Abstract
Double-stranded ribonucleic acid-activated protein kinase (PKR) downregulates translation as a defense mechanism against viral infection. In fish species, PKZ, a PKR-like protein kinase containing left-handed deoxyribonucleic acid (Z-DNA) binding domains, performs a similar role in the antiviral response. To understand the role of PKZ in Z-DNA recognition and innate immune response, we performed structural and functional studies of the Z-DNA binding domain (Zα) of PKZ from Carassius auratus (caZαPKZ). The 1.7-Å resolution crystal structure of caZαPKZ:Z-DNA revealed that caZαPKZ shares the overall fold with other Zα, but has discrete structural features that differentiate its DNA binding mode from others. Functional analyses of caZαPKZ and its mutants revealed that caZαPKZ mediates the fastest B-to-Z transition of DNA among Zα, and the minimal interaction for Z-DNA recognition is mediated by three backbone phosphates and six residues of caZαPKZ. Structure-based mutagenesis and B-to-Z transition assays confirmed that Lys56 located in the β-wing contributes to its fast B-to-Z transition kinetics. Investigation of the DNA binding kinetics of caZαPKZ further revealed that the B-to-Z transition rate is positively correlated with the association rate constant. Taking these results together, we conclude that the positive charge in the β-wing largely affects fast B-to-Z transition activity by enhancing the DNA binding rate.
INTRODUCTION
The regulation of protein synthesis is one of the most important events under stress conditions. The initiation of translation is tightly regulated as the rate limiting phase via the phosphorylation of eukaryotic initiation factor 2α (eIF2α) (1) by various protein kinases including hemeregulated inhibitor kinase, ribonucleic acid (RNA)-dependent protein kinase (PKR), PKR-like endoplasmic reticulum kinase and general control nonrepressed 2 (2,3). These kinases sense different cellular stress signals such as heme depletion, viral infection, Endoplasmic reticulum (ER) stresses and amino acid starvation, and ultimately inhibit protein synthesis and proliferation (2). Among these kinases, PKR plays an important role in the innate immune response against viral infections by recognizing double-stranded RNA (dsRNA) in the cytosol. PKR has a dsRNA binding domain (dsRBD), which consists of two dsRNA binding motifs, and an eIF2α kinase domain at the N- and C-termini, respectively (4). The binding of dsRNA to dsRBD induces the dimerization and autophosphorylation of PKR (5,6), and the activated PKR leads to apoptosis, inflammation and inhibited translation (4).
Functional orthologs of PKR have been identified in several fish species such as Gobiocypris rarus, Salmo salar, Danio rerio, and Carassius auratus (goldfish) (7–10). Since these orthologs contain two left-handed deoxyribonucleic acid (Z-DNA) binding domains (ZBDs) instead of dsRBD of PKR, they were named as protein kinases containing ZBDs (PKZ). Although the sensor domains of PKZ and PKR are different, their kinase domains are well conserved (8). Similar to PKR, PKZ has been known to reduce the overall translational level by phosphorylating Ser51 of eIF2α when it is activated by Z-DNA binding (8). Therefore, PKZ is a functional ortholog of PKR but it recognizes Z-DNA instead of dsRNA.
Left-handed Z-DNA is a higher energy conformation of the double-stranded DNA helix. Unlike canonical right-handed B-DNA, which is common in biological DNAs, Z-DNA adopts the zigzag arrangement of the phosphate backbone as a consequence of alternating anti and syn conformations of glycosidic bonds (11). Thus, alternating purine and pyrimidine sequences energetically favor the Z-DNA formation (12). Z-DNA can be stabilized by reducing the electrostatic repulsion between phosphate groups in the backbone by high ionic strength, positively charged chemicals or modification on bases (13,14). Non-aqueous solvents such as ethanol, methanol and ethylene glycol also stabilize Z-DNA (15). Besides those physicochemical factors, negative supercoiling and Z-DNA binding proteins (ZBPs) induce Z-DNA under physiological conditions (16,17), and the in vivo presence of Z-DNA has been demonstrated by Z-DNA-specific antibodies or ZBPs. So far, four ZBPs have been identified, each of which contains one or two ZBDs (Zα): adenosine deaminase acting on RNA-1 (ADAR1), DNA-dependent activator of interferon (IFN) regulatory factors (DAI, also known as DLM-1, and ZBP-1), protein kinase containing ZBD from fish (PKZ) and viral protein E3L (9,18–20).
Interestingly, all known ZBPs, ADAR1, DAI, E3L and PKZ, are involved in the innate immune response (17,21). Mammalian DAI recognizes cytosolic foreign DNA and consequently activates the inflammatory signal pathway through IFN and NF–κB signaling (22,23). The overexpression of ADAR1 or E3L suppresses the DNA-mediated production of IFN in mouse embryonic fibroblast cells (24). Hence, the competition between ADAR1 and DAI for cytosolic DNA has been suggested as a regulatory mechanism of the innate immune response. However, the distribution of DAI and PKZ is known to vary in different species; that is, fish have only PKZ, while mammals have only DAI, suggesting that PKZ and DAI may perform similar roles as cytosolic DNA sensors.
To date, four crystal structures of ZBDs in complex with Z-DNA are available: Zα domain of human ADAR1 (ZαADAR1) (25), Zα domain of mouse DAI (ZαDAI) (19), Zα domain of Yaba virus (yabZαE3L) (26) and Zβ domain of human DAI (hZβDAI) (27). Zα domains commonly have a winged helix-turn-helix (wHTH) DNA binding motif and recognize Z-DNA in a conformation-specific manner. Accordingly, most of the interactions involve backbone phosphates (17). Despite the similarity in overall structure, the binding modes and activities of Zα domains are quite distinctive (17). For example, it was proposed that the differential Z-DNA binding activities of viral Zα domains are correlated with pathogenicity (28).
caZαPKZ has limited sequence identity with other Zα domains, 28% for hZαADAR1, 20% for hZαDAI, 23% for hZβDAI and 22% for yabZαE3L, respectively. However, the residues involved in DNA backbone recognition are relatively well conserved with few exceptions (Figure 1) (9). It has already been proven that caZαPKZ binds to double-stranded (ds) deoxy-hexanucleotide with three repeats of cytosine-guanine deoxy-dinucleotide (dCdG) in negatively supercoiled DNA and converts ds-(dCdG)3 from B-DNA to Z-DNA in vitro (29,30), but little is known about the detailed Z-DNA binding mode of caZαPKZ. Therefore, structural studies on Z-DNA recognition are essential to understand its binding mode to Z-DNA and its relevance to the biological function of caZαPKZ. In the present study, we determined the crystal structure of caZαPKZ complexed with Z-DNA at 1.7-Å resolution and observed that caZαPKZ mediates B-to-Z transition with the fastest kinetics among the known Zα domains. Structural analyses combined with the functional assay of caZαPKZ mutants provided a basis for its Z-DNA binding mode and higher rate of B-to-Z transition.
Figure 1.
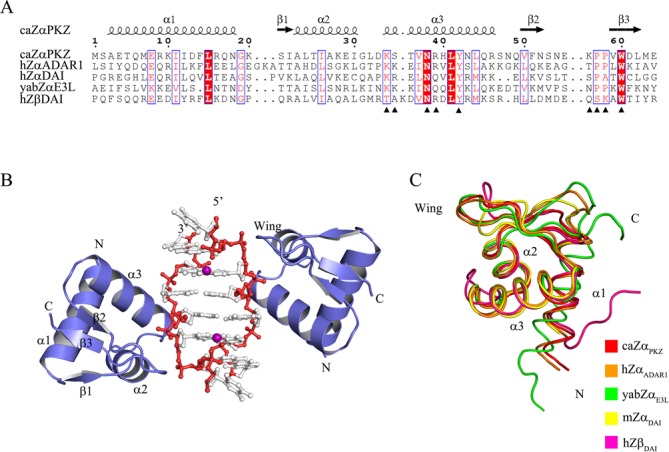
Overall structure of caZαPKZ in complex with ds-dT(dCdG)3 and comparison with other Zα domains. (A) Sequence alignment of caZαPKZ with human ADAR1 (hZαADAR1), human DAI (hZαDAI) and yaba poxvirus E3L (yabZαE3L), and Zβ domain of human DAI (hZβDAI). The secondary structures of caZαPKZ are drawn at the top of the sequences. The residues interacting with the Z-DNA backbone are marked with filled triangles. (B) Overall structure of the caZαPKZ:Z-DNA complex. Double-stranded DNA and two Zα proteins are generated by C2 crystallographic symmetry. Proteins are colored slated blue, and backbones and bases of DNA are colored red and gray, respectively. The manganese ions are shown as purple spheres. (C) Structural comparisons of the Cα traces of caZαPKZ and other Zα domains. Protein structures of caZαPKZ, hZαADAR1 (PDB ID: 1QBJ), yabZαE3L (PDB ID: 1SFU), mZαDAI (PDB ID: 1J75) and hZβDAI (PDB ID: 3EYI) are drawn in red, orange, green, yellow and magenta, respectively.
MATERIALS AND METHODS
Protein preparation
The Zα domains of human DAI (hZαDAI), human ADAR1 (hZαADAR1), E3L homolog of the yaba-like disease virus (yabZαE3L) and C. auratus PKZ (caZαPKZ) were prepared as reported previously (19,25,26,30). The detailed procedures are described in the Supplementary data.
Preparation of duplex oligonucleotide
Deoxyoligonucleotides 5′-dTdCdGdCdGdCdG-3′ [dT(dCdG)3] and 5′-dCdGdCdGdCdGdCdGdCdGdCdG-3′ [(dCdG)6] were purchased from Integrated DNA Technologies (IDT, CA, USA). All oligonucleotides were in 50-mM Tris-HCl pH 8.0, 50-mM NaCl and 1-mM ethylenediaminetetraacetic acid. Duplex deoxyoligonucleotides were prepared by annealing an equimolar mixture of complementary sequences and dialyzing against buffer A. After purification using a MonoQ column (GE Healthcare, NJ, USA), the concentration of the duplex DNA was calculated spectroscopically.
Circular dichroism measurement
The B-to-Z transition was monitored by the circular dichroism (CD) spectrum at 25°C. For each measurement, 7.5 μM of ds-(dCdG)6 in buffer A was used. Wild-type or mutant caZαPKZ was incubated with ds-(dCdG)6 at final concentrations of 0 μM (0×), 7.5 μM (1×), 15 μM (2×), 30 μM (4×), 45 μM (6×) and 60 μM (8×) at 25°C for 1 h. CD spectra were recorded using a Jasco J-810 CD spectrometer between 230 nm and 320 nm at 1-nm intervals averaged over 2 s. The kinetics of the B-to-Z DNA transition of caZαPKZ, hZαADAR1, hZαDAI, yabZαE3L and their mutants were monitored by the time course recording of CD spectra at 255 nm for 2000 s in the presence of 15-μM ds-(dCdG)6.
Structure determination
In our previous study, we confirmed that caZαPKZ induces the B-to-Z transition of the ds-dT(dCdG)3 at 2:1 (protein:DNA) molar ratio and it forms a stable complex with Z-DNA at the same molar ratio (30). We crystallized the caZαPKZ:Z-DNA complex in the C2 space group using 30% PEG 1500 and 15-mM MnCl2 and collected diffraction data at 1.7-Å resolution. The structure of caZαPKZ in complex with Z-DNA was determined by the molecular replacement method. The initial solution was obtained by MOLREP (32) using hZαADAR1 complexed with ds-dT(dCdG)3 (PDB ID: 1QBJ) as a search model. Rigid body refinement of the initial solution and density modification were performed using REFMAC (33) and DM (34), respectively. Iterative model building was done using COOT (35). The final refinement statistics are given in Table 1. The quality of the structure was checked using MolProbity (36). All structural figures were generated using PYMOL (37).
Table 1. Data collection and refinement statistics of caZαPKZ/Z-DNA complex.
| Native | |
|---|---|
| Unit cell parameters | |
| Space group | C2 |
| Unit cell | a = 55.37 Å |
| b = 49.47 Å | |
| c = 29.58 Å | |
| β = 97.22º | |
| Data collection | |
| Beamline | BL4A, PAL |
| Resolution range (Å) | 50.0–1.70 (1.73–1.70)a |
| Redundancy | 3.4 (3.0) |
| Completeness (%) | 97.5 (89.9)a |
| Rmerge (%)b | 6.1 (32.5)a |
| I/σ (I) | 43.3 (4.1)a |
| Refinement | |
| Resolution range (Å) | 27.5–1.70 (1.94–1.70) |
| Number of reflections working sets | 8131 (771) |
| Number of reflections test sets | 382 (33) |
| Number of protein atoms | 504 |
| Number of DNA atoms | 139 |
| Number of waters | 70 |
| Number of metal ions (manganese) | 1 |
| Rwork/Rfree (%)c | 17.2/22.6 (19.2/25.6) |
| RMSD bond lengths/angles (Å) | 0.022/2.226 |
| Average B-factor (Å2) | 25.3(protein)/21.9(DNA)/32.7(water) |
| Ramachandran plot (%)d | 100.0/0.0/0.0 |
aThe values in the parentheses of the resolution range, completeness, Rmerge and I/σ(I) correspond to the last shell.
bRmerge(I) = ∑hkl∑j|[I(hkl)j−I(hkl)]|/[∑hklIhkl] is the jth measurement of the intensity of reflection hkl and <I(hkl)> is the averaged intensity.
cR = ∑hkl∥Fobs|−|Fcalc∥/∑hkl|Fobs|, where Rfree is calculated without an s cutoff for a randomly chosen 5% of reflections, which were not used for structure refinement, and Rwork is calculated for the remaining reflections.
dPercentage of residues in favored/allowed/outlier regions calculated by MolProbity (36).
RESULTS
Overall structure of caZαPKZ in complex with Z-DNA
To understand the difference in the B-to-Z transition rates of Zα domains in relation to their Z-DNA binding modes, we determined the crystal structure of caZαPKZ in complex with dsdT(dCdG)3 oligonucleotide at 1.7-Å resolution with R factor of 17.2% and Rfree of 22.6%. One caZαPKZ molecule bound to a single-stranded DNA was modeled in an asymmetric unit. The current model comprises 62 residues (from Ser2 to Met63) of caZαPKZ, seven nucleotides of single-stranded dT(dCdG)3 [ss-dT(dCdG)3] and one manganese ion. By the C2 symmetry operation, two caZαPKZ molecules bound to both sides of the Z-DNA in a head-to-tail orientation were generated (Figure 1B).
The overall structure of caZαPKZ is well conserved with three β strands and three α helices, like other members of the Zα family (Figure 1). With a wing between the β2 and β3 (β-wing), caZαPKZ adopts the canonical wHTH structure (Figure 1B). Structure comparison revealed that caZαPKZ is a homolog of other known ZBDs: the root-mean-square deviations (RMSDs) from hZαADAR1 (PDB ID: 1QBJ) (25), mZαDAI (PDB ID: 1J75) (19), yabZαE3L (PDB ID: 1SFU) (26) and hZβDAI (PDB ID: 3EYI) (27) are 0.75 Å, 1.57 Å, 1.23 Å and 0.94 Å for 56 Cα atoms, respectively (Figure 1C). The RMSD versus residue plot also indicates that caZαPKZ and hZαADAR1 have no noticeable change throughout the modeled residues. For mZαDAI or yabZαE3L, structural deviations from caZαPKZ were mainly observed near the α1 helix (Figure 1C and Supplementary Figure S1). In contrast, caZαPKZ and hZβDAI showed a noticeable difference in the N-terminal half of α3. The β-wing structure of caZαPKZ is also largely different from that of mZαDAI and hZβDAI (Supplementary Figure S1B). Structural heterogeneity in the α1 helix is expected since this helix is least conserved in sequential and structural aspects. The structural deviation in the α3 helix and the β-wing is directly related to diversity of Z-DNA binding mode since both regions have a major role in recognizing Z-DNA. For example, the conformation movement of α3 helix to 3 (10) helix has been reported in the crystal and nuclear magnetic resonance structures of hZβDAI (27,38).
In the complex structure, dsDNA molecules generated by the C2 symmetry operation adopt a canonical left-handed Z-DNA conformation (Supplementary Table S1). Like other Zα:Z-DNA complexes, all three guanines have the syn conformation, and the three cytosines have the anti conformation. Guanosines 2 and 6 (G2 and G6) adopt 4’-exo sugar pucker instead of 3′-endo sugar pucker, as frequently observed for guanine sugars in Z-DNA (Supplementary Table S2). All seven nucleotides of ds-dT(dCdG)3 including 5′ deoxythymidine (T0) were modeled (Supplementary Figure S2A), whereas the 5′-dT overhang was not modeled due to weak electron density in the structures of other Zα:ds-dT(dCdG)3 complexes. Interestingly, one manganese ion is coordinated with the backbone phosphate (P0), N7 of guanine 2 (G2) and four water molecules with an octahedral coordination geometry (Supplementary Figure S2B).
caZαPKZ has a unique Z-DNA binding mode
The interaction between caZαPKZ and Z-DNA is mediated by five residues in the α3 core and four residues in the β-wing. Among them, Lys34, Asn38 and Arg39 in the α3 core form direct or water-mediated hydrogen bonds with backbone phosphates of Z-DNA, as observed in the hZαADAR1 structure (25) (Figures 1 and 2). Tyr42, which is known as the most critical residue for Z-DNA binding by forming the CH−π stacking with the guanine 4 (G4), is conserved in structural and sequential aspects (Figures 1A and 2B). It is interesting that caZαPKZ has Ser35 in the α3 core, unlike the positively charged Lys or Arg in other Zα domains, which can form electrostatic interaction with the fifth backbone phosphate (P5; Figures 1A and 2). Due to the shorter length and lack of positive charge, Ser35 does not form ionic interactions (Figure 2A and C), but does have hydrogen bonding with the P3 phosphate. The β-wing of caZαPKZ is composed of Lys56, Pro57, Pro58 and Trp60. As in other Zα domains, Trp60 forms a hydrophobic interaction with Tyr42 (19,25,26). The two proline residues, Pro57 and Pro58, interact with P1 and P2 phosphates via hydrophobic interactions. It is notable that caZαPKZ has Lys56, whereas other mammalian Zα domains have a polar residue like Ser or Thr at the corresponding position. This substitution appears to be associated with the unique interaction mode of caZαPKZ, that is, Lys56 interacts not only with P1 but also with P0 (Figure 2A and C), thereby stabilizing T0 and Mn2+ ion (Supplementary Figure S2B). Accordingly, the comparison of charge distribution surfaces of Zα domains reveals that only caZαPKZ has positively charged wing structure (Supplementary Figure S3). Current structural interpretations imply that the Z-DNA binding mode of caZαPKZ is distinguished from those of other Zα domains, which is due to Ser35 and Lys56. Consequently, caZαPKZ recognizes P0–P4 phosphates of Z-DNA, while other Zα domains interact with P1–P5 phosphates (Figure 2).
Figure 2.
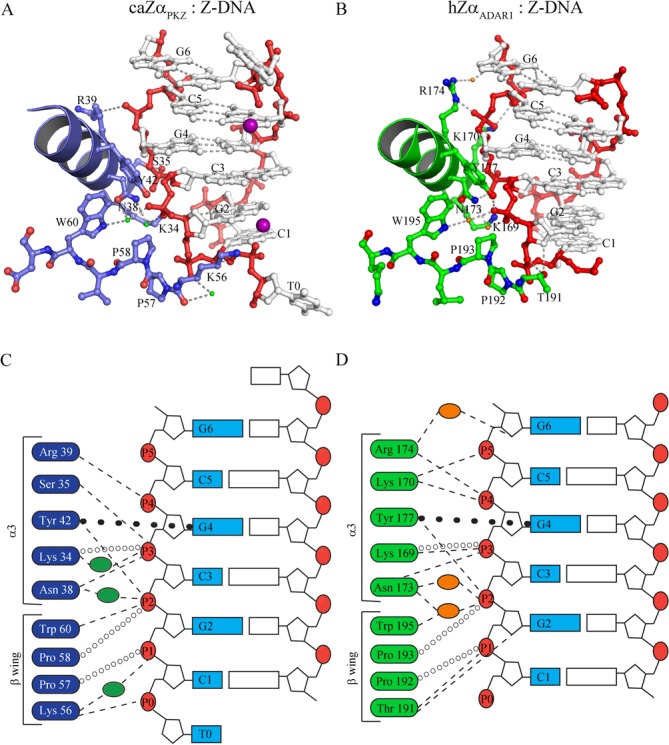
The Z-DNA binding modes of caZαPKZ and hZαADAR1. (A) The binding interface between caZαPKZ and Z-DNA. The DNA binding interface of protein is depicted as a blue ribbon and ball-and-stick models. The residues involved in Z-DNA binding are labeled. The backbone and bases of DNA are drawn as red and gray ball-and-stick models. The manganese atoms are represented by magenta spheres. The water molecules are represented by green dots. (B) The Z-DNA binding interface of hZαADAR1. The DNA binding interface of protein is depicted by a green ribbon and ball-and-stick models. The residues involved in Z-DNA binding are labeled. The backbone and bases of DNA are drawn as red and gray ball-and-stick models. The water molecules are represented by orange dots. The schematic drawings of the DNA binding interfaces of caZαPKZ (C) and hZαADAR1 (D) with Z-DNA. The interaction residues of caZαPKZ and hZαADAR1 are represented in blue (C) and green boxes (D), respectively. The phosphate backbones are numbered from P0 to P5 in red circles. Hydrogen bonds, hydrophobic interactions and π–π interaction are represented by dashed lines, open circles and closed circles, respectively. Green (C) and orange (D) ovals stand for water molecules in the caZαPKZ and hZαADAR1 structures, respectively.
The minimal recognition motif of caZαPKZ for recognizing Z-DNA
It was proposed from the crystal structures of hZαADAR1 in complex with non-CG repeat of Z-DNA that most conserved interactions are mediated by three backbone phosphates, P1, P2 and P3, and five conserved residues, Asn173, Tyr177, Pro193, Pro192 and Trp195 (39). It is clear that caZαPKZ seemingly has a different binding mode to interact with Z-DNA. To elucidate the role of each residue in the DNA binding surface and to identify the minimal DNA recognition motif in caZαPKZ, we mutated the key interacting residues and monitored the B-to-Z transition activity of mutants at various molar ratios of protein/DNA ([P]/[N] ratios) (Supplementary Figure S4). Far-ultraviolet CD spectra (190–240 nm) confirmed that the overall folding of each caZαPKZ mutant was not altered, except for Y42A and W60A mutants (Supplementary Figure S5). Since CD changes at both 255 nm and 292 nm mostly represent the B-to-Z transition, we plotted the ellipticity of DNA at 255 nm and 292 nm in the presence of mutant proteins with various [P]/[N] ratios (0–8) (Supplementary Figure S6) to compare the B-to-Z transition activity of each mutant. CD spectra showed that four molar excess of caZαPKZ can fully convert ds-(dCdG)6 to Z-DNA, and thus the stoichiometry between caZαPKZ and the ds-(dCdG)6 is estimated to be 4:1 (Supplementary Figure S4A). Therefore, to simplify the comparison, the ellipticity changes at 255 and 292 nm from those of B-DNA at the [P]/[N] ratio of 4 were plotted for each mutant (Figure 3). In addition, we introduced the names of backbone phosphates, which are involved in the binding to each mutant in the same plot. Based on their ellipticity change, each mutant was grouped into three. Group 1 included mutants S35A, K56A and R39A and exhibited the largest ellipticity change, suggesting the minimum activity change (Figure 3 and Supplementary Figures S4C, E, G and S6). The double or triple mutants belonging to group 2 showed the intermediate displacement, implying that these mutants are functionally less active. Mutants in group 3 appeared to lose their B-to-Z transition activity, which accounts for the residues essential for the B-to-Z transition. For example, the replacement of either Lys34 or Asn38 to alanine in group 3 resulted in dramatic reduction of the B-to-Z transition (Figure 3 and Supplementary Figure S4B and D). In the caZαPKZ:Z-DNA complex, P2 and P3 form a wide interaction network with many residues in caZαPKZ (Figure 2C). Asn38, Tyr42, Pro58 and Trp60 are the binding partners of P2, while P3 is recognized by Ser35, Lys34, and Asn38 (Figure 2). Accordingly, mutations of residues in this interaction network severely damage protein:DNA interaction. For example, K34A and N38A showed significantly decreased B-to-Z transition activity (Figure 3). These results suggest P2 and P3 interactions are both crucial for protein:Z-DNA interaction. This plot also proved that Tyr42 and Trp60 have the most critical role in DNA binding since the single mutation at those sites almost completely abolished the B-to-Z transition function (Figure 3 and Supplementary Figure S4F and H). Previously, it was suggested that these mutants stabilize Z-DNA conformation by forming a CH–π interaction (25,26). However, our data suggest that they might be also important for the folding of Zα domains since the alanine mutant of Try42 or Trp60 seems to have the altered secondary structures compared with wild-type protein (Supplementary Figure S5).
Figure 3.
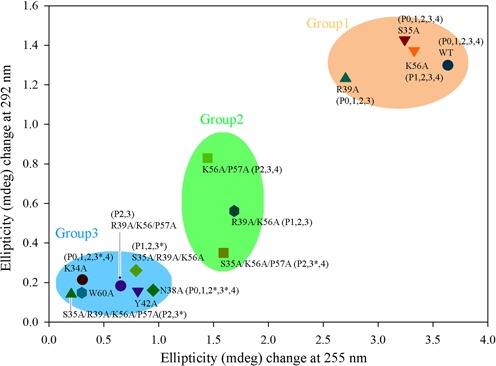
B-to-Z transition induced by caZαPKZ mutants. The Y-axis and X-axis display the ellipticity (mdeg) difference during B-to-Z transition at 292 nm and 255 nm, respectively. The wild-type, K34A, S35A, N38A, R39A, Y42A, K56A, W60A, R39A/K56A, S35A/R39A/K56A, K56A/P57A, S35A/K56A/P57A, R39A/K56A/P57A and S35A/R39A/K56A/P57A mutants of caZαPKZ were tested at a [P]/[N] ratio of 4. The plots are clustered in three groups by using k-means clustering algorithm. The names of phosphate groups inside of parentheses indicate the possible interaction between the residues in the mutants and the backbone phosphates. An asterisk indicates the phosphate groups that lose one of their residue interactions by mutation. In this case, their interactions with proteins partially remain.
Based on the functional study of multisite mutants, we were able to identify the minimal recognition motif of caZαPKZ and interacting phosphates. The K56A/P57A and S35A/K56A/P57A mutants were expected to lose the interaction with P0 and P1, and showed about 50% retention activity relative to the wild-type activity (Figure 3 and Supplementary Figure S7). The R39A/K56A/P57A mutant, which would not interact with P0, P1 and P4, had no detectable B-to-Z transition activity (Figure 3 and Supplementary Figure S7). In addition, the R39A/K56A mutant, in which the interaction with P0 and P4 is abolished, showed about 80% activity (Figure 3 and Supplementary Figure S7). These results indicate that P2 and P3 backbone interactions are not sufficient to stabilize the Z-DNA conformer and suggest that interactions with at least three backbone phosphates, P1, P2 and P3 or P2, P3 and P4, are required. Therefore, the structural geometry of residues involved in either P1, P2 and P3 or P2, P3 and P4 recognition is a likely minimal structural motif of Z-DNA recognition (Figure 4). On the protein side, Lys34, Asn38, Tyr42, Pro58 and Trp60, which are involved in P2 and P3 recognition, Pro57 for P1 binding and Arg39 for P4 binding compose the minimal binding motif for Z-DNA binding (Figure 4).
Figure 4.
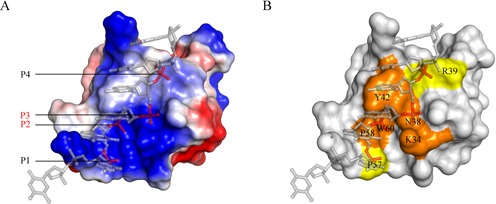
The Z-DNA binding surface of caZαPKZ. (A) Electrostatic distribution of caZαPKZ. Positively charged, negatively charged and hydrophobic regions are depicted in blue, red and white, respectively. P2 and P3 are labeled in red, and P1 and P4 are in black. (B) The Z-DNA binding surface of caZαPKZ. Key residues involved in P2 and P3 binding are colored orange, and P1 or P4 binding residues are shown in yellow.
B-to-Z transition induced by caZαPKZ is faster than that induced by other Zα domains
We examined the B-to-Z transition kinetics by measuring the CD spectra at 255 nm for 2000 s. The B-to-Z transition rate and half transition time of ds-(dCdG)6 induced by caZαPKZ were calculated to be 15.3 (ms−1) and 45.2 s, respectively, by fitting the time course CD using non-linear regression analyses (Figure 5 and Supplementary Table S3). However, B-to-Z transition rates of hZαADAR1, hZαDAI and yabZαE3L were 5.89, 2.51 and 2.39 (ms−1), respectively (Figure 5). Accordingly, the half transition times of hZαADAR1, hZαDAI and yabZαE3L were 117.6, 275.9 and 290.5 s, respectively (Supplementary Table S3). Therefore, the B-to-Z transition rate of caZαPKZ is 2.6 times faster than that of hZαADAR1, and 6.1 and 6.4 times faster than those of hZαDAI and yabZαE3L, respectively.
Figure 5.
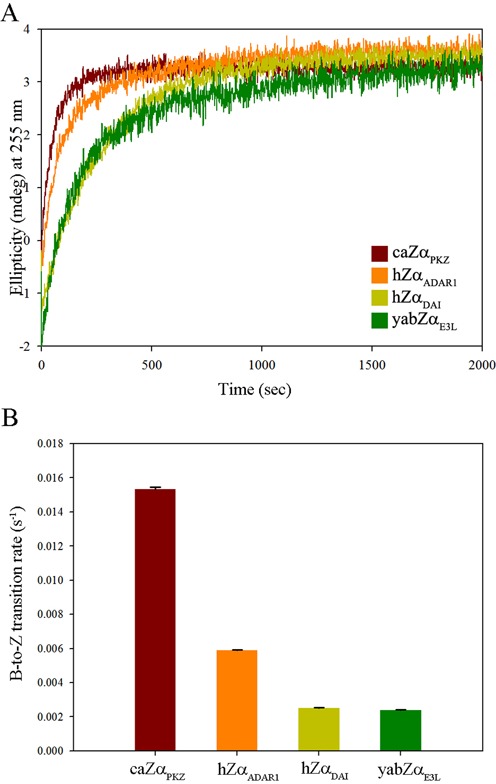
B-to-Z transition kinetics of Zα domains. (A) CD spectra of 15 μM of ds-(dCdG)6 were monitored at 255 nm in the presence of caZαPKZ (brown), hZαADAR1 (orange), hZαDAI (yellow) and yabZαE3L (green). (B) The calculated B-to-Z transition rates (s−1) with error bars from caZαPKZ (brown), hZαADAR1 (orange), hZαDAI (yellow) and yabZαE3L (green).
B-to-Z transition activity of caZαPKZ mutants
The time course B-to-Z transition by caZαPKZ mutants was monitored by CD spectroscopy in order to provide the molecular basis for the high transition rate of caZαPKZ. The replacement of Ser35 or Arg39 in the α3 core had only a marginal effect on the B-to-Z transition rate (Supplementary Figure S7 and Supplementary Table S4). However, mutations in residues in the β-wing significantly affected the B-to-Z transition rate; the calculated half transition time was 239 s for the K56A mutant. The activity of the double-mutant K56A/P57A was more drastically reduced, resulting in a half transition time exceeding 527 s (Supplementary Figure S7 and Supplementary Table S4). These results confirmed that the B-to-Z transition rate is largely affected by the interaction between the β-wing of caZαPKZ and Z-DNA. While residues in β-wing affect the B-to-Z transition rate, K34, N38, Y42 and W60 seem to be essential for B-to-Z transition, since their substitution to Ala almost completely demolished the transition (Figure 3 and Supplementary Figures S4 and S7). We further investigated the roles of Ser35 and Lys56 by measuring the transition activity of S35K and K56T mutants since their corresponding residues in hZαADAR1 are Lys and Thr, respectively, in the sequential and structure alignments (Figures 1A and 2). On the other hand, we also made hZαADAR1 mutants by replacing Lys170 and Thr191 with Ser and Lys, respectively, to make caZαPKZ-mimicking mutants. The time course CD spectra of caZαPKZ mutants (K56T and S35K/K56T), which have no positive charge at the β-wing, showed a decreased B-to-Z transition rate (Figure 6A and B, and Supplementary Table S5). By contrast, the K56R mutant showed a B-to-Z transition rate that was two times faster than that of wild-type caZαPKZ (Figure 6 and Supplementary Table S3). The caZαPKZ-mimicking mutants of hZαADAR1, T191K and K170S/T191K displayed higher B-to-Z transition activity than the wild-type hZαADAR1, while K170S showed the reduced activity (Figure 6C and D, and Supplementary Table S5), although the difference was not as high as that in caZαPKZ. These results confirm that charge–charge interaction between the β-wing and DNA backbone is important for the fast kinetics of Zα. Furthermore, we observed that the B-to-Z transition activity was decreased when the reaction was performed in the presence of NaCl (Figure 7), which further demonstrates that the charge–charge interaction is critical for the B-to-Z transition. The reduction of B-to-Z transition rate is more severe in caZαPKZ than in hZαADAR1, suggesting that the effect of charge–charge interaction on B-to-Z transition activity plays a more critical role in the case of caZαPKZ (Figure 7).
Figure 6.
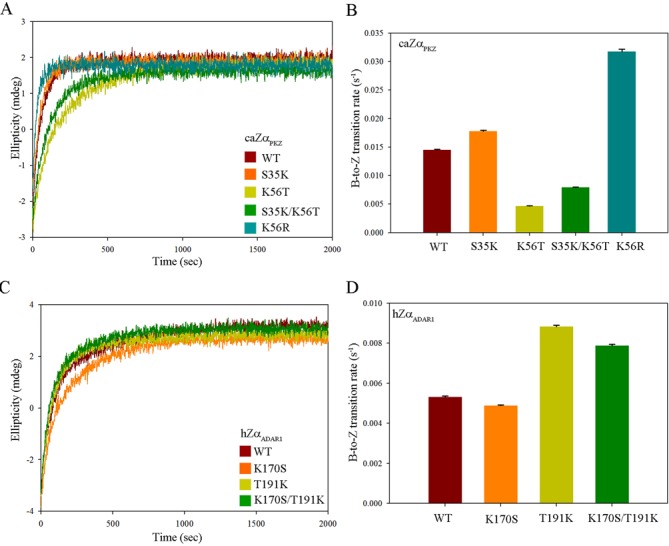
B-to-Z transition activity of caZαPKZ and hZαADAR1. (A) Time course CD spectra of caZαPKZ and S35K, K56T, K56R and S35K/K56T mutants. (B) The calculated B-to-Z transition rates (s−1) of the wild-type, S35K, K56T, S35K/K56T and K56R caZαPKZ are represented by brown, orange, lime, green and turquoise colors, respectively. (C) Time course CD spectra of hZαADAR1 and K170S, T191K and K170S/T191K mutants. (D) The calculated B-to-Z transition rates (s−1) of the wild-type, K170S, T191K and K170S/T191K mutant hZαADAR1 are represented by brown, orange, lime and green colors, respectively.
Figure 7.
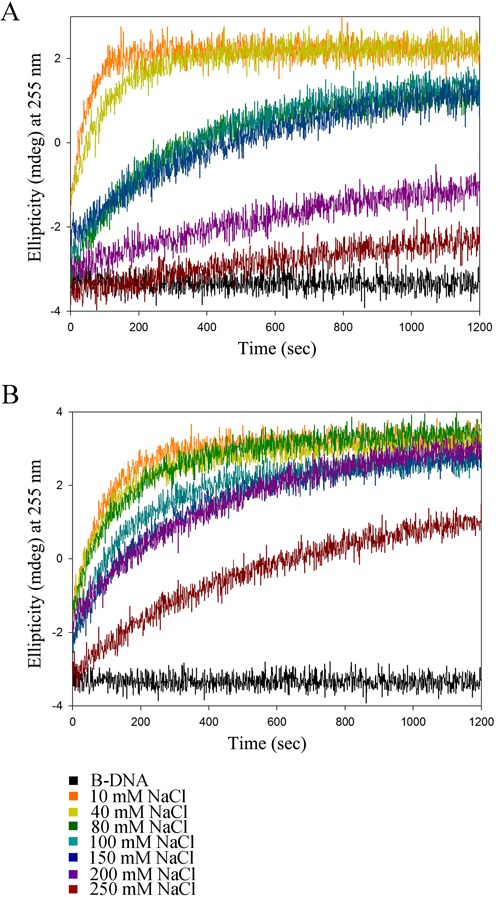
Salt effect on the B-to-Z transition. (A) Time course CD spectra of Z-DNA:caZαPKZ in various NaCl concentrations (10, 40, 80, 100, 150, 200 and 250 mM). The ellipticities (mdeg) monitored at 255 nm are indicated by different colors. (B) Time course CD spectra of Z-DNA:hZαADAR1 in the same conditions with the same color schemes as in Figure 7A.
DNA binding kinetics of ZBPs
By functional studies of the mutants, we confirmed that the positive charge in the β-wing can enhance the B-to-Z transition rate. Given this observation, how does the positive charge in the β-wing affect the B-to-Z transition rate? It was proposed that Zα bind to B-DNA prior to the transition to Z-DNA (40,41), so the association rate of Zα to DNA possibly contributes to the B-to-Z transition rate. Under this hypothesis, it is expected that a positively charged residue can enhance the transition rate by increasing the association of negatively charged DNA molecules to the positively charged protein molecules (42). To further analyze the correlation between the association rate and the B-to-Z transition rate, the kinetic parameters for the binding of the caZαPKZ, hZαADAR1 and their mutants to DNA were calculated from the Bio-Layer Interferometry measurement of the interaction between DNA and Zα and their correlation with the B-to-Z transition rate (s−1) was examined (Supplementary Figures S8, S9 and S10; Supplementary Table S6). We found that B-to-Z transition rates of the wild-type and mutants caZαPKZ are proportional to the association rate constant kon (Supplementary Figure S9E) that is also proportional to the number of positively charged residues in the Z-DNA binding surface. A similar correlation is also found in hZαADAR1, although the correlation coefficient (r) is relatively lower than that of caZαPKZ (Supplementary Figure S10 for hZαADAR1).
DISCUSSION
We have compared the Z-DNA binding structure and B-to-Z transition activity of caZαPKZ with those of other Zα domains. A structural comparison revealed not only the conserved overall binding mode of caZαPKZ but also some distinctive features that appear to be important to its higher B-to-Z transition rate. Unlike other ZBDs, caZαPKZ harbors a positively charged residue in the β-wing, Lys56, which plays a key role in DNA binding by forming a wide interaction with DNA, including bonds with both P1 and P0 (Figure 2A and C, and Supplementary Figure S2B).
Although the overall charge distribution is not conserved among ZBDs, positively charged residues and hydrophobic residues are commonly observed in the α3 core and in the β-wing, respectively, which play essential roles in Z-DNA binding. However, it seems that the positively charged residues have a more significant effect on DNA binding and the B-to-Z transition than hydrophobic residues. There are three basic residues involved in Z-DNA recognition in caZαPKZ (Lys34, Arg38 and Lys56) and hZαADAR1 (Lys169, Lys170 and Arg174) (Figure 1A). However, hZαDAI and yabZαE3L have only two positive residues in the Z-DNA binding surface (Figure 1A). Accordingly, hZαADAR1 induces 2.3 and 2.5 times faster B-to-Z transition than hZαDAI and yabZαE3L, respectively (Figure 5B). In contrast, the B-to-Z transition rate of caZαPKZ, which has one more basic residue in the β-wing than hZαDAI and yabZαE3L, is 6.1 and 6.4 times faster than that of hZαDAI and yabZαE3L, respectively (Figure 5B and Supplementary Table S3). These results suggest that the contribution of positively charged residues during the B-to-Z transition is more significant when they are present in the β-wing than in the α3 helix. Consistently, the K56T mutation in caZαPKZ significantly lowered the transition rate from 14.5 ms−1 to 4.68 ms−1 (Figure 6A and B, and Supplementary Table S5), while the addition of a positively charged residue in the β-wing enhanced the activity of hZαADAR1 from 5.32 ms−1 to 8.83 ms−1 (Figure 6C and D, and Supplementary Table S5). However, the effect of charged residues in the α3 helix seems to be not as significant as the effect of charged residues in the β-wing, although the positive charge in the α3 helix still contributes to an increased B-to-Z transition rate. Furthermore, the S35K mutant showed a higher transition rate than the wild-type caZαPKZ (Figure 6A and B, and Supplementary Table S5), and the K170S mutant showed a slightly decreased B-to-Z transition rate compared to the wild-type hZαADAR1 (Figure 6C and D, and Supplementary Table S5). These results are also consistent with the kinetic study of the viral Zα protein; the positively charged amino acids in the β-wing of ZαE3L enhanced the B-to-Z transition activity (28). Taking these results together, we conclude that the positively charged residue in the β-wing could facilitate the B-to-Z transition (Figure 6). The fastest B-to-Z transition rate of the K56R mutant of caZαPKZ among wild-type and all mutant ZBDs (Figures 5 and 6, and Supplementary Table S3) and the reduction of the transition rate under high salt conditions (Figure 7) also support our conclusion.
From a structural point of view, Arg56 in the model of the K56R mutant of caZαPKZ can fit nicely into the pocket between P0 and P1, suggesting that Arg can form a tight interaction with P0 and P1 groups (Supplementary Figure S11). From the further analyses of their DNA binding kinetics (Supplementary Figures S8–S10), we propose that the increased transition activity is achieved by the enhanced association of ZBDs to DNA through their charge–charge interaction. However, this correlation was not found when different Zα domains were compared (Supplementary Figure S8), which is possibly explained by the fact that the contribution of a positive charge at α3 and the β-wing during the B-to-Z transition is not the same as that shown in Figure 6 and in Supplementary Table S5. Given these results, we conclude that the association rate constant is affected by the presence of a positively charged residue on the DNA binding surface, which eventually enhances the B-to-Z transition rate. Moreover, the contribution of a positive charge in the β-wing is larger than that in the helix α3.
In terms of structure, unlike the other crystal structures of ZBDs complexed with Z-DNA (19,25,31), Mn2+ ion is bound to the caZαPKZ:Z-DNA complex. It was proposed that nickel and cobalt ions can bind to guanine N7 due to the increased solvent accessibility in the Z-DNA conformation (43,44) and the binding of manganese ion to Z-DNA was recently reported (45). However, among all N7 atoms of guanine in caZαPKZ, N7 of G2 appeared to specifically participate in the coordination of the manganese ion. The manganese ion is also coordinated by O1P of C1, which is stabilized by Lys56, so Zα binding is thought to be necessary for coordinating the manganese ion. To investigate the role of manganese ion in the biochemical function of caZαPKZ, the B-to-Z transition rate of caZαPKZ was measured in the presence of 15-mM MnCl2. However, the manganese ion seems to not contribute to the fast B-to-Z transition kinetics of caZαPKZ, since the transition rate decreased from 15.3 (ms−1) to 9.49 (ms−1). hZαADAR1 and hZαDAI showed marginally increased B-to-Z transition kinetics in the presence of manganese ion (Supplementary Figure S12). Therefore, MnCl2 does not seem to be correlated with the fast B-to-Z transition kinetics of caZαPKZ, and thus the binding of manganese ion to caZαPKZ may be considered as a crystallization artifact since the crystallization buffer contained 15-mM MnCl2.
It has been postulated that the interaction of ZBP to Z-DNA is critical for various biological functions, and thus the B-to-Z transition activity of the ZBPs is relevant to the biological functions. DAI binding to the cytosolic dsDNA turns on the innate immune signal by triggering the activation of NF–kB signaling and expression of type-1 IFN (23). However, PKZ works as an innate immune sensor by recognizing poly-d(GC) as Z-DNA and turning off general translation machinery by phosphorylating eukaryotic translation initiation factor 2α (eIF2α) (8,29). Interestingly, in fish species, both PKZ and PKR are known to play an antiviral role as IFN-inducible eIF2α kinases (46). Therefore, we are intrigued by the functional relationship between PKZ and PKR in terms of the antiviral response. A recent study of PKZ and PKR in C. auratus revealed that both proteins work independently, but play a cooperative role in IFN-mediated antiviral response, although antiviral ability of fish PKZ was weaker than that of fish PKR (21). However, the pull-down experiment with poly I:C, a dsRNA mimic, showed that the DNA binding domain of PKZ does not bind to poly I:C, while PKR does bind (21). These results suggest that PKR and PKZ might independently contribute to the innate immune response by recognizing different nucleotide substrates in the initial stage of the innate immune response. Indeed, PKZ-dependent phosphorylation of eIF2α and the subsequent translational shutdown are largely induced not by dsRNA but by Z-DNA (8,29). Since the Z-DNA-induced innate immune responses conducted by DAI could be regulated by competition with other ZBPs such as ADAR1 or E3L, differential Z-DNA binding activity or the B-to-Z transition rate of Zα domains in the innate immune sensors might be a key to controlling the innate immune response. Similarly, since fish virus also harbor the ZBPs (47), it is also expected that PKZ must compete with viral ZBPs to play a role in fish innate immune response during viral infection. Therefore, the fast Z-DNA converting activity of PKZ might be necessary for competing with viral ZBPs. Further studies on the B-to-Z transition activity of Zα domains in cellular environments will clearly define the biological relevance of the structural and kinetic differences between caZαPKZ and other Zα domains. In addition, structural and functional studies on intact ZBPs are also required for better understanding of the Z-DNA-dependent innate immune response.
The current study also provides an insight into a novel ZBD. Z-DNA is known to have various roles in cellular events. Z-DNA formation is necessary for opening chromatin structure and activating transcription of target genes (48,49). Z-DNA is also known to cause genomic instability (50). For the regulation of diverse functions of Z-DNA, more ZBPs with novel ZBDs are expected to mediate various cellular processes associated with Z-DNA. The results from this study combined with those from structural and functional studies of ZαADAR1 confirm that Z-DNA can be recognized by Zα proteins through three backbone phosphates, but the interaction modes for caZαPKZ and ZαADAR1 are not the same in detail (39). Considering the size and charge distribution of the minimum unit of Z-DNA for protein binding and the diversity of the Z-DNA binding mode, it is expected that more ZBPs other than the Zα domain may be present in cells. Consistently, this possibility is partially proven by identifying putative ZBPs that interact with the Z-form of ADAM-12 NRE sequence (51). Therefore, the current study not only reveals a structural requirement for fast B-to-Z transition but also provides insight into the presence of novel ZBPs.
ACCESSION NUMBERS
PDB IDs: 4KMF, 1QBJ, 1J75, 1SFU, 3EYI.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [1–5].
FUNDING
Samsung Science and Technology Foundation [SSTF-BA1301-01 to K.K.K.]; National Research Foundation [2009-0075300 to Y.G.K.]. Funding for open access charge: Samsung Science and Technology Foundation [SSTF-BA1301-01]; NRF [2009-0075300].
Conflict of interest statement. None declared.
Supplementary Material
Footnotes
Correspondence may also be addressed to Yang-Gyun Kim. Email: ygkimmit@skku.edu, Tel: +82 31 299 4563; Fax:+82 31 299 4575
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code is 4KMF).
REFERENCES
- 1.Holcik M., Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 2.Samuel C.E. The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J. Biol. Chem. 1993;268:7603–7606. [PubMed] [Google Scholar]
- 3.Wek R.C., Jiang H.Y., Anthony T.G. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 4.Williams B.R. PKR: a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 5.Wu S., Kaufman R.J. A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J. Biol. Chem. 1997;272:1291–1296. doi: 10.1074/jbc.272.2.1291. [DOI] [PubMed] [Google Scholar]
- 6.Tan S.L., Gale M.J., Jr, Katze M.G. Double-stranded RNA-independent dimerization of interferon-induced protein kinase PKR and inhibition of dimerization by the cellular P58IPK inhibitor. Mol. Cell. Biol. 1998;18:2431–2443. doi: 10.1128/mcb.18.5.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su J., Zhu Z., Wang Y. Molecular cloning, characterization and expression analysis of the PKZ gene in rare minnow Gobiocypris rarus. Fish Shellfish Immunol. 2008;25:106–113. doi: 10.1016/j.fsi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Bergan V., Jagus R., Lauksund S., Kileng O., Robertsen B. The Atlantic salmon Z-DNA binding protein kinase phosphorylates translation initiation factor 2 alpha and constitutes a unique orthologue to the mammalian dsRNA-activated protein kinase R. FEBS J. 2008;275:184–197. doi: 10.1111/j.1742-4658.2007.06188.x. [DOI] [PubMed] [Google Scholar]
- 9.Rothenburg S., Deigendesch N., Dittmar K., Koch-Nolte F., Haag F., Lowenhaupt K., Rich A. A PKR-like eukaryotic initiation factor 2alpha kinase from zebrafish contains Z-DNA binding domains instead of dsRNA binding domains. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1602–1607. doi: 10.1073/pnas.0408714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu C.Y., Zhang Y.B., Huang G.P., Zhang Q.Y., Gui J.F. Molecular cloning and characterisation of a fish PKR-like gene from cultured CAB cells induced by UV-inactivated virus. Fish Shellfish Immunol. 2004;17:353–366. doi: 10.1016/j.fsi.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Rich A. DNA comes in many forms. Gene. 1993;135:99–109. doi: 10.1016/0378-1119(93)90054-7. [DOI] [PubMed] [Google Scholar]
- 12.Johnston B.H. Generation and detection of Z-DNA. Methods Enzymol. 1992;211:127–158. doi: 10.1016/0076-6879(92)11009-8. [DOI] [PubMed] [Google Scholar]
- 13.Rich A., Zhang S. Timeline: Z-DNA: the long road to biological function. Nat. Rev. Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 14.Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B–Z transition in poly(dG-m5dC).poly(dG-m5dC) Proc. Natl. Acad. Sci. U.S.A. 1981;78:1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mamajanov I., Engelhart A.E., Bean H.D., Hud N.V. DNA and RNA in anhydrous media: duplex, triplex, and G-quadruplex secondary structures in a deep eutectic solvent. Angew. Chem. Int. Ed. Engl. 2010;49:6310–6314. doi: 10.1002/anie.201001561. [DOI] [PubMed] [Google Scholar]
- 16.Rahmouni A.R., Wells R.D. Stabilization of Z DNA in vivo by localized supercoiling. Science. 1989;246:358–363. doi: 10.1126/science.2678475. [DOI] [PubMed] [Google Scholar]
- 17.Kim D., Lee Y.H., Hwang H.Y., Kim K.K., Park H.J. Z-DNA binding proteins as targets for structure-based virtual screening. Curr. Drug Targets. 2010;11:335–344. doi: 10.2174/138945010790711905. [DOI] [PubMed] [Google Scholar]
- 18.Herbert A., Lowenhaupt K., Spitzner J., Rich A. Chicken double-stranded RNA adenosine deaminase has apparent specificity for Z-DNA. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7550–7554. doi: 10.1073/pnas.92.16.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwartz T., Behlke J., Lowenhaupt K., Heinemann U., Rich A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 2001;8:761–765. doi: 10.1038/nsb0901-761. [DOI] [PubMed] [Google Scholar]
- 20.Kim Y.G., Muralinath M., Brandt T., Pearcy M., Hauns K., Lowenhaupt K., Jacobs B.L., Rich A. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6974–6979. doi: 10.1073/pnas.0431131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu T.K., Zhang Y.B., Liu Y., Sun F., Gui J.F. Cooperative roles of fish protein kinase containing Z-DNA binding domains and double-stranded RNA-dependent protein kinase in interferon-mediated antiviral response. J. Virol. 2011;85:12769–12780. doi: 10.1128/JVI.05849-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takaoka A., Wang Z., Choi M.K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K., et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 23.Takaoka A., Taniguchi T. Cytosolic DNA recognition for triggering innate immune responses. Adv. Drug Deliv. Rev. 2008;60:847–857. doi: 10.1016/j.addr.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z., Choi M.K., Ban T., Yanai H., Negishi H., Lu Y., Tamura T., Takaoka A., Nishikura K., Taniguchi T. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz T., Rould M.A., Lowenhaupt K., Herbert A., Rich A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 26.Ha S.C., Lokanath N.K., Van Quyen D., Wu C.A., Lowenhaupt K., Rich A., Kim Y.G., Kim K.K. A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zalpha bound to DNA. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14367–14372. doi: 10.1073/pnas.0405586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ha S.C., Kim D., Hwang H.Y., Rich A., Kim Y.G., Kim K.K. The crystal structure of the second Z-DNA binding domain of human DAI (ZBP1) in complex with Z-DNA reveals an unusual binding mode to Z-DNA. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20671–20676. doi: 10.1073/pnas.0810463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quyen D.V., Ha S.C., Lowenhaupt K., Rich A., Kim K.K., Kim Y.G. Characterization of DNA-binding activity of Z alpha domains from poxviruses and the importance of the beta-wing regions in converting B-DNA to Z-DNA. Nucleic Acids Res. 2007;35:7714–7720. doi: 10.1093/nar/gkm748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu C.X., Wang S.J., Lin G., Hu C.Y. The Zalpha domain of PKZ from Carassius auratus can bind to d(GC)(n) in negative supercoils. Fish Shellfish Immunol. 2010;28:783–788. doi: 10.1016/j.fsi.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 30.Kim D., Hwang H.Y., Kim Y.G., Kim K.K. Crystallization and preliminary X-ray crystallographic studies of the Z-DNA-binding domain of a PKR-like kinase (PKZ) in complex with Z-DNA. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 2009;65:267–270. doi: 10.1107/S1744309109002504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ha S.C., Lowenhaupt K., Rich A., Kim Y.G., Kim K.K. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005;437:1183–1186. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- 32.Vagin A., Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Cowtan K. dm: An automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newsl. Protein Crystallogr. 1994;31:34–38. [Google Scholar]
- 35.Emsley P., Cowtan K. Coot: model-building tool for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Chen V.B., Arendall W.B., III, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLano W.L. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 38.Kim K., Khayrutdinov B.I., Lee C.K., Cheong H.K., Kang S.W., Park H., Lee S., Kim Y.G., Jee J., Rich A., et al. Solution structure of the Zbeta domain of human DNA-dependent activator of IFN-regulatory factors and its binding modes to B- and Z-DNAs. Proc. Natl. Acad. Sci. U.S.A. 2011;108:6921–6926. doi: 10.1073/pnas.1014898107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ha S.C., Choi J., Hwang H.Y., Rich A., Kim Y.G., Kim K.K. The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ alpha(ADAR1) Nucleic Acids Res. 2009;37:629–637. doi: 10.1093/nar/gkn976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee Y.M., Kim H.E., Lee E.H., Seo Y.J., Lee A.R., Lee J.H. NMR investigation on the DNA binding and B-Z transition pathway of the Zalpha domain of human ADAR1. Biophys. Chem. 2012;172C:18–25. doi: 10.1016/j.bpc.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Bae S., Kim D., Kim K.K., Kim Y.G., Hohng S. Intrinsic Z-DNA is stabilized by the conformational selection mechanism of Z-DNA-binding proteins. J. Am. Chem. Soc. 2011;133:668–671. doi: 10.1021/ja107498y. [DOI] [PubMed] [Google Scholar]
- 42.von Hippel P.H., Berg O.G. Facilitated target location in biological systems. J. Biol. Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 43.Abrescia N.A., Huynh-Dinh T., Subirana J.A. Nickel-guanine interactions in DNA: crystal structure of nickel-d[CGTGTACACG]2. J. Biol. Inorg. Chem. 2002;7:195–199. doi: 10.1007/s007750100286. [DOI] [PubMed] [Google Scholar]
- 44.Gao Y.G., Sriram M., Wang A.H. Crystallographic studies of metal ion-DNA interactions: different binding modes of cobalt(II), copper(II) and barium(II) to N7 of guanines in Z-DNA and a drug-DNA complex. Nucleic Acids Res. 1993;21:4093–4101. doi: 10.1093/nar/21.17.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mandal P.K., Venkadesh S., Gautham N. Interactions of Mn(2+) with a non-self-complementary Z-type DNA duplex. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 2012;68:1420–1426. doi: 10.1107/S1744309112041759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rothenburg S., Deigendesch N., Dey M., Dever T.E., Tazi L. Double-stranded RNA-activated protein kinase PKR of fishes and amphibians: varying the number of double-stranded RNA binding domains and lineage-specific duplications. BMC Biol. 2008;6:12. doi: 10.1186/1741-7007-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tome A.R., Kus K., Correia S., Paulo L.M., Zacarias S., de Rosa M., Figueiredo D., Parkhouse R.M., Athanasiadis A. Crystal structure of a poxvirus-like zalpha domain from cyprinid herpesvirus 3. J. Virol. 2013;87:3998–4004. doi: 10.1128/JVI.03116-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu R., Liu H., Chen X., Kirby M., Brown P.O., Zhao K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–318. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 49.Xu Y.Z., Thuraisingam T., Marino R., Radzioch D. Recruitment of SWI/SNF complex is required for transcriptional activation of the SLC11A1 gene during macrophage differentiation of HL-60 cells. J. Biol. Chem. 2011;286:12839–12849. doi: 10.1074/jbc.M110.185637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao J., Bacolla A., Wang G., Vasquez K.M. Non-B DNA structure-induced genetic instability and evolution. Cell. Mol. Life Sci. 2010;67:43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ray B.K., Dhar S., Shakya A., Ray A. Z-DNA-forming silencer in the first exon regulates human ADAM-12 gene expression. Proc. Natl. Acad. Sci. U.S.A. 2011;108:103–108. doi: 10.1073/pnas.1008831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


