Abstract
Transcriptional slippage is a class of error in which ribonucleic acid (RNA) polymerase incorporates nucleotides out of register, with respect to the deoxyribonucleic acid (DNA) template. This phenomenon is involved in gene regulation mechanisms and in the development of diverse diseases. The bacteriophage λ N protein reduces transcriptional slippage within actively growing cells and in vitro. N appears to stabilize the RNA/DNA hybrid, particularly at the 5′ end, preventing loss of register between transcript and template. This report provides the first evidence of a protein that directly influences transcriptional slippage, and provides a clue about the molecular mechanism of transcription termination and N-mediated antitermination.
INTRODUCTION
Errors in the transcription process can potentially lead to aberrant gene products and, ultimately, disease. One major class of error, known as transcriptional slippage, can occur during transcription elongation when the nascent ribonucleic acid (RNA) molecule shifts register, backward or forward, with respect to the template deoxyribonucleic acid (DNA) (1). RNA polymerase (RNAP) maintains an ∼8–9 base pair RNA/DNA hybrid during transcription elongation (2). RNAPs depend on this short RNA/DNA hybrid for stability and processivity of ternary elongation complexes (TECs). RNA/DNA hybrids of less than 8-bp display markedly less stability than those that are 8 bp or longer (2–4). If the RNAP shifts backward and resumes transcription, an extra nucleotide will be inserted; conversely, if the complex shifts forward a coded base will be omitted from the transcript. During elongation, transcriptional slippage is typically restricted to DNA sequences containing a single repeated nucleotide, called homopolymeric tracts, generally 8 or more in a stretch (1,5–8). When transcriptional slippage occurs within the coding sequence of a gene, the resulting transcripts can encode disrupted reading frames, changing the sense of all downstream codons (9). This type of error has been implicated in the development of a wide variety of diseases, including colon cancer, non-familial Alzheimer's and Down's syndrome [reviewed in (1) and (10)].
Instances of transcriptional slippage have been observed in many organisms, and certain genetic elements, such as transposons, exploit this phenomenon for use as a regulatory mechanism (5,10,11). Transcription initiation is particularly prone to slippage at short homopolymeric tracts, due in part to the limited length of the RNA/DNA hybrid compared to hybrid length during elongation (12). For example, in Escherichia coli, expression of the pyrBI operon is regulated during initiation by an iterative transcription mechanism that is related to slippage during elongation. In Thermus thermophilus, transcriptional slippage during elongation of dnaX is responsible for producing either the γ- or τ-subunits of the DNA replication machinery, both being produced from the same DNA sequence (13).
Attempts to assess the fidelity of RNAP in vivo have been hampered by the low frequency of transcription errors, the transient nature of the transcript and a comparatively high frequency of translation error (1,14). Early systems used to study transcriptional slippage utilized plasmid-borne constructs containing a lacZ reporter interrupted by homopolymeric tracts of DNA (7). Long stretches of A or T nucleotides (>8 nt) are particularly prone to slippage, possibly due to the weak bonding of the RNA/DNA hybrid during transcription (5,7). A shifted RNA/DNA hybrid in a long repeating tract would be indistinguishable by polymerase from the unshifted RNA/DNA hybrid. Shifts in register may go undetected and uncorrected by RNAP and its associated nucleolytic proofreading functions, e.g. GreA and GreB, because correct pairing of the RNA/DNA hybrid is only structurally monitored up to ∼9 bp (15). Mutant β subunits (encoded by rpoB) of E. coli RNAP exist that increase or decrease the frequency of transcriptional slippage (8). One of the β mutants, P564L, demonstrated increased slippage and showed a defect in the formation of transcription antitermination complexes with the N protein of bacteriophage λ (16–18).
The λ N protein is an extensively studied transcription regulatory factor that prevents both ρ-dependent and intrinsic transcription termination. N also increases the rate of transcription and reduces pausing of RNAP (19,20). The N-antitermination system has served as a model for understanding many regulatory mechanisms that modulate transcription elongation such as the Tat and tar system of human immunodeficiency virus (HIV) and the antitermination system of bacterial ribosomal RNA operons (21–23). The N protein binds to a specific RNA hairpin structure within a regulatory RNA element known as nut (for N utilization) (19,21). Four host proteins (NusA, NusB, NusE and NusG) interact with nut, N and RNAP to generate a termination resistant transcription complex; however, experiments have demonstrated that N alone can modify RNAP when over-expressed in vivo or added in excess of elongation complexes in vitro (24–27).
Given the relationship between slippage and antitermination phenotypes observed in strains carrying the β P564L mutant protein, and the observation that repetitive U-rich sequences are pervasive in intrinsic transcription terminators, we tested if the N-antitermination complex exerts an effect on transcriptional slippage. Here we describe a λ-based genetic assay designed to assess transcriptional slippage by N-modified RNAP. We also use TECs assembled on synthetic scaffolds to probe the effect of N in vitro. We find that (i) N reduces the frequency of transcriptional slippage both in vivo and in vitro and that (ii) N appears to exert its effect on slippage via stabilization of the upstream end of the RNA/DNA hybrid.
MATERIALS AND METHODS
Bacterial strains and plasmids
The N–lacZ slippage reporter strains were constructed using recombineering techniques in the parent strain ZH1041 [W3110 Δ(argF-lac)U169], with the following genetic structure around the λ prophage: gal490*(IS2) pglΔ8 att int-lacZ-int red kil N nutL pL cI857 Δ[cro-bio] (28). Cultures were grown to OD600 ∼0.4, induced at 42°C for 15 min, and prepared for electroporation. These cultures were then electroporated with either overlapping oligonucleotides (Supplementary Table S1) or a single oligonucleotide with 35-bp homology to the N gene, ending at the 33rd codon, a homopolymeric tract, and 35-bp of homology to the lacZ gene, starting at the 16th codon. Recombinants were selected for the loss of the prophage region containing kil and all intervening sequence between N and lacZ, by selecting on L-plates at 42°C, as described in (28). Recombinants were sequenced to confirm the desired genotype. N was expressed from the pZH124 plasmid. This plasmid is a pGB2 derivative (pSC101 replicon), with the coding sequence of N under plac control (28). In control strains, pGB2 alone was used as a vector-only control (29).
β-galactosidase assay
Cultures were grown from three independent colonies per condition overnight in L broth at 30°C, supplemented with 30-μg/ml spectinomycin when plasmids were present. Cultures were diluted 1:100 in fresh L broth and grown 1.5 h at 30°C, without antibiotic in all cases, to an OD600 of ∼0.1. Cultures were then shifted to 42°C shaking water bath and grown for an additional hour, reaching a final OD600 of ∼0.5–0.6. Cultures were transferred to an ice-water bath and chilled for 10 min. One hundred microliters of each culture was assayed for β-galactosidase (β-gal) activity as described by Miller (30).
Processive slippage assay
RNAP and TEC assembly was performed as described by Kireeva and Kashlev (31). Briefly, a 9-nt synthetic RNA (5′-AUC GAG AGG-3′) was annealed to template DNA (5′-TTG GGT TCT CTA TTC GCC TCG TTT TTT TTT CCC TCT CGA TGG CTG TAA GTA TCC TAT ACC-3′) and incubated with histidine-tagged (His-tagged) RNAP, bound to Ni-NTA agarose beads (Qiagen). The non-template strand (5′-GGT ATA GGA TAC TTA CAG CCA TCG AGA GGG AAA AAA AAA CGA GGC GAA TAG AGA ACC CAA-3′) was added, and following a brief incubation period, all complexes were washed free of unbound oligonucleotides. RNA was labeled by extension, adding α-32P guanosine triphosphate (GTP), followed by several wash steps. In samples containing N protein, 100-fold molar excess of N was added and pre-incubated for 10 min. N protein, purified as described by (24), was a kind gift from Stephen Weitzel and Peter von Hippel. To measure the rate of slippage, adenosine triphosphate (ATP) was added at 10 μM, and complexes were allowed to transcribe the A tract for 10, 20, 40, 90 and 180 s, after which they were stopped by the addition of 2× loading buffer containing 7-M urea. Samples were analyzed by denaturing PAGE, visualized by phophorimaging (Molecular Dynamics).
RESULTS
Development of a bacteriophage λ-based transcriptional slippage assay
We employed a genetic reporter to assess transcriptional slippage in a λ-based assay. In this system, transcriptional slippage is monitored by β-gal enzyme activity produced from a defective prophage. The construct consists of a translation fusion of lacZ to the N gene, the first gene transcribed in the λ pL operon (Figure 1). The strong pL promoter can be conditionally controlled by temperature in bacteria expressing the mutant λCI857 repressor protein (28). A homopolymeric tract was placed at the translation fusion junction between N and lacZ. To test for transcriptional slippage within the homopolymeric tracts, we fused the lacZ coding sequence one nucleotide out-of-frame with respect to the start codon of the N protein. Transcriptional slippage events can restore the reading frame of the lacZ gene. Forward slippage of the transcript, coupled with resumed transcription, would lead to the elimination of a coded nucleotide, restoring the reading frame in +1 out-of-frame constructs, whereas backward slippage of the transcript would lead to addition of a nucleotide, restoring the reading frame in −1 out-of-frame constructs.
Figure 1.
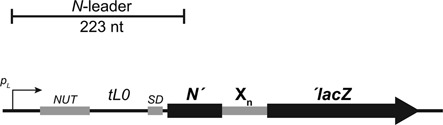
Diagram depicting N leader sequence and N::Xn::lacZ fusion. The 223-nt long leader sequence consists of an N protein binding site (nut) and a weak transcription terminator (tL0). The N::lacZ fusion consists of the Shine–Dalgarno (SD) sequence and the first 33 codons from the N open reading frame, the homopolymeric tract (Xn, where n represents varying numbers of a given nucleotide, X) and the lacZ coding sequence in-frame or out-of-frame (−1 or +1). See Supplementary Figure S1 for controls that demonstrate that no transcription termination or arrest is apparent on homopolymeric A tracts.
We engineered a series of N–lacZ fusions that encode homopolymeric tracts containing 7, 8, 9, 10 and 11 adenines in a row, in both the −1 and +1 reading frames. By convention, we refer to the homopolymeric tracts by the nucleotide that is incorporated into the RNA, for example A9 tracts encode nine As within the RNA. For controls, we also constructed strains that contain a single guanine or uracil interruption within the homopolymeric A tract (A5GA4 and A5UA4). Similar homopolymer interruptions have been reported to be resistant to transcriptional slippage (7,8). Since the A5GA4 maintains the same amino acid composition as A10 constructs, we adopted it as the standard control construct.
Following induction of the pL operon, the A5GA4 fusions in the −1 and +1 frames yielded no significant β-gal activity, consistent with a lack of detectable transcriptional slippage on this construct. In the −1 fusion constructs, A7 shows a small amount of β-gal activity (Table 1), much less than the A8, A9, A10 and A11 constructs. In +1 constructs, both A7 and A8 show limited β-gal activity, less than −1 constructs, but activity increases dramatically on +1 A9, A10 and A11 constructs. In contrast to the out-of-frame constructs, in-frame constructs show decreasing levels of β-gal activity as the length of A tracts increases, explained by greater levels of slippage causing a shift from the correct reading frame to an incorrect reading frame.
Table 1. The effect of homopolymeric tract on β-gal expression.
| β-gal activity (Miller units) | |||
|---|---|---|---|
| Out-of-frame (+1) | Out-of-frame (−1) | In-frame | |
| A7 | 7 (1) | 17 (1) | 2077 (167) |
| A8 | 22 (1) | 180 (16) | 1616 (134) |
| A9 | 180 (15) | 390 (86) | 1260 (43) |
| A10 | 308 (19) | 624 (37) | 1113 (69) |
| A11 | 303 (28) | 539 (46) | 822 (55) |
| U9 | 145 (10) | 307 (28) | 1652 (95) |
| U10 | 431 (33) | 323 (17) | 791 (35) |
| C10 | 9 (1) | 15 (9) | 1246 (76) |
| G10 | 7 (4) | 10 (3) | 1647 (152) |
| A5GA4 | 8 (2) | 7 (1) | 1746 (101) |
| A5UA4 | 3 (1) | 4 (1) | 1698 (100) |
Average β-gal activities are presented in Miller units (bold), with standard error of the mean (SEM) in parentheses, n = ≥3.
We tested for RNAP termination or arrest within the homopolymeric tract by designing a reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) assay that compared the abundance of transcripts containing sequence upstream of the A tract with the abundance of sequence immediately downstream of the A tract. We found no difference in upstream versus downstream RNA levels, further establishing that the homopolymeric tract itself does not cause termination or arrest (Supplementary Figure S1).
In addition to A tracts, we constructed strains that contain U10, C10, or G10 tracts. The out-of-frame U10 tracts yielded activities that were qualitatively similar to A10 tracts, whereas C10 and G10 are comparable to the non-slippery A5GA4 tract (Table 1). These observations are consistent with measurements made in a similar system by others (7). From these data, we conclude that differences in amino acid composition at this site in the fusion result in negligible changes to β-gal enzyme activity, and the C and G tracts are not slippery.
N inhibits transcriptional slippage on homopolymeric A and U tracts
It has been suggested that slippage of the transcript within the elongation complex may contribute to transcription termination (9,32). To test if the N protein has an effect on transcriptional slippage, as well as preventing termination, we expressed N from a medium-copy-number plasmid in reporter strains that contain A7, A8, A9, A10, A11, U9 or U10 homopolymeric tracts. In the in-frame A5GA4 control strain, we found ∼2-fold higher levels of functional β-gal when N protein was expressed from plasmids compared to vector-only controls, 2531 (±87) Miller units versus 1357 (±204) Miller units, respectively. This result reaffirms the presence of a weak terminator (see tL0; Figure 1) in the N-gene leader between nut and the N gene, as described previously (33). These are taken to be the total levels of transcription possible with or without antitermination, respectively. We assayed the fusion constructs containing homopolymeric tracts in −1 and +1 reading frames and normalized all β-gal measurements to the corresponding A5GA4 controls with or without N. There was a consistent decrease in β-gal activity when N was expressed (Table 2), despite the overall increase in transcription observed in A5GA4 controls with N expressed. While both A10 and U10 tracts exhibit extensive transcriptional slippage, N prevented slippage to a greater extent on U tracts, which are known to make the least stable RNA/DNA hybrids (34). The most profound effect of N was observed in the +1 out-of-frame U10 constructs, causing a 7.3-fold decrease in nucleotide omissions (Table 2). N consistently inhibited nucleotide deletions to a greater extent than insertions.
Table 2. Fold effect of N on β-gal activity.
| Fold change in percent slippage | ||
|---|---|---|
| Out-of-frame (+1) | Out-of-frame (−1) | |
| N−/N+ | N−/N+ | |
| A7 | 1.0 | 1.2 |
| A8 | 3.5 | 1.7 |
| A9 | 3.1 | 1.9 |
| A10 | 3.6 | 1.9 |
| A11 | 3.6 | 1.4 |
| U9 | 6.5 | 4.1 |
| U10 | 7.3 | 5.5 |
| A10, nutL7 | 1.0 | 1.0 |
The percent of transcriptional slippage in the out-of-frame constructs was calculated by the formula: [(Miller units An N−/Miller units A5GA4 N−) ÷ (Miller units An N+/Miller units A5GA4 N+)]. A value of 1.0 indicates no change.
The N-leader sequence contains the nut RNA element that is bound by N protein. Antitermination complexes are formed when the N–nut complex binds RNAP, along with additional transcription elongation Nus factors (19). To test the dependence of the anti-slippage phenomenon on the N–nut nucleoprotein complex, we made a mutation that disables the nut site (nutL7) (28). In the A5GA4 control strains, expressing N with nutL7 yielded β-gal activities that were similar to those containing the vector-only control and the wild-type nut sequence (2286 [±75] Miller units versus 1902 [±180] Miller units, P > 0.05, two-tailed t-test). This result demonstrates that nutL7 effectively abolished N's antitermination activity. The β-gal activity is the same with and without N-expressing plasmids in −1 and +1 A10 out-of-frame constructs (Table 2), therefore the inhibition of slippage is also abolished when N cannot bind to nut.
N alone is capable of inhibiting transcriptional slippage in vitro
To further confirm that N protein is capable of inhibiting transcriptional slippage, we assembled purified TECs consisting of RNAP, a synthetic RNA primer, template DNA and non-template DNA oligonucleotides containing an A9 tract and flanking sequence that is unrelated to the in vivo λ constructs (Figure 2A). The TECs did not contain the nut RNA element or Nus elongation factors that are present in the in vivo system. The RNA was annealed one base before the homopolymeric tract, and annealed transcript was labeled at the 3′ end by the addition of α-32P GTP, which also extended the annealed RNA to the beginning of the homopolymeric tract. To commence the slippage assay, ATP alone was added to the TECs, thus only ATP could be incorporated into the RNA transcript. Under this condition, RNAP transcribes to the end of the A tract and continues adding adenines at the end of the A tract by a slippage mechanism (8). The reaction was stopped at selected time points, and the products were separated on a denaturing gel for visualization. This assay measures the length of RNAs over the reaction time course and, therefore, the insertion rate of additional As into the transcript. We observed distributions of transcripts longer than the expected 19 nt, and the length of transcripts steadily increased over the reaction time (Figure 2B). We quantified the bands that were >19 nt in length and assessed their position within the distribution of transcripts at each time point (see arrows in Figure 2B and C). The number of additional nucleotides present in the most abundant transcript was divided by reaction time to provide the rate of slippage. The catalytic rate of slippage was calculated as 0.25 nt/s where no N was present. To test the effect of N on slippage, 100-fold molar excess of N protein was pre-incubated with the complexes, prior to the addition of ATP. High concentrations of N are necessary for antitermination to function in vitro in the absence of nut and Nus factors (24), so similarly high concentrations were used in these experiments. Consistent with in vivo effects, the slippage rate dropped 1.6-fold, compared to reactions lacking N (P < 0.05, two-tailed t-test).
Figure 2.
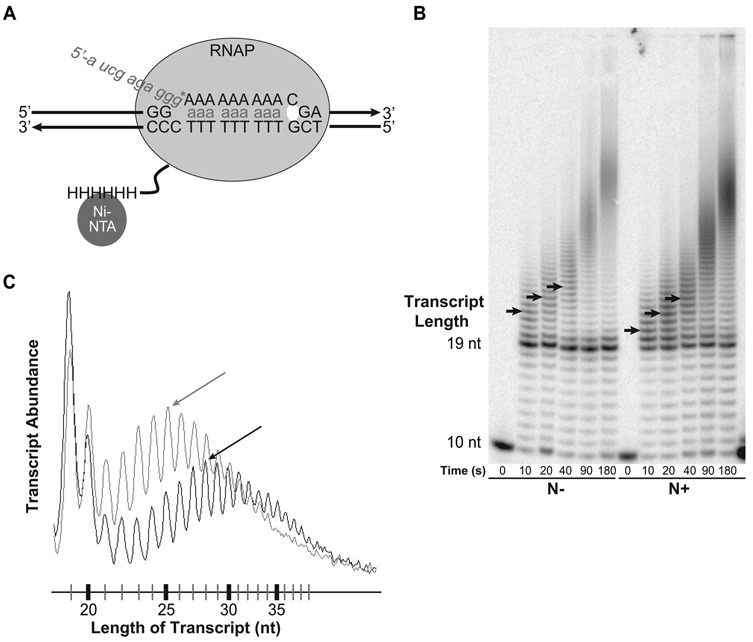
The in vitro transcriptional slippage assay. (A) Diagram depicting the in vitro transcription assay to assess the rate of slippage. His-tagged (HHHHHH) RNAP was immobilized on Ni-NTA functionalized agarose beads. A 9-nt-long RNA was annealed one base upstream of the A9 tract. This RNA/DNA hybrid was added to RNAP that was immobilized on Ni-NTA agarose beads, and the non-template DNA strand was added to complete the assembly of the TEC. α-32P-labeled GTP (G*) was added, elongating the transcript by one nucleotide and radiolabeling the transcript. The complex was washed several times to remove any un-incorporated GTP. To commence the slippage assay, TECs were incubated with ATP or UTP (depending on the homopolymeric tract). The active site is presented as a small circle at the 3′ end of the transcript. (B) A representative denaturing gel from complexes containing A9 homopolymeric tracts. Time points are given in seconds, and N+ and N− conditions are shown. Peak bands corresponding to transcripts >19-nt long are labeled with arrows; note that in the N+ condition peak bands contain fewer nucleotides for all time points. See Supplementary Figure S2 for results of in vitro slippage assay on U11 tracts. See Supplementary Figure S3 for controls that demonstrate that N acts as a positive elongation factor on these in vitro templates. (C) Histograms of samples from in vitro slippage assays enable determination of slippage rates. Transcript abundance, as measured by 32P counts, is presented relative to transcript length. A representative time point (40 s) is shown. Samples containing N are shown in light gray and samples lacking N are shown in black. Arrows indicate the location of the most abundant band.
Similar reactions were carried out on templates containing U11 tracts, and incubating with uridine triphosphate (UTP) instead of ATP. N also reduced slippage on the U11 templates (Supplementary Figure S2). To confirm that N acts as a positive elongation factor under comparable conditions, the elongation rate was compared between N+ and N− reactions in the presence of limited amounts of all four nucleotides. N+ reactions stimulated the rate of nucleotide incorporation >2-fold and decreased pausing (Supplementary Figure S3.), consistent with the effects measured by others (24). We also tested if the NusA elongation factor influences transcriptional slippage. We detected no change in slippage with the addition of NusA (Supplementary Figure S4).
Sequence at the 5′ end of the slippery sequence influences slippage
On A8 tracts, both forward and backward slippage will result in mismatches at the 5′ end of the hybrid, if the RNA/DNA hybrid is restricted to nine nucleotides. The interaction interface between N and RNAP has not been definitively solved; however, there is reason to believe that N interacts near the RNA exit channel (35). For these reasons, we wanted to determine first if slippage, particularly on the shorter A8 tract, is sensitive to nucleotide composition immediately 5′ of the homopolymeric tract, and second, if such an effect is influenced by N. We constructed additional reporter strains, using the in vivo system described above, in which the nucleotide encoded directly upstream of the homopolymeric A8 tract was engineered to contain U, C, or G.
In control experiments without N, we found that β-gal activity in the N–lacZ fusion reporters was highest in the 5′-U constructs, followed by 5′-C then 5′-G (Table 3). This pattern was consistent in both −1 and +1 out-of-frame constructs. These differences may be attributed to relative stability of hybrid base pairs, where greater stability, rG/dC > rC/dG > rU/dA (34), prevents slippage to a larger extent. When N was provided, β-gal activity was reduced in all cases. In −1 out-of-frame constructs, the greatest effect of N was observed on A tracts with a 5′-U; slippage was reduced >3-fold and ∼2-fold in 5′-C and 5′-G reporters, respectively. In +1 out-of-frame constructs, N appears to have a greater effect on 5′-C than on 5′-U. Along with the observation that N inhibits deletions to a greater extent than insertions (Table 2), these results provide evidence that deletions may occur through a different mechanism than insertions and that the deletion process is more sensitive to N's influence.
Table 3. Percent of transcriptional slippage with different nucleotides flanking the 5′ end of the homopolymeric A tract.
| Out-of-frame (+1) | Out-of-frame (−1) | |||||
|---|---|---|---|---|---|---|
| Slippery sequence | N− | N+ | 5′ mispairs | N− | N+ | 5′ mispairs |
| UUaaaaaaaaG | 1.8% (0.2%) | 0.5% (0.1%) | rU/dT | 17.6% (1.7%) | 5.4% (0.8%) | rA/dA |
| UCaaaaaaaaG | 0.9% (0.1%) | 0.2% (0.02%) | rC/dT | 6.7% (0.9%) | 3.7% (0.3%) | rA/dG |
| UGaaaaaaaaG | 0% (0%) | 0.1% (0.2%) | rG/dT | 4.9% (0.4%) | 2.5% (0.3%) | rA/dC |
The percent of transcriptional slippage was calculated by the formula: (Miller units An tract N−/Miller units A5GA4 N−) × 100% or (Miller units An tract N+/Miller units A5GA4 N+)× 100%. Values in parentheses indicate propagated SEM, n = 3. Mispaired nucleotides at the upstream end of the 9-nt RNA/DNA hybrid are shown, assuming the RNA transcript shifts forward relative to the DNA template to restore the +1 reading frame and backward to the −1 reading frame. RNA nucleotides are denoted with the prefix (r) and DNA template nucleotides are denoted with the prefix (d).
DISCUSSION
We have described the development of a new transcriptional slippage assay, specifically designed to determine if the λ antitermination factor N affects the frequency of transcriptional slippage by RNAP. We find that a homopolymeric A tract begins to elicit a large change in β-gal activity at A8, reminiscent of the length of the 8–9-bp RNA/DNA hybrid during transcription elongation (3,4). β-gal functions as a tetramer (36) and multiple functional β-gal subunits can be produced from in-frame transcripts resulting from transcriptional slippage (7). Ribosomal frameshifting could also result in active β-gal subunits; however, the requisite sequences and downstream RNA secondary structures required for ribosomal frameshifting are not predicted in this sequence (13). Moreover, because it generates only one subunit per event, ribosomal frameshifting would be less likely to produce the high local concentration of functional β-gal subunits necessary to make active β-gal tetramers (7,8,13).
We found that modification of RNAP by the N–nut antitermination complex reduces transcriptional slippage in vivo. The greater effect of N on homopolymeric U tracts compared to A tracts supports the hypothesis that stabilization of the hybrid reduces slippage, as rU/dA pairs are more unstable than rA/dT pairs (34). N prevents insertions to a greater extent on A tracts with 5′-U than on 5′-C or 5′-G, suggesting that hybrids that are already reasonably stable are not further stabilized by N to an appreciable extent. Slippage of the transcript on short homopolymeric tracts is also expected to generate mismatches between bases at the upstream end of the RNA/DNA hybrid, which have differential stability (37) and might also contribute to the overall stability of the RNA/DNA hybrid after a slippage event. Stability of mismatches in slippage intermediates does not appear to be an overriding factor in generating insertions or deletions in the transcript. It might be expected that slippage events that result in a relatively stable upstream mismatch in the RNA/DNA hybrid, such as rG/dT (37) and rA/dC (38), would promote more insertions or deletions due to a higher RNA/DNA hybrid stability, compared to less stable mismatches. However, we observed the opposite; there was less slippage in the two contexts that would generate rG/dT or rA/dC mismatches, compared to similar out-of-frame constructs (Table 3).
In vitro, where N was present at a high concentration, N alone was capable of reducing the frequency of insertions at homopolymeric A and U tracts, despite an increase in elongation rate that might have been expected to increase the catalytic rate of slippage. The in vitro experiments were conducted on DNA templates that are unrelated to the λ genetic reporter system, and did not encode the nut element. In vivo, N was expressed from a medium-copy-number plasmid and would not be produced at a high enough concentration to function without nut or Nus factors (39). Indeed, we see no evidence of antitermination or slippage inhibition when nut was disabled in vivo (see nutL7; Table 2). In vitro assays measure the rate of insertion, analogous to the frequency measurements made in the −1 out-of-frame genetic constructs. The 1.6-fold effect is similar in magnitude to the 1.9-fold effect observed on −1 out-of-frame constructs in vivo (Table 2).
N may prevent disruption of the hybrid at the 5′ end, making dissociation of RNA from the DNA template less likely, a step that appears to be important in both termination and slippage (9,32). Loss of stability in the 5′ end of the RNA/DNA hybrid plays a major role in shortening the length of the hybrid, ultimately leading to termination of transcription at intrinsic terminators (4). A shift in the RNA/DNA hybrid at the boundary of a homopolymeric tract will align non-pairing nucleotides with one another at the upstream end of the hybrid, reducing the length of correctly paired nucleotides within the hybrid. Slippage of the transcript might, therefore, also participate in the shortening of the RNA/DNA hybrid and contribute to the termination of transcription. Our findings support the hypothesis that slippage and termination of the elongation complexes are connected phenomena (9,40). It will be interesting to learn if other antitermination systems, such as the Tat/tar in HIV (22), similarly affect transcriptional slippage.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGMENTS
We would like to thank Stephen Weitzel and Peter von Hippel for providing N protein. We thank Yan Ning Zhou, Jessica Law and the members of the Court Lab for many helpful discussions.
FUNDING
Intramural Research Program of the National Institutes of Health at National Cancer Institute's Center for Cancer Research; National Cancer Institute, National Institutes of Health [HHSN261200800001E]. Funding for open access charge: Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Strathern J.N., Jin D.J., Court D.L., Kashlev M. Isolation and characterization of transcription fidelity mutants. Biochim. Biophys. Acta. 2012;1819:694–699. doi: 10.1016/j.bbagrm.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sidorenkov I., Komissarova N., Kashlev M. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol. Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 3.Kireeva M.L., Komissarova N., Waugh D.S., Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J. Biol. Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- 4.Komissarova N., Becker J., Solter S., Kireeva M., Kashlev M. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol. Cell. 2002;10:1151–1162. doi: 10.1016/s1097-2765(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 5.Baranov P.V., Hammer A.W., Zhou J., Gesteland R.F., Atkins J.F. Transcriptional slippage in bacteria: distribution in sequenced genomes and utilization in IS element gene expression. Genome Biol. 2005;6:R25. doi: 10.1186/gb-2005-6-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon A.J., Satory D., Halliday J.A., Herman C. Heritable change caused by transient transcription errors. PLoS Genet. 2013;9:e1003595. doi: 10.1371/journal.pgen.1003595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner L.A., Weiss R.B., Driscoll R., Dunn D.S., Gesteland R.F. Transcriptional slippage occurs during elongation at runs of adenine or thymine in Escherichia coli. Nucleic Acids Res. 1990;18:3529–3535. doi: 10.1093/nar/18.12.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou Y.N., Lubkowska L., Hui M., Court C., Chen S., Court D.L., Strathern J., Jin D.J., Kashlev M. Isolation and characterization of RNA polymerase rpoB mutations that alter transcription slippage during elongation in Escherichia coli. J. Biol. Chem. 2013;288:2700–2710. doi: 10.1074/jbc.M112.429464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macdonald L.E., Zhou Y., McAllister W.T. Termination and slippage by bacteriophage T7 RNA polymerase. J. Mol. Biol. 1993;232:1030–1047. doi: 10.1006/jmbi.1993.1458. [DOI] [PubMed] [Google Scholar]
- 10.Anikin M., Molodtsov V., Temiakov D., McAllister W.T. Transcript slippage and recoding. In: Atkins J. F., Gesteland R. F., Bujnicki J. M., editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. 24th edn. New York, NY: Springer; 2010. pp. 409–432. [Google Scholar]
- 11.Baranov P.V., Fayet O., Hendrix R.W., Atkins J.F. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 2006;22:174–181. doi: 10.1016/j.tig.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Turnbough C.L., Jr Regulation of gene expression by reiterative transcription. Curr. Opin. Microbiol. 2011;14:142–147. doi: 10.1016/j.mib.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen B., Wills N.M., Nelson C., Atkins J.F., Gesteland R.F. Nonlinearity in genetic decoding: homologous DNA replicase genes use alternatives of transcriptional slippage or translational frameshifting. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1683–1688. doi: 10.1073/pnas.97.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummond D.A., Wilke C.O. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vassylyev D.G., Vassylyeva M.N., Perederina A., Tahirov T.H., Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 16.Wilson H.R., Zhou J.G., Yu D., Court D.L. Translation repression by an RNA polymerase elongation complex. Mol. Microbiol. 2004;53:821–828. doi: 10.1111/j.1365-2958.2004.04170.x. [DOI] [PubMed] [Google Scholar]
- 17.Hammer K., Jensen K.F., Poulsen P., Oppenheim A.B., Gottesman M. Isolation of Escherichia coli rpoB mutants resistant to killing by lambda cII protein and altered in pyrE gene attenuation. J. Bacteriol. 1987;169:5289–5297. doi: 10.1128/jb.169.11.5289-5297.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin D.J., Cashel M., Friedman D.I., Nakamura Y., Walter W.A., Gross C.A. Effects of rifampicin resistant rpoB mutations on antitermination and interaction with nusA in Escherichia coli. J. Mol. Biol. 1988;204:247–261. doi: 10.1016/0022-2836(88)90573-6. [DOI] [PubMed] [Google Scholar]
- 19.Roberts J.W., Shankar S., Filter J.J. RNA polymerase elongation factors. Annu. Rev. Microbiol. 2008;62:211–233. doi: 10.1146/annurev.micro.61.080706.093422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mason S.W., Li J., Greenblatt J. Host factor requirements for processive antitermination of transcription and suppression of pausing by the N protein of bacteriophage lambda. J. Biol. Chem. 1992;267:19418–19426. [PubMed] [Google Scholar]
- 21.Das A. Control of transcription termination by RNA-binding proteins. Annu. Rev. Biochem. 1993;62:893–930. doi: 10.1146/annurev.bi.62.070193.004333. [DOI] [PubMed] [Google Scholar]
- 22.Brigati C., Giacca M., Noonan D.M., Albini A. HIV Tat, its TARgets and the control of viral gene expression. FEMS Microbiol. Lett. 2003;220:57–65. doi: 10.1016/S0378-1097(03)00067-3. [DOI] [PubMed] [Google Scholar]
- 23.Li S.C., Squires C.L., Squires C. Antitermination of E. coli rRNA transcription is caused by a control region segment containing lambda nut-like sequences. Cell. 1984;38:851–860. doi: 10.1016/0092-8674(84)90280-0. [DOI] [PubMed] [Google Scholar]
- 24.Rees W.A., Weitzel S.E., Yager T.D., Das A., von Hippel P.H. Bacteriophage lambda N protein alone can induce transcription antitermination in vitro. Proc. Natl. Acad. Sci. U.S.A. 1996;93:342–346. doi: 10.1073/pnas.93.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whalen W., Ghosh B., Das A. NusA protein is necessary and sufficient in vitro for phage lambda N gene product to suppress a rho-independent terminator placed downstream of nutL. Proc. Natl. Acad. Sci. U.S.A. 1988;85:2494–2498. doi: 10.1073/pnas.85.8.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeVito J., Das A. Control of transcription processivity in phage lambda: Nus factors strengthen the termination-resistant state of RNA polymerase induced by N antiterminator. Proc. Natl. Acad. Sci. U.S.A. 1994;91:8660–8664. doi: 10.1073/pnas.91.18.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nudler E., Gusarov I. Analysis of the intrinsic transcription termination mechanism and its control. Methods Enzymol. 2003;371:369–382. doi: 10.1016/S0076-6879(03)71028-3. [DOI] [PubMed] [Google Scholar]
- 28.Wilson H.R., Kameyama L., Zhou J.G., Guarneros G., Court D.L. Translational repression by a transcriptional elongation factor. Genes Dev. 1997;11:2204–2213. doi: 10.1101/gad.11.17.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Churchward G., Belin D., Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984;31:165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- 30.Miller J.H. Experiments in Molecular Genetics. New York, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 31.Kireeva M.L., Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2009;106:8900–8905. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeong S.W., Lang W.H., Reeder R.H. The yeast transcription terminator for RNA polymerase I is designed to prevent polymerase slippage. J. Biol. Chem. 1996;271:16104–16110. doi: 10.1074/jbc.271.27.16104. [DOI] [PubMed] [Google Scholar]
- 33.Wilson H.R., Yu D., Peters H.K., III, Zhou J.G., Court D.L. The global regulator RNase III modulates translation repression by the transcription elongation factor N. EMBO J. 2002;21:4154–4161. doi: 10.1093/emboj/cdf395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Y., Chen C., Russu I.M. Dynamics and stability of individual base pairs in two homologous RNA-DNA hybrids. Biochemistry. 2009;48:3988–3997. doi: 10.1021/bi900070f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheeran A., Babu Suganthan R., Swapna G., Bandey I., Achary M.S., Nagarajaram H.A., Sen R. Escherichia coli RNA polymerase mutations located near the upstream edge of an RNA:DNA hybrid and the beginning of the RNA-exit channel are defective for transcription antitermination by the N protein from lambdoid phage H-19B. J. Mol. Biol. 2005;352:28–43. doi: 10.1016/j.jmb.2005.06.052. [DOI] [PubMed] [Google Scholar]
- 36.Matthews B.W. The structure of E. coli beta-galactosidase. C. R. Biol. 2005;328:549–556. doi: 10.1016/j.crvi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 37.Sugimoto N., Nakano M., Nakano S. Thermodynamics-structure relationship of single mismatches in RNA/DNA duplexes. Biochemistry. 2000;39:11270–11281. doi: 10.1021/bi000819p. [DOI] [PubMed] [Google Scholar]
- 38.Wang W., Hellinga H.W., Beese L.S. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17644–17648. doi: 10.1073/pnas.1114496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lazinski D., Grzadzielska E., Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989;59:207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- 40.Reeder R.H., Lang W.H. Terminating transcription in eukaryotes: lessons learned from RNA polymerase I. Trends Biochem. Sci. 1997;22:473–477. doi: 10.1016/s0968-0004(97)01133-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


