Abstract
The Hedgehog (Hh) pathway depends on primary cilia in vertebrates, but the signaling machinery within cilia remains incompletely defined. We report the identification of a complex between two ciliary proteins, EFCAB7 and IQCE, which positively regulates the Hh pathway. The EFCAB7-IQCE module anchors the EVC-EVC2 complex in a signaling microdomain at the base of cilia. EVC and EVC2 genes are mutated in Ellis van Creveld and Weyers syndromes, characterized by impaired Hh signaling in skeletal, cardiac and orofacial tissues. EFCAB7 binds to a C-terminal disordered region in EVC2 that is deleted in Weyers patients. EFCAB7 depletion mimics the Weyers cellular phenotype— the mis-localization of EVC-EVC2 within cilia and impaired activation of the transcription factor GLI2. Evolutionary analysis suggests that emergence of these complexes might have been important for adaptation of an ancient organelle, the cilium, for an animal-specific signaling network.
Introduction
The Hedgehog (Hh) signaling pathway is orchestrated at primary cilia in vertebrates (Huangfu et al., 2003). Cell biological studies have highlighted the importance of both ciliary compartmentalization and trafficking in regulating Hh signal propagation (Corbit et al., 2005; Haycraft et al., 2005; Rohatgi et al., 2007). In the absence of Hh ligands, the Hh receptor Patched 1 (PTCH1), which suppresses signaling when not bound to its ligand, is localized in and around cilia. Genetic elimination of PTCH1 or its inactivation by Hh ligands results in accumulation of the 7-pass transmembrane (TM) protein Smoothened (SMO) to high levels in the ciliary membrane. SMO activity at cilia promotes transport of GLI and SUFU to the tip of the cilium, allowing the GLI transcription factors to dissociate from SUFU and enter the nucleus to transcribe target genes (Humke et al., 2010; Tukachinsky et al., 2010).
An open question is how SMO (and other 7-pass TM receptors) signal from the ciliary membrane. EVC and EVC2, two homologous Type I single-pass TM proteins that form a complex, have been identified as tissue-specific regulators of Hh signaling. These proteins bind to SMO after it accumulates in cilia in response to Hh ligands (Caparros-Martin et al., 2013; Dorn et al., 2012; Yang et al., 2012). Mutations in the EVC or EVC2 genes cause Ellis van Creveld (EvC) syndrome, characterized by impaired Hh signaling in cardiac, skeletal and orofacial tissues during development (Blair et al., 2011; Galdzicka et al., 2002; Ruiz-Perez et al., 2007; Ruiz-Perez and Goodship, 2009; Ruiz-Perez et al., 2000; Ruiz-Perez et al., 2003). Localization of these proteins to the EvC zone, a distinct compartment at the base of primary cilia, is critical for their function in Hh signaling. The importance of this precise compartmentalization was demonstrated by the analysis of a dominant EVC2 allele identified in patients with Weyers Acrofacial Dysostosis (Weyers), a skeletal ciliopathy characterized by phenotypes similar to that of EvC syndrome (Weyers, 1952). The Weyers EVC2 allele encodes a truncated protein that lacks the C-terminal 43 amino acids (a.a.) and is distributed along the entire ciliary membrane rather than being restricted to the EvC zone (Caparros-Martin et al., 2013; Dorn et al., 2012; Valencia et al., 2009; Ye et al., 2006). This mutant protein (hereafter called EVC2ΔW) is a dominant inhibitor of Hh signaling, explaining the dominant mode of inheritance seen in Weyers families (Valencia et al., 2009). These observations suggested that a SMO signaling complex assembles at the EvC zone in cilia.
We have isolated a protein complex that restricts EVC and EVC2 at the base of cilia and consequently promotes Hh signaling. In the absence of the complex, EVC and EVC2 are instead dispersed throughout the ciliary membrane. While SMO still accumulates in cilia in response to Hh ligands, it fails to transmit the signal downstream to activate GLI2. Interestingly, SMO remains competent to regulate repressor forms of GLI3 (GLI3R), suggesting an unexpected bifurcation in signaling downstream of SMO. These data suggest that signaling by ciliary receptors may be organized by scaffolds that assemble in specific ciliary compartments.
Results
EFCAB7 and IQCE are EVC2-interacting proteins
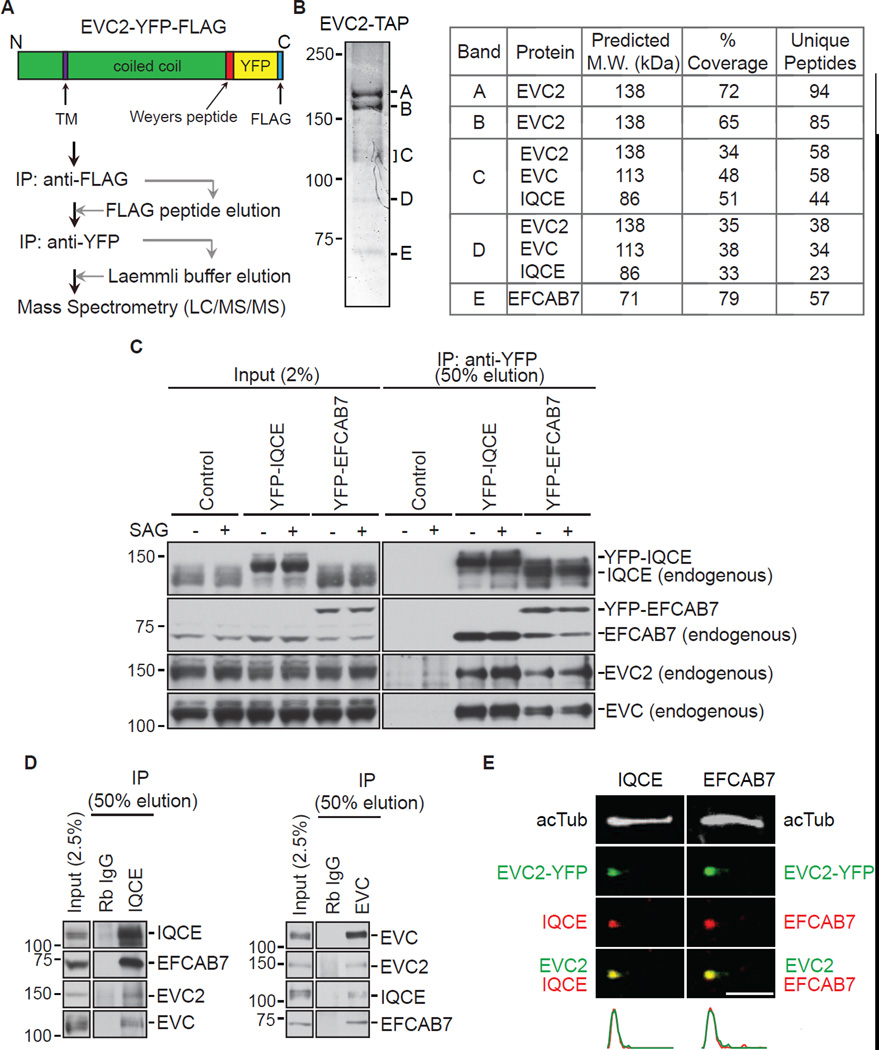
We used tandem affinity purification (TAP) followed by mass spectrometry to identify EVC2-interacting proteins from NIH/3T3 cells stably expressing EVC2 fused to a dual Yellow Fluorescent Protein (YFP)-FLAG tag (EVC2-YFP-FLAG; Figure 1A). In addition to EVC, previously known to form a complex with EVC2, two other proteins co-purified with the EVC2 bait: IQ-domain containing protein E (IQCE; NP_083109) and EF-hand calcium-binding domain-containing protein 7 (EFCAB7; NP_663524.1) (Figure 1B). While the predicted molecular weight of IQCE is 86 kDa, both endogenous IQCE (Figures 1C and 1D) and an epitope-tagged version of IQCE (Figure 2C) consistently fractionated anomalously above the 100 kDa marker on SDS-PAGE gels. IQCE and EFCAB7 had been previously detected in cilia proteomic surveys (Ishikawa et al., 2012; Ostrowski et al., 2002).
Figure 1. Identification of EVC2 binding proteins.
(A) EVC2 binding partners were identified with a tandem affinity approach using NIH/3T3 cells stably expressing EVC2-YFP-FLAG. (B) A Coomassie-stained 8% SDS-PAGE gel (left) showing the five protein bands isolated after the procedure shown in (A). The proteins contained in each band were identified by mass spectrometry, as summarized in the table on the right. (C) Immunoblots showing endogenous EVC2 and EVC isolated on anti-YFP beads incubated with extracts of NIH/3T3 cells stably expressing YFP-IQCE, YFP-EFCAB7, or neither protein (control). Cells were treated with SAG (24 h) to activate Hh signaling or left untreated. (D) The four endogenous components of the EvC complex from NIH/3T3 extracts could be isolated on anti-IQCE beads (left) or anti-EVC beads (right). IPs using beads coated with Rabbit IgG (Rb IgG) served as a control. (E) Endogenous IQCE and EFCAB7 (red) co-localize with EVC2-YFP (green; anti-YFP) at the base of primary cilia labeled with the axonemal marker acetylated tubulin (white; acTub). Plots below the images indicate normalized fluorescent intensities for each of the channels along the cilium from base (left) to tip (right). Scale bars: 2 µm. See also Figure S1.
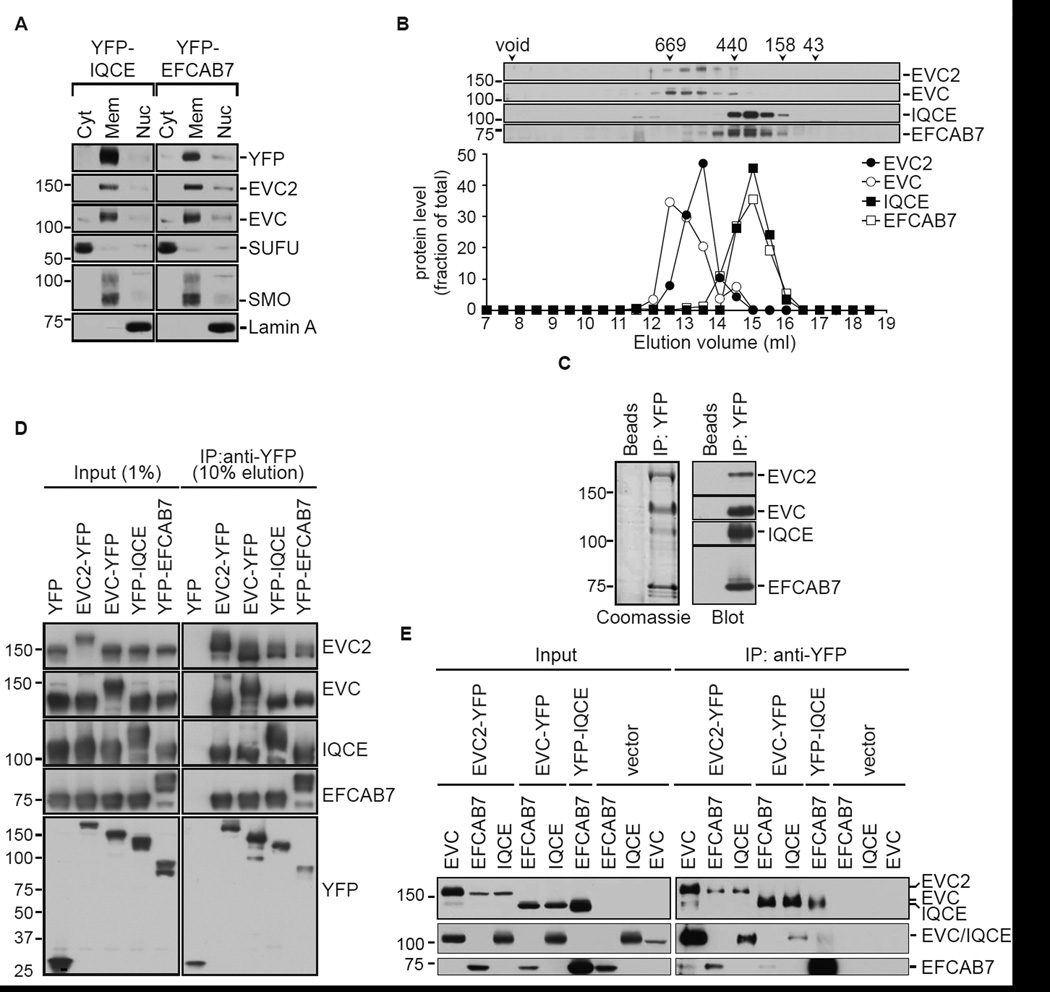
Figure 2. Architecture of the EvC complex.
(A) Subcellular distribution of EvC complex components in NIH/3T3 cells stably expressing YFP-IQCE or YFP-EFCAB7. SUFU, SMO and Lamin A served as markers for the cytoplasmic (Cyt), membrane (Mem) and nuclear (Nuc) fractions, respectively. (B) Fractionation of endogenous EvC complex components on a Superose 6 gel-filtration column. The immunoblot (top) and graph (below) depict the amount protein found in each of the column fractions. The numbers above the immunoblots denote the molecular weights (in kDa) and peak elution positions of a set of standard proteins. (C) A Coomasie-stained gel (left) and immunoblot (right) shows isolation of an intact EvC complex on anti-YFP beads but not on control beads from extracts of 293T cells simultaneously transfected with constructs encoding YFP-tagged EVC2 along with EVC, IQCE and EFCAB7. (D) All four EvC complex subunits could be isolated from transfected 293T cells (see C) regardless of which subunit was used as the YFP-tagged bait. Above each lane, the subunit used as the YFP-tagged bait is denoted; the remaining three subunits were tagged with HA or FLAG. (E) Binary interactions between the four different subunits of the EvC complex were tested after transfecting the proteins in sets of two into 293T cells with YFP-tagged bait proteins and HA-tagged prey proteins. See also Figure S1.
These results suggested that EVC, EVC2, EFCAB7 and IQCE formed a protein complex (hereafter called the EvC complex) in cells. Immunoprecipitation (IP) of YFP-tagged EFCAB7 and IQCE from NIH/3T3 cells stably expressing each protein on anti-YFP beads also captured endogenous versions of the three other proteins in the EvC complex (Figure 1C). These interactions were not altered when Hh signaling was stimulated by SAG, a direct SMO agonist. Using antibodies to isolate IQCE and EVC, we also detected the presence of a completely endogenous EvC complex (Figure 1D). Importantly, both endogenous and YFP-tagged EFCAB7 and IQCE co-localized with EVC2 at the base of cilia in the EvC zone (Figures 1E, S1A and S1B). The levels of EFCAB7 and IQCE at the EvC zone did not change after activation of Hh signaling (Figure S1C). The subcellular localization and biochemical interactions of EFCAB7 and IQCE suggested that they were likely to regulate the EVC-EVC2 complex and consequently Hh signal transduction.
The architecture of the EvC complex
While EVC and EVC2 are single pass, Type I TM proteins with extracellular N-termini and intracellular C-termini, EFCAB7 and IQCE are not predicted to have any TM segments. However, sub-cellular fractionation revealed that both proteins were membrane associated to same degree as the integral membrane proteins EVC, EVC2 and SMO (Figure 2A). To investigate the stability of the endogenous EvC complex, detergent extracts of membranes isolated from NIH/3T3 cells were subjected to gel filtration chromatography (Figure 2B) and glycerol gradient sedimentation (Figure S1D). By both hydrodynamic methods, the complex did not fractionate as a stable, unitary species but rather as two distinct sub-complexes, EVC-EVC2 and EFCAB7-IQCE.
To dissect the pattern of interactions between the components of the EvC complex, we reconstituted its formation in HEK 293T cells. Co-expression of YFP-EVC2 with EVC, IQCE, and EFCAB7 in 293T cells allowed us to isolate the entire tetrameric complex on anti-YFP beads, as visualized by Coomasie staining or immunoblotting (Figure 2C). We repeated this isolation in four combinations by moving the YFP tag iteratively to each of the four subunits of the complex (Figure 2D). Regardless of which subunit was isolated on anti-YFP beads, the remaining three subunits were co-precipitated. This data is most consistent with the model that EFCAB7, IQCE, EVC and EVC2 form a complex when co-expressed in cells.
Using the 293T reconstitution system, we tested all possible binary interactions between each of the four subunits of the EvC complex (Figure 2E), allowing us to construct a provisional map of the interactions between the components. EVC2 appeared to play the most central role in complex assembly, interacting with EVC, EFCAB7 and IQCE. Interestingly, EVC2-EVC and EFCAB7-IQCE expression levels were clearly correlated, being significantly higher when co-expressed with the interaction partner within each sub-complex (Figure 2E, left input panel). This pattern of co-regulation was consistent with the hydrodynamic characterization of the EvC complex (Figure 2B).
Taken together, these data suggest that the EvC complex is built from two stable sub-complexes, EVC-EVC2 and EFCAB7-IQCE, which in turn associate through EVC2-mediated interactions.
The Weyers peptide in EVC2 links the EVC-EVC2 and EFCAB7-IQCE subcomplexes
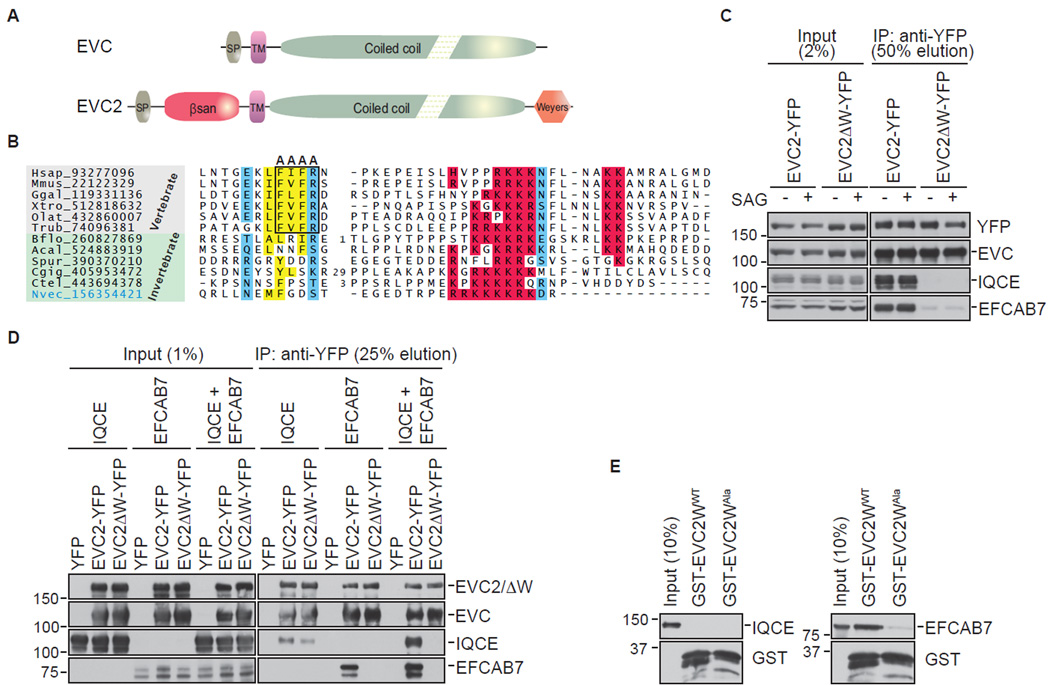
EVC and EVC2 are paralogous proteins that share a single TM region followed by a long cytoplasmic region that is predicted to adopt a coiled-coil conformation (Figure 3A) (Blair et al., 2011). Whereas EVC has a very short extracellular segment, EVC2 has a globular extracellular domain that is likely to adopt a β-sandwich fold that may contact ligands in the extracellular space (see supplemental experimental procedures). In the intracellular region, beyond the shared coiled-coil region, EVC2 has a distinct C-terminal disordered region, which in vertebrates contains a well-conserved solitary hydrophobic tetrapeptide motif followed by an arginine (“IFVFR” in the mouse) (Figure 3B). Strikingly, this disordered region (hereafter called the W-peptide) is the same region that is deleted in a truncated EVC2 protein (EVC2ΔW) encoded by the EVC2 allele found in families with Weyers syndrome. When this 43 a.a. W-peptide is deleted, as in Weyers patients, or the tetrapeptide motif contained within this peptide is mutated, EVC2 becomes a dominant inhibitor of Hh signaling and fails to be restricted to the EvC zone, instead being distributed along the length of the ciliary membrane (Caparros-Martin et al., 2013; Dorn et al., 2012). Both sequence conservation and mutational analysis of this disordered W-peptide in EVC2 points to its importance in mediating a protein interaction that is important for EvC zone localization and Hh signaling in vertebrates.
Figure 3. The Weyers peptide of EVC2 links the EVC-EVC2 and EFCAB7-IQCE sub-complexes.
(A) Domain composition of EVC and EVC2. Abbreviations: SP, signal peptide; TM, TM segment; βsan, β-sandwich domain; Weyers, Weyers peptide. (B) Multiple sequence alignment of the Weyers peptide in EVC2 with basic residues highlighted in red, hydrophobic residues in yellow, and polar residues in blue. The alanine mutations that lead to the delocalization and dominant-negative function of EVC2 are boxed. (C) Immunoblots showing the amount of endogenous EVC, IQCE, and EFCAB7 isolated on anti-YFP beads from NIH/3T3 cells stably expressing EVC2-YFP or EVC2ΔW-YFP and treated ± SAG (24 h). (D) Interaction between the EVC-EVC2 or the EVC-EVC2ΔW complex and IQCE alone, EFCAB7 alone or both IQCE and EFCAB7 together was assessed after transient transfections in 293T cells with YFP-tagged bait proteins and HA-tagged prey proteins; all transfections also included EVC. (E) Binding of in vitro translated, full-length IQCE or EFCAB7 to GST fused to the C-terminal 88 amino acids of EVC2 (GST-EVC2WWT) or a mutant fragment bearing alanine mutations in four conserved amino acids within the Weyers peptide (GST-EVC2WAla) shown in (B).
To investigate a role for the W-peptide in assembly of the EvC complex, we isolated EVC2-YFP and EVC2ΔW-YFP from stable cell lines (Dorn et al., 2012) expressing each of these proteins and tested their association with endogenous EVC, IQCE and EFCAB7 (Figure 3C). EVC bound equally well to both EVC2 and EVC2ΔW; however, EVC2ΔW failed to interact with either EFCAB7 or IQCE. Experiments in the 293T reconstitution system confirmed that the W-peptide was required for the interaction between the EVC-EVC2 and IQCE-EFCAB7 sub-complexes (Figure 3D). Both EFCAB7 alone and the EFCAB7-IQCE complex interacted with EVC2-EVC, but not with EVC2ΔW-EVC.
To determine if the W-peptide was sufficient to bind to EFCAB7 or IQCE, we fused the C-terminal 88 amino acids of EVC2 to Glutathione S-Transferase (GST-EVC2WWT). As a stringent control, a variant GST-EVC2W protein (GST-EVC2WAla) carrying alanine mutations (FVFR→AAAA) in the conserved tetrapeptide motif was also prepared (boxed in Figure 3B). Both proteins were expressed as soluble proteins in bacteria, purified, immobilized on glutathione beads and tested for binding to in vitro translated full-length IQCE or EFCAB7. Neither GST-EVC2WWT nor GST-EVC2WAla could bind to IQCE; however, EFCAB7 bound selectively to GST-EVC2WWT (Figure 3E).
The interaction between the W-peptide, including the tetrapeptide motif within this domain, and EFCAB7 likely plays an important role in regulating the Hh signaling function and EvC zone localization of EVC2. A deletion of the W-peptide, as seen in patients with Weyers syndrome, would be predicted to compromise the assembly of the EvC complex.
Mapping the interactions between EVC2, EFCAB7 and IQCE
Given that EFCAB7 and IQCE were previously unstudied proteins, we undertook a detailed sequence analysis to identify both recognizable and novel domains (Figures 4A, 4B, S2 and S3).
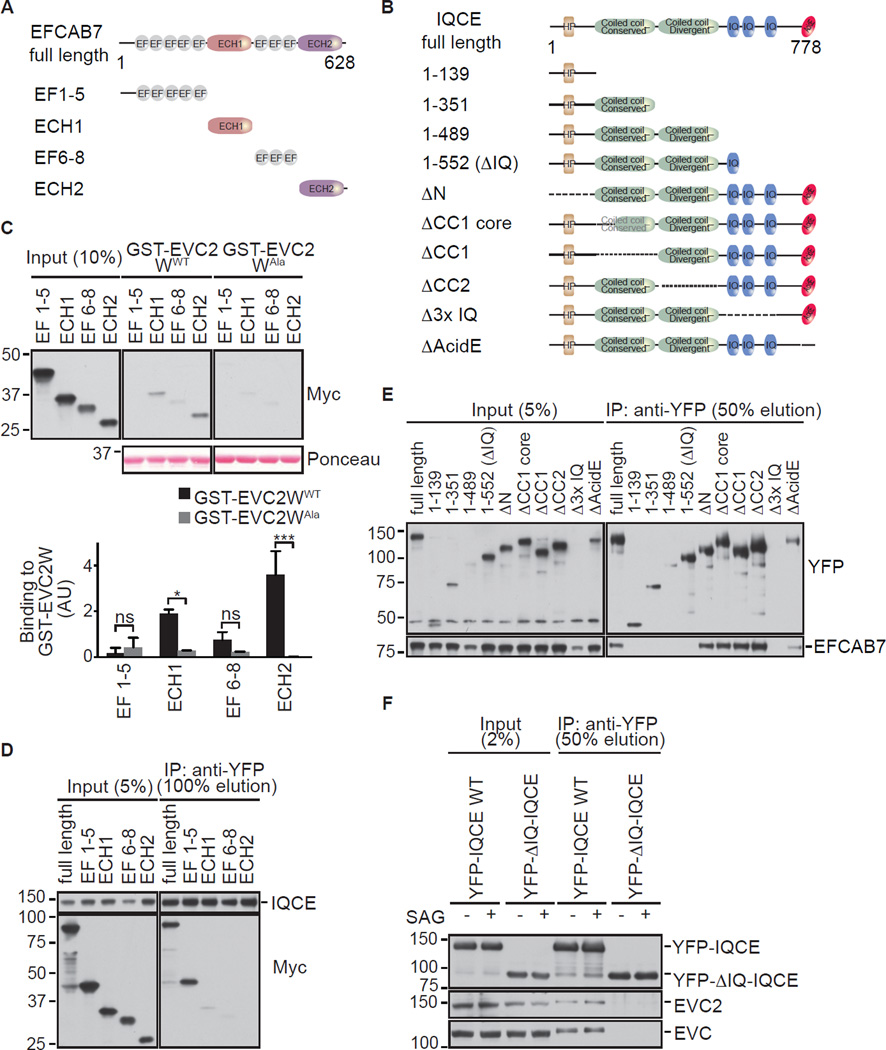
Figure 4. Mapping the interaction between EFCAB7 and IQCE.
(A) and (B) Domain compositions of EFCAB7, IQCE and the fragments of each protein used for interaction studies. Abbreviations: EF, EF-hand domain; ECH, EFCAB7-Calpain Homology domain; HP, hydrophobic peptide; IQ, IQ calmodulin-binding motif; AcidE, acidic peptide. See sequence alignments in Figures S2 and S3. (C) An Immunoblot and graph (mean ± SD, n=2, one-way ANOVA with *=p<0.05 and ***=p<0.001) showing the amount of in vitro translated EFCAB7 fragments captured by the wild-type (WT) GST-Weyers peptide fusion or the Alanine (Ala) mutant (see Figure 3E). (D) Binding of Myc-tagged EFCAB7 fragments to YFP-tagged, full-length IQCE assessed by co-in vitro translation of both proteins followed by an anti-YFP IP. (E) Binding of YFP-tagged IQCE fragments to Myc-tagged, full-length EFCAB7 was assessed as in (D) with an anti-YFP IP. (F) Immunoblots showing the amount of endogenous EVC and EVC2 that co-precipitated with YFP-tagged full-length IQCE or a deletion mutant (1–522 or ΔIQ) from NIH/3T3 cells stably expressing each protein and treated ± SAG (24 h).
EFCAB7 was annotated as a protein containing EF-hand domains. Using sensitive sequence profiles with the RPS-BLAST program and profile-profile comparisons with the HHpred program, we established that EFCAB7 possesses at least 8 EF-hand domains arranged in two distinct runs: an N-terminal set of 5 EF-Hands (EF1–5), followed by an uncharacterized globular domain, followed by a second run of at least 3 EF-Hands (EF6–8) followed by another globular domain (Figure 4A). At least two EF-hand domains in the second set (EF6–8) preserved conserved acidic residues that are required for binding to calcium. Profile searches using the PSI-BLAST program demonstrated that the two uncharacterized globular domains in EFCAB7 were related to conserved β-sandwich domains found in calpains, prompting us to name them EFCAB7-calpain homology 1 and 2 (ECH1 and ECH2) (Figures 4A and S2).
Likewise we systematically analyzed the structure of the IQCE protein: its core contains a coiled coil region, the N-terminal part of which is strongly conserved whereas the C-terminal part is rapidly evolving (Figure 4B and Figure S3). This coiled coil region is followed by 3 tandem α-helical IQ motifs that are often determinants for interaction with EF-hand domains, suggesting a mechanism for the interaction between IQCE and EFCAB7. The coiled-coiled core is flanked on the N- and C- terminal sides by disordered regions. Embedded within the N-terminal disordered segment is a conserved hydrophobic segment (HP) predicted to adopt an extended conformation. The C-terminal disordered region contains a stretch enriched in acidic residues and serines (AcidE). These N- and C- terminal conserved motifs in IQCE may mediate interactions of IQCE with other binding partners.
To map EFCAB7 interactions with both the W-peptide (Figure 4C) and IQCE (Figure 4D), we split the protein into 4 domains, EF1–5, ECH1, EF6–8, and ECH2, guided by the sequence analysis described above (Figure 4A). When these individual domains were produced by in vitro translation (IVT) and tested for their interaction with GST-EVC2W immobilized on beads, the ECH2 domain showed the strongest and most specific interaction (Figure 4C). On the other hand, EF1–5, the first cluster of EF hands, was the only fragment that bound to full-length IQCE (Figure 4D). Finally, to map the interaction domain in IQCE, a series of truncation mutants of IQCE (Figure 4E) were tested for their ability to bind to full-length EFCAB7. Full-length IQCE, along with a series of truncation mutants that carry deletions of the N-terminal and coiled-coil domains of IQCE, bound to EFCAB7. However, C-terminal truncation mutants that deleted the IQ motifs generally abrogated binding. The smallest truncation mutation that was expressed but failed to bind EFCAB7 (a.a. 1–552; hereafter ΔIQ-IQCE) still retained IQ1, suggesting that IQ2 and IQ3 are most important for the interaction with EFCAB7.
Taken together, the interaction mapping experiments suggested that EFCAB7 functioned as an adaptor to link the EVC-EVC2 complex to IQCE, using ECH2 to engage the W-peptide in EVC2 and EF1–5 to bind to IQ domains in IQCE. Consistent with this model, ΔIQ-IQCE, a truncation mutant of IQCE that cannot bind to EFCAB7 (1–552 truncation in Figure 4E), failed to interact with the EVC-EVC2 complex when stably expressed in cells (Figure 4F).
EFCAB7 and IQCE are positive regulators of Hh signaling
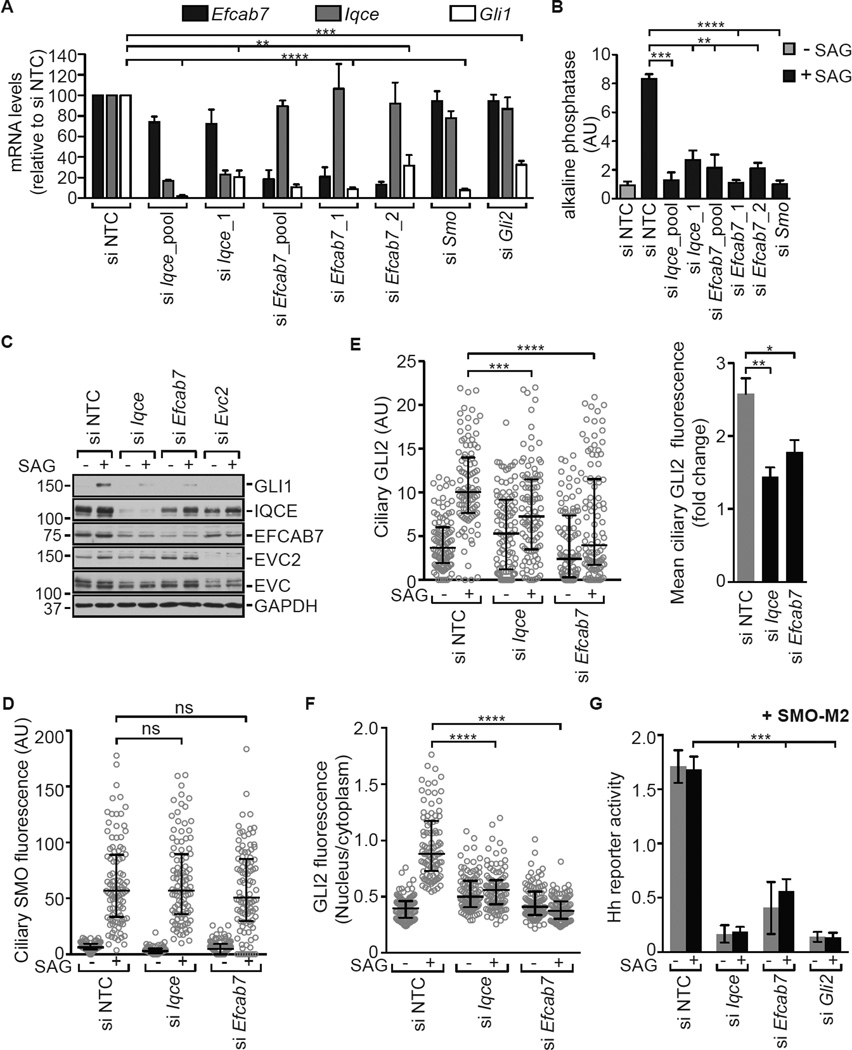
EVC2 and EVC are tissue-specific positive regulators of Hh signaling. Since the EVC-EVC2 complex plays a role in skeletal development, we depleted IQCE and EFCAB7 using siRNA in C3H10T1/2 cells, mesenchymal cells that differentiate into osteoblasts when exposed to Hh ligands (Kinto et al., 1997). Depletion of either EFCAB7 or IQCE inhibited both the induction of Gli1, a direct Hh target gene, and the induction of alkaline phosphatase, a marker of osteoblast differentiation (Figures 5A and 5B). This same block in Hh signaling was also seen when EFCAB7 and IQCE were depleted in a different cell type, NIH/3T3 cells, commonly used to study Hh signal transduction (Figures 5C and S4A). Importantly, depletion of either protein did not significantly alter the frequency of ciliation (Figure S4B). Two distinct siRNAs against EFCAB7 produced the same phenotype (Figures 5A and 5B), and the effect seen with a single IQCE siRNA could be rescued with a siRNA-resistant IQCE transcript (Figure S4C).
Figure 5. EFCAB7 and IQCE are positive regulators of the Hh signaling pathway.
(A) Quantitative reverse-transcription PCR (qRT-PCR) was used to measure levels (mean ± SD, n=3, unpaired student’s t-test) of Iqce, Efcab7, and Gli1 transcripts in C3H10T1/2 cells transfected with a non-targeting control (NTC) siRNA or siRNAs against the indicated transcripts. All cells were treated with SAG (36 h) and Gli1 served as a metric for signaling. (B) The SAG-mediated differentiation (36 h) of C3H10T1/2 cells into osteoblasts was assayed by the increase in alkaline phosphatase activity (mean ± SD, n=3, unpaired student’s t-test) after siRNA-mediated depletion of IQCE, EFCAB7, or SMO. (C) Immunoblots showing the effect of IQCE, EFCAB7, and EVC2 depletion on GLI1 induction in NIH/3T3 cells after treatment with SAG (24 h). (D, E, F) Accumulation of endogenous SMO in cilia (D), GLI2 at the cilia tip (E), and GLI2 in the nucleus of NIH/3T3 cells (n=100) treated (4–6 h) with SAG. Each point represents a measurement from a single cell, black brackets depict median fluorescence and interquartile ranges, and the Kruskal-Wallis test is used to test significance. In (E), the right panel shows the fold-change in mean (± SD, n=4, Mann-Whitney test) GLI2 fluorescence at the cilia tip after treatment with SAG. (G) Activation of a Hh reporter (mean ± SD, n=3, unpaired student’s t-test) by the SMO-M2 mutant was assessed in NIH/3T3 cells after depletion of IQCE, EFCAB7 or GLI2. In all panels, statistical significance is depicted as follows: p<0.05 (*), p<0.01 (**), p<0.001 (***), p<0.0001 (****) and p>0.05 (not significant-ns). See also Figure S4.
When EFCAB7 and IQCE were depleted, SMO accumulated in cilia normally in response to SAG (Figure 5D), but there was a defect in the activation of GLI2, the major Hh transcriptional activator. Both GLI2 accumulation at the tips of primary cilia and GLI2 translocation into the nucleus were significantly reduced (Figures 5E and 5F). This suggested that the EFCAB7-IQCE complex functioned at step between SMO and the GLI proteins. Consistent with this notion, depletion of both IQCE and EFCAB7 significant impaired signaling by SMO-M2, a constitutively active, oncogenic mutant of SMO (Figure 5G). EVC and EVC2 have also been implicated in regulating the SMO→GLI step in Hh signaling (Dorn et al., 2012).
Genetic analysis of the EvC complex
We used CRISPR/Cas9-based genome editing to investigate the consequences of completely eliminating the function of each of the two modules that comprise the EvC complex in NIH/3T3 cells (Cong et al., 2013). We first disrupted both alleles of the Efcab7 gene, since biochemical studies (Figure 4) had suggested that EFCAB7 was the critical link between IQCE and the EVC-EVC2 complex. Efcab7−/− cells were poorly responsive to signaling induced by the direct SMO agonist SAG (Figure 6A) or the native ligand Shh (Figure S4D). Efcab7−/− cells had no defects in the length of cilia (Figure S4E), the frequency of ciliation (Figure S4F) or in the localization of cilia markers (Figure S4G). In Efcab7−/− cells treated with SAG, SMO accumulated in cilia (Figure S4H), but GLI2 showed a defect in nuclear translocation (Figures S4I and S4J).
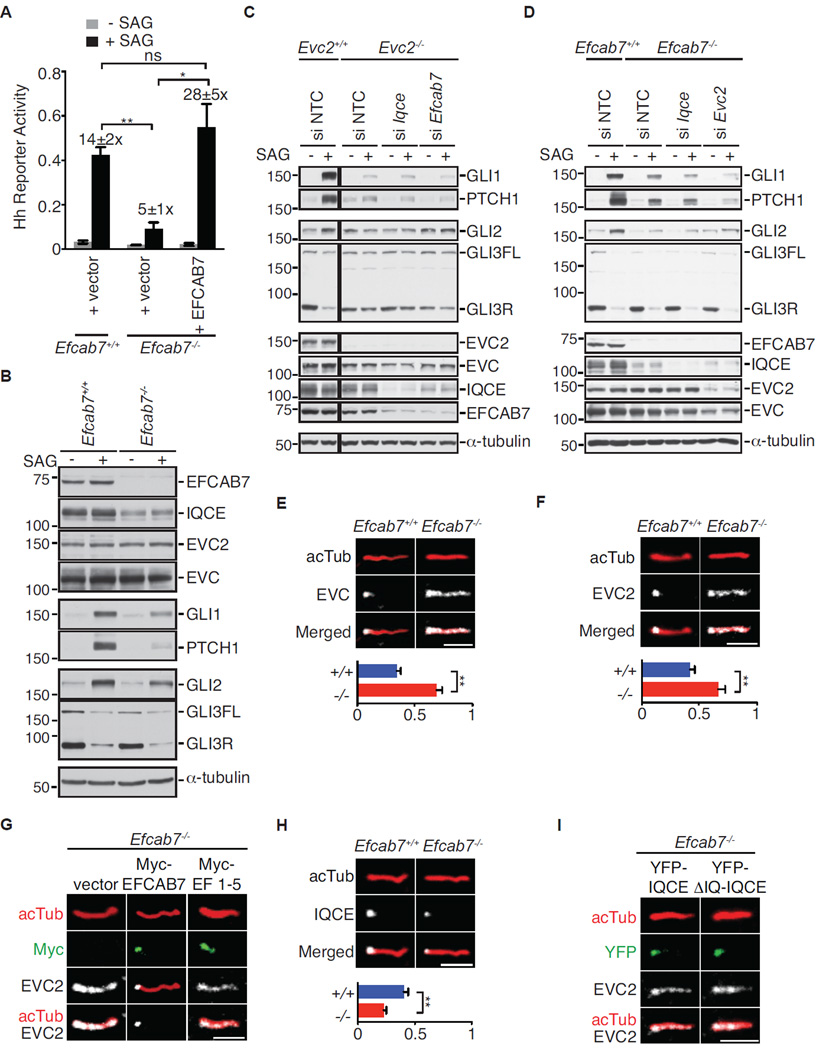
Figure 6. EFCAB7 tethers the EVC-EVC2 complex at base of primary cilia.
(A) SAG-induced (24 h) Hh reporter activity in wild-type NIH/3T3 cells, Efcab7−/− cells, and Efcab7−/− cells rescued with transient expression of YFP-EFCAB7. Numbers above the bars show the fold-induction in reporter activity seen after SAG treatment. Mean ± S.E.M. (n=3) reporter activity is depicted with significance tested using an unpaired student’s t-test; p<0.05 (*) and p<0.01 (**). (B) Immunoblots of extracts from Efcab7+/+ and Efcab7−/− cells treated ± SAG (24 h) were used to assess levels of the indicated EvC complex and Hh pathway components. α-tubulin served as the loading control, GLI1 and PTCH1 are Hh target genes, and GLI3FL and GLI3R point to the full-length GLI3 and its repressor fragment, respectively. (C, D) Immunoblots of extracts from Evc2−/− (C) and Efcab7−/− (D) cells further depleted of the indicated EvC complex components were used to assess levels of various proteins as in (B). (E, F) Localization of endogenous EVC and EVC2 (both in white) within individual cilia, identified by axonemal anti-acetylated tubulin staining (acTub, red). The graphs below show the fraction of acTub staining that co-localizes with EVC or EVC2 using a Mander’s coefficient. The mean (±SD, Mann-Whitney test with **=p<0.01) co-localization fraction was determined from 5 independent images, each containing ~50 cells each. (G) Localization of endogenous EVC2 in Efcab7−/− cells transfected with Myc-tagged full-length EFCAB7 or an EFCAB7 fragment (EF1–5) that cannot bind to EVC2 (see Figure 4C). (H) Localization of endogenous IQCE at the cilium in cells with or without EFCAB7; co-localization measurement below is depicted as in (E). (I) Localization of transiently transfected YFP-tagged, full-length IQCE or its ΔIQ truncation mutant (1–552 a.a; Figure 4B) in Efcab7−/− cells. Scale bars are 2 µm in all images. See also Figures S4 and S5.
Overall, the results obtained using CRISPR/Cas9-mediated gene disruption were consistent with those obtained using siRNA-mediated protein depletion. However, an important difference between these two techniques is that only the former allows complete elimination of gene function. Efcab7−/− cells had undetectable amounts of EFCAB7 and severely reduced amounts of IQCE (Figure 6B), consistent with our previous observations that levels of these two complexed proteins are tightly co-regulated in cells (Figures 1C, 2E, and 5C). In contrast, EVC and EVC2 levels were unaltered. Efcab7−/− cells remained capable of transducing low-level Hh signals, as shown by the slight induction of both a Hh reporter gene (Figure 6A) and the endogenous target genes Ptch1 and Gli1 (Figure 6B). To understand the basis of this residual signaling, we examined two Hh transcription factors-- GLI2, the major activator, and GLI3, the major repressor. When SMO is activated, GLI3R production is blocked and GLI2 is converted into a transcriptional activator that enters the nucleus to activate target genes. Two markers of GLI2 activation, nuclear translocation and stabilization (Humke et al., 2010; Pan et al., 2006), were both substantially decreased in Efcab7−/− cells (Figures 6B, S4I and S4J). However, GLI3R regulation was unaffected− GLI3R was produced in the basal state and extinguished normally in response to SAG (Figure 6B). The Hh signaling phenotypes seen in Efcab7−/− cells were similar to those observed in Iqce−/− cells independently generated using CRISPR/Cas9 (Figure S5A). Thus, the requirement for EFCAB7 and IQCE seems to be selective for GLI2 over GLI3R regulation.
The discordance between GLI2 and GLI3 regulation was unexpected since previous work had suggested that EVC and EVC2 regulated both transcription factors (Caparros-Martin et al., 2013; Dorn et al., 2012). Therefore, we knocked-out Evc2 in the same NIH/3T3 cell line used to disrupt Efcab7 and Iqce (Figures 6C and S5B). As reported previously, loss of EVC2 blocked the SAG-induced activation of Gli1 and Ptch1 (Figure 6C and S5B). However, unlike Efcab7−/− cells, there was a clear defect in the ability of Hh signaling to extinguish GLI3R levels.
To clarify the relationship between the EFCAB7-IQCE and EVC-EVC2 sub-complexes, we used siRNA to deplete IQCE and EFCAB7 in Evc2−/− cells (Figure 6C) or IQCE and EVC2 in Efcab7−/− cells (Figure 6D)— analogous to a double mutant analysis. Hh target gene induction in Evc2−/− cells was not further impaired by the depletion of EFCAB7 or IQCE (Figure 6C). However, the depletion of EVC2 (but not IQCE) in Efcab7−/− cells further reduced the Hh-stimulated induction of Gli1 or Ptch1 (Figure 6D). SAG-induced reduction in GLI3R was not affected, likely because the depletion of EVC2 by siRNA was incomplete.
Thus, EFCAB7-IQCE and EVC-EVC2 sub-complexes are not redundant. The former selectively regulates GLI activator functions, while the latter regulates both GLI activator and repressor functions.
An assembly pathway for the EvC complex at the base of primary cilia
The derivation of an Efcab7−/− cell line allowed us to address an important question-- which component of the EvC complex specifies localization in the EvC zone at the cilia base? In Efcab7−/− cells, as predicted from our biochemical mapping experiments, EVC and EVC2 were not restricted to the EvC zone but rather were distributed along the entire ciliary membrane (Figures 6E and 6F). This altered localization is reminiscent of the phenotype seen in cells expressing the Weyers protein EVC2ΔW (Caparros-Martin et al., 2013; Dorn et al., 2012), which lacks the W-peptide and hence cannot bind to EFCAB7. Reintroduction of full-length EFCAB7 restored the EvC zone localization of EVC2 (Figures 6G and S5C) and EVC (Figure S5D). The EF1–5 fragment of EFCAB7, which can bind to IQCE (Figure 4D) but lacks the ECH2 domain that interacts with EVC2, was itself correctly localized in the EvC zone but failed to restrict EVC2 to the base of cilia (Figures 6G and S5C).
In contrast to EVC and EVC2, the localization of endogenous IQCE at the cilia base was unaffected in Efcab7−/− cells, though levels of IQCE at cilia were lower as expected from the immunoblot in Figure 6B (Figure 6H). Overexpressed YFP-IQCE also localized appropriately to the base of cilia in Efcab7−/− cells (Figure 6I), as did ΔIQ-IQCE, a truncation mutant IQCE that cannot bind to EFCAB7, EVC or EVC2. Moreover, the localization of both IQCE and EFCAB7 at the EvC Zone was unaltered in Evc2−/− cells, which also lacked EVC at cilia (Figure S5E). Taken together, these data are consistent with the view that EvC zone localization of this complex is driven by the EFCAB7-IQCE complex. The ΔIQ-IQCE fragment of IQCE contains the primary targeting signal for the EvC zone. While EVC and EVC2 can accumulate in cilia independently of EFCAB7, they require EFCAB7 to be restricted to the EvC zone. This provides an illustration of how distinct, separable mechanisms can regulate ciliary trafficking and compartmentalization.
Evolutionary history of the EvC complex
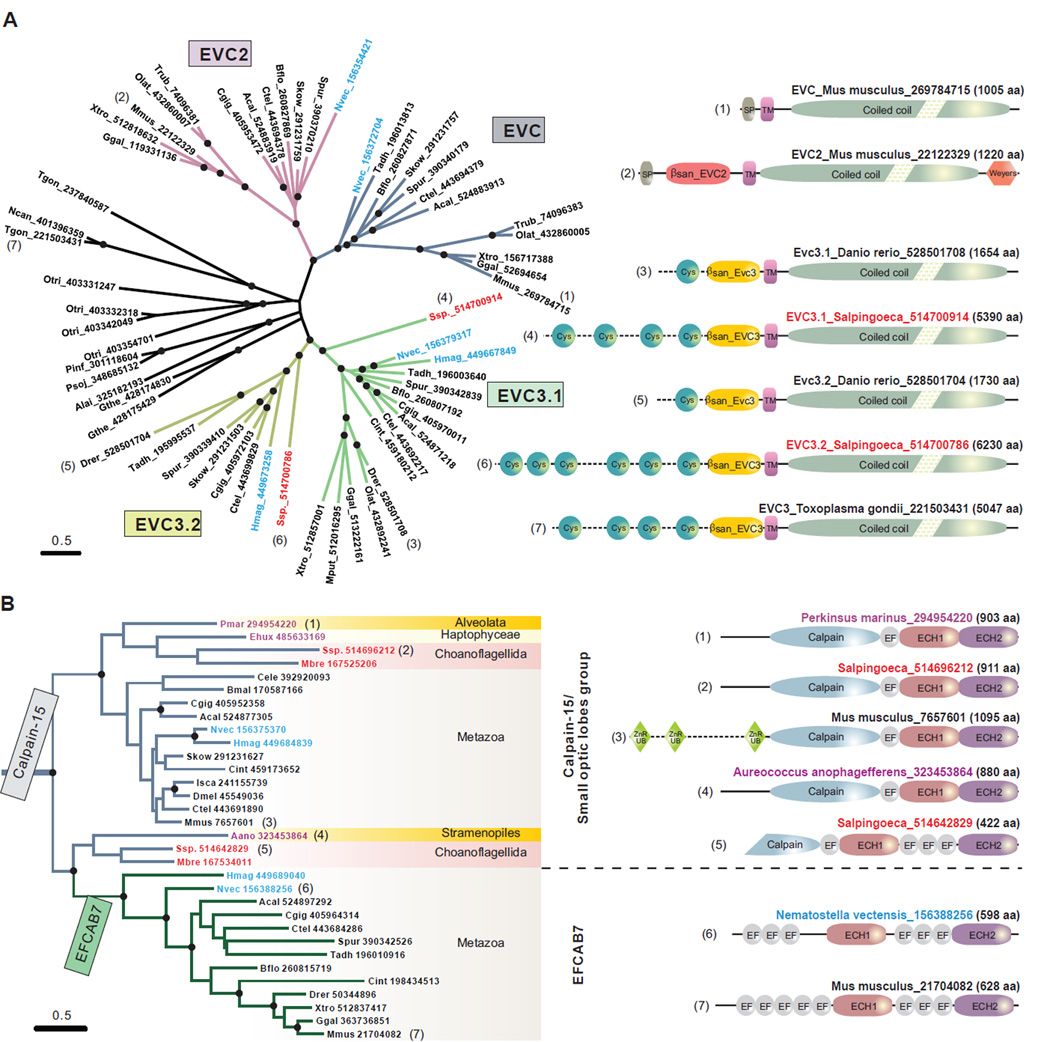
The cilium is an ancient organelle that can be traced back to the last eukaryotic common ancestor (Zhang and Aravind, 2012). Canonical Hh signaling is currently restricted to eumetazoans (i.e. all animals excluding ctenophores, sponges, and placozoans), raising the question of how this ancient organelle was co-opted for a newly emergent signaling pathway in animals. Given the role of the EvC complex in ciliary Hh signaling, we systematically investigated the evolutionary history of both the integral membrane and intracellular components. Our analysis identified a novel clade of proteins related to EVC and EVC2 that we named EVC3 (e.g. Zebrafish; gi: 528501708 and 528501704). EVC3, like EVC and EVC2, had a characteristic single TM region followed by a long cytoplasmic coiled-coil region (Figure 7A). However, its extracellular region is much longer; in addition to a β-sandwich, EVC3 has a variable number of repeats of a cysteine-rich domain that likely form disulfide-bonded structures (Figure 7A). In terms of phyletic patterns, both EVC and EVC2 are restricted to metazoans and are found in the basal most animal lineages such as cnidarians. Curiously, EVC and EVC2 appear to have been lost secondarily in certain animals, such as zebrafish (a pseudogene of evc can be identified on chromosome 14), even though they are conserved in other fishes. EVC3 shows a more widespread pattern; in addition to metazoans, it is also present in their sister group the choanoflagellates, and earlier-branching eukaryotes such as ciliates, apicomplexans, cryptophyte algae and various stramenopiles (Figure 7A). EVC3 has been recently lost in certain mammals including humans where a pseudogene remnant (gi: 119578130) can be identified on chromosome 22. However, zebrafish, which has lost EVC and EVC2, possesses two EVC3 paralogs. Importantly, EVC3 is present only in eukaryotes that possess cilia at some stage of their life cycle, suggesting that it could potentially be a novel ciliary membrane component.
Figure 7. Evolutionary analysis of EvC complex components.
(A) Evolution and domain architectures of the EvC family proteins, EVC, EVC2, EVC3.1 and EVC3.2. An un-rooted phylogenetic tree is presented with nodes supported at 90% bootstrap shown with black circles. Each primary metazoan branch is highlighted in different color. The proteins are represented by their species abbreviations followed by their GenBank Identifiers (GIs). The basal animal species, Nematostella vectensis and Hydra magnipapillata, are highlighted in blue, while the sister clade of animals, Choanoflagellida, in red. Domain architectures are shown for selected proteins (right); the number next to each domain map is also used to mark that protein in the tree. (B) Origin of EFCAB7 from the Calpain-15 family and associated domain architecture changes. Basal animal species are highlighted in blue; the animal-sister clade, Choanoflagellida, in red; and basal eukaryotic species in purple. Domain abbreviations: SP, signal peptide; TM, transmembrane region; Weyers, Weyers peptide; Cys, Cys-rich motif; βsan_EVC2 and βsan_EVC3, the β-sandwich domains in the EVC2 and EVC3 subfamilies; Calpain, the papain-like peptidase domain; EF, EF-hand domain; ECH1 and ECH2, EFCAB7-Calpain Homology domains 1 and 2.
Species abbreviations: Aano, Aureococcus anophagefferens; Acal, Aplysia californica; Alai, Albugo laibachii; Bflo, Branchiostoma floridae; Bmal, Brugia malayi; Cele, Caenorhabditis elegans; Cgig, Crassostrea gigas; Cint, Ciona intestinalis; Ctel, Capitella teleta; Dmel, Drosophila melanogaster; Drer, Danio rerio; Ehux, Emiliania huxleyi; Ggal, Gallus gallus; Gthe, Guillardia theta; Hmag, Hydra magnipapillata; Isca, Ixodes scapularis; Mbre, Monosiga brevicollis; Mmus, Mus musculus; Mput, Mustela putorius; Ncan, Neospora caninum; Nvec, Nematostella vectensis; Olat, Oryzias latipes; Otri, Oxytricha trifallax; Pinf, Phytophthora infestans; Pmar, Perkinsus marinus; Psoj, Phytophthora sojae; Skow, Saccoglossus kowalevskii; Spur, Strongylocentrotus purpuratus; Ssp., Salpingoeca sp.; Tadh, Trichoplax adhaerens; Tgon, Toxoplasma gondii; Trub, Takifugu rubripes; Xtro, Xenopus tropicalis.
A phylogenetic analysis of the common conserved region shared by these proteins shows that EVC and EVC2 are sister groups. Their divergence predates the radiation of Eumetazoa because they are present in the placozoan Trichoplax, a basal animal lineage (Figure 7A). However, EVC and EVC2 are absent in the currently available sponge genome. EVC3 appears to have undergone a parallel duplication in metazoans into two paralogs we term EVC3.1 and EVC3.2. This, along with the pattern reciprocal gene loss in mammals and zebrafish raises the possibility that EVC3.1-EVC3.2 comprise a distinct ciliary protein complex, which may have some functional overlap with EVC-EVC2. The tree and phyletic patterns suggest that EVC3 was the original representative, with EVC and EVC2 being derived from the former early in metazoan evolution through loss of the extracellular cysteine-rich domains.
The phyletic pattern of EFCAB7 closely mirrors that of EVC and EVC2, being found only in Placozoa and Eumetazoa. However, calpains with homologous domains are found more widely across the eukaryotic tree with representatives from more earlier-branching groups such as choanflagellates, alveolates and stramenopiles. A phylogenetic tree of the ECH2 domain shows that the EFCAB7 version is nested within those from the calpain-15 clade (Figure 7B). Importantly, those closest to EFCAB7 are a specific subset of choanoflagellate (sister group of Metazoa) calpain-15 proteins with similar organization of EF-hands and ECH1/ECH2 domains as observed in EFCAB7 (Figure 7B). This suggests that EFCAB7 probably arose as a breakaway from an ancestral calpain-15 (Sorimachi et al., 1997) prior to the metazoan radiation. IQCE, like EVC3 but unlike EVC, EVC2 and EFCAB7, is also found outside of Metazoa in their closest sister group the choanoflagellates. Thus, both EVC-EVC2 and the EFCAB7-IQCE sub-complexes can be confidently inferred as being present in the common ancestor of placozoans and eumetazoans. However, an ancestral version of the EFCAB7-IQCE sub-complex with IQCE and a calpain-like protein might have predated the origin of metazoans.
Discussion
Important molecular insights into the regulation of EVC proteins came from the study of dominant alleles of EVC2, initially isolated from families with Weyers syndrome (Valencia et al., 2009; Ye et al., 2006). These alleles encode a protein (EVC2ΔW) lacking the C-terminal 43 a.a. W-peptide. Loss of this peptide had two major consequences— it endowed the protein with the ability to inhibit Hh signaling and it led to a specific change in the sub-ciliary localization of EVC and EVC2 (Caparros-Martin et al., 2013; Dorn et al., 2012; Valencia et al., 2009). Instead of being tightly restricted to the base of the cilium in the EvC zone, the EVC-EVC2ΔW complex was instead distributed along the entire length of the cilium. Notably, the EVC-EVC2ΔW complex still bound SMO efficiently (Dorn et al., 2012), suggesting that the W-peptide mediated unknown protein interactions that were essential for transmission of the ciliary Hh signal.
Motivated by these observations about the importance of the W-peptide for Hh signaling and skeletal development, we have identified a previously unstudied complex composed of two ciliary proteins, EFCAB7 and IQCE, that engages EVC-EVC2 through an interaction with the W-peptide. EFCAB7 and IQCE are positive regulators of Hh signaling. Guided by detailed domain mapping, genetic perturbations, and deep phylogenetic analysis of the four proteins in this module, we elucidated the architecture of the EvC complex at the base of primary cilia (see graphical abstract). TM proteins often recruit cytoplasmic proteins in signal transduction; however, in the case of the EvC complex, the arrangement is reversed. IQCE, a protein without TM domains, is sufficient to localize correctly at the base of cilia in the EvC zone, and we believe it is the major determinant of localization in this unique ciliary compartment through yet to be identified protein or lipid factors. EFCAB7 is best viewed as an adaptor that then recruits the EVC-EVC2 complex by interactions between a β-sandwich domain and the W-peptide of EVC2. Support for our model comes from the observation that depletion of EFCAB7 phenocopies the effect of deleting the W-peptide—Hh signaling is impaired and the EVC and EVC2 proteins fail to be restricted to the EvC zone.
Further research is required to understand the mechanism by which the EvC complex regulates Hh signaling. The EVC-EVC2 complex interacts with SMO in a signal-dependent manner but we have not been able to detect an interaction with downstream Hh signaling components such as PKA, SUFU or the GLI proteins. One possibility is that this complex serves as a scaffold to concentrate SMO in a distinct ciliary compartment, the EvC zone, for efficient activation or signal transmission (Dorn et al., 2012). A scaffolding role that determines the amplitude of signaling is suggested by the fact that even the total loss of EVC, EVC2 or EFCAB7 does not completely interrupt signaling (as a loss of SMO does) and still allows a low level of target gene induction (Figure 6). This might also explain why the loss of EVC and EVC2 has an effect in some tissues and not in others, suggesting a context-specific or level-specific role. It is interesting to note that G-protein coupled receptors, the superfamily to which SMO belongs, are often regulated by scaffolding proteins that play tissue-specific roles in modulating the amplitude or speed of signaling (reviewed in Ritter and Hall, 2009).
Our results suggest that the Hh pathway may be bifurcated in cilia downstream of SMO to allow independent regulation of the repressor and activator functions of the GLI proteins. The presence of EVC-EVC2 in cilia, irrespective of EvC zone restriction, was sufficient for Hh signaling to regulate levels of GLI3R. However, restriction of EVC-EVC2 to the cilia base (mediated by EFCAB7-IQCE) was required for Hh signaling to promote the activation of GLI2. One possibility is that the EFCAB7-IQCE module, and consequently the EvC zone, is most relevant for high-level Hh signaling. The relative importance of Gli activator and repressor functions seems to vary depending on the tissue during development, supporting the view that Hh signaling may be bifurcated at this step. This phenotype is reminiscent of mice lacking the ciliary small GTPase ARL13B, whose loss did not impact GLI3R levels but instead left GLI2 in a constitutive state of low-level activity; however, we did not observe any changes in the localization of ARL13B in cells lacking EFCAB7 (Figure S4G) (Caspary et al., 2007).
Finally, evolutionary analysis of the EvC complex provides some interesting insights into how ciliary complexes and Hh signaling may have become coupled. The components of this complex show a degree of phyletic correlation with the distribution of the Hh signaling pathway and the pervasive presence of primary cilia (Figure S6). Choanoflagellates and sponges possess HINT and N-terminal hedgehog-like peptidase domains fused to cadherin repeats (the “Hedgeling” proteins) but they lack canonical hedgehog signaling; likewise they entirely or partly lack EvC complex components. Placozoans appear to lack Hh signaling but possess a full-fledged EvC complex. In Eumetazoa the EvC complex is present right from the basal-most lineage (Cnidaria), which possesses a canonical Hh signaling pathway. Within Eumetazoa, the loss of the EvC complex is correlated with loss of Hh signaling and reduced expression of primary cilia. Thus, the EvC complex is entirely lost (Figure S6) in Drosophila, where cilia are limited to sensory neurons and spermatozoa, and in C.elegans, where again cilia are limited to sensory neurons and Hh signaling is absent. Together, these observations suggest that the EvC complex arose primarily in the context of a ciliary function and subsequently developed a connection to Hh signaling within Eumetazoa.
The linkage between Hh signaling and cilia is best characterized in vertebrates. We examined the conservation of various domains in EvC complex proteins within vertebrates, compared to conservation across animals. EVC2 has a distinctive C-terminal tail corresponding to the W-peptide in all metazoans where it is present, but the hydrophobic tetrapeptide motif within this segment, experimentally shown to be critical for the interaction with EFCAB7, EvC zone localization and Hh signaling, is strongly conserved only in vertebrates (Figure 3B). The C-terminal 3 strands of the EFCAB7 ECH2 domain, which interacts with the tetrapeptide motif, are also rather divergent in sequence outside vertebrates, implying co-evolution of ECH2 and EVC2. In addition, the β-sandwich extracellular domain of EVC2, while present in all metazoans, is strongly conserved in vertebrates but is highly variable in sequence in other animals. The relatively strong constraints on both the extracellular and intracellular parts of EVC2 in vertebrates might have arisen from selective pressures resulting from the need for interactions unique to signaling events in vertebrates. In this regard, we note that the major phenotypes seen when EVC and EVC2 are lost are found in the skeletal system, a vertebrate-specific innovation.
Experimental Procedures
Methods for Hh signaling assays, microscopy-based cilia trafficking assays, quantitative image analysis, western blotting, immunoprecipitation and qRT-PCR have been described previously (Dorn et al., 2012) and are also described in detail in the supplemental experimental procedures.
Constructs
All EvC complex constructs used the mouse sequence. Constructs encoding EVC, EVC2, EVC2ΔW (a.a. 1–1176), and SMO-M2 were described previously (Dorn et al., 2012). For this study, mouse EVC was also cloned with an N-terminal 3xHA tag. Full-length IQCE and EFCAB7 (or fragments thereof) were cloned with N-terminal YFP, 3xHA tag, or 6xMyc tags. pCS2+ or pCS-DEST (Addgene 22423) vectors were used for in vitro translation or transient expression in mammalian cells, pEF5/FRT/V5-DEST (Life Technologies) for transient mammalian expression or the construction of stable Flp-In cell lines, pLENTI CMV Puro DEST (Addgene 17452) for lentiviral production and pGex-FA vector (GE Life Sciences) or pMal-FA (New England Biolabs) for bacterial production of recombinant protein. Mutations were introduced by inverse PCR or the QuikChange method (Agilent) to generate N-terminal tagged versions of IQCE 1–139 (a.a. 1- 139), IQCE 1–351 (a.a. 1–351), IQCE 1–489 (a.a. 1–489), IQCE 1–552 (ΔIQ-IQCE; a.a. 1–552), IQCE ΔN (a.a. 1–139 deleted), IQCE ΔCC1 core (a.a. 142–211 deleted), IQCE ΔCC1 (a.a. 140–351 deleted), IQCE ΔCC2 (a.a. 362–489 deleted), IQCE Δ3× IQ (a.a. 530–635 deleted), IQCE ΔAcidE (a.a. 749–778 deleted), EFCAB7 EF 1–5 (a.a. 1–248), EFCAB7 ECH1 (a.a. 249–396), EFCAB7 EF 6–8 (a.a. 400–540), EFCAB7 ECH2 (a.a. 540–628), EVC2WWT (a.a. 1133–1220), and EVC2WAla (a.a. 1133–1220 with mutations in FVFR motif).
Antibodies
Rabbit polyclonal antibodies against the N-terminal (1–166) and C-terminal (637–778) domains of mouse IQCE and full-length mouse EFCAB7 were generated in rabbits (Cocalico Biologicals) and affinity purified prior to use.
Cell Lines
NIH/3T3, C3H10T1/2 and HEK293T cells were obtained from ATCC, and Flp-In NIH/3T3 cells for stable cell lines construction from Life Technologies. Stable cell lines expressing tagged EVC, EVC2, EFCAB7 and IQCE (or their mutants) were produced by site-specific recombination into a single site in the genome using the Flp-In system (Life Technologies).
Tandem affinity purification and Mass spectrometry
One hundred 15 cm plates of NIH/3T3 cells stably expressing EVC2-YFP-FLAG were serum starved for 24 h and then treated with 100 nM SAG for 2 h. Cells were washed twice with ice-cold PBS followed by 10 mM HEPES pH 7.4 and swollen in the same hypotonic buffer for 10 min. The buffer was removed carefully and cells were scraped with SEAT buffer (10 mM triethanolamine/acetic acid, 1 mM EDTA pH 8.0, 250 mM sucrose, protease inhibitor cocktail) to a final volume of 100 ml and homogenized with twenty strokes of a dounce homogenizer. The sample was spun at 900×g for 8 min to collect the post-nuclear supernatant (PNS). The PNS was spun at 142,000×g for 60 min at 4 °C to harvest a membrane pellet, which was then resuspended by douncing in a membrane extraction buffer (MEB) containing 50 mM Tris-HCl pH-7.4, 150 mM NaCl, 1% DDM, 0.1% CHS, 1 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 0.2 mM DTT, 10% glycerol, 1 mM NaF, 1 mM Na3VO4, 1 µM microcystin-LR, and 1× protease inhibitor cocktail. After incubation for 1h at 4°C, the detergent extract was clarified by centrifugation at 140,000×g for 60 min at 4°C and then incubated with Anti-FLAG-M2 conjugated agarose beads (Sigma) for 16 h at 4°C. Beads were washed once with MEB followed by a single wash each in wash buffer A (MEB containing 0.1% DDM, 0.1% CHS, and 300 mM NaCl) and wash buffer B (MEB containing 0.1% DDM, 0.1% CHS). Bound protein complexes were eluted with 0.2 mg/ml FLAG peptide (Sigma) for 4 h at 4°C followed by an additional round of elution at 22°C for 1 h. The eluates were pooled and subjected to a second round of immunoprecipitation with GFP binding protein (GBP) covalently conjugated to carboxylic acid decorated Dynabeads M-270 for 2 h at 4°C. GBP beads were washed five times with buffer B and the bound complexes were eluted in 5× Laemmli buffer at 37°C for 30 min. The eluted sample was subjected to SDS-PAGE on a 8% Tris-Glycine gel followed by GelCode Blue staining (Thermo Scientific) to visualize protein bands. Protein bands were excised, digested with trypsin and analyzed (MS Bioworks) using a nano LC/MS/MS with a NanoAcquity HPLC system (Waters) interfaced to an Orbitrap Velos Pro (Thermo Scientific). The data were processed with the Mascot Server (Matrix Science) and the Mascot DAT files were parsed into the Scaffold software for validation, filtering and to create a non-redundant list per sample. Data were filtered using a minimum protein value of 90%, a minimum peptide value of 50% (Prophet scores) and requiring at least two unique peptides per protein.
In vitro binding assays
For GST pulldown assays, 10 µg of purified GST-EVC2WWT and GST-EVC2WAla were immobilized on glutathione sepharose 4B beads and tested for their ability to pull-down IQCE and EFCAB7 expressed using the TNT coupled wheat germ extract system (Promega). For interactions between EFCAB7 and IQCE, protein fragments whose interaction was being tested were in vitro translated together, followed by immunoprecipitation of one of the fragments.
Generation of NIH/3T3 cell lines carrying null mutations in EvC complex subunits
Efcab7−/−, Evc2−/− and Iqce−/− cells were generated using the CRIPSR/Cas9 genome editing strategy using Addgene plasmid number 42230 and the guide sequences 5’- aactgaggcgtcaacagtt-3’ (Efcab7), 5’-gatatttcaaaaatgctcac-3’ (Evc2), 5’-ggcgatctctgaagacggca-3’ (Iqce). (Cong et al., 2013). The targeting CRISPR plasmid was co-transfected with a YFP-containing plasmid using X-tremeGENE 9 and single cells expressing YFP were sorted into 96 well plates using FACSAria II (BD). Single clones that had lost expression of the targeted protein were identified using immunoblotting. The frequencies at which NIH/3T3 clones bearing null mutations in both alleles of Efcab7, Iqce, and Evc2 were retrieved were 5%, 1% and 5% respectively.
Statistical analysis
Statistical analysis for fluorescence intensity comparisons between two or multiple groups was performed by Mann-Whitney test or the Kruskal-Wallis non-parametric ANOVA test, respectively. The statistical significance between the means in Hh reporter assays was determined by a two-tailed, unpaired Student’s t test.
Supplementary Material
Highlights.
-
⇨
EFCAB7 and IQCE are positive regulators of Hedgehog signaling.
-
⇨
EFCAB7-IQCE complex tethers the EVC-EVC2 complex to the base of cilia.
-
⇨
Depletion of EFCAB7 mimics the cellular phenotype seen in a Hh-related ciliopathy.
-
⇨
The evolution of these proteins might have adapted cilia for Hh signaling.
Acknowledgements
KD purified the EvC complex and identified EFCAB7 and IQCE. GP, CH, KD and PS characterized the assembly, localization and signaling properties of these proteins. DZ and LA performed the sequence and evolutionary analyses that guided experiments. RR, LA, and GP wrote the paper, with input from all authors. We thank A. Lebensohn for purification of the GBP protein used in all the anti-GFP IP experiments and for help with the CRISPR/Cas9 system, G. Luchetti for insightful discussions and EvC complex experiments in 293T cells, E. Nigg (anti- CEP164) and J. Eggenschwiler (anti-GLI2) for reagents, and X. Ge for help with IF. This work was supported by the March of Dimes Foundation (6-FY13-104), the NIH (DP2GM105448), the Pew Foundation and the Sontag Foundation. DZ and LA are supported by the intramural funds of the National Institutes of Health, Department of Health and Human Services.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blair HJ, Tompson S, Liu YN, Campbell J, MacArthur K, Ponting CP, Ruiz-Perez VL, Goodship JA. Evc2 is a positive modulator of Hedgehog signalling that interacts with Evc at the cilia membrane and is also found in the nucleus. BMC Biol. 2011;9:14. doi: 10.1186/1741-7007-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparros-Martin JA, Valencia M, Reytor E, Pacheco M, Fernandez M, Perez-Aytes A, Gean E, Lapunzina P, Peters H, Goodship JA, et al. The ciliary Evc/Evc2 complex interacts with Smo and controls Hedgehog pathway activity in chondrocytes by regulating Sufu/Gli3 dissociation and Gli3 trafficking in primary cilia. Hum Mol Genet. 2013;22:124–139. doi: 10.1093/hmg/dds409. [DOI] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Dorn KV, Hughes CE, Rohatgi R. A Smoothened-Evc2 complex transduces the Hedgehog signal at primary cilia. Dev Cell. 2012;23:823–835. doi: 10.1016/j.devcel.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdzicka M, Patnala S, Hirshman MG, Cai JF, Nitowsky H, Egeland JA, Ginns EI. A new gene, EVC2, is mutated in Ellis-van Creveld syndrome. Mol Genet Metab. 2002;77:291–295. doi: 10.1016/s1096-7192(02)00178-6. [DOI] [PubMed] [Google Scholar]
- Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010;24:670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Thompson J, Yates JR, 3rd, Marshall WF. Proteomic analysis of mammalian primary cilia. Curr Biol. 2012;22:414–419. doi: 10.1016/j.cub.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinto N, Iwamoto M, Enomoto-Iwamoto M, Noji S, Ohuchi H, Yoshioka H, Kataoka H, Wada Y, Yuhao G, Takahashi HE, et al. Fibroblasts expressing Sonic hedgehog induce osteoblast differentiation and ectopic bone formation. FEBS letters. 1997;404:319–323. doi: 10.1016/s0014-5793(97)00014-8. [DOI] [PubMed] [Google Scholar]
- Ostrowski LE, Blackburn K, Radde KM, Moyer MB, Schlatzer DM, Moseley A, Boucher RC. A proteomic analysis of human cilia: identification of novel components. Mol Cell Proteomics. 2002;1:451–465. doi: 10.1074/mcp.m200037-mcp200. [DOI] [PubMed] [Google Scholar]
- Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol Cell Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter SL, Hall RA. Fine-tuning of GPCR activity by receptor-interacting proteins. Nature reviews Molecular cell biology. 2009;10:819–830. doi: 10.1038/nrm2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Ruiz-Perez VL, Blair HJ, Rodriguez-Andres ME, Blanco MJ, Wilson A, Liu YN, Miles C, Peters H, Goodship JA. Evc is a positive mediator of Ihh-regulated bone growth that localises at the base of chondrocyte cilia. Development. 2007;134:2903–2912. doi: 10.1242/dev.007542. [DOI] [PubMed] [Google Scholar]
- Ruiz-Perez VL, Goodship JA. Ellis-van Creveld syndrome and Weyers acrodental dysostosis are caused by cilia-mediated diminished response to hedgehog ligands. Am J Med Genet C Semin Med Genet. 2009;151C:341–351. doi: 10.1002/ajmg.c.30226. [DOI] [PubMed] [Google Scholar]
- Ruiz-Perez VL, Ide SE, Strom TM, Lorenz B, Wilson D, Woods K, King L, Francomano C, Freisinger P, Spranger S, et al. Mutations in a new gene in Ellis-van Creveld syndrome and Weyers acrodental dysostosis. Nat Genet. 2000;24:283–286. doi: 10.1038/73508. [DOI] [PubMed] [Google Scholar]
- Ruiz-Perez VL, Tompson SW, Blair HJ, Espinoza-Valdez C, Lapunzina P, Silva EO, Hamel B, Gibbs JL, Young ID, Wright MJ, et al. Mutations in two nonhomologous genes in a head-to-head configuration cause Ellis-van Creveld syndrome. Am J Hum Genet. 2003;72:728–732. doi: 10.1086/368063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorimachi H, Ishiura S, Suzuki K. Structure and physiological function of calpains. Biochem J. 1997;328(Pt 3):721–732. doi: 10.1042/bj3280721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukachinsky H, Lopez LV, Salic A. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol. 2010;191:415–428. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia M, Lapunzina P, Lim D, Zannolli R, Bartholdi D, Wollnik B, Al-Ajlouni O, Eid SS, Cox H, Buoni S, et al. Widening the mutation spectrum of EVC and EVC2: ectopic expression of Weyer variants in NIH 3T3 fibroblasts disrupts Hedgehog signaling. Hum Mutat. 2009;30:1667–1675. doi: 10.1002/humu.21117. [DOI] [PubMed] [Google Scholar]
- Weyers H. A correlated abnormality of the mandible and extremities (dysostosis acrofacialis) Fortschritte auf dem Gebiete der Rontgenstrahlen. 1952;77:562–567. [PubMed] [Google Scholar]
- Yang C, Chen W, Chen Y, Jiang J. Smoothened transduces Hedgehog signal by forming a complex with Evc/Evc2. Cell research. 2012;22:1593–1604. doi: 10.1038/cr.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Song G, Fan M, Shi L, Jabs EW, Huang S, Guo R, Bian Z. A novel heterozygous deletion in the EVC2 gene causes Weyers acrofacial dysostosis. Hum Genet. 2006;119:199–205. doi: 10.1007/s00439-005-0129-2. [DOI] [PubMed] [Google Scholar]
- Zhang D, Aravind L. Novel transglutaminase-like peptidase and C2 domains elucidate the structure, biogenesis and evolution of the ciliary compartment. Cell Cycle. 2012;11:3861–3875. doi: 10.4161/cc.22068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.