, we have provided important data that are crucial for further understanding the development and functions of ILC2s during type 2 immune responses.
Previous reports suggest that ILC2s can be identified by a combination of several cell surface markers, including T1/ST2 (IL-33R), IL-25R, ICOS, Kit, Sca-1 and KLRG-1 in addition to IL-2Rα and IL-7Rα (Halim et al., 2012b; Hoyler et al., 2012; Moro et al., 2010; Neill et al., 2010; Price et al., 2010; Saenz et al., 2010; Wong et al., 2012). We have shown that GATA3 regulates most of these molecules. However, the expression of IL-7Rα and Sca-1 is only modestly reduced, and KLRG-1 is not affected by Gata3 inactivation. In addition to known ILC2 cell surface markers, we also identified other cell surface molecules including IL-9R, CCR8, Cysteinyl leukotriene receptor 1 (Cysltr1), 5-hydroxytryptamine (serotonin) receptor 1B (Htr1b) and TNFR superfamily members 4-1BB and RANK, whose expression in ILC2s is GATA3 dependent. Cysltr1 is expressed by ILC2s and leukotriene D4 is able to induce the expression of IL-5, IL-13 as well as IL-4(Doherty et al., 2013). Therefore, GATA3 may indirectly affect type 2 cytokine production in vivo by regulating two stimulating receptors, T1/ST2 and Cysltr1. Overall, our data indicate that while GATA3 regulates many of the distinguishing cell surface markers on ILC2s, other transcription factors are also involved. Identification of other factors that regulate KLRG1 expression may add important insights into the transcription factor networks controlling the development and maturation of ILC2s.
GATA3 plays critical roles in the differentiation and maintenance of both Th2 cells and ILC2s, and thus is important for mediating type 2 immune responses at different stages. Through genome-wide profiling of the genes that are regulated by GATA3 in ILC2s, we find a substantial overlap of the genes that are regulated by GATA3 in Th2 cells and in ILC2s. Most strikingly, these commonly regulated genes include many key effector cytokines and receptors, such as Il13, Il5, Areg, Il10, Il24, Lif, Il1rl1, Il9r and Ccr8. Except for Il10 and Il24that are expressed only in activated ILC2s, all other key cytokines and receptors are also regulated by GATA3 in “naïve” ILC2s at the steady state. IL-4 is regulated by GATA3 in “naïve” ILC2s but not cultured ILC2s, consistent with our previous findings in T cells that GATA3 is important for the induction of IL-4 during Th2 cell differentiation but plays a minor role in IL-4 production in already developed Th2 cells. Thus, our data indicate that ILC2s resemble the innate counterpart of Th2 cells at a genomic scale and that GATA3 regulates the key functions shared by ILC2s and Th2 cells, possibly through a similar mechanism.
GATA3 is thought to be critical for controlling the cell fate of ILC2 but not other ILCs (Hoyler et al., 2012). The discrepancy between this report and our study can be explained by the timing of Gata3 inactivation. In that study, Id2-driven Cre-ERT2 was used to conditionally delete Gata3 by tamoxifen in otherwise normal mice. Because all the ILCs have already developed in these mice before Gata3 inactivation and the half-life of ILCs may be longer than 3 weeks, the specific effect on ILC2s 3 weeks after GATA3 removal only confirms that GATA3 is critical for the maintenance of ILC2s but not other ILCs. Indeed, we were also able to show that tamoxifen-induced Gata3 deletion in Gata3fl/fl-CreERT2 mice abolished ILC2s but had no effect on RORγt+ ILCs once they had developed. Therefore, acute Gata3 ablation after ILCs have developed obscures the role we have revealed here for GATA3 during development of ILCs. Vav-Cre-mediated gene inactivation disrupts Gata3 gene before ILCs have developed, allowing us to conclude that GATA3 is critical for the development of all IL-7Rα+ ILCs including ILC2s.
All the known subsets of ILCs and NK cells depend on Id2 expression for their development (Moro et al., 2010; Yokota et al., 1999). While E4BP4 (also known as NF-IL3) is critical for the development of NK cells (Gascoyne et al., 2009; Kamizono et al., 2009), no severe ILC development defect has been reported in mice deficient in E4BP4. For the development of IL-7Rα+ ILC subsets, RORα is a transcription factor specifically required for inducing ILC2s(Halim et al., 2012b; Wong et al., 2012) but not RORγt-expressing ILCs(Halim et al., 2012b). On the other hand, RORγt is only needed for the development of RORγt-expressing ILCs(Eberl et al., 2004a; Halim et al., 2012b) but not ILC2s(Halim et al., 2012b; Moro et al., 2010). Up to now, the transcription factor critical for the development of all the IL-7Rα+ ILC lineages but not NK cells has not been identified. Here we report that GATA3 is such a transcription factor. GATA3 does not seem to regulate Id2, RORa or ROR t, suggesting that GATA3 may collaborate with Id2 for ILC development in general, with RORa for ILC2 development, and with RORγt for ILC3 development. Our data indicate that different IL-7Rα+ ILC subsets may develop through a GATA3-dependent mechanism from a common progenitor. Thus, a hematopoietic branch point, with E4BP4 committing progenitors to the NK cell lineage and GATA3 leading to the development of IL-7Rα+ ILCs, may exist. Therefore, it is reasonable to consider that IL-7Rα+ ILC subsets resemble innate version of CD4+ T helper subsets, whereas conventional NK cells, which have higher cytotoxic activity than other IFNγ-expressing ILCs, represent innate version of CD8+ T cells.
Our study show that there is an excellent symmetry in ILC and T helper cell development: while GATA3 is regarded as the master regulator for Th2 cells, it is also critical for the maintenance and functions of ILC2s; while GATA3 is critical for the development of all CD4+ T cells(Ho et al., 2009), it is also indispensable for the development of all IL-7Rα+ ILC subsets. GATA3, therefore, plays parallel roles in establishing and regulating both adaptive and innate lymphocyte populations.
Experimental Procedures
Mice
Gata3fl/fl mice on C57BL/6 background were previously described(Yagi et al., 2010). These mice were bred with either CreERT2(Taconic line 10471) or Vav-Cre transgenic mice (JAX line 8610)on a C57BL/6 background to generate Gata3fl/fl-CreERT2 or Gata3fl/fl-Vav-Cre lines. C57BL/6 mice were ordered from Taconic. CD45.1 congenic mice (Line 7) and Rag2−/ −Il2rg−/ − mice (Line 4111) were from Taconic-NIAID contract. All the mice were bred and/or maintained in the NIAID specific pathogen free animal facility and the experiments were done when mice were 8 to 16 weeks of age under protocols approved by the NIAID Animal Care and Use Committee.
Citrobacter rodentium infections
C. rodentium (formerly Citrobacter freundii, biotype 4280) strain DBS100 was prepared by selecting a single colony and culturing in LB broth for 8 hours. Mice were inoculated with approximately 1 × 1010 CFU in 200 μL of PBS via oral gavage. Cells from large intestinal lamina propria were prepared as previously described(Sun et al., 2007). CFU were determined via serial dilutions on MacConkey’s agar from overnight cultures of homogenized fecal pellets.
Cell preparation
Single cell suspensions were prepared directly from fetal livers or different lymphoid organs of mice including lymph nodes, spleen and bone marrow. Cells from small intestinal lamina propria and intraepithelial lymphocytes (IELs) were prepared as previously described (Sun et al., 2007). To prepare cells from tissues such as lung, mice were perfused with PBS before organs were harvested. The lung was cut into small pieces and digested with DNaseI (Roche) and Liberase (Roche) for 30 min at 37°C. Single cell suspension from digested lung was subjected to Percoll-density gradient centrifugation. In some experiments, recombinant murine IL-25 (from either eBioscience or PeproTech, 0.4 ug per mouse) in PBS was injected intraperitoneally into mice for three consecutive days before mesenteric lymph nodes were harvested. In other experiments, tamoxifen (Sigma) was injected intraperitoneally into mice every other day for 5 times (5mg tamoxifen in 150 ul corn oil per mouse per injection). In mixed bone marrow chimera experiments, bone marrow cells from Gata3fl/fl or Gata3fl/fl-Vav-Cre mice were mixed with bone marrow cells from CD45.1 congenic mice (Line 7) at 1:1 ratio and injected (10 million per mouse) into sublethally irradiated (450 Rad) Rag2−/−Il2rg−/− mice (Line 4111). Cells were stained with a cocktail of antibodies to various lineage markers, including antibodies to CD3, CD5, CD45R, CD11b, CD11c, NK1.1, Gr-1, TER119, FcεRI and TCRγ/δ to identify lineage negative (Lin−) cells. Lin− cells were sorted using a FACS Aria and then cultured with IL-7 (10ng/ml), IL-33 (10ng/ml) and IL-25 (10ng/ml) for ~ 1 week. In some experiments, cells were treated with 100 nM 4-hydroxytamoxifen (4-OHT) after 5 days of culture to delete Gata3 gene from Gata3fl/fl-CreERT2 cells.
Flow cytometry (FACS) analysis
Cell surface molecules and cytokine intracellular staining was performed as previously described (Zhu et al., 2004). Staining for transcription factors was carried out with Foxp3 Staining Buffer Set (eBioscience) according to the manufacturer’s instructions. Flow cytometry data were collected with LSR II (BD Biosciences) and results were analyzed using FlowJo software (Tree Star). Antibodies specific for mouse CD5 (53–7.3), CD45R (RA3-6B2), Gr-1 (RB6-8C5), NK1.1 (PK136), TCRγ/δ (UC7-13D5) and an erythroid cell marker (TER119), were purchased from BioLegend; antibodies specific for mouse CD3 (2C11), CD4 (RM4-5), CD11b (M1/70), CD11c (N418), CD127 (A7R34), CD45.1 (A20), CD45.2 (104), FcεRI (MAR-1), IL-13 (eBio13A), KLRG1 (2F1), NKp46 (29A1.4), RORγt (AFKJS-9), Sca-1 (D7), Flt3 (A2F10), α4β7 (DATK32) and c-Kit (2B8) were purchased from eBiosciences; antibodies specific for Thy1.2 (53–2.1) and GATA3 (L50-823) were purchased from BD Biosciences; and antibodies for IL-33R (DJ8) were purchased from MD Bioproducts.
RNA-Seq and data analysis
Lin− cells were sorted from Gata3fl/fl-CreERT2 mice that had been injected with IL-25 for 3 days. After 5 days culture in IL-7, IL-25 and IL-33, cells were either treated with 4-OHT or ethanol as a control for 2 days. Gata3-sufficient ILC2s were sorted from ethanol treated samples. Gata3-deficient cells were sorted from 4-OHT treated samples excluding residual T1/ST2-expressing cells. In other experiments, Gata3fl/fl or Gata3fl/fl-CreERT2 mice were injected with tamoxifen (5mg/mouse). Two to three days later, Lin−Thy1+CD127+KLRG1+ (mostly ILC2s) and Lin−Thy1+CD127+KLRG1− (mostly ILC3s) were sorted from small intestine lamina propria. RNA-Seq experiments were performed. Briefly, 100 ng of total RNA was amplified using the Ovation RNA-Seq system V2 kit (NuGEN). The resulting dsDNA was sonicated to 200–400 bp. 250 ng of sonicated DNA was used to prepare sequencing libraries for multiplex sequencing (Illumina) and 50 bp reads were generated by the NHLBI Sequencing and Genomics core. Sequencing reads were mapped to the mm9 genome. Gene expression levels were measured by RPKM (Reads Per Kilobase of exon per Million reads in library) (Mortazavi et al., 2008). Differentially expressed genes were identified by edgeR (Robinson et al., 2010) with criteria of false discovery rate (FDR)< 0.001, fold change (FC)> 2 and RPKM >3 in either Gata3-sufficient or deficient samples. Data are available in the Gene Expression Omnibus (GEO) database under the accession number GSE47851.
Supplementary Material
Highlights.
GATA3 is critical for the development of all IL-7Rα-expressing ILCs.
Gata3 conditional deficient mice fail to develop lymph node structure.
GATA3 is essential for the maintenance of ILC2s but not ILC3s.
Genome wide analysis indicates that GATA3 regulates Th2-related genes in ILC2s.
Acknowledgments
We thank Drs. Ronald Germain, Dragana Jankovic, William Paul and John O’Shea for their critical reading of our manuscript; Julie Edwards for her excellent assistance in cell sorting; the NHLBI DNA Sequencing Core facility for sequencing the RNA-Seq libraries; Naofumi Takemoto, Hidehiro Yamane, George Punkosdy, Michelle Crank, Amina Metidji, Liying Guoand Giuseppe Sciume for their helpful discussions. The work is supported by the Division of Intramural Research, NIAID and NHLBI, National Institutes of Health, USA.
Footnotes
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bernink JH, Peters CP, Munneke M, te Velde AA, Meijer SL, Weijer K, Hreggvidsdottir HS, Heinsbroek SE, Legrand N, Buskens CJ, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nature immunology. 2013;14:221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, Powrie F. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YJ, Kim HY, Albacker LA, Baumgarth N, McKenzie AN, Smith DE, Dekruyff RH, Umetsu DT. Innate lymphoid cells mediate influenza induced airway hyper-reactivity independently of adaptive immunity. Nature immunology. 2011;12:631–638. doi: 10.1038/ni.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrier M, Sawa S, Eberl G. Notch, Id2, and RORgammat sequentially orchestrate the fetal development of lymphoid tissue inducer cells. The Journal of experimental medicine. 2012;209:729–740. doi: 10.1084/jem.20111594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty TA, Khorram N, Lund S, Mehta AK, Croft M, Broide DH. Lung type 2 innate lymphoid cells express cysteinyl leukotriene receptor 1, which regulates TH2 cytokine production. J Allergy Clin Immunol. 2013;132:205–213. doi: 10.1016/j.jaci.2013.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littman DR. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nature immunology. 2004a;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi YW, Littman DR. An essential function for the nuclear receptor ROR gamma(t)in the generation of fetal lymphoid tissue inducer cells. Nature immunology. 2004b;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- Fallon PG, Ballantyne SJ, Mangan NE, Barlow JL, Dasvarma A, Hewett DR, McIlgorm A, Jolin HE, McKenzie AN. Identification of an interleukin (IL)-25-dependent cell population that provides IL-4, IL-5, and IL-13 at the onset of helminth expulsion. The Journal of experimental medicine. 2006;203:1105–1116. doi: 10.1084/jem.20051615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, Cella M, Colonna M. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity. 2013;38:769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa J, Moro K, Motomura Y, Okamoto K, Zhu J, Takayanagi H, Kubo M, Koyasu S. Critical role of p38 and GATA3 in natural helper cell function. J Immunol. 2013;191:1818–1826. doi: 10.4049/jimmunol.1300379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJ. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nature immunology. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- Halim TY, Krauss RH, Sun AC, Takei F. Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity. 2012a;36:451–463. doi: 10.1016/j.immuni.2011.12.020. [DOI] [PubMed] [Google Scholar]
- Halim TY, MacLaren A, Romanish MT, Gold MJ, McNagny KM, Takei F. Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity. 2012b;37:463– 474. doi: 10.1016/j.immuni.2012.06.012. [DOI] [PubMed] [Google Scholar]
- Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nature reviews immunology. 2009;9:125–135. doi: 10.1038/nri2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, Voehringer D, Busslinger M, Diefenbach A. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, Grosveld G, Akashi K, Lind EF, Haight JP, Ohashi PS, et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. The Journal of experimental medicine. 2009;206:2977–2986. doi: 10.1084/jem.20092176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Vahedi G, Hirahara K, Singleton K, O’Shea JJ. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annual review of immunology. 2012;30:707–731. doi: 10.1146/annurev-immunol-020711-075058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein Wolterink RG, Serafini N, van Nimwegen M, Vosshenrich CA, de Bruijn MJ, Fonseca Pereira D, Veiga Fernandes H, Hendriks RW, Di Santo JP. Essential, dose-dependent role for the transcription factor Gata3 in the 26 development of IL-5+ and IL-13+ type 2 innate lymphoid cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10240–10245. doi: 10.1073/pnas.1217158110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d’Hargues Y, Goppert N, Croxford AL, Waisman A, Tanriver Y, Diefenbach A. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, Mantovani A, Kopan R, Bradfield CA, Newberry RD, Colonna M. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nature immunology. 2012;13:144– 151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HE, Reinhardt RL, Bando JK, Sullivan BM, Ho IC, Locksley RM. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nature immunology. 2012;13:58–66. doi: 10.1038/ni.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjosberg J, Bernink J, Golebski K, Karrich JJ, Peters CP, Blom B, te Velde AA, Fokkens WJ, van Drunen CM, Spits H. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity. 2012;37:649–659. doi: 10.1016/j.immuni.2012.08.015. [DOI] [PubMed] [Google Scholar]
- Mjosberg JM, Trifari S, Crellin NK, Peters CP, van Drunen CM, Piet B, Fokkens WJ, Cupedo T, Spits H. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nature immunology. 2011;12:1055–1062. doi: 10.1038/ni.2104. [DOI] [PubMed] [Google Scholar]
- Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, Angelosanto JM, Laidlaw BJ, Yang CY, Sathaliyawala T, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nature immunology. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, Furusawa J, Ohtani M, Fujii H, Koyasu S. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature. 2010;463:540–544. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5:621– 628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TK, Bucks C, Kane CM, Fallon PG, Pannell R, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- Possot C, Schmutz S, Chea S, Boucontet L, Louise A, Cumano A, Golub R. Notch signaling is necessary for adult, but not fetal, development of RORgammat(+) innate lymphoid cells. Nature immunology. 2011;12:949–958. doi: 10.1038/ni.2105. [DOI] [PubMed] [Google Scholar]
- Powell N, Walker AW, Stolarczyk E, Canavan JB, Gokmen MR, Marks E, Jackson I, Hashim A, Curtis MA, Jenner RG, et al. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity. 2012;37:674–684. doi: 10.1016/j.immuni.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AE, Liang HE, Sullivan BM, Reinhardt RL, Eisley CJ, Erle DJ, Locksley RM. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11489–11494. doi: 10.1073/pnas.1003988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz SA, Siracusa MC, Monticelli LA, Ziegler CG, Kim BS, Brestoff JR, Peterson LW, Wherry EJ, Goldrath AW, Bhandoola A, Artis D. IL-25 simultaneously elicits distinct populations of innate lymphoid cells and multipotent progenitor type 2 (MPPtype2) cells. The Journal of experimental medicine. 2013;210:1823–1837. doi: 10.1084/jem.20122332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz SA, Siracusa MC, Perrigoue JG, Spencer SP, Urban JF, Jr, Tocker JE, Budelsky AL, Kleinschek MA, Kastelein RA, Kambayashi T, et al. IL25 elicits a multipotent progenitor cell population that promotes T(H)2 cytokine responses. Nature. 2010;464:1362–1366. doi: 10.1038/nature08901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Sciume G, Hirahara K, Takahashi H, Laurence A, Villarino AV, Singleton KL, Spencer SP, Wilhelm C, Poholek AC, Vahedi G, et al. Distinct requirements for T-bet in gut innate lymphoid cells. The Journal of experimental medicine. 2012;209:2331–2338. doi: 10.1084/jem.20122097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Artis D. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity. 2012;37:601–610. doi: 10.1016/j.immuni.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, et al. Innate lymphoid cells - a proposal for uniform nomenclature. Nature reviews immunology. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- Spits H, Cupedo T. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annual review of immunology. 2012;30:647–675. doi: 10.1146/annurev-immunol-020711-075053. [DOI] [PubMed] [Google Scholar]
- Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nature immunology. 2011;12:21–27. doi: 10.1038/ni.1962. [DOI] [PubMed] [Google Scholar]
- Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. The Journal of experimental medicine. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, Mebius RE, Littman DR. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science (New York, N Y. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T, Flach M, Bengsch B, Thimme R, Holscher C, et al. Regulated expression of nuclear 28 receptor RORgammat confers distinct functional fates to NK cell receptor expressing RORgammat(+) innate lymphocytes. Immunity. 2010;33:736–751. doi: 10.1016/j.immuni.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshenrich CA, Garcia-Ojeda ME, Samson-Villeger SI, Pasqualetto V, Enault L, Richard-Le Goff O, Corcuff E, Guy-Grand D, Rocha B, Cumano A, et al. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nature immunology. 2006;7:1217–1224. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- Walker JA, Barlow JL, McKenzie AN. Innate lymphoid cells - how did we miss them? Nature reviews immunology. 2013;13:75–87. doi: 10.1038/nri3349. [DOI] [PubMed] [Google Scholar]
- Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SH, Walker JA, Jolin HE, Drynan LF, Hams E, Camelo A, Barlow JL, Neill DR, Panova V, Koch U, et al. Transcription factor RORalpha is critical for nuocyte development. Nature immunology. 2012;13:229–236. doi: 10.1038/ni.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Junttila IS, Wei G, Urban JF, Jr, Zhao K, Paul WE, Zhu J. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity. 2010;32:507–517. doi: 10.1016/j.immuni.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Zhu J, Paul WE. An updated view on transcription factor GATA3-mediated regulation of Th1 and Th2 cell differentiation. Int Immunol. 2011;23:415–420. doi: 10.1093/intimm/dxr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Monticelli LA, Saenz SA, Chi AW, Sonnenberg GF, Tang J, De Obaldia ME, Bailis W, Bryson JL, Toscano K, et al. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity. 2013;38:694–704. doi: 10.1016/j.immuni.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, Gruss P. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF, Jr, Guo L, Paul WE. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nature immunology. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annual review of immunology. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.