Abstract
Conjugation of DNA through a type IV secretion system (T4SS) drives horizontal gene transfer. Yet little is known on the diversity of these nanomachines. We previously found that T4SS can be divided in eight classes based on the phylogeny of the only ubiquitous protein of T4SS (VirB4). Here, we use an ab initio approach to identify protein families systematically and specifically associated with VirB4 in each class. We built profiles for these proteins and used them to scan 2262 genomes for the presence of T4SS. Our analysis led to the identification of thousands of occurrences of 116 protein families for a total of 1623 T4SS. Importantly, we could identify almost always in our profiles the essential genes of well-studied T4SS. This allowed us to build a database with the largest number of T4SS described to date. Using profile–profile alignments, we reveal many new cases of homology between components of distant classes of T4SS. We mapped these similarities on the T4SS phylogenetic tree and thus obtained the patterns of acquisition and loss of these protein families in the history of T4SS. The identification of the key VirB4-associated proteins paves the way toward experimental analysis of poorly characterized T4SS classes.
INTRODUCTION
Prokaryotes have the ability to adapt quickly by acquiring genes from other prokaryotes (1–3). Conjugation, which is one of the major mechanisms of gene transfer, requires cell-to-cell contact and is able to deliver the whole genome of one cell into another. Conjugation-specific proteins are found in all major taxa of prokaryotes, even though experimental evidence is still mostly restricted to Proteobacteria and Firmicutes (4–8). The most frequent mechanism of DNA conjugation involves the passage of single-stranded DNA (ssDNA) from the donor cell to the recipient, upon which replication re-establishes double-stranded (dsDNA) copies in each cell (7). This mechanism relies on three major components: a relaxosome, a coupling protein (T4CP) and a type IV secretion system (T4SS). The relaxosome includes a protein essential for conjugation—the relaxase (MOB)—that nicks the dsDNA and binds the resulting ssDNA at the origin of transfer (see (7,9) for reviews). The relaxase bound to the ssDNA molecule is coupled to a T4SS by a T4CP and translocated through the donor membrane(s) to the cytoplasm of the recipient. Two different coupling proteins have been identified: VirD4 and TcpA. TcpA is found within certain systems of Firmicutes, and is more closely related to FtsK, a protein involved in chromosome segregation, than to VirD4 (10,11). VirD4 is associated with the vast majority of T4SS and probably originated from an ssDNA translocase (12). Some mobile genetic elements encode a relaxase and occasionally a T4CP but no T4SS. These elements are very abundant in bacterial genomes and are called ‘mobilizable’ because they use a T4SS encoded in trans. Most T4SS are thought to be involved in conjugation (nucleoprotein secretion), but some are specialized in protein secretion, allowing the delivery of effector proteins to the cytosol of eukaryotic organisms. These T4SS typically lack a relaxase (MOBless T4SS), but require a T4CP (see (5) for exceptions). Several systems are able to deliver both the DNA-bound relaxase and other protein effectors (13–15). There are also examples of T4SS involved in DNA import (the ComB system of Helicobacter pylori (16)) or in DNA secretion (the GGI system of Neisseria gonorrhoeae (17)) associated with natural transformation. T4SS are thus remarkably flexible nanomachines adapted to translocate large macromolecules through multiple cell membranes.
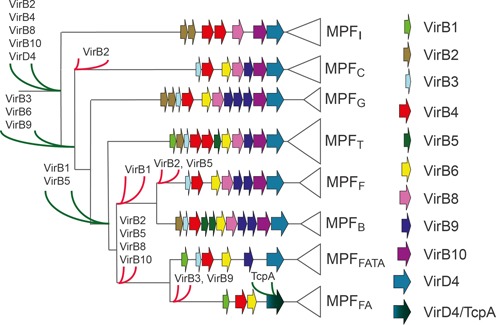
The plasticity of T4SS results in a diversity of systems, most of which are yet poorly characterized. One complication in the study of T4SS is the lack of a standard nomenclature: genes of similar names are not necessarily homologs and homologs do not have necessarily the same names. Here, we follow the convention that mating-pair formation (MPF) genes are labeled with the name of the associated mobile element, and protein profiles are labeled with the name of the MPF class. For example, TraBF is the protein TraB encoded by plasmid F and TraBMPFF is the protein profile for the protein family including TraBF of MPFF. The only exception concerns the VirB proteins, which by default concern the T-DNA transfer virB system of Agrobacterium tumefaciens, and whose names are in general used without ambiguity (i.e. similar names correspond to homologs). We use the VirB system, composed of 11 genes from VirB1 to VirB11, as a model because it is by far the best characterized T4SS (9,18) (Figure 1). The core secretion channel complex of the VirB T4SS (including VirB7, VirB9 and VirB10) that spans the periplasm and both cell membranes is thought to be the first to assembly. VirB10 lines the inner surface of the core complex chambers, and spans the whole length of the core complex, forming the bottom ring of the inner membrane layer. The protein VirB9 forms the outer sheath of the core complex that interacts and is stabilized by VirB7, a small lipoprotein (9,19). The three proteins VirB3, VirB6 and VirB8 are thought to join the core complex to produce the inner-membrane pore (20). VirB6 is a polytopic membrane protein with a number of transmembrane domains (TMDs) and a large central periplasmic loop (21). The role of VirB3 is not yet clear, even if the protein is essential for pilus assembly and substrate translocation. In some systems, VirB3 is fused to VirB4, which suggests a strong functional link between the two (22). Three AAA+ ATPases (VirB4, VirB11 and the T4CP) join the assembled complex, one of which (VirB4) is implicated in energizing pilus biogenesis (23). The pilus is composed of a major and a minor pilin (respectively VirB2 and VirB5). Finally, VirB1 is a non-essential transglycosylase that degrades peptidoglycan and thus facilitates T4SS assembly across the cell wall (24,25).
Figure 1.
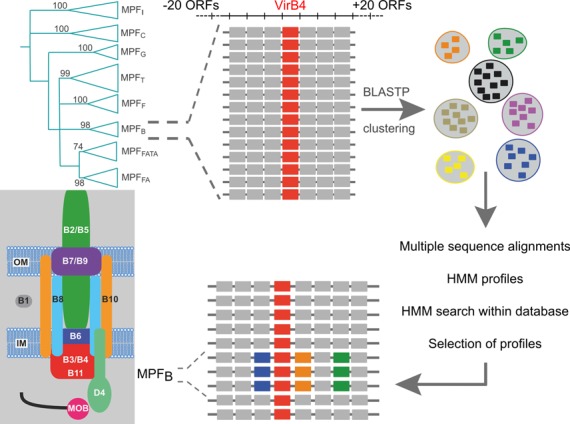
Procedure used to create mating-pair formation (MPF) protein profiles. We used all the genes found within a frame of −20/+20 genes around the VirB4 proteins of each clade (named MPFI, MPFC, MPFG, MPFT, MPFF, MPFB, MPFFATA and MPFFA—see text for details) of its phylogeny (11). Numbers on the phylogeny correspond to bootstrap values, from (11). With these seven datasets of proteins (one for each class, except for MPFT for which we already had the protein profiles) we performed all-versus-all BLASTP and used the scores to build protein families. We made protein alignments of these families, and kept the ones that give hits within the class. Thumbnail: scheme of the virB system.
In a recent study, we found that the phylogeny of VirB4, the only ubiquitous protein with recognizable homologs in all known T4SS, is divided in eight large robust clades that correspond to eight MPF classes (11). Based on these results we proposed an evolution-aware classification of T4SS that shows strong associations with prokaryote's systematics and, to some extent, to the structure of the cell envelope (11). The association between T4SS composition and cell envelope structure has been recently reviewed (20). It should be stressed that within a given MPF class there is also co-evolution between the composition of the membrane and the T4SS (26). Four MPF classes encompass the conjugation systems of Proteobacteria and closely related taxa: VirB-like systems are the most numerous (MPFT); F-like systems are particularly abundant in plasmids of γ-Proteobacteria (MPFF); R64-like systems are much rarer (MPFI); and ICEHin1056-like systems are almost exclusively found as integrative elements (ICE) in Proteobacteria (MPFG) (6,8,27). Two MPF classes are much less well known and include systems present only in Cyanobacteria (MPFC) or in Bacteroidetes (MPFB). Finally, we delimited two different MPF classes in monoderms (organisms devoid of an outer membrane). One class is found in Firmicutes and Actinobacteria (MPFFA) whereas the other is also found in Tenericutes and Archaea (MPFFATA).
T4SS are very diverse: 10 of 11 virB genes are essential (24), whereas the conjugative system of R64 is encoded by 49 genes of which 23 are essential for solid mating plus 12 for liquid mating (28). The exact number of genes and their essentiality for T4SS assembly or function is unknown in many classes of T4SS. Some systems of Firmicutes are thought to lack pili altogether (4), e.g. the T4SS of the pCF10 plasmid of Enterococcus faecalis, the pGO1 plasmid or the ICE TnGBS of Streptococcus encode adhesins that stabilize the mating process. In taxa such as Archaea, Actinobacteria or Cyanobacteria, there are very few reports on the mechanisms of ssDNA conjugation, and none, to the best of our knowledge, on the T4SS structure and composition. Yet these taxa encode homologs of VirB4 and VirD4 in their mobile genetic elements suggesting the presence of T4SS-mediated conjugation (8).
Here, we took advantage of our previous dataset of VirB4 homologs to characterize all MPF classes. The repertoires, diversity and evolution of relaxases have been recently reported (29). Therefore, we focused our attention on T4SS. To establish the repertoire of protein families typical of each class of T4SS, we detected the families of genes systematically associated and co-localized with VirB4 in the eight MPF classes. With these families we built protein profiles that we used to detect and classify T4SS. This resulted in a much more complete database of T4SS than those currently available. Using profile–profile alignments we identified distant homologies between protein families of different MPF classes thereby providing the first large-scale systematic analysis of protein homology between all different MPF classes. Finally, we built a web resource, called CONJdb, that presents all our data in a searchable and comprehensive manner.
MATERIALS AND METHODS
Data
Data on complete prokaryotic chromosomes and plasmids were taken from GenBank Refseq (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/, last accessed February 2013). The dataset of 2262 complete prokaryotic genomes comprised 2393 chromosomes and 1813 plasmids. We used the annotations of the GenBank files, having removed all pseudogenes and proteins with inner stop codons. Some protein profiles were taken and improved from our previous work (8).
Construction of protein profiles
Figure 1 describes the procedure used to construct the protein profiles. We searched for genes encoding TraU/VirB4 in the genomes using HMMER (see below). We then gathered the 20 genes on each side of each traU/virB4 gene. This resulted in seven sets of proteins, one for each T4SS class (except for the previously analyzed MPFT). We made all-against-all BLASTP searches in each set (default settings) and used the output to build protein clusters using SiLiX (30) (identity ≥30% and overlap >50%). We carried out a multiple alignment of the proteins in each cluster with more than five proteins using MUSCLE (31) (default parameters). We used the multiple alignments to build phylogenetic trees using PHYML (32). With these two pieces of evidence we removed the very few cases of extreme divergence, the proteins that were too short and the proteins that were too long (typically false positives, fusions or fissions of proteins motivated by sequencing errors or pseudogenization). Then, we re-built multiple alignments of the selected proteins with MUSCLE, checked manually the alignments and trimmed them to remove poorly aligned regions at the edges, if relevant. Finally, we used HMMER 3.0 (33) to build protein profiles from the manually curated multiple alignments.
It should be pointed out that homologous proteins with very little sequence similarity might escape the BLASTP detection used in the clustering procedure if a protein does not find a single hit in a family. However, the use of a method allowing detection of these homologs would probably not help at this stage, since the protein profiles must be built from proteins providing reliable multiple alignments. For a few very divergent proteins this may lead to their exclusion from the analysis. For families aligning poorly with other sub-families, our method will provide several independent profiles. In this case, the profile–profile alignment procedure will identify the homology between families.
Identification and analysis of MPF protein profiles
To identify the profiles corresponding to MPF proteins within all the protein profiles, we performed hidden Markov model (HMM) searches on the genome data. We used HMMER to identify components of the T4SS. We kept the hits showing an i-e-value < 0.01 and a coverage of the protein profile higher than 50%. Some T4SS genes may be more than 20 genes apart from the VirB4 homolog (even if we could not find a single such occurrence in the model systems). The proteins encoded by these genes will not be used to build the protein profiles. Yet these proteins will be identified by these profiles when we scan the genomes. We will keep them for further analysis when more than 50% of the hits of the protein family are located in a neighborhood of −20/+20 genes around a traU/virB4 gene, i.e. as long as the elements of the protein family are not systematically distant from VirB4. All genes regarded as essential components of the T4SS were in this situation. Then, we applied the following procedure on the remaining HMM profiles. We compared all pairs of profiles with each other and with the ones of the Pfam 26 database (13 672 protein families) using HHsearch (34) (p < 0.001 threshold) (35). As common usage in comparative genomics, the pairs of significant hits were regarded as homologs. We cannot totally exclude the possibility that the very low similarity between some families could be due to convergence. We removed from further analysis the profiles whose Pfam annotation was clearly related to functions other than conjugation. We inspected the co-localization patterns of the hits of the remaining profiles. We removed protein profiles that gave hits systematically distant (more than five ORFs) from the others. The resulting set of profiles was used to scan the genomes and build CONJdb.
RESULTS
Characterization of T4SS protein families
We built a procedure to identify T4SS based on two well-established features (5). (i) Genes encoding the T4SS are generally grouped together in one or a few operons. (ii) VirB4 is the only protein family identified in all functional T4SS. We identified the occurrences of virB4 in 2269 complete prokaryotic genomes using a previously defined protein profile (see the Materials and Methods section). These proteins were found in all major taxa. We then fetched the genes in the genomic neighborhood of each of the 1623 virB4 genes. Pairwise similarity searches followed by clustering and curation resulted in 652 families. The multiple alignments of each family were used to build protein profiles (HMMs), which were applied to scan the genomic data. Some T4SS components may be more than 20 genes apart from virB4. This is the case of T4SS encoded in several loci scattered in the genomes of Rickettsiales (36). These proteins were not used to build the protein profiles. Nevertheless, they were subsequently identified in the step of genome scanning with the protein profiles. They are therefore included in the analysis of T4SS. Our method has no phylogenetic bias, i.e. we do not a priori restrict protein families associated with a VirB4 class to a given taxonomic group. However, most studied T4SS are from Proteobacteria and Firmicutes and this may lead to two inevitable implicit biases in the analysis. Firstly, analogous, not homologous, proteins may fill the same function in different taxa and might be missed if there are few representatives of the taxa or if these genes are not encoded systematically close to virB4. For example, we have reported that we probably miss relaxases from Archaea and Actinobacteria (11). Secondly, proteins evolving too rapidly produce smaller protein families that might miss representatives from the T4SS more evolutionarily distant from the model systems. Candidates for such functions can be fetched using other protein features like peptide signals or TMDs (see below). In spite of this, we found T4SS in nearly all taxa for which there is a significant number of genomes. Profile–profile comparisons also showed homology between many of these profiles between MPF classes and between taxa (see below). Therefore, we believe to have uncovered the majority of protein families systematically associated with T4SS.
Some of the new profiles are not specific for T4SS because they match genes lacking a neighboring virB4 more than 50% of the times. Most of these non-specific profiles match proteins typically encoded by mobile genetic elements, like primases or zinc-finger proteins, which are not directly associated with conjugation or T4SS (Supplementary Table S1). These profiles may help the characterization of the genetic context of the T4SS but they are of little use to the computational study of these systems. In our genome scans we ignored them, except when experimental data showed their implication in conjugation. This was the case of relaxases and T4CP. Some profiles match proteins encoded by genes systematically neighboring virB4. These profiles are specific to T4SS, i.e. they contribute to their accurate identification. In the vast majority of cases, they also match proteins from one single T4SS class (Figure 2) and can therefore be used to class T4SS. Out of the 1623 VirB4 hits, 93% co-localized with other T4SS-associated profiles in Proteobacteria and 80% in other taxa. Hence, in the vast majority of cases, VirB4 is indeed associated with T4SS. Exceptions may be due to ongoing genetic degradation of conjugation systems, to the existence of unknown classes of T4SS and/or to co-option of VirB4 for other functions. A T4SS is assigned to a given class if the direct neighborhood of VirB4 harbors the hits of at least three protein profiles from this class. Some systems cannot be classed this way because they contain less than three hits, or no hits at all, of MPF protein profiles. We call them MPFO (‘O’ for ‘others’). They are mostly (75%) T4SS loci lacking a neighboring relaxase, thus probably devoted to protein secretion or ongoing genetic degradation.
Figure 2.
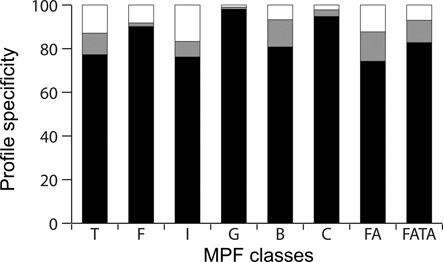
Specificity of the profiles obtained for the different mating-pair formation (MPF) classes named I, C, G, T, F, B, FATA and FA (see text for details). Black corresponds to the percentage of hits found within −20/+20 genes around a virB4 of the corresponding class. Gray corresponds to the percentage of proteins found within −20/+20 genes neighborhood of a virB4 of another class. White corresponds to the percentage of proteins that were not found to be associated with a VirB4.
The observation that profiles typically match T4SS of one single class means that homologous proteins are more similar within than between T4SS classes. This can be explained either by lack of homology between components of different T4SS classes or by tight co-evolution between these proteins and VirB4. The latter hypothesis applies to at least certain components that have known homologs between T4SS classes (e.g. VirB6 or VirB1) (20,37). Hence, homologous components exist but are not usually exchanged between classes. We used profile–profile comparisons to systematically detect significant sequence similarity between protein families of different T4SS classes (34). These protein families are similar in sequence and thus they are likely to correspond to bona fide homologs. Nevertheless, since divergence between the proteins precludes the use of phylogenetic methods to test homology, we cannot exclude the possibility of convergent evolution. Our analysis pinpointed 57 relations of similarity between protein families associated with different classes of T4SS (Figure 3), in addition to the relations between VirB4, T4CP and MOB homologs. In the following sections, we describe the protein families identified in the different T4SS (Figure 4). We then compare these among themselves and with our previously defined set of profiles for the VirB system (11).
Figure 3.
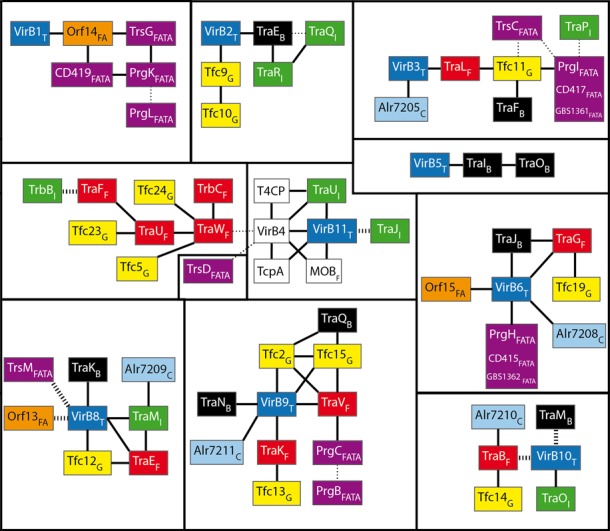
Relationships of homology between protein families of different mating-pair formation (MPF) classes. Subscript letters correspond to the I, C, G, T, F, B, FATA and FA MPF classes (see text for details). Black lines represent direct relationships, i.e. an HHsearch P-value < 0.001. Dotted wide lines correspond to relationships that have been established by structure or sequence similarity, but not by profile alignment. Dotted thin lines represent less certain relationships given by profile alignments: the HHsearch score suggests a relation of homology, but the two proteins exhibit different features (e.g., domain organization, protein length or presence of specific motifs). White squares represent profiles matching many classes (e.g. VirB4). The color scheme used for the boxes correspond to the MPF classes: blue for MPFT, red for MPFF, green for MPFI, yellow for MPFG, cyan for MPFC, black for MPFB, orange for MPFFA and purple for MPFFATA.
Figure 4.
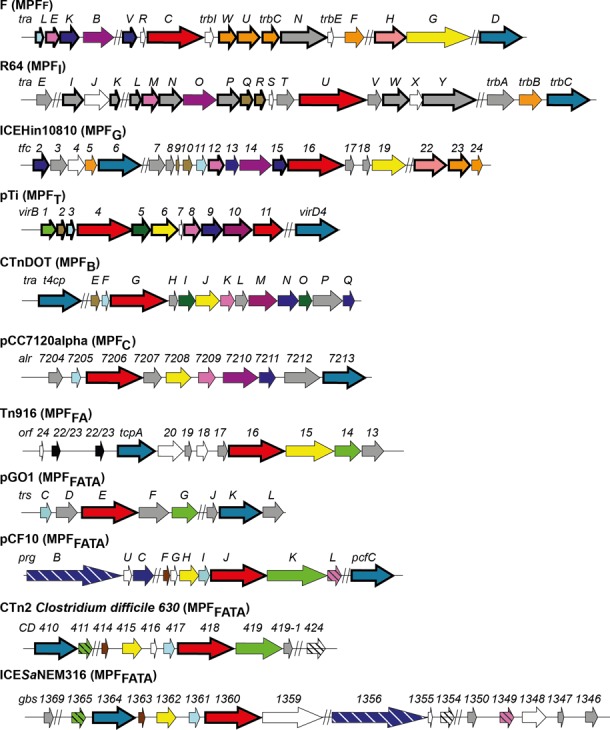
Representation of the different mating-pair formation (MPF) classes. The length of the arrows is proportional to the mean length of the corresponding genes. Bold arrows represent genes for which the corresponding protein profile was already available (6,8,11). White arrows represent genes for which we did not obtain a profile. Gray arrows represent genes for which we built a protein profile lacking homologs in other classes. A given color (except white and gray) corresponds to a single family of homologs.
MPFF
We used the F plasmid T4SS as the model of MPFF. This system is composed of 18 proteins, some homologous to VirB components (reviewed in (38)). We built profiles for the three components of the core complex of the T4SS—TraBF, TraVF and TraKF (39)—that display structural similarities to the VirB10, VirB9 and VirB7 complex (40). While these proteins do seem to have analogous roles, only the profile–profile alignments of VirB9 and TraKMPFF and TraVMPFF were significant (P-values of 7.6×10−6 and 8.3×10−05 respectively). Profile–profile comparisons of VirB10 and TraBMPFF are not significant, whereas they both exhibit the same PFAM domain (PF03743, with e-values of 4.8×10−56 and 1.3×10−39 respectively). This is because our long profiles (respectively 375 and 431 positions) only align at the region common to the much smaller profile PF03743 (187 positions). The inner-membrane pore is thought to be composed of TraGF in interaction with TraLF and TraEF (homolog to VirB3 and VirB8 respectively). The N-terminal region of TraGF is homolog to VirB6 while the C-terminal part is involved in mating-pair stabilization (41). TraNF interacts with OmpA and LPS moieties during conjugation, resulting in mating-pair stabilization (41,42). Its profile did not match any other profile. TraAF is the only pilin of MPFF (43,44), but its rapid evolution precluded the definition of a protein profile for the family. We built four different profiles for the periplasmic proteins TraWF, TraUF, TraFF and TrbCF (45), all homologous according to our profile–profile alignments (Figure 3). The inactivation of these proteins leads to shortened pili (46,47). We also built a profile for TraHF that is thought to participate in pilus extension (44,45). Overall, we have obtained 12 protein profiles for MPFF matching all known essential genes, except TraQF (pilin chaperone) and the fast-evolving TraAF pilin (48,49) (and TraXF but this is dispensable (50)). Interestingly, these three latter genes interact physically (51,52).
MPFI
We used the IncI plasmid R64 conjugative system as the model for MPFI. We built 16 protein profiles for MPFI, few of which have homologs in other classes. The profile TraOMPFI is homologous to VirB10 and thus probably part of the core complex (53). TraMMPFI is homologous to TraEMPFF/VirB8, and thus is probably part of the inner scaffold. TraJR64 is homologous to VirB11 (BlastP e-value <10−14), as reported (54). TraQMPFI and TraRMPFI are homologs and profile–profile comparisons show an indirect relation of homology with the pilin VirB2 (Figure 3). We also built profiles for proteins required for conjugation both in liquid and in surfaces that are encoded outside of the main T4SS operon: TrbAMPFI and TrbBMPFI (28) and for TraEMPFI whose function is still unknown (55). TrbBMPFI is homolog to TraFMPFF as previously suggested (44). We could not obtain profiles for TraJR64, TraHR64, TraSR64 and TraXR64 that are not essential for conjugation (28,55). We also could not build a specific protein profile for the SogLR64 and SogSR64 proteins; they have been reported to be essential for conjugation, but not directly involved in DNA transfer (55). Except these ones, the set of profiles for MPFI includes all proteins for which the corresponding gene disruption completely abolishes transfer (55).
MPFG
MPFG were originally described from ICEHin1056 from Haemophilus influenzae (27,56). Here, we used the very closely related ICEHin10810 element as a model because it is included in a complete genome sequence. The T4SS is encoded in the 24-genes operon tfc. We could find no studies on the structure or assembly of this T4SS, but comparative analyses identified 13 genes present in many homologous elements and a number of essential genes (27,56,57). We built profiles for 18 different proteins of MPFG (Figure 4). We were unable to retrieve profiles for Tfc1ICEHin10810, Tfc4ICEHin10810, Tfc20ICEHin10810 and Tfc21ICEHin10810; the first two seem non-essential for conjugation (27). The core complex of MPFG might resemble the one of MPFF since Tfc13MPFG, Tfc14MPFG and Tfc15MPFG (as well as Tfc2MPFG) are respectively homologs to TraKMPFF, TraBMPFF and TraVMPFF. Profile–profile alignments revealed similarities between MPFG and the inner scaffold of MPFT and MPFF with Tfc11MPFG, Tfc18MPFG and Tfc12MPFG being homologous to VirB3/TraLMPFF, VirB6/TraGMPFF and VirB8/TraEMPFF proteins. MPFG, like MPFI, exhibits two VirB2-like pilins, Tfc9MPFG and Tfc10MPFG. Interestingly, MPFG contains TraWMPFF, TraUMPFF, TrbCMPFF and TraFMPFF homologs, although they were thought to be specific to MPFF (38). Deletion of Tfc24, the homolog of TraWMPFF, results in decreased transcription of many of the T4SS genes (27). We could also find the only detected homolog to TraHMPFF among all T4SS classes: Tfc22MPFG. Overall, our set includes 18 protein profiles including nearly all T4SS essential proteins, except Tfc20ICEHin10810 and Tfc21ICEHin10810. Profile–profile alignments show striking homologies between MPFG and MPFF in spite of their evolutionary distance and the use of very different pili by the two systems.
MPFB
Bacteroides thetaiotaomicron CTnDOT ICE encodes the MPFB model system in a 17-genes operon: traA-traQCTnDOT (58). Only one of these proteins (TraGCTnDOT) was previously found to be homologous to components of the T4SS of Proteobacteria (VirB4 protein) (59). The genes traA-traDCTnDOT seem more variable than the rest of the system when compared with the closely related element CTnERL (60); they might thus be non-essential or evolve so fast that sequence similarity between distant elements is lost. The proteins encoded by traG-NCTnDOT are reportedly essential for conjugation whereas traO-QCTnDOT might have a regulatory role (58,60). We obtained profiles for all proteins TraE-TraQCTnDOT. The vast majority (99%) of T4SS that we detected with these profiles is encoded in genomes of the Bacteroidetes phylum. Profile–profile comparisons show that MPFB has at least two VirB9 homologs, TraQMPFB and TraNMPFB. TraMMPFB matches the VirB10 PFAM domain, but not our VirB10 protein profile. As for the homology between VirB10 and TraBF, this is because the PFAM domain is much shorter and includes more distant homologs. The comparisons also revealed that the most-conserved proteins of the inner scaffold (VirB3, VirB6 and VirB8) have homologs in MPFB (respectively TraFMPFB, TraJMPFB and TraKMPFB). The TraECTnDOT and TraICTnDOT proteins are thought to be pilins since we find them to be homologous to VirB2 and VirB5 in our analysis. TraOMPFB is homologous to TraIMPFB, suggesting that MPFB might have the peculiarity of encoding three different pilins. We were thus able to define 12 protein profiles for MPFB, including most known essential genes of CTnDOT, of which eight have homologs in other systems. This analysis suggests that this class of T4SS strongly resembles some proteobacterial T4SSs. Hopefully, this will facilitate further studies on these systems.
MPFC
There is some circumstantial evidence of conjugation in Cyanobacteria (61), and we have previously identified VirB4 homologs in genomes of this taxa (11). Yet we were unable to find experimental or computational studies on cyanobacterial conjugative systems. We built eight protein profiles that were highly MPFC-specific (>95% hitting genomes of Cyanobacteria). These hits are systematically associated with VirB4 and form compact genetic loci. We used the alpha plasmid of Nostoc sp. PCC 7120 as a model for MPFC. Profile–profile comparisons show that MPFC have two proteins homologous to the core complex in other systems (a VirB9/TraVMPFF and a VirB10/TraBMPFF homolog). The MPFC inner scaffold might resemble those of MPFT and MPFF since we found homologs to VirB3/TraLMPFF, VirB6/TraKMPFF and VirB8/TraEMPFF (Figure 3). Overall, among the eight new MPFC protein profiles, five show homologs with components of MPF from Proteobacteria.
MPFFA
The 12-gene model conjugative locus of Tn916 is the best-described MPFFA system. This class is mostly found in Firmicutes and Actinobacteria (62,63). Tn916, ICEBs1 of Bacillus subtilis and the plasmid pCW3 of Clostridium perfringens, all use TcpA as the coupling protein instead of VirD4 and encode a peculiar relaxase (orf20MPFFA, a MOBT) related to rolling-circle replication initiators from plasmids and phages (29,64). The presence of a relaxase within the MPF region is unique among the systems we considered in this work. These systems do not encode cell surface adhesins (5). We built seven specific profiles for this system: the TcpA distant homolog of VirD4, and six putative components of the T4SS (Figure 4). Orf22MPFFA and Orf23MPFFA are the only pair of protein families from a single system for which we could only build one single profile that systematically matches both proteins. As expected, given the absence of an outer membrane, our analysis suggests that MPFFA lacks homologs to the components of the core complex found in proteobacterial systems. Orf15MPFFA is the only profile homologous to components of the inner scaffold (VirB6/TraGMPFF) that we could identify in this T4SS class. The structure of TcpCpCW3, a close homolog to Orf13Tn916, has remarkable similarities with the one of VirB8 even though sequence similarity is not significant (65). Hence, MPFFA might have two components homologous to the inner scaffold of MPFT—Orf15MPFFA and Orf13MPFFA—and these proteins have been shown to interact (66). MPFFA also has a component homologous to VirB1: Orf14MPFFA. Its homolog TcpGpCW3 has a hydrolase-like activity on C. perfringens peptidoglycan (67), which is consistent with a VirB1-like role in MPFT. We could not build a profile for Orf18Tn916 matching our specificity criteria (less that 50% of the hits neighbored virB4). This gene encodes an anti-restriction protein that is probably not part of the typical MPFFA (68). The last gene of the operon, orf24Tn916, is often found in other MPFFA systems isolated from the rest of the operon (69), or even not mentioned as part of the conjugative machinery (70). It is thus probably not an essential component of MPFFA.
MPFFATA
This class includes systems from an extremely diverse group of Prokaryotes (Firmicutes, Actinobacteria, Tenericutes and Archaea). These monoderms have very diverse cell envelopes—no cell wall in Tenericutes, different lipids in Archaea, thick cell walls in Firmicutes and Actinobacteria—and this may have accelerated their diversification. This might explain why we could not create a single set of protein profiles for all the systems in the class. The lack of experimental studies in most MPFFATA systems further complicated our task. Nevertheless, we identified proteins associated with four sub-classes of T4SS for which conjugative systems have been studied experimentally. These sub-classes correspond to monophyletic sub-groups in the MPFFATA VirB4 phylogeny. Some of the components have highly similar homologs between sub-classes.
The 13-genes operon encoding the conjugative system of the pGO1 plasmid from Staphylococcus aureus (71,72) was used as a model for a sub-class only found among plasmids of Firmicutes. We built six profiles specific to genes encoded in the operon. The TrsIpGO1 and TrsMpGO1 profiles were not specific, with respectively only 7 and 2% of the hits associated to a VirB4. The three other sub-classes were modeled from the prg/pcf system of plasmid pCF10 from Enterococcus faecalis (73,74), from the ICE CTn2 from Clostridium difficile (75) and from an ICE of the Streptococcus agalactiae NEM316 genome (ICESaNEM316) of the ICESa2603 family (76,77). We did not use ICESa2603 itself as a model because it lacks a T4CP and therefore it does not strictly fit our definition of an ICE. These three MPFFATA sub-classes share a core of three proteins (in addition to the VirB4 and VirD4 homologs): PrgFpCF10 is homologous to CD414CTn2 and GBS1363ICESaNEM316; PrgIpCF10 is homologous to CD417CTn2 and GBS1361ICESaNEM316; PrgHpCF10 is homologous to CD415CTn2 and GBS1362ICESaNEM316. Thus, these nine proteins are recovered by only three different profiles. ICESaNEM316-like systems are only found within some Streptococcus, and never on plasmids. This element also shares an additional profile with the Prg/Pcf system, namely PrgLpCF10, which corresponds to GBS1349ICESaNEM316.
Profile–profile comparisons showed no protein profile in MPFFATA with homologies to the T4SS core complex. The exception is the homology between PrgCpCF10 and VirB9 at the N-terminus of the latter, where VirB9 interacts with the inner membrane. The VirB9 region interacting with the outer membrane has no discernible homologs in MPFFATA. On the other hand, we found some homologs to components of the inner-membrane pore complex: VirB3 homologs (TrsCpGO1, PrgIpCF10/CD417CTn2/GBS1361ICESaNEM316) and VirB6 homologs (PrgHpCF10/CD415CTn2/GBS1362ICESaNEM316). Besides, TraMpIP501, homolog of TrsMpGO1for which we could not build a profile, is structurally similar to VirB8 (78). All the MPFFATA systems that we describe except ICESaNEM316 encode a VirB1 homolog (TrsGpGO1, PrgKpCF10, CD419CTn2). The peptidoglycan-degrading activity of TraGpIP501 and PrgKpCF10, homologs of TrsGpGO1, has been shown, confirming the relationship with VirB1 (79,80). We found no homologs to T4SS pilins in MPFFATA. This it not unexpected, since these systems are thought to encode adhesins to stabilize the mating process (see the Introduction section).
DISCUSSION
Homology between MPF classes
Although profiles of one MPF class typically do not match proteins from other classes, profile–profile alignments revealed a number of homologs between classes. Importantly, our analyses revealed networks of homology between profiles of different MPF classes (Figure 3). Some of these homologies had previously been noticed in comparisons between pairs of T4SS (4,5,38,60). Our analysis generalizes these results in a common methodological setup.
Some components of the core complex of T4SS (VirB7, VirB9 and VirB10) are conserved among diderms. This is most notably the case for VirB9 (Figure 3), which even has two homologs in MPFF and MPFB and three in MPFG. VirB10 has homologs in all MPF classes of diderms, including MPFI. VirB7 is a small fast-evolving lipoprotein, which may justify why we could not find its homologs in other classes. Several profiles from other systems are annotated as lipoproteins and could thus be VirB7 analogs (Supplementary Table S2). Monoderms lack an outer membrane and thus have few homologs to the core complex of diderms. Remarkably, PrgCMPFFATA shares some homology with VirB9. This protein has no specific attributed function (81) but displays cell wall anchor motifs and features of other monoderm surface proteins (repeat regions enriched with proline and negatively charged residues) (5). Part of the VirB10 protein forms the outer membrane pore (40) and this may explain its absence from monoderms.
The three proteins of the inner-membrane pore (VirB3, VirB6 and VirB8) are found in almost all MPF classes. VirB3 has homologs in every class except MPFI and MPFFA. The VirB3 partner ATPase (VirB4) has a distant homolog in MPFI, so the presence of an unrecognized VirB3 homolog within MPFI cannot be excluded. We checked for the possibility that TraU could carry region homologs to VirB3 and VirB4, since VirB3 and VirB4 fusions are known to occur in some members of MPFT (82). TraU homologs are larger than average VirB4s. However, we could not find VirB3 signatures in the TraU homologs. VirB3 proteins typically exhibit two TMDs (83). Detection of TMD confirmed this for VirB3 and its homologs with the exception of TraLMPFF (Supplementary Table S2). VirB6 has recognizable homologs in every MPF class except MPFI. This key component of the T4SS has between 30 and 35 kDa and a high number of TMDs (21). In MPFI, we find a protein family (TraYMPFI) with >30 kDa and typically more than four TMDs (nine domains in TraYR64). This is a good candidate for an analog of VirB6. This hypothesis is reinforced by the fact that TraYR64 is the partner of ExcA in R64 entry exclusion (84), an interaction also observed between VirB6 and Eex (85). All the other homologs of VirB6 present at least three TMDs (Supplementary Table S2). VirB8 is a bitopic protein that shows recognizable homologs in all systems of diderms. Interactions of VirB8 have been reported with nearly all other components of the T4SS (86). The differences between monoderms and diderms in terms of the external structure of the T4SS may explain why Orf13MPFFA (including TcpCpCW3), a protein highly similar in structure, has no significant sequence similarity with VirB8 (65). PrgLMPFFATA might also be an analog of VirB8 (20). All the homologs of VirB8, including Orf13MPFFA and the putative analog PrgLMPFFATA, seem to be bitopic since they exhibit one single TMD (Supplementary Table S2). The major pilin (VirB2) has homologs in several MPF classes of diderms, with the exception of MPFF and MPFC. Unsurprisingly, monoderms, which are not known to have pili, lack homologs of VirB2. Finally, VirB1, a non-essential cell wall hydrolase found in MPFT, has homologs only in the two classes of monoderms. These homologous hydrolases are larger than VirB1 and they have an N-terminus anchored in the inner membrane possibly as an adaptation to the absence of an outer membrane and the thicker cell wall of many monoderms (20,87).
Two of the ATPases of the VirB system (VirB4 and VirD4) are widespread. VirB4 is thought to be ubiquitous in T4SS and our analysis relied on this hypothesis. Importantly, very few large (> = 4) clusters of profiles lack VirB4 (9%). Absence of VirB4 might be due to pseudogenization of the conjugation system or sequencing errors. Accordingly, some of these large clusters matching a large number of profiles for a given class include a pseudogene of VirB4. VirD4 is also nearly ubiquitous in T4SS loci. This protein is only replaced by a different AAA+ ATPase (TcpAFA) in a sub-class of MPFFA (10,11,88). On the other hand, VirB11, once thought to be the most frequent ATPase of T4SS (89), is rarely found outside MPFT (19%). Its homolog in MPFI (TraJMPFI) was shown to be distantly related and closer to PilT, the ATPase involved in the retraction of type IV pili (90).
Among the 104 protein profiles used in this study, 34 have no homologs. These ‘orphan’ profiles are not equally distributed (gray arrows in Figure 4). Only one is found in MPFF, whereas they represent most of the profiles of MPFI. This matches the high complexity of MPFI and its early divergence of MPFI and MPFC from the remaining classes (11). Some of these ‘orphan’ profiles might be distant homologs of proteins present in other systems that passed unnoticed in our sequence-based analysis. For example, we found no homologs among pilins, which evolve very fast in sequence, of different classes. Structural data, when it becomes available for the different classes, will help highlighting these cases and complete the networks of homology of Figure 3.
Evolution of the T4SS gene repertoires
The analysis of the total number of relationships of homology between profiles shows that MPFT is the class with the largest number of homologs in other classes (23 links) (Figure 5). MPFF and MPFG are highly connected among themselves suggesting some homology between these classes at least at the level of the core complex and inner-membrane pore, in spite of not being neighbors in the VirB4 phylogeny (11) (Figure 1). MPFC have a number of components homologous with other MPF classes organized in loci resembling the VirB operon, in spite of the large evolutionary distance between MPFT and MPFC in the VirB4 phylogeny (11). MPFB has more homologs with MPFF (two VirB9) and MPFT (homology to the pilins VirB2 and VirB5). Interestingly, MPFI is the least connected class. This is consistent with its position at the base of the VirB4 phylogeny and its use of a distant homolog of VirB4, TraU (11). MPFI also has the peculiarity of encoding two types of pili, one of which being necessary for liquid mating and homologous to the type 4 pili (55,91). The systems of monoderms, and particularly MPFFA, also have few homologs reinforcing the claims that they have fewer components than the other T4SS (20,92), possibly as a result of adaptation to monodermy.
Figure 5.
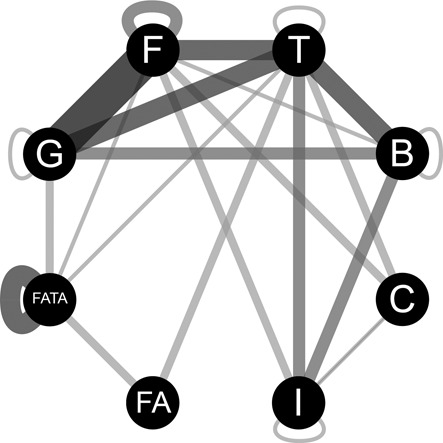
Pairwise relations of homology between the protein profiles of the different mating-pair formation (MPF) classes named I, C, G, T, F, B, FATA and FA as in (11). The width of the links is proportional to the number of couples of profiles, between two MPF classes or within a single MPF class, that have an HHsearch score below 0.001.
We mapped the patterns of the presence/absence of VirB homologs in the phylogenetic trees and drew the most parsimonious scenario for the recruitment of the different components to the T4SS (Figure 6). The scenarios for fast-evolving proteins must be taken with care because lost sequence similarity may lead to an under-estimation of the proteins already present in the last common ancestor of all T4SS. Nevertheless, our analysis suggests that a large number of components of the VirB systems were present early in the history of T4SS.
Figure 6.
Representation of the presence/absence of the most-conserved protein families along the VirB4 phylogeny as presented in Figure 1 and in (11). Green shapes represent inferred protein gains, whereas red shapes represent protein losses. The colors of the arrows (as shown on the right) correspond to those in Figure 4. The bicolor VirD4/TcpA arrow means that some MPFFA systems use VirD4 as coupling protein, whereas others use TcpA.
Webservers
We had previously made available a web site (now called CONJscan, http://mobyle.pasteur.fr/cgi-bin/portal.py#forms::CONJscan-T4SSscan or the short URL http://bit.ly/CONJscan) that allows searching for T4SS and conjugation-related protein profiles. In the present work, we increased the number of profiles available for search from 45 to 116. Our previously identified T4SS and relaxases were distributed as a simple text file. This made its analysis difficult with no intuitive or user-friendly way to search or filter the results according to different criteria. We have now created a web site, CONJdb, which allows searching and browsing our extended dataset thanks to a graphical interface. Our web site also implements the classification scheme. Finally, the graphical interface allows to performs searches by bacterial species, by T4SS class, or according to the presence or absence of T4SS components, namely the T4SS, the coupling protein and the relaxase. This should facilitate the discrimination of T4SS dedicated to conjugation from T4SS dedicated to protein secretion. We are working on the development of a stand-alone application that will be distributed and should be of use to analyze large metagenomic datasets with local computational resources. In the future, CONJdb will be regularly updated and linked with other genomic databases. The web site is accessible at http://conjdb.web.pasteur.fr.
The present release of CONJdb has the results of the analysis of 2393 chromosomes and 1813 plasmids for a total of 2262 complete prokaryotic genomes. Plasmids sequenced without the corresponding chromosome were not included. We detected 947 conjugative systems (up from 515), 1181 mobilizable elements (up from 595) and 646 T4SS lacking nearby relaxases (up from 243), for a total of 1623 T4SS. The most recent and comprehensive T4SS database to date, SecReT4 (93), contains 811 T4SS present on 638 different replicons. SecReT4 does not contain Cyanobacteria or Archaea T4SS. Another tool, AtlasT4SS (94), contains information from 70 genomes (58 Bacteria, 1 Archaea and 11 plasmids) and contains 134 clusters of orthologs. Importantly, CONJdb includes information on the T4SS class and on the putative T4SS function. Finally, because proteins are annotated using HMM profiles, instead of blast-like searches in a large databank of sequences, CONJscan and CONJdb provide standard sequence annotations. We hope this study and these web resources will be useful for scientists studying T4SSs and particularly for those engaging in the study of poorly known ones.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Economy [BFU2011-26608]; European Seventh Framework Program [282004/FP7-HEALTH.2011, 612146/FP7-ICT-2013]; European Research Council Grant [EVOMOBILOME no. 281605]. Source of open access funding: European Research Council grant to the PI.
Supplementary Material
REFERENCES
- 1.Ochman H., Lerat E., Daubin V. Examining bacterial species under the specter of gene transfer and exchange. Proc. Natl. Acad. Sci. U.S.A. 2005;102((Suppl. 1)):6595–6599. doi: 10.1073/pnas.0502035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de la Cruz F., Davies J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 2000;8:128–133. doi: 10.1016/s0966-842x(00)01703-0. [DOI] [PubMed] [Google Scholar]
- 3.Gogarten J.P., Doolittle W.F., Lawrence J.G. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 4.Grohmann E., Muth G., Espinosa M. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alvarez-Martinez C.E., Christie P.J. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smillie C., Garcillan-Barcia M.P., Francia M.V., Rocha E.P., de la Cruz F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010;74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de la Cruz F., Frost L.S., Meyer R.J., Zechner E. Conjugative DNA metabolism in gram-negative bacteria. FEMS Microbiol. Rev. 2010;34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- 8.Guglielmini J., Quintais L., Garcillan-Barcia M.P., de la Cruz F., Rocha E.P. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011;7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fronzes R., Christie P.J., Waksman G. The structural biology of type IV secretion systems. Nature Rev. 2009;7:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsons J.A., Bannam T.L., Devenish R.J., Rood J.I. TcpA, an FtsK/SpoIIIE homolog, is essential for transfer of the conjugative plasmid pCW3 in Clostridium perfringens. J. Bacteriol. 2007;189:7782–7790. doi: 10.1128/JB.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guglielmini J., de la Cruz F., Rocha E.P. Evolution of conjugation and type IV secretion systems. Mol. Biol. Evol. 2013;30:315–331. doi: 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tato I., Zunzunegui S., de la Cruz F., Cabezon E. TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA-dependent ATPase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8156–8161. doi: 10.1073/pnas.0503402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christie P.J. Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochim. Biophys. Acta. 2004;1694:219–234. doi: 10.1016/j.bbamcr.2004.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagai H., Kubori T. Type IVB secretion systems of Legionella and other gram-negative bacteria. Front. Microbiol. 2011;2(136) doi: 10.3389/fmicb.2011.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroder G., Schuelein R., Quebatte M., Dehio C. Conjugative DNA transfer into human cells by the VirB/VirD4 type IV secretion system of the bacterial pathogen Bartonella henselae. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14643–14648. doi: 10.1073/pnas.1019074108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofreuter D., Odenbreit S., Haas R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 2001;41:379–391. doi: 10.1046/j.1365-2958.2001.02502.x. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton H.L., Dominguez N.M., Schwartz K.J., Hackett K.T., Dillard J.P. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol. Microbiol. 2005;55:1704–1721. doi: 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
- 18.Thompson D.V., Melchers L.S., Idler K.B., Schilperoort R.A., Hooykaas P.J. Analysis of the complete nucleotide sequence of the Agrobacterium tumefaciens virB operon. Nucleic Acids Res. 1988;16:4621–4636. doi: 10.1093/nar/16.10.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivera-Calzada A., Fronzes R., Savva C.G., Chandran V., Lian P.W., Laeremans T., Pardon E., Steyaert J., Remaut H., Waksman G., et al. Structure of a bacterial type IV secretion core complex at subnanometre resolution. EMBO J. 2013;32:1195–1204. doi: 10.1038/emboj.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhatty M., Laverde Gomez J.A., Christie P.J. The expanding bacterial type IV secretion lexicon. Res. Microbiol. 2013;164:620–639. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Judd P.K., Mahli D., Das A. Molecular characterization of the Agrobacterium tumefaciens DNA transfer protein VirB6. Microbiology (Reading, England) 2005;151:3483–3492. doi: 10.1099/mic.0.28337-0. [DOI] [PubMed] [Google Scholar]
- 22.Mossey P., Hudacek A., Das A. Agrobacterium tumefaciens type IV secretion protein VirB3 is an inner membrane protein and requires VirB4, VirB7, and VirB8 for stabilization. J. Bacteriol. 2010;192:2830–2838. doi: 10.1128/JB.01331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerr J.E., Christie P.J. Evidence for VirB4-mediated dislocation of membrane-integrated VirB2 pilin during biogenesis of the Agrobacterium VirB/VirD4 type IV secretion system. J. Bacteriol. 2010;192:4923–4934. doi: 10.1128/JB.00557-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger B.R., Christie P.J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bayer M., Iberer R., Bischof K., Rassi E., Stabentheiner E., Zellnig G., Koraimann G. Functional and mutational analysis of p19, a DNA transfer protein with muramidase activity. J. Bacteriol. 2001;183:3176–3183. doi: 10.1128/JB.183.10.3176-3183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gillespie J.J., Brayton K.A., Williams K.P., Diaz M.A., Brown W.C., Azad A.F., Sobral B.W. Phylogenomics reveals a diverse Rickettsiales type IV secretion system. Infect. Immun. 2010;78:1809–1823. doi: 10.1128/IAI.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juhas M., Crook D.W., Dimopoulou I.D., Lunter G., Harding R.M., Ferguson D.J., Hood D.W. Novel type IV secretion system involved in propagation of genomic islands. J. Bacteriol. 2007;189:761–771. doi: 10.1128/JB.01327-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komano T., Yoshida T., Narahara K., Furuya N. The transfer region of IncI1 plasmid R64: similarities between R64 tra and legionella icm/dot genes. Mol. Microbiol. 2000;35:1348–1359. doi: 10.1046/j.1365-2958.2000.01769.x. [DOI] [PubMed] [Google Scholar]
- 29.Garcillan-Barcia M.P., Francia M.V., de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 2009;33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 30.Miele V., Penel S., Duret L. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics. 2011;12(116) doi: 10.1186/1471-2105-12-116. doi:10.1186/1471-2105-12-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gascuel O., Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 33.Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics (Oxford, England) 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 35.Finn R.D., Tate J., Mistry J., Coggill P.C., Sammut S.J., Hotz H.R., Ceric G., Forslund K., Eddy S.R., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillespie J.J., Ammerman N.C., Dreher-Lesnick S.M., Rahman M.S., Worley M.J., Setubal J.C., Sobral B.S., Azad A.F. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS ONE. 2009;4:e4833. doi: 10.1371/journal.pone.0004833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abajy M.Y., Kopec J., Schiwon K., Burzynski M., Doring M., Bohn C., Grohmann E. A type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in gram-positive bacteria. J. Bacteriol. 2007;189:2487–2496. doi: 10.1128/JB.01491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arutyunov D., Frost L.S. F conjugation: back to the beginning. Plasmid. 2013;70:18–32. doi: 10.1016/j.plasmid.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Harris R.L., Hombs V., Silverman P.M. Evidence that F-plasmid proteins TraV, TraK and TraB assemble into an envelope-spanning structure in Escherichia coli. Mol. Microbiol. 2001;42:757–766. doi: 10.1046/j.1365-2958.2001.02667.x. [DOI] [PubMed] [Google Scholar]
- 40.Chandran V., Fronzes R., Duquerroy S., Cronin N., Navaza J., Waksman G. Structure of the outer membrane complex of a type IV secretion system. Nature. 2009;462:1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klimke W.A., Frost L.S. Genetic analysis of the role of the transfer gene, traN, of the F and R100–1 plasmids in mating pair stabilization during conjugation. J. Bacteriol. 1998;180:4036–4043. doi: 10.1128/jb.180.16.4036-4043.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klimke W.A., Rypien C.D., Klinger B., Kennedy R.A., Rodriguez-Maillard J.M., Frost L.S. The mating pair stabilization protein, TraN, of the F plasmid is an outer-membrane protein with two regions that are important for its function in conjugation. Microbiology (Reading, England) 2005;151:3527–3540. doi: 10.1099/mic.0.28025-0. [DOI] [PubMed] [Google Scholar]
- 43.Haase J., Lurz R., Grahn A.M., Bamford D.H., Lanka E. Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage propagation, and pilus production require the same Tra2 core components of a proposed DNA transport complex. J. Bacteriol. 1995;177:4779–4791. doi: 10.1128/jb.177.16.4779-4791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawley T.D., Klimke W.A., Gubbins M.J., Frost L.S. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 2003;224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 45.Arutyunov D., Arenson B., Manchak J., Frost L.S. F plasmid TraF and TraH are components of an outer membrane complex involved in conjugation. J. Bacteriol. 2010;192:1730–1734. doi: 10.1128/JB.00726-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Firth N., Ippen-Ihler K., Skurray R. Structure and function of the F factor and mechanisms of conjugation. In: In: F. C. Neidhardt J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (eds), editor. Escherichia coli and Salmonella: cellular and molecular biology. 2nd edn. Washington, DC: ASM Press; 1996. pp. 2377–2401. [Google Scholar]
- 47.Moore D., Maneewannakul K., Maneewannakul S., Wu J.H., Ippen-Ihler K., Bradley D.E. Characterization of the F-plasmid conjugative transfer gene traU. J. Bacteriol. 1990;172:4263–4270. doi: 10.1128/jb.172.8.4263-4270.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kathir P., Ippen-Ihler K. Construction and characterization of derivatives carrying insertion mutations in F plasmid transfer region genes, trbA, artA, traQ, and trbB. Plasmid. 1991;26:40–54. doi: 10.1016/0147-619x(91)90035-u. [DOI] [PubMed] [Google Scholar]
- 49.Anthony K.G., Klimke W.A., Manchak J., Frost L.S. Comparison of proteins involved in pilus synthesis and mating pair stabilization from the related plasmids F and R100–1: insights into the mechanism of conjugation. J. Bacteriol. 1999;181:5149–5159. doi: 10.1128/jb.181.17.5149-5159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maneewannakul K., Maneewannakul S., Ippen-Ihler K. Characterization of traX, the F plasmid locus required for acetylation of F-pilin subunits. J. Bacteriol. 1995;177:2957–2964. doi: 10.1128/jb.177.11.2957-2964.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moore D., Hamilton C.M., Maneewannakul K., Mintz Y., Frost L.S., Ippen-Ihler K. The Escherichia coli K-12 F plasmid gene traX is required for acetylation of F pilin. J. Bacteriol. 1993;175:1375–1383. doi: 10.1128/jb.175.5.1375-1383.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris R.L., Sholl K.A., Conrad M.N., Dresser M.E., Silverman P.M. Interaction between the F plasmid TraA (F-pilin) and TraQ proteins. Mol. Microbiol. 1999;34:780–791. doi: 10.1046/j.1365-2958.1999.01640.x. [DOI] [PubMed] [Google Scholar]
- 53.Vincent C.D., Friedman J.R., Jeong K.C., Buford E.C., Miller J.L., Vogel J.P. Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol. 2006;62:1278–1291. doi: 10.1111/j.1365-2958.2006.05446.x. [DOI] [PubMed] [Google Scholar]
- 54.Segal G., Feldman M., Zusman T. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol. Rev. 2005;29:65–81. doi: 10.1016/j.femsre.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 55.Sampei G., Furuya N., Tachibana K., Saitou Y., Suzuki T., Mizobuchi K., Komano T. Complete genome sequence of the incompatibility group I1 plasmid R64. Plasmid. 2010;64:92–103. doi: 10.1016/j.plasmid.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 56.Juhas M., Power P.M., Harding R.M., Ferguson D.J., Dimopoulou I.D., Elamin A.R., Mohd-Zain Z., Hood D.W., Adegbola R., Erwin A., et al. Sequence and functional analyses of Haemophilus spp. genomic islands. Genome Biol. 2007;8(R237) doi: 10.1186/gb-2007-8-11-r237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seth-Smith H.M., Fookes M.C., Okoro C.K., Baker S., Harris S.R., Scott P., Pickard D., Quail M.A., Churcher C., Sanders M., et al. Structure, diversity, and mobility of the Salmonella pathogenicity island 7 family of integrative and conjugative elements within Enterobacteriaceae. J. Bacteriol. 2012;194:1494–1504. doi: 10.1128/JB.06403-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bonheyo G.T., Hund B.D., Shoemaker N.B., Salyers A.A. Transfer region of a Bacteroides conjugative transposon contains regulatory as well as structural genes. Plasmid. 2001;46:202–209. doi: 10.1006/plas.2001.1545. [DOI] [PubMed] [Google Scholar]
- 59.Franco A.A. The Bacteroides fragilis pathogenicity island is contained in a putative novel conjugative transposon. J. Bacteriol. 2004;186:6077–6092. doi: 10.1128/JB.186.18.6077-6092.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonheyo G., Graham D., Shoemaker N.B., Salyers A.A. Transfer region of a bacteroides conjugative transposon, CTnDOT. Plasmid. 2001;45:41–51. doi: 10.1006/plas.2000.1495. [DOI] [PubMed] [Google Scholar]
- 61.Muro-Pastor A.M., Kuritz T., Flores E., Herrero A., Wolk C.P. Transfer of a genetic marker from a megaplasmid of Anabaena sp. strain PCC 7120 to a megaplasmid of a different Anabaena strain. J. Bacteriol. 1994;176:1093–1098. doi: 10.1128/jb.176.4.1093-1098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roberts A.P., Mullany P. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 2009;17:251–258. doi: 10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 63.Senghas E., Jones J.M., Yamamoto M., Gawron-Burke C., Clewell D.B. Genetic organization of the bacterial conjugative transposon Tn916. J. Bacteriol. 1988;170:245–249. doi: 10.1128/jb.170.1.245-249.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rocco J.M., Churchward G. The integrase of the conjugative transposon Tn916 directs strand- and sequence-specific cleavage of the origin of conjugal transfer, oriT, by the endonuclease Orf20. J. Bacteriol. 2006;188:2207–2213. doi: 10.1128/JB.188.6.2207-2213.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Porter C.J., Bantwal R., Bannam T.L., Rosado C.J., Pearce M.C., Adams V., Lyras D., Whisstock J.C., Rood J.I. The conjugation protein TcpC from Clostridium perfringens is structurally related to the type IV secretion system protein VirB8 from Gram-negative bacteria. Mol. Microbiol. 2012;83:275–288. doi: 10.1111/j.1365-2958.2011.07930.x. [DOI] [PubMed] [Google Scholar]
- 66.Teng W.L., Bannam T.L., Parsons J.A., Rood J.I. Functional characterization and localization of the TcpH conjugation protein from Clostridium perfringens. J. Bacteriol. 2008;190:5075–5086. doi: 10.1128/JB.00386-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bantwal R., Bannam T.L., Porter C.J., Quinsey N.S., Lyras D., Adams V., Rood J.I. The peptidoglycan hydrolase TcpG is required for efficient conjugative transfer of pCW3 in Clostridium perfringens. Plasmid. 2012;67:139–147. doi: 10.1016/j.plasmid.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 68.Serfiotis-Mitsa D., Roberts G.A., Cooper L.P., White J.H., Nutley M., Cooper A., Blakely G.W., Dryden D.T. The Orf18 gene product from conjugative transposon Tn916 is an ArdA antirestriction protein that inhibits type I DNA restriction-modification systems. J. Mol. Biol. 2008;383:970–981. doi: 10.1016/j.jmb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 69.Bertsch D., Uruty A., Anderegg J., Lacroix C., Perreten V., Meile L. Tn6198, a novel transposon containing the trimethoprim resistance gene dfrG embedded into a Tn916 element in Listeria monocytogenes. J. Antimicrob. Chemother. 2013;68:986–991. doi: 10.1093/jac/dks531. [DOI] [PubMed] [Google Scholar]
- 70.de Vries L.E., Christensen H., Skov R.L., Aarestrup F.M., Agerso Y. Diversity of the tetracycline resistance gene tet(M) and identification of Tn916- and Tn5801-like (Tn6014) transposons in Staphylococcus aureus from humans and animals. J. Antimicrob. Chemother. 2009;64:490–500. doi: 10.1093/jac/dkp214. [DOI] [PubMed] [Google Scholar]
- 71.Morton T.M., Eaton D.M., Johnston J.L., Archer G.L. DNA sequence and units of transcription of the conjugative transfer gene complex (trs) of Staphylococcus aureus plasmid pGO1. J. Bacteriol. 1993;175:4436–4447. doi: 10.1128/jb.175.14.4436-4447.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Caryl J.A., O’Neill A.J. Complete nucleotide sequence of pGO1, the prototype conjugative plasmid from the Staphylococci. Plasmid. 2009;62:35–38. doi: 10.1016/j.plasmid.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 73.Chen Y., Zhang X., Manias D., Yeo H.J., Dunny G.M., Christie P.J. Enterococcus faecalis PcfC, a spatially localized substrate receptor for type IV secretion of the pCF10 transfer intermediate. J. Bacteriol. 2008;190:3632–3645. doi: 10.1128/JB.01999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li F., Alvarez-Martinez C., Chen Y., Choi K.J., Yeo H.J., Christie P.J. Enterococcus faecalis PrgJ, a VirB4-like ATPase, mediates pCF10 conjugative transfer through substrate binding. J. Bacteriol. 2012;194:4041–4051. doi: 10.1128/JB.00648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brouwer M.S., Warburton P.J., Roberts A.P., Mullany P., Allan E. Genetic organisation, mobility and predicted functions of genes on integrated, mobile genetic elements in sequenced strains of Clostridium difficile. PLoS ONE. 2011;6:e23014. doi: 10.1371/journal.pone.0023014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davies M.R., Shera J., Van Domselaar G.H., Sriprakash K.S., McMillan D.J. A novel integrative conjugative element mediates genetic transfer from group G Streptococcus to other {beta}-hemolytic Streptococci. J. Bacteriol. 2009;191:2257–2265. doi: 10.1128/JB.01624-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tettelin H., Masignani V., Cieslewicz M.J., Eisen J.A., Peterson S., Wessels M.R., Paulsen I.T., Nelson K.E., Margarit I., Read T.D., et al. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12391–12396. doi: 10.1073/pnas.182380799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goessweiner-Mohr N., Grumet L., Arends K., Pavkov-Keller T., Gruber C.C., Gruber K., Birner-Gruenberger R., Kropec-Huebner A., Huebner J., Grohmann E., et al. The 2.5 A structure of the enterococcus conjugation protein TraM resembles VirB8 type IV secretion proteins. J. Biol. Chem. 2013;288:2018–2028. doi: 10.1074/jbc.M112.428847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arends K., Celik E.K., Probst I., Goessweiner-Mohr N., Fercher C., Grumet L., Soellue C., Abajy M.Y., Sakinc T., Broszat M., et al. TraG encoded by the pIP501 type IV secretion system is a two-domain peptidoglycan-degrading enzyme essential for conjugative transfer. J. Bacteriol. 2013;195:4436–4444. doi: 10.1128/JB.02263-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Laverde Gomez J.A., Bhatty M., Christie P.J. PrgK, a multidomain peptidoglycan hydrolase, is essential for conjugative transfer of the pheromone responsive plasmid pCF10. J. Bacteriol. 2013;196:527–539. doi: 10.1128/JB.00950-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dunny G.M. The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007;362:1185–1193. doi: 10.1098/rstb.2007.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arechaga I., Pena A., Zunzunegui S., del Carmen Fernandez-Alonso M., Rivas G., de la Cruz F. ATPase activity and oligomeric state of TrwK, the VirB4 homologue of the plasmid R388 type IV secretion system. J. Bacteriol. 2008;190:5472–5479. doi: 10.1128/JB.00321-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Christie P.J., Whitaker N., Gonzalez-Rivera C. Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta. 2014 doi: 10.1016/j.bbamcr.2013.12.019. doi:10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sakuma T., Tazumi S., Furuya N., Komano T. ExcA proteins of IncI1 plasmid R64 and IncIgamma plasmid R621a recognize different segments of their cognate TraY proteins in entry exclusion. Plasmid. 2013;69:138–145. doi: 10.1016/j.plasmid.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 85.Garcillan-Barcia M.P., de la Cruz F. Why is entry exclusion an essential feature of conjugative plasmids. Plasmid. 2008;60:1–18. doi: 10.1016/j.plasmid.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 86.Baron C. VirB8: a conserved type IV secretion system assembly factor and drug target. Biochem. Cell Biol. 2006;84:890–899. doi: 10.1139/o06-148. [DOI] [PubMed] [Google Scholar]
- 87.Laverde Gomez J.A., Bhatty M., Christie P.J. PrgK, a multidomain peptidoglycan hydrolase, is essential for conjugative transfer of the pheromone-responsive plasmid pCF10. J. Bacteriol. 2014;196:527–539. doi: 10.1128/JB.00950-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Steen J.A., Bannam T.L., Teng W.L., Devenish R.J., Rood J.I. The putative coupling protein TcpA interacts with other pCW3-encoded proteins to form an essential part of the conjugation complex. J. Bacteriol. 2009;191:2926–2933. doi: 10.1128/JB.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cao T.B., Saier M.H., Jr Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: organismal distribution, structural constraints and evolutionary conclusions. Microbiology (Reading, England) 2001;147:3201–3214. doi: 10.1099/00221287-147-12-3201. [DOI] [PubMed] [Google Scholar]
- 90.Planet P.J., Kachlany S.C., DeSalle R., Figurski D.H. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2503–2508. doi: 10.1073/pnas.051436598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim S.R., Komano T. The plasmid R64 thin pilus identified as a type IV pilus. J. Bacteriol. 1997;179:3594–3603. doi: 10.1128/jb.179.11.3594-3603.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang W., Rong C., Chen C., Gao G.F. Type-IVC secretion system: a novel subclass of type IV secretion system (T4SS) common existing in gram-positive genus Streptococcus. PLoS ONE. 2012;7:e46390. doi: 10.1371/journal.pone.0046390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bi D., Liu L., Tai C., Deng Z., Rajakumar K., Ou H.Y. SecReT4: a web-based bacterial type IV secretion system resource. Nucleic Acids Res. 2013;41:D660–D665. doi: 10.1093/nar/gks1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Souza R.C., del Rosario Quispe Saji G., Costa M.O., Netto D.S., Lima N.C., Klein C.C., Vasconcelos A.T., Nicolas M.F. AtlasT4SS: a curated database for type IV secretion systems. BMC Microbiol. 2012;12:172. doi: 10.1186/1471-2180-12-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


