Abstract
Peripheral ameloblastoma (PA) in gingiva is rare and often confused with oral basal cell carcinoma (OBCC). The tissues of one case of PA and one case of OBCC with the same mandibular molar area affected were compared via an immunohistochemical examination using 50 antisera. The PA and OBCC showed similar proliferation of basaloid epithelial strands, but toluidine blue staining revealed that the PA had pinkish juxta-epithelial myxoid tissue, whereas the OBCC was infiltrated by many mast cells. Immunohistochemical comparisons showed that the PA was strongly positive for ameloblastin, KL1, p63, carcinoembryonic antigen, focal adhesion kinase, and cathepsin K, and slightly positive for amelogenin, Krox-25, E-cadherin, and PTCH1, whereas the OBCC was not. On the other hand, the OBCC was strongly positive for EpCam, matrix metalloprotease (MMP)-1, α1-antitrypsin, cytokeratin-7, p53, survivin, pAKT1, transforming growth factor-β1, NRAS, TGase-1, and tumor nescrosis factor-α, and consistently positive for β-catenin, MMP-2, cathepsin G, TGase-2, SOS-1, sonic hedgehog, and the β-defensins-1, -2, -3, while the PA was not. These data suggest that the tumorigeneses of PA and OBCC differ, and that PAs undergo odontogenic differentiation and generate oncogenic signals for infiltrative growth and bone resorption, whereas OBCCs undergo basaloid epidermal differentiation as a result of growth factor/cytokine-related oncogenic signals.
Keywords: Peripheral ameloblastoma; Carcinoma, basal cell; Immunohistochemistry
The differential diagnosis of peripheral ameloblastoma (PA) and oral basal cell carcinoma (OBCC) is difficult based on histological observation alone, because both tumors exhibit similar basal cell proliferation forming epithelial strands. It is well known that PAs usually arises from the gingival epithelium,1,2 and that OBCCs rarely occurs in the gingival epithelium.3,4 The cases examined in the present study had similar gingival ulcerations localized in the mandibular molar area and were both male, though their ages differed (PA, 61 years; OBCC, 33 years).
PA is an exophytic growth in the tooth-bearing areas of the jaw, and is likely fibrous epulis.5 In most cases, superficial bone erosion manifesting as cupping or saucerization is observed during surgery. PA accounts for 2% to 10% of all ameloblastomas, and overall average patient age is 52.1 years.2 Thus, PA occurs at a significantly higher age than intraosseous ameloblastoma (37.4 years).6 As to the location of PA, its maxilla/mandible ratio is 1:2.6, and the mandibular premolar region accounts for 32.6% of all sites.7 Pathogenetically, two major sources of PA have been proposed: remnants of dental lamina and oral surface epithelium. Histologically, PA consists of proliferating odontogenic epithelium exhibiting the same histomorphological cell types and patterns as those seen in intraosseous ameloblastoma.2,8
Basal cell carcinoma (BCC) is a common neoplasm of the skin with a well-known variable morphology and a wide incident age range. The most common nodular type BCC consists of islands and nests of uniform basaloid cells, which often exhibit stromal retraction artifacts, mucin within stroma, and associated mononuclear infiltrates.9,10 Morpheaform, infiltrating, and basosquamous BCCs have been associated with aggressive behaviors, including perineural invasion, whereas adenoid and adamantinoid BCCs are generally considered to be no more aggressive than nodular BCCs. Adenoid BCC shares histologic features with PA, and sometimes grows infiltratively into the deep subcutis, and thus, its differential diagnosis is obligatory.11,12 The present case of BCC occurred in the gingival epithelium and showed features of nodular, infiltrative, and adenoid patterns of tumor growth. Therefore it was simply described as an OBCC.
In the present study, the cases of PA and OBCC showed similar histological features, nevertheless the proliferating patterns and cytological characteristics of their tumor epithelia differed slightly. Accordingly, they were diagnosed primarily based on histological observations. The immunohistochemical study was conducted to compare protein expressions in the PA and OBCC.
CASE REPORT
A case of PA (S2009-75) in a 61-year-old male patient exhibited an ulcerative lesion in the right mandibular molar area and diffuse osteolytic depression of alveolar bone by plain radiography. In addition, a case of OBCC (S2012-109) in a 33-year-old male patient presented as an ulcerative lesion in the gingiva of the right mandibular molar area, involving only regional periodontium by plain radiography. The primary gingival lesions in both cases were examined pathologically. The usage of biopsy specimens (S2009-75 and S2012-109) filed in the Department of Oral Pathology, Gangneung-Wonju National University Dental Hospital was approved by the Institutional Review Board (IRB2013-2).
Multiple serial microsections of the PA and OBCC excised tissue samples were immunohistochemically analyzed with the three-layered indirect immunostaining method using 50 antisera. The antisera were selected for important signaling pathways, including odontogenic differentiation, cellular proliferation, apoptosis, growth factors, tumor oncogenesis, inflammation, and angiogenesis (Table 1). All immunohistochemical (IHC) results were confirmed by repeat IHC staining, two times for clear IHC reactions and three times for obscure IHC reactions. The IHC images were obtained from similar representative areas of the PA and OBCC microsections, and immunoreactions for the PA and OBCC were compared.
Table 1.
Antibodies used in this study
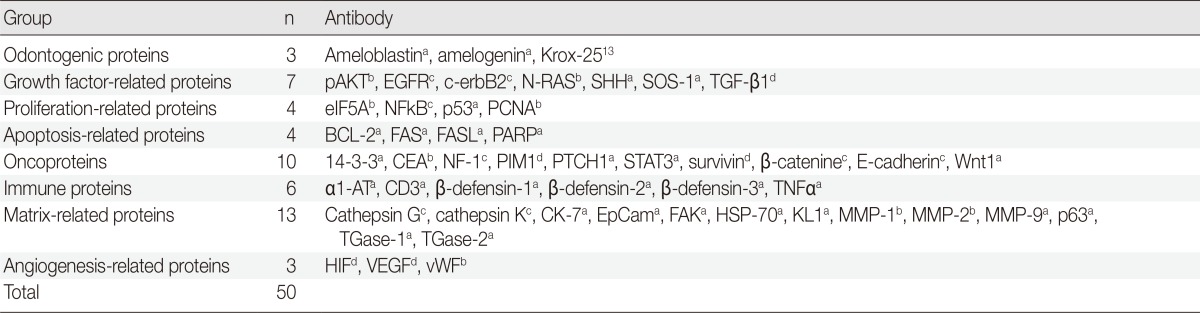
pAKT1, v-akt murine thymoma viral oncogene homolog 1, phosphorylated at Thr 308; EGFR, epithelial growth factor receptor; SHH, sonic hedgehog; SOS-1, Son of sevenless-1; TGF-β1, transforming growth factor-β1, eIF5A, eukaryotic translation initiation factor 5A; NFkB, nuclear factor kappa-light-chain-enhancer of activated B cells; PCNA, proliferating cell nuclear antigen; BCL-2, B-cell leukemia/lymphoma-2; FASL, FAS (CD95/Apo1) ligand; PARP, poly-ADP ribose polymerase; CEA, carcinoembryonic antigen; NF-1, neurofibromin-1; PIM1, pivotal integration site 1; PTCH1, patched homologue 1; STAT3, signal transducer and activator of transcription-3; α1-AT, α1-antitrypsin; TNFα, tumor nescrosis factor-α; CK, cytokeratin; EpCam, epithelial cell adhesion molecule, Ber-EP4; FAK, focal adhesion kinase; HSP-70, heat shock protein-70; KL1, pan cytokeratin 1, 2, 5, 6, 7, 8, 11, 14, 16, 17, 18; MMP, matrix metalloprotease; TGase, transglutaminase; HIF, hypoxia inducible factor; VEGF, vascular endothelial growth factor; vWF, von Willebrand factor.
aSanta Cruz Biotechnology, Santa Cruz, CA, USA; bNeomarkers, Fremont, CA, USA; cDAKO, Glostrup, Denmark; dZymed, San Francisco, CA, USA.
Histological observations
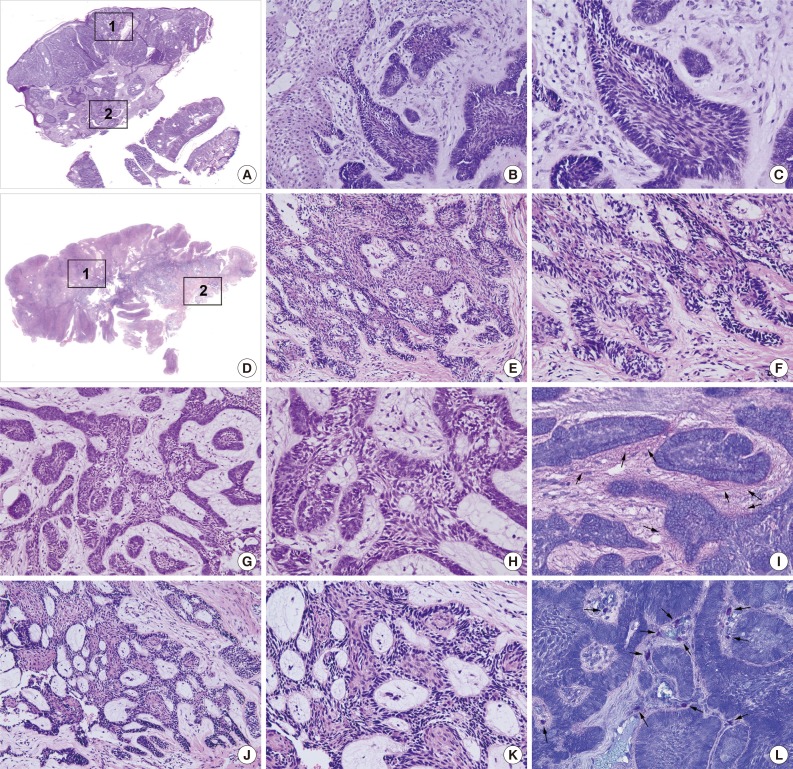
The PA and OBCC both showed basaloid epithelial strands proliferating in the subepithelial area of the gingiva and gradually infiltrating into deep connective tissue (Fig. 1A, D). In the PA, epithelial follicles and strands directly proliferated from the basal layer of epithelium, which continuously grew into underlying connective tissue. Tumor epithelium was composed of a palisading basal layer composed of reversely polarized tall columnar cells (Fig. 1B, C), and formed epithelial bridges in a plexiform fashion (Fig. 1G, H).
Fig. 1.
Photomicrographs, hematoxylin and eosin stain. (A-C, G-I) Peripheral ameloblastoma (PA). (D-F, J-L) Oral basal cell carcinoma (OBCC). (B, E, G, J) Low magnification. (C, F, H, K) High magnification. 1: Subepithelial area (B, C, E, F). 2: Deep connective tissue area (G, H, J, K). (B, C) Subepithelial area in PA. (G, H) Deep connective tissue in PA, note the palisading basal layer cells. (E, F) Subepithelial area in OBCC. (J, K) Deep connective tissue in OBCC, note the proliferating basal layer cells. (I, L) Toluidine blue stain. (I) Juxta-epithelial pink staining (arrows). (L) Many mast cells (arrows) in the stromal tissue.
The OBCC also showed tumor epithelial strands and cords connected with the basal layer of epithelium (Fig. 1E, F) that subsequently grew into adjacent fibrous connective tissue. Tumor epithelia were anastomosed with proliferating basal cells containing elongated nuclei (Fig. 1J, K).
Toluidine blue staining in the PA showed pinkish juxta-epithelial myxoid stromal tissue (Fig. 1I), whereas the OBCC tissues were infiltrated by many mast cells (Fig. 1L).
IHC comparison
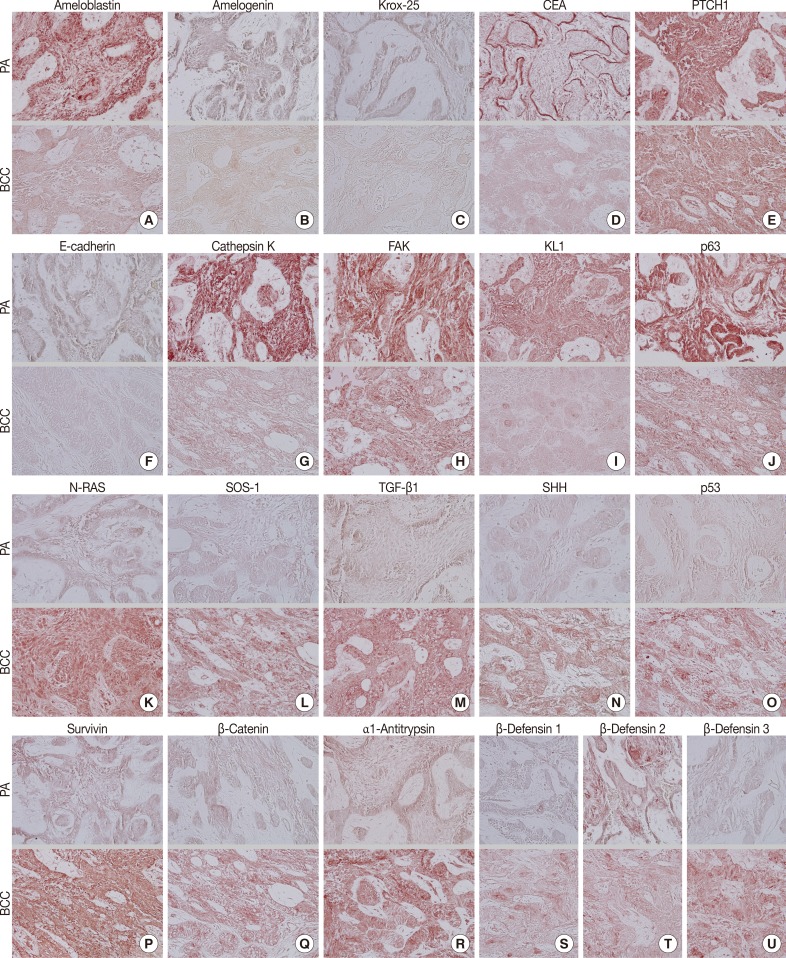
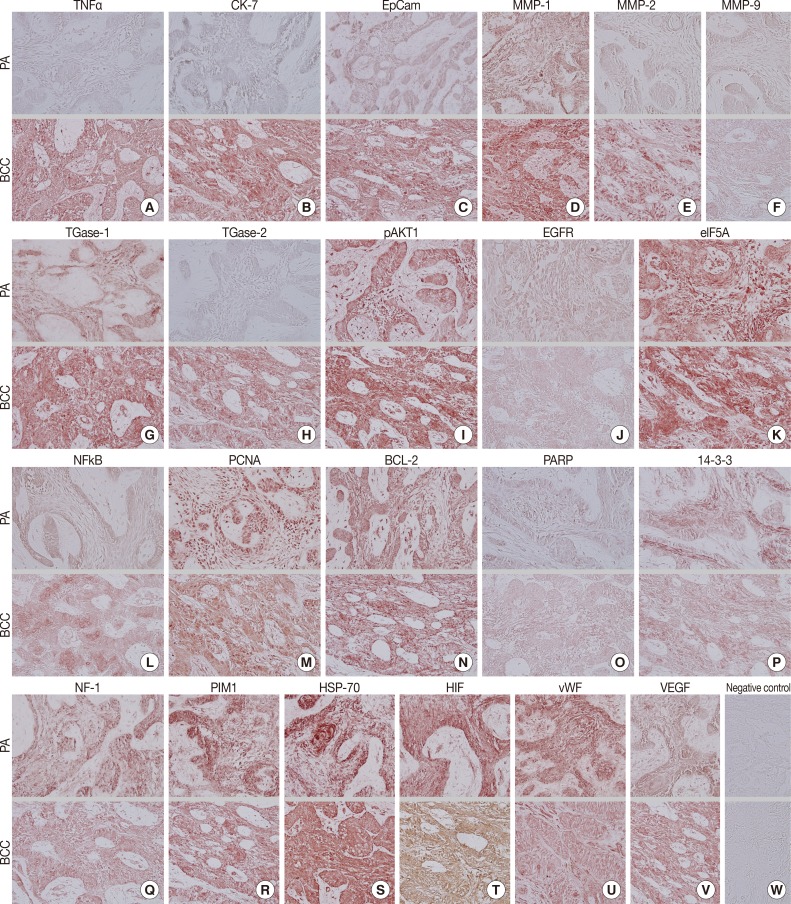
During comparative IHC screening, the PA exhibited strong positive reactions for ameloblastin, KL1, p63, carcinoembryonic antigen (CEA), focal adhesion kinase (FAK), and cathepsin K, and slight positive reactions for amelogenin, Krox-25, E-cadherin, whereas the OBCC did not. On the other hand, the OBCC exhibited strong positive reactions for epithelial cell adhesion molecule (EpCam), matrix metalloprotease (MMP)-1, α1-antitrypsin, cytokeratin (CK)-7, p53, survivin, transforming growth factor-β1 (TGF-β1), N-RAS, v-akt murine thymoma viral oncogene homolog 1, phosphorylated at Thr 308 (pAKT1), transglutaminase (TGase)-1, and tumor nescrosis factor-α (TNFα), and consistent positive reactions for β-catenin, MMP-2, cathepsin G, TGase-2, Son of sevenless-1 (SOS-1), sonic hedgehog (SHH), β-defensin-1, -2, -3, while the PA did not. Immunoreactions for proliferating cell nuclear antigen (PCNA), nuclear factor kappa-light-chain-enhancer of activated B cells (NFkB), MMP-9, eukaryotic translation initiation factor 5A (eIF5A), B-cell leukemia/lymphoma-2 (BCL-2), poly-ADP ribose polymerase (PARP), pivotal integration site 1 (PIM1), neurofibromin-1 (NF-1), heat shock protein-70 (HSP-70), 14-3-3, hypoxia inducible factor (HIF), von Willebrand factor (vWF), and vascular endothelial growth factor (VEGF) were similarly positive in the PA and OBCC tissues (Table 2, Figs. 2, 3).
Table 2.
IHC array comparison between PA and OBCC
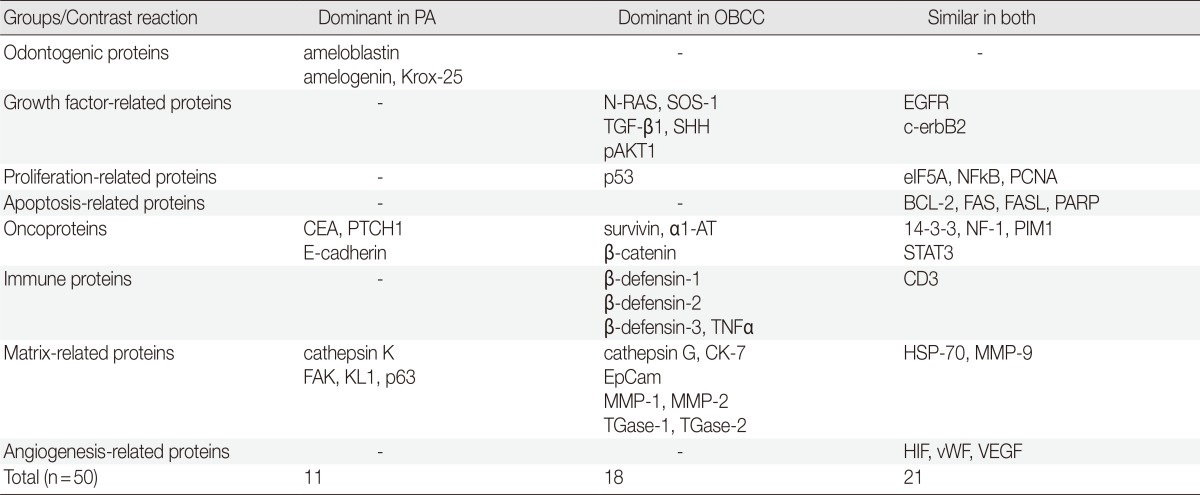
PA, peripheral ameloblastoma; OBCC, oral basal cell carcinoma; SOS-1, Son of sevenless-1; EGFR, epithelial growth factor receptor; TGF-β1, transforming growth factor-β1; SHH, sonic hedgehog; pAKT1, v-akt murine thymoma viral oncogene homolog 1, phosphorylated at Thr 308; eIF5A, eukaryotic translation initiation factor 5A; NFkB, nuclear factor kappa-light-chain-enhancer of activated B cells; PCNA, proliferating cell nuclear antigen; BCL-2, B-cell leukemia/lymphoma-2; FASL, FAS (CD95/Apo1) ligand; PARP, poly-ADP ribose polymerase; CEA, carcinoembryonic antigen; PTCH1, patched homologue 1; α1-AT, α1-antitrypsin; NF-1, neurofibromin-1; PIM1, pivotal integration site 1; STAT3, signal transducer and activator of transcription-3; TNFα; tumor nescrosis factor-α; CK-7, cytokeratin 7; HSP-70, heat shock protein-70; MMP, matrix metalloprotease; EpCam, epithelial cell adhesion molecule, Ber-EP4; TGase, transglutaminase; HIF, hypoxia inducible factor; vWF, von Willebrand factor; VEGF, vascular endothelial growth factor.
Fig. 2.
Photomicrographs of immunohistochemical staining results, no background stain. (A-T, upper panels) Peripheral ameloblastoma (PA). (A-T, lower panels) Oral basal cell carcinoma (OBCC). (A) Ameloblastin. (B) Amelogenin. (C) Krox-25. (D) Carcinoembryonic antigen (CEA). (E) Patched homologue 1 (PTCH1). (F) E-Cadherin. (G) Cathepsin K. (H) Focal adhesion kinase (FAK). (I) KL1. (J) p63. (K) N-RAS. (L) Son of sevenless-1 (SOS-1). (M) Transforming growth factor-β1 (TGF-β1). (N) Sonic hedgehog (SHH). (O) p53. (P) Survivin. (Q) β-Catenin. (R) α1-Antitrypsin. (S) β-Defensin 1. (T) β-Defensin 2. (U) β-Defensin 3.
Fig. 3.
Photomicrographs of immunohistochemical stainings, no background stain. (A-V, upper panels) Peripheral ameloblastoma (PA). (A-V, lower panels) Oral basal cell carcinoma (OBCC). (A) Tumor nescrosis factor-α (TNFα). (B) Cytokeratin-7 (CK-7). (C) Epithelial cell adhesion molecule, Ber-EP4 (EpCam). (D) Matrix metalloprotease (MMP)-1. (E) MMP-2. (F) MMP-9. (G) Transglutaminase (TGase)-1. (H) TGase-2. (I) v-akt murine thymoma viral oncogene homolog 1, phosphorylated at Thr 308 (pAKT1). (J) Epithelial growth factor receptor (EGFR). (K) Eukaryotic translation initiation factor 5A (eIF5A). (L) Nuclear factor kappa-light-chain-enhancer of activated B cells (NFkB). (M) Proliferating cell nuclear antigen (PCNA). (N) B-cell leukemia/lymphoma-2 (BCL-2). (O) Poly-ADP ribose polymerase (PARP). (P) 14-3-3. (Q) Neurofibromin-1 (NF-1). (R) Pivotal integration site 1 (PIM1). (S) Heat shock protein-70 (HSP-70). (T) Hypoxia inducible factor (HIF). (U) von Willebrand factor (vWF). (V) Vascular endothelial growth factor (VEGF). (W) Negative control.
In terms of protein functions, the biomarkers of odontogenic epithelium (ameloblastin, amelogenin, Krox-25) (Fig. 2A-C) were conspicuously positive in the PA but rare in the OBCC. Tumor epithelia of the PA and OBCC showed contrasting cytodifferentiation characterized by positivity for KL1 (Fig. 2I) in the PA, but positivity for CK-7 and EpCam (Fig. 3B, C) in the OBCC. Furthermore, a transformation-related oncoprotein, p63, created by interactions between mesenchyme and epithelium (Fig. 2J); a biomarker of infiltrative growth (FAK) (Fig. 2H); a biomarker of bone resorption (cathepsin K) (Fig. 2G); and a biomarker of epithelial adhesion (E-cadherin) (Fig. 2F) were more strongly positive in the PA than in the OBCC. In particular, a cell surface-anchored glycoprotein (CEA) (Fig. 2D) was strongly positive in some tumor epithelium in the PA, at which juxta-epithelial myxoid stroma accumulated, but was rarely positive in the OBCC. In addition, a tumor suppressor protein functioning in the formation of embryonic structures and tumorigenesis (patched homologue 1 [PTCH1]) (Fig. 2E) was more strongly positive in the epithelium of the PA than in the epithelium of the OBCC.
Oncoproteins relevant to tumor progression (p53, survivin, β-catenin) (Fig. 2O-Q) were strongly positive in the OBCC, but weak in the PA. The cytokine-related proteins (β-defensin-1, -2, -3, TNFα, cathepsin G) (Figs. 2S, 2T, 2U, 3A) (cathepsin G, data not shown), the growth factor-related signaling proteins (TGF-β1, N-RAS, SOS-1) (Fig. 2K-M), the cross-linking enzymes for keratinocyte differentiation (TGase-1 and TGase-2) (Fig. 3G, H), and the serine/threonine-specific protein kinase signaling cell survival (pAKT1) (Fig. 3I) were more strongly positive in the OBCC than in the PA. The signaling molecule for developmental organogenesis (SHH) (Fig. 2N) was strongly positive in the epithelium of the OBCC, but not in that of the PA.
Biomarkers of cellular proliferation (PCNA) (Fig. 3M), DNA transcription (NFkB) (Fig. 3L), protein translation (eIF5A) (Fig. 3K), and angiogenesis (HIF, vWF, VEGF) (Fig. 3T, U, V) were consistently positive in the PA and OBCC.
Additionally, a biomarker of T cell receptor complex (CD3; data not shown), a ubiquitous heat shock protein (HSP-70) (Fig. 3S), an antiapoptotic protein (BCL-2) (Fig. 3N), and a proto-oncoprotein signaling for cell cycle progression, apoptosis, and transcriptional activation (PIM1) (Fig. 3R) were strongly positive in the PA and OBCC. A vital regulatory protein for diverse cellular functions (14-3-3) (Fig. 3P), and a tumor suppressor protein which is a negative regulator of the RAS signal transduction pathway (NF-1) (Fig. 3Q), were diffusely and slightly positive in both the PA and OBCC. The cellular apoptosis-related proteins (PARP, FAS, FAS [CD95/Apo1] ligand [FASL]) (Fig. 3O) (FAS and FASL; data not shown) and the epidermal growth factor receptor proteins (EGFR, c-erbB2) (Fig. 3J) (c-erbB2; data not shown) were weakly positive in the PA and OBCC. A transcription factor in response to cytokine and growth factors (signal transducer and activator of transcription-3 [STAT3]) and a signaling protein for developmental structures and oncogenesis (Wnt1) were rarely positive in the tumor epithelium of the PA or OBCC (data not shown). On the other hand, negative control staining done without primary antibody showed no background reaction (Fig. 3W).
DISCUSSION
PA is classified as an odontogenic tumor with the histopathological characteristics of intraosseous ameloblastomas, and mainly occurs in soft tissues covering the teeth and tooth-bearing tissues.1,14 Histologically, PAs are composed of islands and strands of columnar-appearing basaloid epithelial cells. A PA may demonstrate continuity with the surface epithelium, may have an associated mononuclear infiltrate, and may produce keratin pearls.14 However, no consistent histological features can easily differentiate OBCCs and PAs, which explains why it has been frequently been asserted that PAs are simply BCCs of the gingival epithelium.1
The two described cases of PA and OBCC occurred at the same area of the right mandibular molar area in male patients of different ages (61 and 31 years old, respectively). Histologically, the PA exhibited typical odontogenic differentiation mimicking dental lamina or enamel epithelium connected with a basal layer of gingival epithelium. The basal layer cells of the PA were well polarized in a reverse manner and arranged in a palisading fashion, whereas the OBCC showed proliferating epithelial strands and cords with undifferentiated basal cells, which had elongated, spindle shaped hyperchromatic nuclei. These findings suggest that the OBCC featured cellular malignancy and the PA did not.
The present study also demonstrates that the PA had abundant juxta-epithelial myxoid tissue, which was stained pink by the metachromasia of toluidine blue, implying the presence of the active ectomesenchymal interaction usually found in odontogenic tissue. The OBCC tissues were infiltrated by many mast cells found in toluidine blue stain, suggesting that the tumor growth of the OBCC is affected by pro-inflammatory reactions.
Continued progress in the elucidation of molecular signaling pathways makes it possible to explain normal developmental processes, organogenesis, tumorigenesis, and malignant transformation by the differential expressions of functional proteins. Several high throughput technologies such as DNA microarray and proteomics have been developed to detect the expressional changes of multiple genes/proteins. Nevertheless, IHC studies are still the preferred method for observing protein expressions in specific cells, and thus the present study used an IHC-based approach to compare protein expressions in the PA and OBCC.
The IHC reactions of the known differential biomarkers (EpCam and KL1) of PA and OBCC were found to be identical to previous reports.15 That is, EpCam was exclusively positive in the OBCC and KL1 was exclusively positive in the PA. However, this study also found that the odontogenic proteins amelogenin, ameloblastin, and Krox-25 were strongly positive in the PA but scarce in the OBCC. Therefore, the differential diagnosis of the present cases was more certain than the primary diagnosis based on histological observation alone.
Proteins associated with embryonal processes, epithelial adhesion, ectomesenchymal interaction, cellular migration, and bone resorption (CEA, E-cadherin, p63, FAK, cathepsin K, respectively) were more strongly positive in the PA than in the OBCC. These findings suggest that the PA has the characteristic properties of an active ectomesenchymal interaction and infiltrative growth into underlying bone, and that OBCC does not.16,17
Immunostaining of PTCH1 was strongly positive in the PA but weak in the OBCC, whereas the signaling molecule SHH (a ligand of PTCH1 receptor) was common in the OBCC but rare in the PA. These findings suggest that the pathogenesis of ameloblastoma is related to that of the OBCC.18,19,20
In the present study, the OBCC also exhibited dominant expression of growth factor-related proteins (N-RAS, SOS-1, TGF-β1), whereas the PA did not. In particular, the inflammation-related proteins, i.e., cytokine (TNFα), innate immunity proteins (β-defensin-1, -2, -3, and α1-antitrypsin), and matrix proteases (MMP-1, MMP-3, and MMP-9) were strongly positive in the OBCC but weak in the PA. These findings indicate that the tumorigenesis of OBCC is more affected by gingival inflammation compared to PA, and the production of cleft-like ulcerated lesions in OBCCs are caused by active inflammatory reactions.
The OBCC showed stronger reactions for p53, survivin, and pAKT1 than PA, suggestive of malignant transformation, and greater nuclear β-catenin overexpression indicative of a more aggressive behavior.
The cytodifferentiation of the OBCC was also distinct from that of the PA as indicated by the stronger immunostainings of CK-7 and TGase-1, -2 in the OBCC. This finding suggests that the OBCC forms solid epithelial tumor tissue with transitional or glandular structures.
On the other hand, many biomarkers of cellular proliferation, DNA transcription, protein translation, apoptosis, and angiogenesis were similarly positive in the PA and OBCC, which indicates that the two diseases have similar potential for tumor growth and propagation.
In conclusion, the present IHC-based, comparative study screened for differentially expressed proteins in the PA and OBCC using 50 antisera, and found that EpCam was exclusively expressed in the OBCC and that KL1 was dominantly expressed in the PA, as previously reported.15 Furthermore, immunostainings of ameloblastin, amelogenin, Krox-25, CEA, E-cadherin, p63, FAK, and cathepsin K were consistently positive in the PA, and N-RAS, TGF-β1, survivin, α1-antitrypsin, TNFα, and MMP-1 were consistently positive in the OBCC. These findings suggest that the above-mentioned proteins should be viewed as candidate biomarkers for the differential diagnosis of PA and OBCC. Although further investigations to elucidate characteristic protein expressions in PA and OBCC are required, the present IHC comparison identifies proteins differentially expressed in the cellular signaling pathways of cytodifferentiation and tumorigenesis in PA and OBCC. The present findings demonstrate that PA is clearly distinguishable from OBCC by IHC.
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Gardner DG. Peripheral ameloblastoma: a study of 21 cases, including 5 reported as basal cell carcinoma of the gingiva. Cancer. 1977;39:1625–1633. doi: 10.1002/1097-0142(197704)39:4<1625::aid-cncr2820390437>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 2.Philipsen HP, Reichart PA, Nikai H, Takata T, Kudo Y. Peripheral ameloblastoma: biological profile based on 160 cases from the literature. Oral Oncol. 2001;37:17–27. doi: 10.1016/s1368-8375(00)00064-6. [DOI] [PubMed] [Google Scholar]
- 3.Koutlas IG, Koch CA, Vickers RA, Brouwers FM, Vortmeyer AO. An unusual ostensible example of intraoral basal cell carcinoma. J Cutan Pathol. 2009;36:464–470. doi: 10.1111/j.1600-0560.2008.01059.x. [DOI] [PubMed] [Google Scholar]
- 4.Nishimura N, Sakurai K, Noguchi K, Urade M. Keratotic basal cell carcinoma of the upper gingiva with cervical lymph node metastasis: report of a case. J Oral Maxillofac Surg. 2001;59:677–680. doi: 10.1053/joms.2001.23401. [DOI] [PubMed] [Google Scholar]
- 5.Gardner DG, Baker DC. The relationship of the canine acanthomatous epulis to ameloblastoma. J Comp Pathol. 1993;108:47–55. doi: 10.1016/s0021-9975(08)80226-5. [DOI] [PubMed] [Google Scholar]
- 6.Bologna-Molina R, Mosqueda-Taylor A, de Almeida-Oslei P, Toral-Rizo V, Martínez-Mata G. Peripheral desmoplastic ameloblastoma: histopathological and immunohistochemical profile of a case. Med Oral Patol Oral Cir Bucal. 2010;15:e846–e849. doi: 10.4317/medoral.15.e846. [DOI] [PubMed] [Google Scholar]
- 7.López-Jornet P, Bermejo-Fenoll A. Peripheral ameloblastoma of the gingiva: the importance of diagnosis. J Clin Periodontol. 2005;32:12–15. doi: 10.1111/j.1600-051X.2004.00627.x. [DOI] [PubMed] [Google Scholar]
- 8.Vigneswaran N, Whitaker SB, Budnick SD, Waldron CA. Expression patterns of epithelial differentiation antigens and lectin-binding sites in ameloblastomas: a comparison with basal cell carcinomas. Hum Pathol. 1993;24:49–57. doi: 10.1016/0046-8177(93)90062-l. [DOI] [PubMed] [Google Scholar]
- 9.Park HR, Han YI, Sol MY, Lee SK. Nevoid basal cell carcinoma syndrome: report of a case. Korean J Pathol. 1995;29:263–267. [Google Scholar]
- 10.Song KY, Choi YH, Kim MK, Lee KK, Ham EK. Pathological analysis of the basal cell carcinoma. Korean J Pathol. 1994;28:160–167. [Google Scholar]
- 11.Golda N, Wheeland RG, Thomas TL, Trout C. Adenoid Basal cell carcinoma simulating ameloblastoma. Dermatol Surg. 2009;35:1410–1413. doi: 10.1111/j.1524-4725.2009.01249.x. [DOI] [PubMed] [Google Scholar]
- 12.Fresko D, Lazarus SS. Oral carcinoma in situ. Its progression to squamous, basosquamous, and basal-cell carcinoma. Arch Pathol Lab Med. 1981;105:15–19. [PubMed] [Google Scholar]
- 13.Lee SK, Kim YS, Lee SS, et al. Molecular cloning, chromosomal mapping, and characteristic expression in tooth organ of rat and mouse Krox-25. Genomics. 2004;83:243–253. doi: 10.1016/j.ygeno.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Lowe L, Rapini RP. Newer variants and simulants of basal cell carcinoma. J Dermatol Surg Oncol. 1991;17:641–648. doi: 10.1111/j.1524-4725.1991.tb01312.x. [DOI] [PubMed] [Google Scholar]
- 15.Kato N, Endo Y, Tamura G, Motoyama T. Ameloblastoma with basal cell carcinoma-like feature emerging as a nasal polyp. Pathol Int. 1999;49:747–751. doi: 10.1046/j.1440-1827.1999.00932.x. [DOI] [PubMed] [Google Scholar]
- 16.Ishida M, Kojima F, Okabe H. Cathepsin K expression in basal cell carcinoma. J Eur Acad Dermatol Venereol. 2013;27:e128–e130. doi: 10.1111/j.1468-3083.2011.04436.x. [DOI] [PubMed] [Google Scholar]
- 17.Zheng Z, Kye Y, Zhang X, Kim A, Kim I. Expression of p63, bcl-2, bcl-6 and p16 in basal cell carcinoma and squamous cell carcinoma of the skin. Korean J Pathol. 2005;39:91–98. [Google Scholar]
- 18.Ponti G, Pollio A, Mignogna MD, et al. Unicystic ameloblastoma associated with the novel K729M PTCH1 mutation in a patient with nevoid basal cell carcinoma (Gorlin) syndrome. Cancer Genet. 2012;205:177–181. doi: 10.1016/j.cancergen.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 19.Jorgensen TJ, Ruczinski I, Yao Shugart Y, et al. A population-based study of hedgehog pathway gene variants in relation to the dual risk of basal cell carcinoma plus another cancer. Cancer Epidemiol. 2012;36:e288–e293. doi: 10.1016/j.canep.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vangelista G. An unusual neoplasm of the stomach: basal cell epithelioma with adamantinoid evolution. Arch Ital Mal Appar Dig. 1955;21:139–150. [PubMed] [Google Scholar]