Abstract
The streptokinase (SK) is emerging as an important thrombolytic therapy agent in the treatment of patients suffering from cardiovascular diseases. We reported highly effective renaturation of a SK from S. pyogeness DT7 overexpressed in E. coli, purification, and biochemical characterization. A gene coding for the SK was cloned from S. pyogeness DT7. Because accumulation of active SK is toxic to the host cells, we have expressed it in the form of inclusion bodies. The mature protein was overexpressed in E. coli BL21 DE3/pESK under the control of the strong promoter tac induced by IPTG with a level of 60% of the total cell proteins. The activity of the rSK, renatured in phosphate buffer supplemented with Triton X-100 and glycerol, was covered with up to 41 folds of its initial activity. The purified of protein was identified with MALDI-TOF mass spectrometry through four peptide fragments, which showed 100% identification to the corresponding peptides of the putative SK from GenBank. Due to overexpression and highly effective renaturation of large amounts of inclusion bodies, the recombinant E. coli BL21 DE3/pESK system could be potentially applied for large-scale production of SK used in the therapy of acute myocardial infarction.
1. Introduction
Streptokinase (EC 3.4.99.22) (SK), a commercially important nonprotease, binds stoichiometrically to both circulating and thrombus-bound plasminogen (Plg) to generate SK-plasminogen activator complex. Cleavage of plasminogen in zymogen form at an Arg-Val bond generates plasmin, an active enzyme that degrades fibrin component of thrombin [1]. Due to this property, the streptokinase has been widely used in the therapy of acute myocardial infarction for its strong activity in dissolving blood clots [2].
Most group A, C, and G β-hemolytic streptococci isolated from human hosts secrete streptokinase with molecular mass of 47 kDa, which convert the plasminogen to the serine protease plasmin. However, due to low SK production yields from natural host and its pathogenicity, so research interest has shifted to cloning and expression of SK in hyperproductive and safe heterologous host systems. Therefore, sk genes have been cloned and expressed in different expression systems including Bacillus subtilis [3], Streptococcus sanguis [4], Streptomyces lividans [5, 6], Schizosaccharomyces pombe [7], Pichia pastoris [1, 8], Lactococcus lactis [9], and Escherichia coli [10, 11]. However, there are some disadvantages of producing recombinant proteins in Pichia pastoris due to high glycosylation level [12] or in Lactococcus lactis due to low cell density [9].
Escherichia coli is the most commonly used host for the production of recombinant proteins, both in research and industry [13]. High-level expression of recombinant proteins in the form of a soluble intracellular product, secretory product, or as insoluble inclusion bodies depends on promoter system, host-vector interactions, sequence, and characteristics of recombinant products and the effect of the expressed foreign protein on host cell physiology [14].
The expression of SK as inclusion bodies by E. coli systems is shown to be useful for obtaining large amounts of protein, provided that renaturation is effective and recovery of active protein is high. Thus, the purpose of this study was firstly to overproduce the recombinant streptokinase in E. coli BL21 (DE3) and simultaneously to refold effectively the large amount of the recombinant streptokinase as inclusion bodies overexpressed by E. coli BL21 (DE3) under the control of the promoter T7. Only both objectives were gained; then the recombinant E. coli overproducing SK as inclusion bodies can become a potential strain for industrial SK production.
2. Materials and Methods
2.1. Chemicals and Reagents
DNA cloning kit, RNase A, restriction enzymes (BamHI, NotI, and EcoRI), T4-ligase, and Proteinase K were purchased from Fermentas (Thermo Fisher Scientific Inc., Waltham, USA). The DNA Extraction Kit was from Qiagen (Venlo, Netherlands). Protein Extraction Kit and ProBond resin were supplied by Invitrogen Corp. (Carlsbad, CA, USA). Human plasminogen from MP Biomedicals (Santa Ana, USA); SK, N (p-tosyl) gly-pro-lys-4-nitro anilide acetate salt (AAS), SDS from Sigma Aldrich Co. (St. Luis, USA); Plasminogen, Tween 20 and Tween 80 from BioBasic Inc. (NY, USA); Triton X-100 and EDTA from Merck (Darmstadt, Germany). All other reagents were of analytical grade unless otherwise stated.
2.2. Plasmids, Bacterial Strains, and Culture Conditions
The bacterial strain Streptococcus pyogenes DT7 (GQ247718) isolated from a patient at the Army Hospital No. 103 (Hanoi, Vietnam) was used as the source of the streptokinase (sk) gene. Escherichia coli DH5α (F−, ø80dlacZΔM15, Δ(lacZYA-argF) U169, deoR, recA1, endA1, hsdR17(rK−, mK+), phoA, supE44, λ–, thi-1, gyrA96, relA1) and the vector pJET1.2/blunt (Fermentas, Thermo Fisher Scientific Inc., Waltham, USA) were used for DNA manipulations and amplification. Escherichia coli BL21 (DE3) cells (F – ompT gal dcm lon hsdS B (r B − m B −) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) and pET22b+ vector (Novagen, Merck KGaA, Darmstadt, Germany) were used for expression of SK. LB medium (Luria-Bertani) containing 1% (w/v) bacto tryptone; 0.5% (w/v) yeast extract; 1% (w/v) NaCl; pH 7–7.5 was used for cultivation of E. coli DH5α and BL21 (DE3). LB agar contained additionally 2% (w/v) agar and 100 μg ampicillin/mL.
2.3. DNA Manipulations
Genomic and plasmid DNA isolation was carried out by methods which have been previously described [15]. DNA fragments and PCR products were excised from a 0.8% agarose gel and purified by a gel extraction kit (Qiagen, Venlo, The Netherlands) according to the manufacturer's instructions. DNA sequencing was performed on an ABI PRISM 3100 Avant Genetic Analyzer (Applied Biosystems Inc., Foster City, USA). E. coli DH5α and BL21 were transformed using heat shock method that has been previously described [15].
2.4. DNA Amplification and Plasmid Construction
The putative sk-coding DNA fragment was amplified from S. pyogenes DT7 genomic DNA by PCR with Taq DNA polymerase. Based on the nucleotide sequence of the sk gene from S. pyogenes strain (GenBank: Z48617), 3 oligonucleotides, mSKF GGC GGATCC CATATG ATTGCTGGACCTG, and SKF: GCC CAT GGG CAA AAA TTA CTT AT and SKR GCC TCG AGT TTG TCB TTA GGG TT were designed as primers for introduction of the underlined BamHI and XhoI restriction sites, respectively. The PCR mixture contained 2.5 μL 10x PCR buffer; 2 μL of 2.5 mM dNTP; 2.5 μL of 25 mM MgCl2; 0.5 μL genomic DNA (50–100 ng); 0.25 μL 5 unit Taq polymerase, and 1 μL each primer (10 pmol), supplemented with 15.25 μL distillated water to a final volume of 25 μL. The thermocycler conditions were as follows: 95°C/4′; 30 cycles of (95°C/30′′, 52°C/45′′, 72°C/45′′); 72°C/10′. The PCR products amplified from the genomic DNA with the primer pair SKF and SKR were inserted into the cloning vector pJET1.2/blunt, resulting in pJSK. DNA sequencing was performed on ABI PRISM 3100 Avant Genetic Analyzer. Sequence alignments were constructed and analyzed using the program MegAlign DNAStar. It was followed by ligation of the BamHI-XhoI digested PCR products (with the primer pair mSKF and SKR) with pET22b+ linearized by the same enzymes, resulting in pESK under the control of the T7-promoter induced by isopropyl-β-D-thiogalactopyranoside (IPTG) and possessing the ampicillin marker. The streptokinase rSKhis encoded by the plasmid pESK contains the mature streptokinase fused with the 6x histidine-tag and no native leader sequence.
2.5. rSK Expression
The transformant E. coli BL21/pESK was cultivated overnight in 5 mL of LB medium containing 5 μL of 100 mg/mL ampicillin at 37°C on an orbital shaker at 200 rpm. Overnight culture (2 mL) was inoculated in a 1-liter Erlenmeyer flask containing 200 mL of LB broth and 200 μL of 100 mg/mL ampicillin. The culture was grown at 37°C with agitation at 200 rpm and until an optical density (OD) at 600 nm reached 0.6 (for approximately 2.5 h); then 200 μL of 100 mM IPTG was added. The culture was continuously incubated at 37°C with agitation at 200 rpm for 3–6 h induction. Cells were harvested by centrifugation at 6000 rpm for 10 min at 4°C. Wet weight cells were used for protein purification.
2.6. Purification of Streptokinase
The fusion form rSKhis carrying a C-terminal 6xHis tag was expressed in E. coli BL21. To purify rSK, 100 mg wet weight cells from a 120 mL culture in LB medium were harvested by centrifugation and suspended in 10 mL of guanidine lysis buffer containing 6 M guanidine hydrochloride, 20 mM sodium phosphate, 500 mM NaCl, and pH 7.8. The cell suspension was sonificated (three bursts of 1 min each at 1 min interval). After 30–60 min incubation in ice with slight shaking, the cell lysate was centrifuged at 13000 rpm and 4°C for 25 min to remove cell debris. A volume of 8 mL cell lysate was applied to a Ni-NTA column (Invitrogen Corp., Carlsbad, USA) containing 2 mL resin which was equilibrated with denaturing binding buffer and incubated for 45 min at room temperature with gentle hand shaking for several times. The column was washed with 4 times of 8 mL denaturing wash buffer. The bound protein was eluated with 8 mL of denaturing eluation buffer. Then 6 mL of the enzyme extract was applied to a Bio-gel column (2,6 × 6 cm) with elution of 50 mM Tris-HCl buffer (pH 8) at a flow rate of 25 mL/h and then washed with the same buffer.
2.7. Streptokinase Renaturation
The pool of purified SK fragments were renaturated using 50 mM phosphate buffer pH 7 supplemented with 10% (w/v) glycerol and different detergents (0.5% (w/v) Triton X-100, 1% (w/v) Tween 20, 1.5% (w/v) Tween 80) [11]. Diluted cell lysate (1 : 200) and purified rSK (1 : 100) in renaturation buffer were incubated at 37°C for 1 h and 4°C for 6 h. The residual activity was then determined as described below.
2.8. Streptokinase Assay
To estimate the activity of the purified rSK, 10 μL purified protein solution was added to 10 μL of 50 mM Tris buffer pH 7.5 containing 0.05 unit of human plasminogen and incubated at 37°C for 30 min. The color reaction was developed by the addition of 40 μL of 1 mM AAS solution and incubated at 37°C for 15 min. The reaction was stopped by the addition of 10 μL of 0.4 N acetic acid. The absorbance was read at 405 nm against a blank containing human plasminogen, Tris buffer, and AAS but without rSK solution. The activity was estimated using standard SK (Sigma Aldrich Co., St. Luis, USA). One unit (U) of rSK was defined as one unit of standard SK, which liquefies a standard clot of fibrinogen, plasminogen, and thrombin at 37°C and pH 7.5 for 10 min.
2.9. Protein Electrophoresis and Quantification
The homogeneity and molecular mass of the streptokinase were determined by 12.5% SDS polyacrylamide gel electrophoresis [16] with Biometra equipment (Göttingen, Germany). Proteins were visualized by staining with Coomassie Brilliant Blue R-250 or with 0.1% (w/v) of silver nitrate. Protein concentrations were measured by Bradford assay with the bovine serum albumin as standard [17].
2.10. MALDI-TOF Mass Spectrometry
The rSK was identified by MALDI-TOF mass spectrometry as previously described [18]. The predicted protein band on SDS-PAGE was cut out and the target protein was digested by trypsin treatment into small peptide fragments. The mixture of peptides was analyzed on nano-LC liquid chromatography and ionized by the ESI (electrospray ionization). The mass spectra were obtained by QSTAR XL mass spectrometer (Applied Biosystems, MDS SCIEX, Canada) with a nano-ESI ion source. Protein fragments were identified by the Mascot v1.8 Search Software from the database (NCBInr, SwissProt). Peptide fragments showing ion scores above 42 were identified uniquely or high-similarly with P < 0.05.
2.11. Biochemical Characterization of rSK
The pH and temperature optimum of rSK were determined by measuring the activity as described above using 100 mM potassium phosphate buffer (pH 5.5–7.5) and 100 mM Tris-HCl buffer (pH 7.5–10) at 37°C, and in the temperature range of 4 to 60°C using 100 mM potassium phosphate buffer, pH 7.5, respectively.
For the determination of temperature and pH stability, the purified rSK, 0.1 μg for each reaction, was preincubated in 100 mM potassium phosphate buffer pH 7 at different temperatures 4–60°C for 0–96 h, and pH range from 4 to 9.5 (pH 4-5, 100 mM potassium acetate buffer; pH 5.5–7.5, 100 mM potassium phosphate buffer; and pH 7.5–9.5, 100 mM Tris-HCl) at 37°C for 0–48 h, respectively. The residual activity was then determined.
Effect of surfactants on the activity of rSK was check by mixture of 0.4 unit purified rSK and substrate and supplemented with either Triton X-100, Tween 20, or Tween 80, each at a final concentration of 0.5, 1.0, 1.5, and 2.0% (w/v) in appropriate buffer pH 7 and incubated at 37°C for 60 min. The residual activity of rSK was determined as described above.
The effect of additives on the activity of the purified rSK was investigated by incubating the mixture of 0.4 unit of the purified rSK and either of Ag+, Ca2+, Co2+, Cu2+, Fe2+, K+, Mn2+, Ni2+, Zn2+, or EDTA, at a final concentration of 1, 3, and 5 mM. The reaction mixtures were incubated at 28°C for 60 min. The residual activity of rSK was then measured as shown above. All measurements were carried out in triplicate with the resulting values being the mean of the cumulative data obtained.
3. Results and Discussion
3.1. Gene Cloning and Analysis
The recombinant plasmid pTSK with inserted sk gene was sequenced and aligned with sequences from GenBank using DNAstar. Nucleotide sequence of sk gene from S. pyogenes DT7 exhibited 84.4% to 99.6% identities with sequences from Streptococcus pyogenes groups of A, C, and G strains in GenBank (CP000262, CP000261, M19347, AM903378, and AY234136). The putative amino acid sequence of the gene sk showed 77.9 to 99.3% identities with the corresponding amino acid sequences from the abovementioned Streptococcus pyogenes strains. The sequence was deposited in the GenBank with an accession number of ACG50170.
3.2. Expression and Purification of SK
The DNA fragment (1245 bps) encoding the mature streptokinase (SK) truncated 26 N-terminal amino acids from S. pyogenes DT7 was inserted into pET22b+ vector at the BamHI and XhoI sites resulting in the recombinant plasmid pESK. The transformant E. coli BL21/pESK was grown in LB medium for the SK production. After IPTG induction, the cells were collected and used for purification and renaturation. The expression level of rSK as inclusion bodies by E. coli BL21/pESK was 60% of the total proteins (Figure 1(a), lane 1) using Dolphin 1D software. This level was as high as that (65%) reported by Zhang et al. (1999) [19] and more than two to four times as high as those (25%) reported by [20], (20%) by [21], and 15% by [22].
Figure 1.
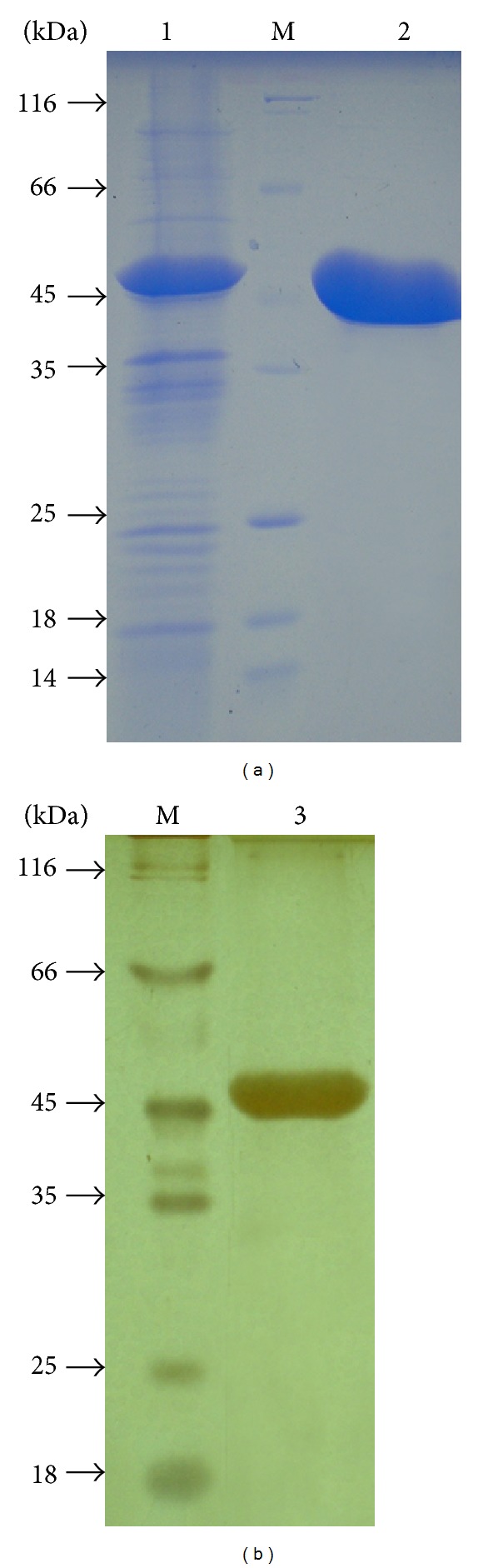
SDS-PAGE of the purified rSK by ProBond Resin. Lane 1, E. coli BL21/pESK cell lysate; Lane 2, purified rSK stained by using Coomassie Brilliant Blue R250; Lane 3, purified rSK stained by using silver nitrate; Lane M, molecular standards indicated in kDa.
3.3. Renaturation of Streptokinase
The cell lysate was renatured by using various surfactants including Triton X-100, Tween 20, and Tween 80 each or in combination with glycerol. Triton X-100 was known as detergent to dissolves and refolding aggregated protein. In absence of surfactants, rSK exhibited the same activity (182–189 U/mL) with or without glycerol (Table 1). The addition of surfactants increased the rSK activity obviously to 3.5–3.8 folds without glycerol, but steeply to 25.2–30.6-folds in combination with glycerol at 37°C for 60 min, even to 36.1–41.7 folds at 4°C for 6 h. At lower temperature (4°C), the enzyme activity was recovered better than at higher temperature (37°C), increased by 26–43%. The combination of glycerol at the concentration of 10% (w/v) and Triton X-100 at the concentration of 0.5% (w/v) recovered the highest activity of rSK and reached 7,591 U/mL at 4°C (Table 1). The renaturation of the purified rSK with 10% glycerol containing 0.5% Triton X-100 at 4°C for 6 h and at 37°C for 1 h recovered the enzyme activity of 28.6 and 36.5 folds, respectively (Table 2), corresponding to the specific activity of 10,312.5, and 11,264.2 U/mg protein. The reason the renaturation efficiency in this study was much higher than that reported by Cherish Babu et al. (2008). At the same conditions for treatment, the enzyme activity was recovered with only 9.7 folds in comparison to control [11].
Table 1.
Effect of surfactant, glycerol, and temperature on the renaturation of cell lysate E. coli/pESK.
| Parameter | SK activity (U/mL) | ||
|---|---|---|---|
| at 37°C for 1 h | at 37°C for 1 h + 10% glycerol | at 4°C for 6 h + 10% glycerol | |
| 0.5% Triton X-100 | 643.4 ± 3.6 | 5574.0 ± 36.2 | 7591.3 ± 45.2 |
| 1% Tween 20 | 657.6 ± 5.7 | 4592.8 ± 27.1 | 6587.7 ± 27.1 |
| 1.5% Tween 80 | 691.6 ± 6.6 | 5226.8 ± 50.6 | 6578.7 ± 63.3 |
| No surfactant | 182.1 ± 1.8 | 184.3 ± 3.6 | 189.4 ± 3.6 |
Table 2.
The renaturation of purified rRSK.
| Fraction number | rSK activity (U/mg) | ||
|---|---|---|---|
| at 37°C for 1 h | at 4°C for 6 h + 0.5% Triton X-100 and 10% glycerol | at 37°C for 1 h + 0.5% Triton X-100 and 10% glycerol | |
| 1 | 69.1 ± 1.0 | 1622.4 ± 29.3 | 2519.0 ± 15.7 |
| 2 | 360.5 ± 4.8 | 10312.5 ± 55.2 | 11264.2 ± 27.6 |
| 3 | 199.9 ± 1.4 | 2039.3 ± 21.1 | 3862.5 ± 15.2 |
| 4 | 55.3 ± −0.3 | 1516.9 ± 18.9 | 1907.6 ± 11.8 |
3.4. Purification of Recombinant SK
rSK from S. pyogenes DT7 overexpressed by E. coli BL21/pESK cells was purified through affinity chromatography column of Ni2+-ProBond resin to the homogeneity on SDS-PAGE with a molecular mass of approximately 47 kDa (Figure 1, lane 2). The purified rSak gained a specific activity of 10,336 U/mg proteins with a purification factor of 2.56 and a yield of 52% (Table 3). The solution containing rSK protein was loaded onto Biogel P-100 packed column for fractionating and obtained with a purity of 95.7% and specific activity of 11,558 U/mg (Figure 1, lane 3).
Table 3.
Purification steps of the streptokinase from E. coli/pESK.
| Total proteins | Purified proteins | Specific activity (U/mg) of | Yield (%) | Purification factor | |
|---|---|---|---|---|---|
| supernatant of cell lysate | purified rSK | ||||
| 90375 mg | 21789 mg | 4568 | 10336 | 52% | 2.26 |
3.5. Identification of Recombinant SK
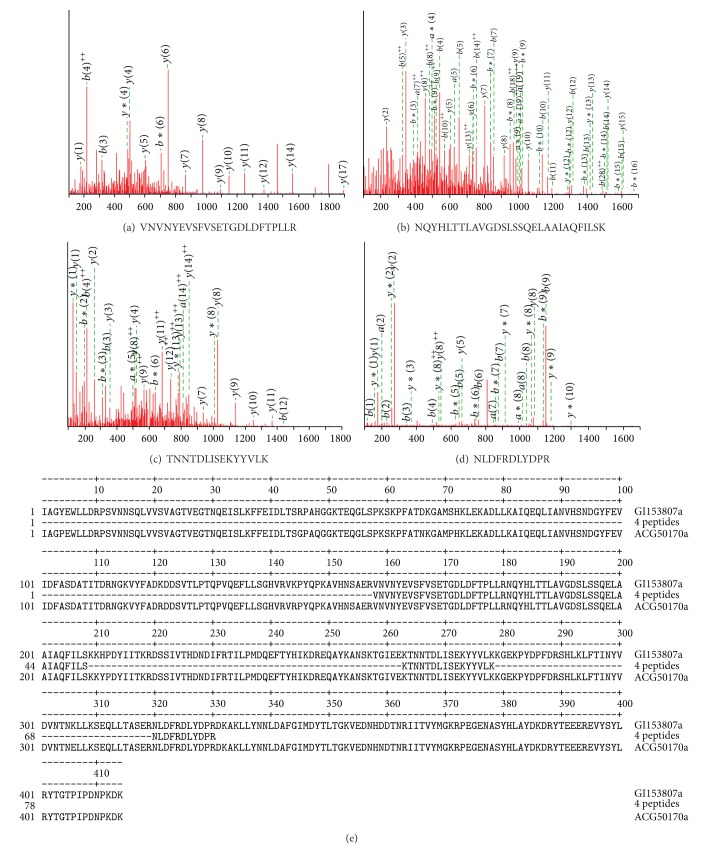
The single protein on SDS-PAGE (Figure 1, lane 3) was cut out from the gel and used for LC-ESI-MS/MS analysis of mass spectrum database by using Mascot v1.8 program. The total score of SK identification was 203 to 509 and matched peptides were 29 to 39 fragments. Four peptide fragments of the purified enzyme identified by MALDI-TOF mass spectrometry agreed with those of the streptokinase found in GenBank gi|153807, streptokinase (S. pyogenes) VNVNYEVSFVSETGDLDFTPLLR (position 158–180) (Figure 2(a)), NQYHLTTLAVGDSLSSQELAAIAQFILSK (position 181–209) (Figure 2(b)), TNNTDLISEKYYVLK (position 263–278) (Figure 2(c)), NLDFRDLYDPR (position 320–330) (Figure 2(d)), corresponding to a monoisotopic mass of 2613.3, 3117.63, 1799.93, and 1422.69 Da and to m/z ion scores of 102, 111, 73, and 51, respectively. Whereas the peptide fragments showing ion scores above 42 were identified uniquely or highly similarly to P < 0.05. These peptides of the recombinant streptokinase expressed by E. coli/pESK showed 100% identity with the corresponding fragments of the putative streptokinase protein from S. pyogenes (gi|153807) (Figure 2(e)).
Figure 2.
Monoisotopic mass of three neutral identified peptides. (a) VNVNYEVSFVSETGDLDFTPLLR position 158–180 (a); (b) NQYHLTTLAVGDSLSSQELAAIAQFILSK position 181–208; (c) TNNTDLISEKYYVLK position 263–279; (d) NLDFRDLYDPR position 320–330 found in gi: 153807, streptokinase from Streptococcus pyogenes (GenBank, AAA26973) corresponding to ion scores of 102, 111, 73, and 51 with P < 0.05, respectively. (e) Alignment of four neutral identified peptides (4 peptides) with streptokinase from Streptococcus pyogenes AAA26973 (gi153807) and rSK from S. pyogenes DT07 (ACG50170).
3.6. Temperature and pH Optimum
The temperature and pH optimum for the reaction of SK-plasmin were observed at 37°C and pH 7 (Figures 3(a) and 3(b)). The enzyme showed over 80% activity at the temperature range from 25 to 45°C and pH 6.7–7.5 (for 100 mM potassium phosphate buffer) and pH 8.5–10 (for Tris-HCl buffer) in comparison with the optimum activity. The temperature optimum for the SK-plasmin reaction was in agreement with that from other reports. Rajagopalan et al. (1987) reported that the reactions of α2-macroglobulin (α2M) with plasmin or streptokinase-plasmin (ogen) (SkP1) was markedly temperature-dependent and initial rates of reaction at 0 and 24°C were only 3 and 40% of the rate of 37°C, respectively [23]. Mumme et al. (1993) reported that the highest fibrinolysis activity with streptokinase was obtained at 40°C, with lower activities having been recorded at both higher and lower temperatures [24]. The optimum temperature and pH of streptokinase from β-haemolytic streptococci were 27–37°C and 7 [25]. Another thrombolytic agent, closely related to the streptokinase, staphylokinase (Sak) from Staphylococcus aureus exhibited the same profile. The native Sak from S. aureus V8 showed the pH optimum at pH 7.5 and 8.5 [26]. The temperature and pH optimum for Sak from S. aureus QT08 expressed in E. coli and P. pastoris were observed at 30–37°C, pH 7, and pH 9 [27] and pH 7,5, and pH 8.5 [28], respectively.
Figure 3.

Temperature (a) and pH (b) optimum of rSK from S. pyogenes DT07.
Why the streptokinases and staphylokinases shared a common property that the optimum temperature was not more than 40°C and pH optimum exhibited 2 peaks? because the fibrinolytic activity of streptokinase originates in its ability to activate blood plasminogen to plasmin, the enzyme that degrades fibrin cloth through its specific lysine binding site [29]. The temperature optimum for the human plasmin was at 37°C [30] and the optimal pH value for the human plasmin and that for SK or Sak were significantly different.
3.7. Temperature and pH Stability
The streptokinase from S. pyogenes DT7 was stable up to 37°C and retained more than 80% of its initial activity after incubation for 9 h and more than 50% after incubation for 96 h (Figure 4(a)). The enzyme exhibited more stability at pH 7 than at pH 9 and retained more than 73% of its initial activity after incubation at pH 7 for 24 h, whereas it retained only more than 65% of its initial activity after incubation at pH 9 for 8 h (Figure 4(b)). K. Vesterberg and O. Vesterberg (1972) also reported that the concentrated material containing Sak from S. aureus V8 was stable at refrigerator temperature over a pH range of 3.0–8.5. Sak from S. aureus QT08 expressed in E. coli and P. pastoris was stable at a temperature range from 25°C to 50°C, and at a pH range from 7 to 9 after incubation for 2 h with a residual activity of more than 70% [26, 28]. The results depicted in Figure 4(b) indicating that there were two sharp peak, one at pH 7.0 and the other one at pH 9.0 with the activity of 100% and 98%, respectively. The experiments of the optimal pH value for the high level activity of rSK were rather complicated since two reactions happened continuously in the same reaction mixture: at first, the activation reaction of plasminogen to plasmin was activated by rSK, and second, the digestion process of AAS was catalyzed by plasmin. The optimal pH value for human plasmin and that for SK was significantly different; therefore, this could cause the appearance of second peak activity. The data depicted in Figure 4(b) showing that the second peak activity at pH value of 9,0 might therefore be due to optimal pH for the plasmin activity in Tris-HCl buffer. Similarly, these observations were also reported by K. Vesterberg and O. Vesterberg (1972) in which staphylokinase was a plasminogen activator.
Figure 4.
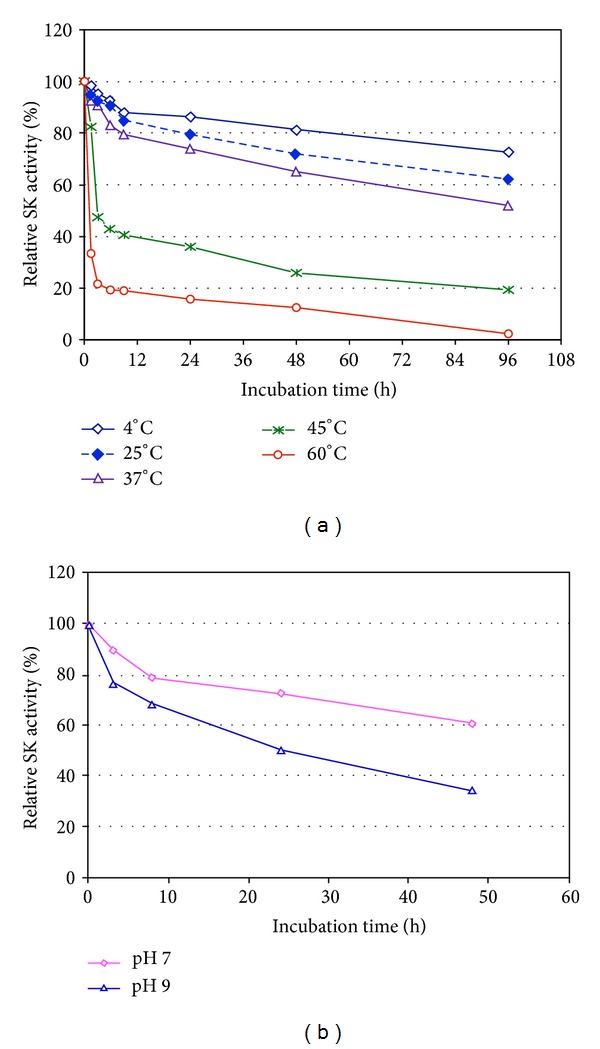
Temperature (a) and pH (b) stability of rSK from S. pyogenes DT07.
3.8. Effect of Surfactants
The addition of either Tween 80, Tween 20, or Triton X-100 at the final concentration of 0.5–2% (w/v) in reaction mixture showed an activation of the streptokinase from S. pyogenes DT07 up to 150% of its original activity. The enzyme activity increased up to 154% after incubation for 24 h but deeply decreased to 18% after longer incubation for 48 h (Table 4). Similarly, Cherish Babu et al. (2008) reported that rSK was treated with guanidine and then supplemented with Triton X-100 that enhanced the activity of rSK.
Table 4.
Effect of surfactants on streptokinase activity.
| Detergent | Residual activity (%) at the concentration (%) | |||
|---|---|---|---|---|
| 0.5 | 1.0 | 1.5 | 2.0 | |
| Tween 20 | 137.5 ± 2.6 | 138.3 ± 1.8 | 135.0 ± 2.3 | 136.5 ± 2.6 |
| Tween 80 | 119.1 ± 2.8 | 125.9 ± 2.6 | 142.2 ± 1.5 | 115.4 ± 2.2 |
| Triton X-100 | 150.3 ± 1.9 | 140.6 ± 3.6 | 143.6 ± 0.9 | 126.6 ± 2.0 |
|
| ||||
| Relative activity (%) after incubation for (h) | ||||
| 0 | 8 | 24 | 48 | |
|
| ||||
| Tween 20 | 117.6 ± 1.5 | 149.2 ± 1.3 | 154.7 ± 1.2 | 21.9 ± 0.2 |
| Tween 80 | 121.9 ± 1.9 | 151.9 ± 0.7 | 154.4 ± 1.1 | 27.6 ± 0.2 |
| Triton X-100 | 119.3 ± 1.6 | 162.6 ± 1.3 | 15.4 ± 0.1 | 18.1 ± 0.2 |
3.9. Effect of Metal Ions and EDTA
In the present study, effect of various additives on the purified rSK activity was investigated. The addition of EDTA and metal ions showed a clear effect on the streptokinase activity. EDTA, Mn2+, and K+ inhibited the enzyme partially whereas Ag+, Ca2+, and Co2+ exhibited a strong inhibition. But the metal ions Cu2+, Fe2+, Ni2+, and Zn2+ at a final concentration of 1 mM completely inhibited the streptokinase (Table 5). In previous studies, it was also observed that the addition of Zn2+ and Cu2+ almost completely inhibited the activity of the recombinant staphylokinase from Staphylococcus aureus QT08 [27] and the native staphylokinase from S. aureus V8 [31], another thrombolytic agent, closely related to the streptokinase. Why the streptokinases and staphylokinases shared a common property that addition of Zn2+ and Cu2+ resulted in almost completely inhibition of activities? Because the plasmin completely lost its activity when it was incubated with Zn2+ and Cu2+ [32, 33].
Table 5.
Effect of metal ions on streptokinase activity.
| Additive | Residual activity (%) at the concentration (mM) | ||
|---|---|---|---|
| 1 | 3 | 5 | |
| AgNO2 | 16.1 ± 0.1 | n.d. | n.d. |
| CaCl2 | 14.5 ± 0.2 | n.d. | n.d. |
| CoCl2 | 15.6 ± 0.1 | n.d. | n.d. |
| EDTA | 50.4 ± 0.2 | 70.5 ± 0.3 | 88.2 ± 0.4 |
| KCl | 18.5 ± 0.1 | 25.2 ± 0.2 | 59.6 ± 0.5 |
| MnSO4 | 40.5 ± 0.4 | 54.2 ± 0.3 | 68.4 ± 0.5 |
4. Conclusion
SK is a promising blood-clot dissolving agent for the treatment of patients suffering from a heart attack. It would be desirable to produce high yield of protein with high activity for thrombolytic therapy. In the present study, a sk gene from Streptococcus pyogenes DT7 was overexpressed in E. coli with a level of 60% of total proteins which is highest yield of any rSK expressed in E. coli till date. A simple renaturation system dramatically covered the rSK activity with 41 folds, which was not reported before. Overproduction of rSK in E. coli in combination with a simple and highly effective renaturation made the recombinant E. coli become a potential strain for industrial SK production.
Acknowledgments
This study was supported by the Ministry of Science and Technology with the project KC10.28/06-10 “Production of Recombinant Streptokinase and Tissue Plasminogen Activator Used for Therapy” and Vietnam Academy of Science and Technology (2008).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Vellanki RN, Potumarthi R, Mangamoori LN. Constitutive expression and optimization of nutrients for streptokinase production by Pichia pastoris using statistical methods. Applied Biochemistry and Biotechnology. 2009;158(1):25–40. doi: 10.1007/s12010-008-8315-z. [DOI] [PubMed] [Google Scholar]
- 2.Kunadian V, Gibson CM. Thrombolytics and myocardial infarction. Cardiovascular Therapeutics. 2012;30(2):e81–e88. doi: 10.1111/j.1755-5922.2010.00239.x. [DOI] [PubMed] [Google Scholar]
- 3.Klessen C, Malke H. Expression of the streptokinase gene from Streptococcus equisimilis in Bacillus subtilis . Journal of Basic Microbiology. 1986;26(2):75–81. doi: 10.1002/jobm.3620260203. [DOI] [PubMed] [Google Scholar]
- 4.Malke H, Gerlach D, Kohler W, Ferretti JJ. Expression of a streptokinase gene from Streptococcus equisimilis in Streptococcus sanguis . Molecular and General Genetics. 1984;196(2):360–363. doi: 10.1007/BF00328072. [DOI] [PubMed] [Google Scholar]
- 5.Pimienta E, Ayala JC, Rodríguez C, et al. Recombinant production of Streptococcus equisimilis streptokinase by Streptomyces lividans . Microbial Cell Factories. 2007;6, article 20 doi: 10.1186/1475-2859-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim M-R, Choeng Y-H, Chi W-J, Kang D-K, Hong S-K. Heterologous production of streptokinase in secretory form in Streptomyces lividans and in nonsecretory form in Escherichia coli . Journal of Microbiology and Biotechnology. 2010;20(1):132–137. [PubMed] [Google Scholar]
- 7.Kumar R, Singh J. Expression and secretion of a prokaryotic protein streptokinase without glycosylation and degradation in Schizosaccharomyces pombe . Yeast. 2004;21(16):1343–1358. doi: 10.1002/yea.1184. [DOI] [PubMed] [Google Scholar]
- 8.Pratap J, Dikshit KL. Effect of signal peptide changes on the extracellular processing of streptokinase from Echerichia coli: requirement for secondary structure at the cleavage junction. Molecular and General Genetics. 1998;258(4):326–333. doi: 10.1007/s004380050738. [DOI] [PubMed] [Google Scholar]
- 9.Sriraman K, Jayaraman G. Enhancement of recombinant streptokinase production in Lactococcus lactis by suppression of acid tolerance response. Applied Microbiology and Biotechnology. 2006;72(6):1202–1209. doi: 10.1007/s00253-006-0410-x. [DOI] [PubMed] [Google Scholar]
- 10.Malke H, Ferretti JJ. Streptokinase: cloning, expression, and excretion by Escherichia coli . Proceedings of the National Academy of Sciences of the United States of America. 1984;81(11):3557–3561. doi: 10.1073/pnas.81.11.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherish Babu PV, Srinivas VK, Krishna Mohan V, Krishna E. Renaturation, purification and characterization of streptokinase expressed as inclusion body in recombinant E. coli . Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences. 2008;861(2):218–226. doi: 10.1016/j.jchromb.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Pratap J, Rajamohan G, Dikshit KL. Characteristics of glycosylated streptokinase secreted from Pichia pastoris: enhanced resistance of SK to proteolysis by glycosylation. Applied Microbiology and Biotechnology. 2000;53(4):469–475. doi: 10.1007/s002530051643. [DOI] [PubMed] [Google Scholar]
- 13.Baneyx F. Recombinant protein expression in Escherichia coli . Current Opinion in Biotechnology. 1999;10(5):411–421. doi: 10.1016/s0958-1669(99)00003-8. [DOI] [PubMed] [Google Scholar]
- 14.Balagurunathan B, Jayaraman G. Theoretical and experimental investigation of chaperone effects on soluble recombinant proteins in Escherichia coli: effect of free DnaK level on temperature-induced recombinant streptokinase production. Systems and Synthetic Biology. 2008;2(1-2):27–48. doi: 10.1007/s11693-009-9021-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quyen DT, Dao TT, Nguyen SLT. A novel esterase from Ralstonia sp. M1: gene cloning, sequencing, high-level expression and characterization. Protein Expression and Purification. 2007;51(2):133–140. doi: 10.1016/j.pep.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Analytical Biochemistry. 1976;72(1-2):248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 18.Vu TTH, Quyen DT, Dao TT, Nguyen SLT. Cloning, high-level expression, purification, and properties of a novel endo-β-1,4-mannanase from Bacillus subtilis G1 in Pichia pastoris . Journal of Microbiology and Biotechnology. 2012;22(3):331–338. doi: 10.4014/jmb.1106.06052. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X-W, Sun T, Huang X-N, Liu X, Gu D-X, Tang Z-Q. Recombinant streptokinase production by fed-batch cultivation of Escherichia coli . Enzyme and Microbial Technology. 1999;24(10):647–650. [Google Scholar]
- 20.Pérez N, Urrutia E, Camino J, et al. Hydrophobic interaction chromatography applied to purification of recombinant streptokinase. Minerva Biotecnologica. 1998;10(4):174–177. [Google Scholar]
- 21.Balagurunathan B, Ramchandra NS, Jayaraman G. Enhancement of stability of recombinant streptokinase by intracellular expression and single step purification by hydrophobic interaction chromatography. Biochemical Engineering Journal. 2008;39(1):84–90. [Google Scholar]
- 22.Pupo E, Baghbaderani BA, Lugo V, Fernández J, Páez R, Torréns I. Two streptokinase genes are expressed with different solubility in Escherichia coli W3110. Biotechnology Letters. 1999;21(12):1119–1123. [Google Scholar]
- 23.Rajagopalan S, Gonias SL, Pizzo SV. The temperature-dependent reaction between alpha 2-macroglobulin and streptokinase-plasmin(ogen) complex. The Journal of Biological Chemistry. 1987;262(8):3660–3664. [PubMed] [Google Scholar]
- 24.Mumme A, Kemen M, Homann H-H, Zumtobel V. Temperature-dependent fibrinolysis with streptokinase. Deutsche Medizinische Wochenschrift. 1993;118(44):1594–1596. doi: 10.1055/s-2008-1059489. [DOI] [PubMed] [Google Scholar]
- 25.Dubey R, Kumar J, Agrawala D, Char T, Pusp P. Isolation, production, purification, assay and characterization of fibrinolytic enzymes (Nattokinase, Streptokinase and Urokinase) from bacterial sources. African Journal of Biotechnology. 2011;10(8):1408–1420. [Google Scholar]
- 26.Vesterberg K, Vesterberg O. Studies of staphylokinase. Journal of Medical Microbiology. 1972;5(4):441–450. doi: 10.1099/00222615-5-4-441. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen THT, Quyen DT. Cloning, high-level expression, purification and characterization of a staphylokinase variant SakφC from Staphylococcus aureus QT08 in Escherichia coli BL21. African Journal of Biotechnology. 2012;6:2129–2136. [Google Scholar]
- 28.Nguyen THT, Quyen DT. High-level expression, purification and properties of a fully active even glycosylated staphylokinase variant SakφC from Staphylococcus aureus QT08 in Pichia pastoris . African Journal of Microbiology Research. 2012;11:5995–6003. [Google Scholar]
- 29.Rodriguez P, Fuentes P, Barro M, et al. Structural domains of streptokinase involved in the interaction with plasminogen. European Journal of Biochemistry. 1995;229(1):83–90. doi: 10.1111/j.1432-1033.1995.tb20441.x. [DOI] [PubMed] [Google Scholar]
- 30.Lu H, Soria C, Cramer EM, et al. Temperature dependence of plasmin-induced activation or inhibition of human platelets. Blood. 1991;77(5):996–1005. [PubMed] [Google Scholar]
- 31.Vesterberg K, Vesterberg O. Studies of staphylokinase. Journal of Medical Microbiology. 1972;5(4):441–450. doi: 10.1099/00222615-5-4-441. [DOI] [PubMed] [Google Scholar]
- 32.Sokolovskaya LI, Slominskii AY, Volkov GL. Induction of catalytic activity of plasminogen by monoclonal antibody IV-Ic in the presence of divalent metal cations and α2-antiplasmin. Biochemistry. 2006;71(6):627–633. doi: 10.1134/s000629790606006x. [DOI] [PubMed] [Google Scholar]
- 33.Nowak P, Zgirski A. Effects of metal ions on activity of plasmin. Biological Trace Element Research. 2003;93(1-3):87–94. doi: 10.1385/BTER:93:1-3:87. [DOI] [PubMed] [Google Scholar]