Abstract
The transcription factor Msn2 mediates a significant proportion of the environmental stress response, in which a common cohort of genes changes expression in a stereotypic fashion upon exposure to any of a wide variety of stresses. We have applied genome-wide chromatin immunoprecipitation and nucleosome profiling to determine where Msn2 binds under stressful conditions and how that binding affects, and is affected by, nucleosome positioning. We concurrently determined the effect of Msn2 activity on gene expression following stress and demonstrated that Msn2 stimulates both activation and repression. We found that some genes responded to both intermittent and continuous Msn2 nuclear occupancy while others responded only to continuous occupancy. Finally, these studies document a dynamic interplay between nucleosomes and Msn2 such that nucleosomes can restrict access of Msn2 to its canonical binding sites while Msn2 can promote reposition, expulsion and recruitment of nucleosomes to alter gene expression. This interplay may allow the cell to discriminate between different types of stress signaling.
INTRODUCTION
Regulation of eukaryotic gene expression involves a complex interplay among transcription factors, core transcriptional machinery and the chromatin template on which these factors operate. A number of studies over the last several years have documented that the chromatin structure across a cell's genome remains well defined and remarkably static under all conditions (1–3). Generally, well-positioned nucleosomes bracket the promoter region of most genes to maintain a nucleosome-depleted region (NDR) upstream of the transcriptional start site of the gene, with nucleosomes assuming a well-ordered periodic array extending into the coding region with periodicity diminishing with increasing distance from the promoter (1,4–6). This chromatin structure serves an instructive role in transcription factor binding, with factors able to bind to their cognate sites lying within the NDR but unable to bind to those sites occluded by nucleosomes in other regions (3,7,8). Against this backdrop of static chromatin structure, nucleosome depletion around the NDR is in some cases associated with transcriptional activation and nucleosome recruitment to the NDR associated with transcriptional repression (9–12). This local reorganization depends on the action of chromatin remodeling factors that slide, evict or recruit nucleosomes (2,11,13–19). These rearrangements also occur in concert with transcription factor binding and transcriptional reprogramming, although the causal nature of those relations is not entirely clear. To address this question, we have examined transcriptional reprogramming and nucleosome rearrangements associated with the yeast stress response.
All cells mount a rapid adaptive response to a new and stressful environment and that response generally includes substantial transcriptional reprogramming. The transcriptional response of yeast cells to any of a wide variety of stresses, including heat shock, oxidative agents, nutrient depletion and hypo- and hyperosmolarity, comprises a stereotypic repression and induction of the same large number of genes independent of the particular type of stress, referred to as the environmental stress response (ESR), as well as changes specific to the individual stress (20,21). This transcriptional reprogramming has minimal impact on the survival of cells to the eliciting stressful condition but does serve to protect cells to subsequent stresses (22–25).
The structurally related, stress-responsive transcription factors, Msn2 and Msn4, mediate a major component of the ESR (21,26). These two transcription factors reside in the cytoplasm in unstressed cells, due to active export from the nucleus by the Msn5 exportin machinery and to restricted import as a consequence of protein kinase A (PKA) catalyzed phosphorylation and inhibition of the nuclear import signals on the proteins (27–30). A number of microfluidics-based single-cell time lapse studies have documented that acute stress causes rapid cycling of Msn2 and Msn4 into and out of the nucleus, due to inhibition of PKA and activation of protein phosphatase 1 and the Snf1 adenosine-monophosphate (AMP)-activated kinase (31–35). Cells exhibit idiosyncratic patterns of Msn2 nuclear cycling such that genetically identical cells under the same stress condition show markedly different patterns of cycling. Moreover, different stresses elicit different classes of nuclear localization patterns (34,35). How Msn2 cycling relates to the transcriptional output from Msn2 remains to be resolved, although recent results suggest that different promoters respond to Msn2 cycling in different ways (36).
Once in the nucleus, Msn2 can bind to stress response elements (STREs) within the genome to alter transcription of genes neighboring the sites (26,37). Approximately 3000 STREs reside upstream of yeast genes, but only a fraction of these serve as binding sites for Msn2. Many of these sites are likely occluded by positioned nucleosomes that prevent access to Msn2 (38). Moreover, since each cell contains only 100–200 Msn2 molecules (39), formation of stable Msn2 complexes with a large number of STREs within a single cell's genome would not be possible. One explanation for the rapid dynamics of Msn2 localization may be to facilitate sampling of multiple sites by individual Msn2 molecules. Whether different stresses affect the selection of different subsets of sites—either by modifying Msn2's deoxyribonucleic acid (DNA) binding recognition region or by altering the accessibility of different sites—has not been extensively explored.
Several previous studies have addressed the localization of Msn2 binding on a genome-wide basis. Venters et al. (40) examined global Msn2 binding in response to heat shock in the context of a much larger study to map the majority of transcription factor binding sites in yeast. More recently, Huebert et al. (41) mapped the location of Msn2 binding over time over the entire genome following treatment of cell with peroxide and correlated that binding with genome-wide changes in nucleosome positioning. Here we examine the binding of Msn2 to genomic sites in response to a nutritional stress. We also measure the global nucleosome architecture before and after application of the stress both in the presence and absence of Msn2 in order to address the extent to which Msn2 binding influences and is influenced by nucleosomes. Finally, we assess the sufficiency and necessity of Msn2 binding on changes in expression of each associated gene to determine the effect on transcription elicited by Msn2 binding. Our results document an extensive interplay between nucleosome binding and Msn2 binding such that nucleosome occlusion can restrict Msn2 binding in some conditions but in other cases Msn2 binding leads to repositioning of occluding nucleosomes. These results point to a complex choreography between general and specific transcription factors in order to mount a coherent transcriptional program. In a companion paper (Elfving et al. 2014, submitted), we also examine the role of Mediator in this process.
MATERIALS AND METHODS
Yeast strain growth and construction
Standard methods were used to grow, maintain and construct strains (42). Strains used in this study are listed in Table 1.
Table 1. Strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| Y2864 | MATα gal1::HIS3 ade2–1 can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 | (46) |
| Y3513 | MATα msn2::KanMX4 msn4::KanMX4 gal1::HIS3 ade2–1 can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 (isogenic to Y2864) | This study |
| Y4105 | MATaleu2Δ0 ura3Δ0 MSN2-GFP | ” |
| Y4108 | MATaleu2Δ0 ura3Δ0 KanMX4-PZ4EV-MSN2-GFP | ” |
| Y4015 | MATa prototroph | (44) |
| Y4016 | MATaMSN2–8xGly-13xMyc-KanMX4 (isogenic to Y4015) | This study |
| Y4131 | MATaleu2Δ::PACT1-Z4EV-NatMX KanMX4-PZ4EV-MSN2-GFP ybr032w::LEU2 | ” |
| Y4132 | MATaleu2Δ::PACT1-Z4EV-NatMX KanMX4-PZ4EV-MSN26A-GFP ybr032w::LEU2 | ” |
| Y4108 | MATaleu2Δ KanMX-PZ4EV-MSN2-GFP | ” |
The myc-tagged Msn2 strain was made by replacing the stop codon in the corresponding ORF (YMR037C) with a G8–13xMyc-KanMx6 construct. This construct was made by polymerase chain reaction (PCR) from the template plasmid pFA6a-13Myc-kanMX6. The flexible G8–linker was introduced in order to improve in vivo function of the epitope-tagged protein as suggested by Sabourin et al. (43). The construct was transformed using standard methods into strain FY4 (Y4015), a prototroph in the S288C background (44).
The estradiol inducible MSN2 strains were constructed essentially as described in (45). First, MSN2-GFP was integrated into the MSN2 locus of strain Y4098 (MATa msn2::URA3 leu2Δ0 ura3Δ0) as described (35) to generate strain Y4105. MSN2S288A,S582A,S620A,S625A,S633A,S686A-GFP, designated MSN26A, was constructed from the MSN2-GFP LEU2 plasmid B2802 using the QuikChange® II XL Site-directed mutagenesis kit (Agilent) and also integrated into strain Y4098 to generate strain Y4107. The Z4EV promoter was inserted upstream of MSN2-GFP or MSN26A-GFP locus in these two strains by PCR amplification of the Z4EV promoter from plasmid pMN10 (45). The resulting strains, Y4108 and Y4109, were crossed with strain DBY12416 (MATα leu2-PACT1-Z4EV ybr034w::LEU2) (45) and prototrophic segregants from each cross carrying the Z4EV transcription factor and the Z4EV-driven MSN2 locus were retained as Y4131 and Y4132.
Growth protocols
Glucose downshift
Yeast cells were grown at 30°C in Synthetic Complete (SC) + 2% glucose media and maintained in mid log phase for at least 24 h by dilution. When cultures reached a density of (3–4) × 106 cells/ml, cells were collected by filtration (Stericup-GP, 0.22-μm filtering systems, Millipore Corporation, Billerica, MA, USA), washed and resuspended in SC + 3% glycerol media and then subjected to sample preparation at the desired time points. Zero-minute samples were taken before this procedure.
Estradiol induction
Cells were grown at 30°C in SD + 2% glucose to a cell density of 4 × 106 cells/ml and estradiol added to a final concentration of 10 μM. Samples (5 ml) were harvested at specified times and ribonucleic acid (RNA) extracted as described below.
Nucleosomal DNA sample preparation
Mononucleosomal DNA from the wild-type parent strain and the msn2Δmsn4Δ strain was isolated as previously described (3). Briefly, for each sample, a 650-ml culture was formaldehyde crosslinked, converted to spheroplasts, then resuspended in NP buffer, and micrococcal nuclease (Sigma-Aldrich, St. Louis, MO, USA) was added. DNA–protein cross-links were reversed by incubation at 65°C for at least 4 h. DNA was then purified by PCR clean-up kit (Qiagen, Valencia, CA, USA), and the sample was analyzed by gel electrophoresis to ensure that the extent of digestion did not vary significantly from sample to sample. Data were obtained from single samples for each time point, with greater than 1.3 × 107 unique 2 × 100-bp reads per sample.
mRNA sample preparation and transcript analysis
Messenger RNA (mRNA) was isolated and hybridized to Agilent yeast microarrays as described in (46). Briefly, 5-ml cultures were collected on filters and snap frozen in liquid nitrogen. Total RNA was extracted using the Qiagen RNeasy Mini kit, including the additional DNase I digestion step. Chromosomal RNA (cRNA) for microarray hybridization was synthesized following the standard protocol of the Agilent Low RNA Input Linear Amplification kit (Agilent Technologies). cRNA was extracted using the Qiagen RNeasy Mini kit and hybridized to Agilent Yeast Gene Expression Microarray (8 × 15K G4813A) slides and scanned at 5-μm resolution. Data were extracted using Agilent Feature Extraction software version 9.5 with Linear Lowess dye normalization and no background subtraction and were submitted to the Princeton University Microarray database for storage and analysis.
For estradiol induction experiments, time course fold change in transcript levels was fit to a Hill plot by optimization of n, f0, K and Vmax for each gene for the equation f(t) = f0 + Vmax·tn/(Kn + tn). Delay times were determined by extrapolation of the derivative of this function at f(t) = Vmax/2 to the x-axis intercept.
Chromatin preparation for chromatin immunoprecipitation
Chromatin extract production was adapted from (47), with some modifications. Briefly, 45-ml yeast cultures prior to or post the glucose downshift procedure were crosslinked with formaldehyde (0.8% final concentration) for 10 min and quenched with glycine for 5 min. Cells were harvested by centrifugation, 3000 X g, 4°C, 5 min, and washed with cold buffer (50-mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.5, 140-mM NaCl), resuspended in 400-μl cold ChIP lysis buffer (50-mM HEPES pH 7.5, 140-mM NaCl, 1-mM ethylenediaminetetraacetic acid (EDTA), 1% Triton, 0.1% sodium deoxycholate, 1-mM phenylmethylsulfonyl fluoride and a Roche complete protease inhibitor tablet) and snap frozen in liquid nitrogen. Samples were thawed in 37°C water bath, put on ice, and cold glass beads were added to 1 mm below meniscus. Cells were disrupted with a Fast Prep-24 (MP Biomedicals) bead beating system on setting 5.5 m/s 3 × 40 s in a 4°C cold room. The resulting cell lysates were centrifuged at 20 800 x g, 4°C, 30 min. The supernatants were removed, and the pellets were resuspended in 100-μl ChIP lysis buffer and placed in 120-μl Covaris tubes for sonication shearing. Chromatin was sheared to an average fragment size of 350 bp using a Covaris E220 system. The sheared chromatin samples were transferred to an Eppendorf tube and sample volume adjusted to 200 μl (by adding ChIP lysis buffer) and centrifuged at 10000 x g, 4°C, 5 min. The pellets were the ‘insoluble fraction’ and the supernatants were transferred to a new Eppendorf tube and centrifuged again, 10 000 x g, 4°C, 15 min. The final supernatants were the chromatin extract used for ChIP.
Chromatin immunoprecipitation
For each ChIP, 2.5-μl anti-myc (Clontech, clone 9E10, cat#631206) or anti-Pol II C-terminal domain (Pol II 8WG16 Monoclonal Antibody, Covance) antibody was added to 15-μl resuspended protein G Dynabeads (Invitrogen), coupled according to the Dynabeads manual and washed and resuspended in 233-μl lysis buffer per sample. Sixty-seven microliter chromatin extract was incubated with the antibody-bound beads (total volume 300 μl) with rotation for 4 h at room temperature (RT). The beads were then collected with the magnet and washed (resuspended and nutated 4 min, RT) with 0.5-ml sodium dodecyl sulphate (SDS) buffer (50-mM HEPES pH 7.5, 140-mM NaCl, 1-mM EDTA and 0.025% SDS). Beads were subsequently washed with 0.5-ml high salt buffer (50-mM HEPES pH 7.5, 500-mM NaCl, 1-mM EDTA), followed by 0.5-ml tris-lithium (TL) buffer (20-mM Tris-Cl pH 7.5, 140-mM NaCl, 250-mM LiCl, 1-mM EDTA), followed by two washes in 0.5-ml tris-EDTA (TE) buffer (20-mM Tris-Cl pH 7.5, 0.1-mM EDTA). Washed beads were resuspended for elution in 72-μl TE + 1% SDS buffer (20-mM Tris-Cl pH 7.5, 0.1-mM EDTA, 1% SDS), vortexed and heated in a 65°C water bath, 2 min. The beads were vortexed well again and supernatants were taken from the beads. Twenty-five microliter was used for western blots and 45 μl was taken to reverse crosslink at 65°C, 14 h.
Reverse crosslinking and purification of DNA
Input DNA (2-μl chromatin extract (input DNA) and 118-μl TE + 1% SDS buffer) and ChIP DNA (45-μl ChIP eluate + 75-μl TE + 1% SDS buffer) were incubated at 65°C, 14 h for reverse crosslinking. Reverse crosslinked samples were purified on Qiagen PCR purification columns, eluted in 2 × 35-μl Qiagen Elution buffer and kept frozen until library construction.
Library construction
ChIP-DNA was amplified using the LM-PCR method described in Agilent Yeast ChIP-on-chip analysis protocol version 9.2, May 2007 and subjected to the Illumina TruSeq paired-end sequencing protocol.
Sequence analysis
Paired-end sequences were mapped to the Saccharomyces cerevisiae reference genome s288c Saccharomyces Genome Database (SGD) version r64-1-1_20110203, using Bowtie for Illumina (version 1.1.2) with seed length 22 and a maximum permitted total of quality scores of 70 at mismatched read positions, also allowing a maximum of two mismatches in the seed. Twenty samples each for the Msn2 ChIP at 0 and 20 min were combined to yield 2 × 105 total reads for each time point. Alignments that mapped to more than one position on the reference genome were randomly distributed between the reportable alignments. To eliminate PCR amplification artifacts, precise duplicates of paired-end ChIP read alignments mapping to a genomic position were excluded from analysis. Resulting sequence positions were then subjected to further analysis in MATLAB. Occupancy at each base pair position across the genome for both nucleosomes and ChIP profiles was determined by summing the total number of unique sequence reads at that position and then normalizing the summed values such that the average occupancy per bp for each experiment equals 1 over each chromosome. Peaks of Msn2 binding were identified either as those with a maximal peak intensity 6-fold above the average binding over the chromosome in which it is located or as those with z-score greater than 5 for the integrated area of binding in the 250-bp region around a binding maximum. Visualizations were performed using MATLAB standard bioinformatics methods. The positions of STRE elements were obtained from SGD (http://www.yeastgenome.org/cgi-bin/PATMATCH/nph-patmatch). Functional analysis of groups of genes was performed using the Gene Ontology Term Finder from SGD.
Accession numbers and deposition of microarray data
Read data for the ChIP-Seq and MNase-Seq experiments are publically available at NCBI SRA with the accession number SRP033438. Microarray data are publicly available at http://puma.princeton.edu/cgi-bin/publication/viewPublication.pl?pub_no=559 and as a processed spreadsheet in Supplementary Table S2.
RESULTS
Msn2 binds to a limited number of sites in vivo
To explore the relation between transcription factor binding, transcriptional changes and nucleosome repositioning, we determined the global binding pattern of Msn2 by chromatin immunoprecipitation and DNA sequencing of the precipitated fragments (ChIP-Seq) prior to and 20 min after transition of cells from growth on glucose to growth on glycerol, a condition that induces the ESR. We performed ChIP-Seq using anti-Myc antibodies on a strain in which MSN2 was replaced with MSN2 tagged with 13 copies of the Myc epitope attached to the carboxy terminus of the protein and expressed under its own promoter. The Myc-tagged version of the protein showed normal nuclear localization and transcriptional activation in response to both hydrogen peroxide and glucose downshift conditions (Elfving et al. 2014, submitted). We obtained 3–4-fold average sequence coverage over the entire genome for both time points and 180 reads over the most abundant unique binding site at the 20 min time point. To assess the interplay of nucleosome remodeling and Msn2 binding, we concurrently mapped genome-wide nucleosome positions prior to and 20 min after the glucose-to-glycerol switch in an MSN2 MSN4 strain and in an isogenic msn2 msn4 strain by sequencing size-selected DNA fragments following micrococcal nuclease treatment of cross-linked chromatin. We obtained >100-fold sequence coverage of the entire genome for both strains at each time point.
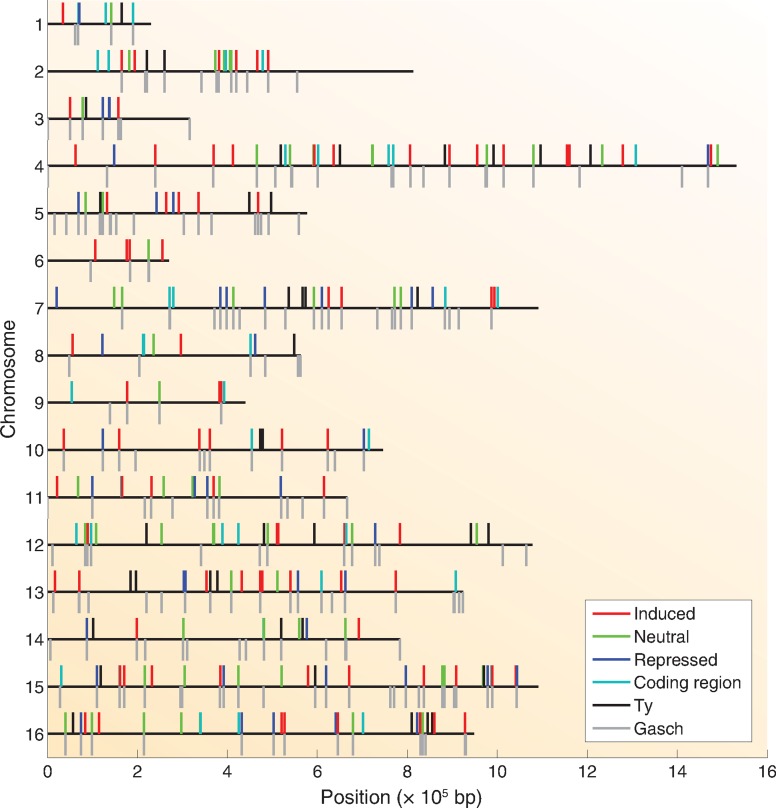
ChIP-Seq identified few Msn2 binding sites prior to the carbon source downshift and a large number after the downshift. We computationally identified sites of Msn2 binding as described in the Materials and Methods section. The positions of the major Msn2 binding sites are shown in Figure 1. We hand annotated each of the peaks to identify the genomic features associated with each site. This process yielded 273 distinct and robust peaks of bound Msn2, distributed over 269 genes, 20 min after the glucose downshift. The positions of these sites, the associated gene or genomic feature and the relative abundance of Msn2 at these sites prior to and after the glucose downshift are listed in Supplementary Table S1. The majority (192) of those sites corresponded to promoter regions with the remaining peaks mapping solely to gene coding sequences (36 peaks) or to Ty elements (40 peaks), with the latter displaying a very characteristic pattern of Msn2 distribution based on the manner in which sequence reads were apportioned to repeat sequences. Because Ty elements are not readily distinguished by sequence, we could not determine whether all Ty elements bind Msn2 at equal levels or whether some have greater affinity than others. Most of the coding sequences registering significant Msn2 binding were expressed at high levels, as measured by PolII occupancy (Supplementary Table S1), consistent with the growing appreciation that highly expressed genes are retrieved inadvertently as artifacts of the ChIP protocol (48). In fact, more than half of the 50 most highly expressed genes were recovered in the Msn2 ChIP experiment (P < 10−100). Moreover, most coding regions to which Msn2 bound lack an STRE, the binding motif for Msn2. In sum, our experiments identified a number of sites for Msn2 binding following glucose downshift, with a majority of the robust sites mapping to promoter regions and a smaller number mapping to Ty elements and to coding domains of highly expressed genes.
Figure 1.
Msn2 binding sites. The relative positions of the 268 Msn2 binding sites determined in this study are indicated by short vertical lines above each chromosome (horizontal black lines). The vertical lines below the chromosomes denote the 212 most robust binding sites identified in (41) following exposure of cells to hydrogen peroxide. The binding sites identified in the current study are denoted by color specified in the legend as residing solely in the coding region of a gene (coding region), over a transposable element (Ty) or in the promoter of a gene induced, repressed or unaffected (neutral) by Msn2. See Supplementary Table S1 for a detailed description of each site.
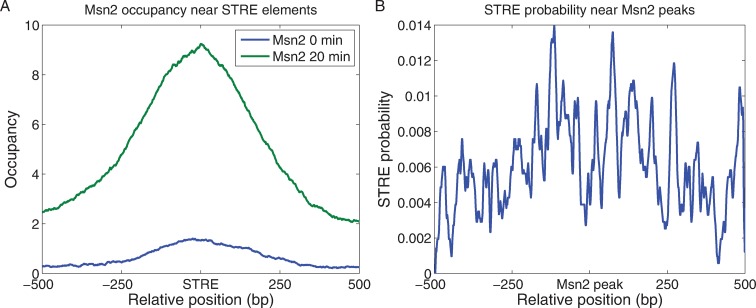
Previous in vivo and in vitro studies have identified a canonical binding site for Msn2, the STRE with a sequence RGGGG (26,37,49–51). As shown in Figure 2A, Msn2 binding is significantly enriched around STREs. Moreover, the probability of an STRE residing close to a peak of Msn2 binding is significantly greater than that expected for a random distribution of STREs relative to binding sites (Figure 2B). However, while approximately 11,500 STRE sequences are present in yeast genome, with 3160 lying in the promoters of almost 2000 genes, Msn2 fails to bind most of these sites. Reasons for this selective binding to only a subset of STREs are addressed below. The probability of Msn2 binding to a promoter was correlated with the number of STREs within that promoter (Supplementary Figure S1). This may result from cooperative binding of Msn2 to adjacent STREs, particularly since the fraction of multiple STRE-containing promoters that are bound by Msn2 is higher than that predicted assuming independent interaction of Msn2 with each STRE within a promoter (Supplementary Figure S1). We also observed low but detectable levels of Msn2 binding over STREs prior to the nutrient downshift. This is consistent with the observation that, while Msn2 resides predominantly in the cytoplasm in the absence of stress, Msn2 is not completely excluded from the nucleus under those conditions (35). Moreover, even under robust growth conditions, Msn2 exhibits random nuclear bursting, such that Msn2 congregates in the nucleus for a short period of time in a small number of individual cells. Thus, the low level of Msn2 binding we observe on average in unstressed cells may represent robust Msn2 binding in a very small number of cells in the population.
Figure 2.
Msn2 binds near STREs. (A) The average Msn2 occupancy at a particular site is plotted as a function of distance of that site from each of the 3150 promoter STREs (RGGGG motifs), before and 20 min after a glucose-to-glycerol downshift. (B) The likelihood of an STRE residing at a particular site is plotted as a function of distance of that site from each of the peaks of Msn2 binding identified in this study and listed in Supplementary Table S1. The values are calculated as the fraction of Msn2 peaks that contain an STRE at the indicated position, averaged over a 20 bp moving window. By comparison, the probability of finding STRE at a random site is less than 9 × 10−4.
We compared the binding sites identified in our experiment with those observed by Huebert et al. following treatment of cells with hydrogen peroxide (41). This study reported 1289 sites that were statistically enriched in the treated samples. Plotting the 212 sites with the highest reported peak values from the Huebert et al. experiment relative to the positions of the most robust sites from our experiment indicates a significant overlap in the two sets of data (Figure 1). In fact, the peaks of 113 sites (42%) identified in our experiment lie within 500 bp of a peak among the 212 most abundant sites identified by Huebert et al. (P < 10−250). Moreover, 76% of the peaks identified in our experiment coincide with the location of one of the significant sites identified by Huebert et al. Thus, the vast majority of de novo Msn2 binding sites are recapitulated under different stress conditions, consistent with a relative uniform transcriptional response of cells to stress, independent of the nature of the initiating stress.
Stress-specific binding of Msn2 to promoters reflected both common and condition-specific responses to stress. While most of the Msn2-binding sites identified upon nutrient downshift overlap those identified following oxidative stress, we noted that a number of stress-induced Msn2 binding sites identified after oxidative stress were absent upon nutrient downshift, and vice versa. Those genes in whose promoters Msn2 bound following oxidative stress but not after glucose downshift were enriched for telomeric sequences and for those involved specifically in response to treatment with oxidizing agents (13/69; P = 10−4). Similarly, those genes in whose promoters Msn2 bound following glucose downshift but not in response to oxidative agents were highly enriched in those involved in carbohydrate metabolism (P = 4 × 10−5). Thus, in addition to binding to promoters of genes involved in a common stress response, Msn2 binds specifically to a subset of genes associated with response to the particular initiating stress. We discuss below possible mechanistic bases for this discrimination.
Msn2 mediates both gene activation and gene repression
We assessed the effect of Msn2 on transcriptional reprogramming under nutrient downshift conditions in several ways. First, we determined the level of Pol II associated with all coding regions prior to and 20 min after glucose to glycerol transition by performing ChIP-Seq using antibodies targeting the C-terminal repeat domain of the Rpb1 subunit of Pol II. In addition, we examined global transcript levels by microarray analysis of both an MSN2 MSN4 and an msn2 msn4 strain pre- and post-transition. These data provide information on the extent to which transcriptional changes that occur following the carbon source downshift are dependent on the Msn transcription factors. Finally, to identify those transcriptional changes that occur specifically in response to activation of Msn2, we measured global transcriptional changes following ectopic induction of Msn2. This was accomplished using a hybrid zinc-finger transcription factor recently described, termed Z4EV (45). Specifically, we used a strain in which MSN2 expression was driven by a modified GAL1 promoter in which four repeats of a zinc-finger DNA binding protein recognition sequence replaced the Gal4 binding sites. The strain expressed the Z4EV fusion protein, consisting of the zinc-finger binding protein targeting the modified GAL1 promoter as well as the VP16 activation domain and an estrogen receptor fragment encompassing the Hsp90 and estrogen binding domains. Z4EV resides in the cytoplasm in the absence of estradiol, due to binding Hsp90, and rapidly dissociates from Hsp90 following estradiol addition, resulting in relocation to the nucleus and induction of transcription of genes with a upstream activator sequence (UAS) containing the specific zinc-finger binding motifs. Since this sequence does not normally exist in the yeast genome, the only gene induced by estradiol treatment is the one engineered to be linked to that sequence. Accordingly, estradiol induces production only of Msn2 in this strain. While the induced Msn2 resides predominantly in the cytoplasm upon estradiol treatment, sufficient nuclear localization occurs, through partitioning in all cells and bursting in some cells, to allow Msn2 to exert its transcriptional effects (see below). We conclude that any gene rapidly induced or repressed by estradiol treatment must be under the direct regulation of Msn2.
Using the transcript data from the experiments described above, we could identify those genes whose expression was directly affected by Msn2 in response to the glucose downshift. The strong correlation between transcript level changes and the changes in Pol II occupancy over the corresponding coding regions following nutrient downshift confirmed that transcript level changes were a consequence of changes in transcriptional activation rather than post-transcriptional processes (Supplementary Figure S2). We identified those genes activated by Msn2 as those that showed increased transcript levels upon estradiol treatment of the Z4EV strain described above as well as diminished induction, or more substantial repression, of transcript levels in the msn2 msn4 strain versus the MSN2 MSN4 strain after the glucose downshift. Similarly, we identified genes repressed by Msn2 as those whose transcript levels fell upon estradiol treatment of the Z4EV strain and exhibited higher transcript levels in the msn2 msn4 strain versus the MSN2 MSN4 strain after glucose downshift. These independent measures of sufficiency and necessity of Msn2 activity on gene expression were reasonably consistent (Supplementary Table S1). Moreover, approximately two-thirds of genes exhibiting Msn2-dependent regulation by the above criteria showed Msn2 promoter binding in response to a glucose downshift, consistent with the hypothesis that Msn2 binding directly affected expression of the corresponding gene and that most genes whose transcription modulation are Msn2-dependent are directly regulated by Msn2 binding. Finally, a significant number of genes (86/192) to whose promoter Msn2 bound after the glucose downshift showed Msn2-dependent transcriptional activation, consistent with Msn2's reported role as a transcriptional activator. These were enriched in genes involved in energy reserve metabolism (P = 3 × 10−11), oxidation-reduction processes (P = 5 × 10−10) and glycogen (P = 7 × 10−10) and trehalose (P = 4 × 10−7) metabolism. However, a significant number of genes (44/192) to whose promoter Msn2 bound exhibited Msn2-dependent transcriptional repression following glucose downshift or during induction of Msn2. These were enriched in genes involved in glucose catabolism (P = 10−4). The remaining genes to which Msn2 bound were either Ty elements or coding regions noted above or showed no Msn2-dependent change in expression. These results indicate that Msn2 functions both as a transcriptional activator and a transcriptional repressor. The basis of this dual activity is discussed below.
Msn2 elicits different patterns of gene regulation kinetics
Evaluation of the transcriptional consequences of activating Msn2 using the Z4EV system revealed several unexpected aspects of Msn2 regulation. First, a number of genes significantly changed expression following Z4EV induction of Msn2 but did not exhibit significant Msn2 binding in the ChIP-Seq analysis or display Msn2-dependent transcriptional changes following the glucose downshift. The induced genes in this set generally contained one or more STREs in their upstream intergenic regions. For instance, 85 of the 100 most induced genes following estradiol treatment of the Z4EV strain contained one or more upstream STREs, although only 40 of these showed significant binding of Msn2 by ChIP-Seq in the glucose downshift experiment. This suggests a hierarchy of STRE binding affinities such that lower affinity sites are bound only when Msn2 is expressed at higher levels but that such sites can mediate activation under that condition. A number of these promoters exhibited Msn2-dependent nucleosome remodeling (see below) during the glucose downshift, indicating that even low level or transient Msn2 binding can affect local chromatin structure. Those genes repressed following Msn2 induction by Z4EV are highly enriched for ribosome biogenesis genes (P = 10−24) but only ∼10% show Msn2 binding by ChIP-Seq and most (60%) do not contain STREs in their promoters. This suggests that much of the repression is an indirect effect of Msn2 induction.
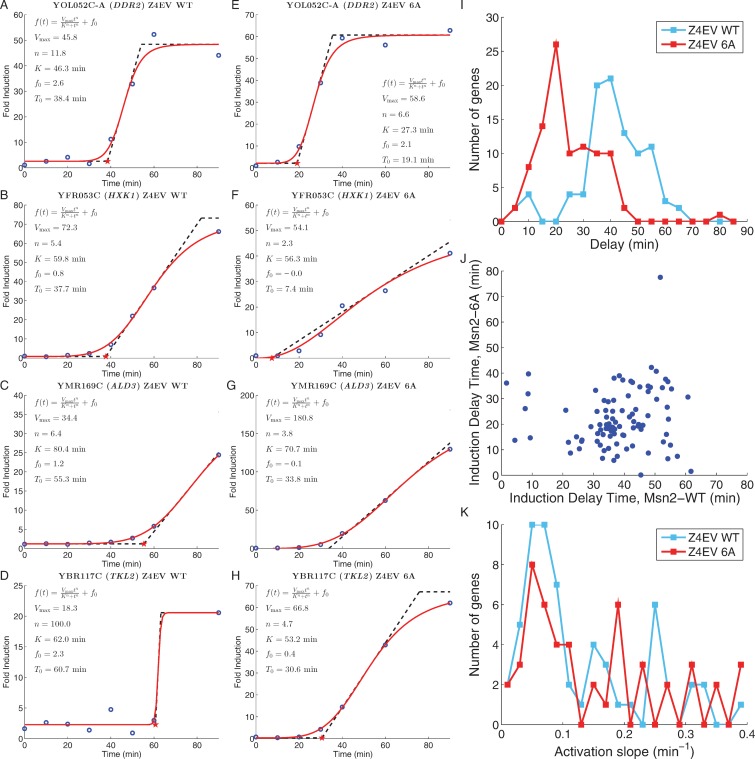
The Z4EV induction data also revealed unexpected diversity in the kinetics of activation of Msn2 responsive genes. Genes induced upon activation of Msn2 in the Z4EV strain begin to accumulate transcripts following estradiol addition but only after a delay. As illustration, the induction kinetics of several genes are shown in Figure 3A–D. The duration of this delay showed a broad distribution among the induced genes, with the majority of genes initiating transcript accumulation 35–45 min after estradiol addition while a significant minority of the genes initiated accumulation greater than 45 min following addition (Figure 3I and J). Since the Z4EV cells in which Msn2 was induced by estradiol were unstressed, Msn2 remained predominantly in the cytoplasm, with only occasional random bursts of nuclear occupancy in individual cells (Supplementary Movie S1 and Supplementary Figure S3A). To test whether this limited nuclear occupancy contributed to the variable lag in transcript accumulation, we performed the transcription study using the Z4EV strain to drive expression of a mutant Msn2, designated Msn26A, in which all six PKA phosphorylation sites in the protein were converted to alanines. Since PKA-mediated phosphorylation is responsible for retaining Msn2 in the cytoplasm and Msn26A could not be phosphorylated by PKA, Msn26A induced by estradiol treatment immediately entered the nucleus in all cells (Supplementary Movie S2 and Supplementary Figure S3B). Notably, the duration of the delay in transcript accumulation was reduced for most genes to 15–25 min (Figure 3E–J), without a significant change in the rate of transcript accumulation once induction began (Figure 3K). Repressed genes exhibited a similar pattern of kinetics (data not shown). Thus, Msn2 responsive genes fall into roughly two categories based on their activation kinetics in conditions of limited Msn2 nuclear occupancy: rapid responders and slow responders. Notably, this dichotomy is lost under conditions of high Msn2 nuclear occupancy, ruling out the possibility that the two classes simply reflect direct versus indirect targets of Msn2 regulation. Rather, Msn2-regulated genes exhibit distinctly different patterns of response to wild-type Msn2 activation.
Figure 3.
Different genes exhibit different induction kinetics in response to Msn2. (A–D) Induction kinetics for four Msn2-inducible genes following estradiol addition to strains containing wild-type MSN2 under control of the hybrid Z4EV transcription factor. Blue circles: fold increase in gene transcript levels relative to that immediately prior to estradiol addition; solid red line: best fit of the data for each gene to the function f(t) = f0 + Vmax · tn/(Kn + tn); dashed line: tangent line to the curve at f(t) = Vmax/2, whose extrapolation to the x-axis provides the measure of the time delay in response to Msn2 induction. (E–H) Induction kinetics for the genes shown in (A–D) following estradiol addition to strains containing MSN26A under control of the hybrid Z4EV transcription factor. (I) Histogram of time delay values for the 96 genes induced more than 2-fold in both the MSN2 wild-type and MSN26A strains and whose induction values are reasonably fit by the Hill curve. Blue line: delay values in the MSN2 wild-type strain; red line: delay values in the MSN26A mutant strain. (J) Scatter plot of the delay time for each gene in I in the MSN26A strain relative to that in the MSN2 wild-type strain. (K) Histogram of rates of induction, i.e. the slope of the tangent line to the fitted curve at f(t) = Vmax/2, for the 96 genes in (I). Blue line: delay values in the MSN2 wild-type strain; red line: delay values in the MSN26A mutant strain.
Msn2 can promote repositioning of nucleosomes following nutritional stress
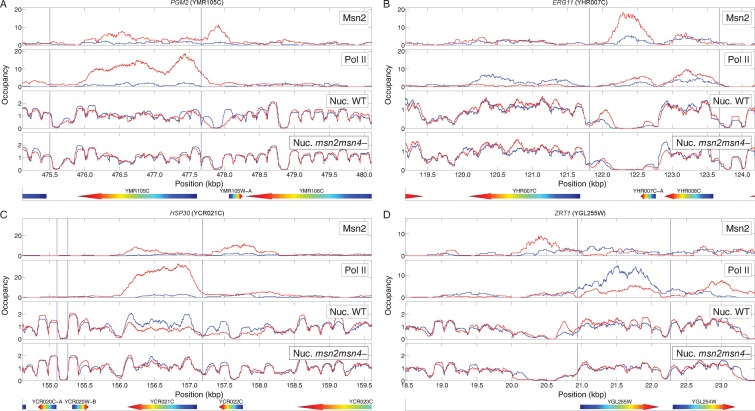
Consistent with our previous observations (3), we find that nutrient downshift results in relatively limited nucleosome repositioning, in spite of the significant transcriptional reprogramming (Supplementary Figure S4). However, by determining global nucleosome positions before and after glucose downshift in both an MSN2 MSN4 and an msn2 msn4 strain, we were able to identify promoters in which expression change was associated with nucleosome repositioning and determine whether that repositioning was dependent on Msn2 and/or Msn4. In this manner, we identified a number of genes in which transcriptional activation, Msn2 binding and nucleosome depletion from the promoter were coincident and in which nucleosome depletion was dependent on Msn2 and/or Msn4. Two examples of genes, PGM2 and HSP30, that show this pattern are profiled in Figure 4A and C. Similarly, we identified several genes at which transcriptional repression, Msn2 binding and nucleosome acquisition are coincident and in which nucleosome acquisition is dependent on Msn2 (Figure 4B and D). In sum, 72% (62/86) of induced genes regulated by Msn2 show nucleosome depletion and 60% of those show complete or partial dependence of the nucleosome depletion on Msn2 and/or Msn4. Similarly, 50% (22/44) of repressed genes regulated by Msn2 show nucleosome remodeling, primarily nucleosome acquisition, and 45% of those show dependence of nucleosome remodeling on Msn2 and/or Msn4. From these studies, we conclude that a significant function of Msn2 is to expose promoter regions during gene activation and to occlude promoter regions during gene repression. Thus, Msn2 is not simply a passive respondent to remodeling promoted by other agents but plays an active role in restructuring the NDR during transcriptional reprogramming.
Figure 4.
Msn2 promotes nucleosome repositioning over gene promoters. Shown are binding profiles of Msn2 (row 1) and PolII (row 2) as well as the nucleosome profiles (rows 3 and 4) over four different Msn2-regulated genes and the surrounding regions along the genome. Each panel shows a binding profile before the glucose-to-glycerol switch (blue lines) and 20 min after the transition (red lines). The nucleosome profiles were obtained for both an MSN2 MSN4 (row 3) and an msn2 msn4 strain (row 4). Two of the genes, PGM2 and HSP30, are induced by Msn2 following the nutrient downshift, while the other two, ERG11 and ZRT1, are repressed. In order to be able to compare the occupancy profiles obtained from different experiments with different sequencing coverage, we normalized all profiles such that the average occupancy across each chromosome in each experiment equals 1.
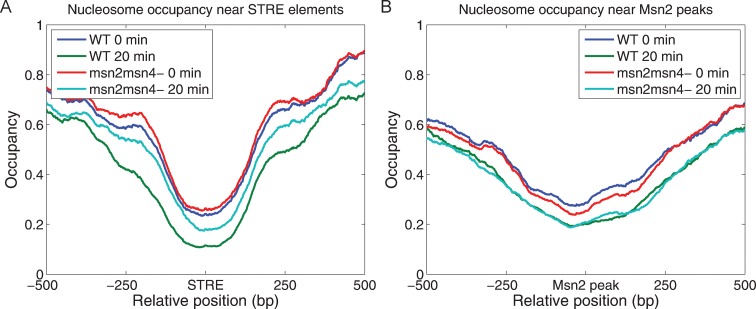
The Msn2/Msn4-dependent reorganization of nucleosomes noted above could be observed on a global scale. We calculated the nucleosome occupancy around all promoter STRE sites before and after the nutrient downshift in both the wild-type and msn2 msn4 strains. As evident from the plot of the average nucleosome occupancy as a function of distance from every STRE under these four conditions, nutrient downshift results in reduction of nucleosome density over these sites (Figure 5A). This is consistent with an overall increase in expression of stress-responsive genes following a downshift, and observed correlation between reduced occupancy of nucleosomes in promoters and increased transcription. However, the reduction in nucleosome density of STREs was approximately twice as large in the wild-type strain compared to the msn2 msn4 strain. We observed similar results by examining nucleosome density around sites of Msn2 binding (Figure 5B). These observations demonstrate that Msn proteins play an active role in reducing nucleosome occupancy during transcriptional activation.
Figure 5.
Msn2 reduces the average nucleosome occupancy near its binding sites following nutrient downshift. The distribution of nucleosome occupancy around STREs (A) and Msn2 binding sites (B), as defined in the legend to Figure 2, are shown before (blue line) and after (green line) the glucose-to-glycerol downshift in wild-type cells and before (red line) and after (cyan line) the glucose-to-glycerol downshift in msn2 msn4 mutant cells.
Nucleosomes restrict access of Msn2 to STREs
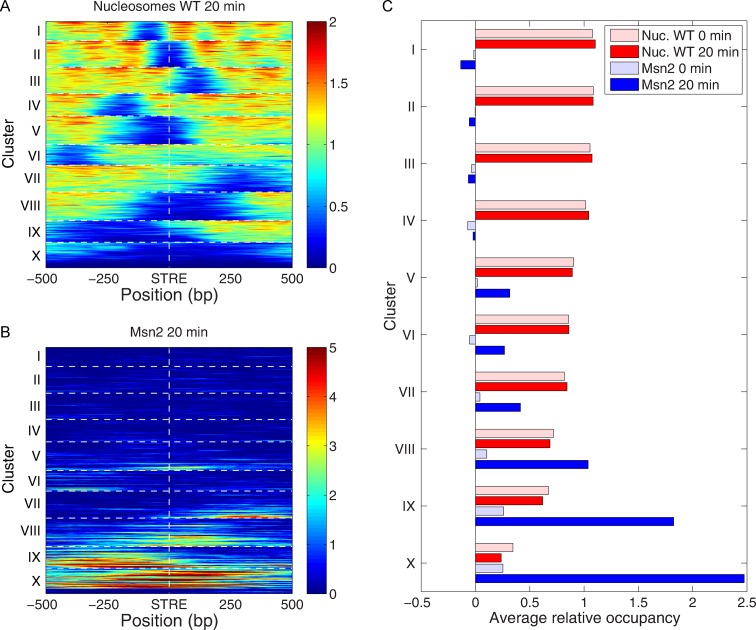
While our data described above demonstrate that Msn2 binding can alter adjacent nucleosome occupancy, we find that nucleosome position significantly restricts accessibility of Msn2 to its canonical binding sites. First, while most STREs in the genome reside in coding regions, Msn2 did not bind to any of these sites, likely because they are occluded by well-positioned nucleosomes. Second, we used k-means clustering to categorize patterns of nucleosome occupancy around the 3150 transcription start site proximal STREs prior to the nutrient downshift (Figure 6A and C). These patterns range from cases in which nucleosomes sit directly over the STRE to cases in which the STRE resides in an NDR centered on the STRE to cases in which the STRE resides in an extended NDR. We then determined to which of these STREs Msn2 bound following the nutrient downshift. As evident in Figure 6B and C, Msn2 bound almost exclusively only to those STREs residing in extended NDRs. Moreover, the extent of Msn2 binding was essentially inversely proportional to the nucleosome density around the STRE. Surprisingly, even those STREs lying in open chromatin domains of limited dimension were not substrates for Msn2 binding (cluster 2, for example). Thus, at the global scale, nucleosome occupancy restricts Msn2 binding.
Figure 6.
Msn2 binds to STREs in nucleosome free promoters. All STREs residing upstream of gene transcription start sites were organized by k-means clustering (k = 10) on the basis of the nucleosome occupancy profiles over the 1 kb regions centered on STRE at 20 min following the glucose-to-glycerol nutrient downshift. The clustered nucleosome occupancy profiles are presented as a heat map (A). A heat map of the binding profiles of Msn2 at 20 min following the nutrient downshift is shown in (B), with the same gene order as in (A). The average nucleosome and Msn2 occupancy in the 1kb regions centered on STRE elements for each cluster, before and 20 min after the switch, are shown in (C).
We also find a local effect of nucleosome occupancy on Msn2 binding. In Figure 7 we plot the density of STRE elements within the footprint of well-positioned nucleosomes across the genome. Superimposed on that plot is the average binding of Msn2 to STREs at the indicated position following nutrient downshift. Quite evident from this diagram, STREs are enriched under the nucleosome umbrella, perhaps reflecting the guanosine/cytosine bias in nucleosomal positioning sequences. In contrast, Msn2 binding is uniformly low within the 90-bp inner core of the positioned nucleosomes, significantly higher in the nucleosome adjacent region and proportionately increasing with increasing distance from the inner core of the nucleosome footprint. These results suggest that well-positioned nucleosomes prevent access of Msn2 to its cognate binding sites when those sites reside under the core of the nucleosome. However, Msn2 can gain access to its cognate sites that lie under the edges of the positioned nucleosomes. As discussed below, this observation is consistent with dynamic partial unwrapping of DNA bound to nucleosomes and competition between nucleosome binding and transcription factor binding to specific target sequences.
Figure 7.
Msn2 and STRE distribution relative to the well-positioned nucleosomes. We selected well-positioned nucleosome, with dyad positions characterized by at least five sequencing reads. The blue line shows the average Msn2 occupancy relative to the dyad locations of the well-positioned nucleosomes. The green line shows the probability of finding a nearby STRE element.
DISCUSSION
Stress-specific and stress-non-specific Msn2 binding
We have mapped Msn2 binding sites across the genome following a nutrient downshift, which elicits the ESR. Some of the binding sites correspond to structural features such as transposable elements or transcribed regions of highly expressed genes. The former are repeated sequences, which prevents assignment of binding to specific elements, so we cannot determine from our data whether all Ty elements or only a subset binds Msn2. However, binding to Ty elements is consistent with stress activation of Ty transposition (52,53) and suggests that Msn2 may play a role in stimulating such transposition. The latter structural feature—transcribed coding regions—may be an artifact of the ChIP-Seq protocol, since antibodies to many unrelated proteins have been reported to retrieve these same sequences (48,54,55). We also find that anti-Myc antibodies to multiple different tagged proteins in this experiment retrieved these sequences (data not shown). The majority of the binding sites correspond to promoters of transcribed genes. Moreover, most of these binding sites lie within several hundred base pairs of one or more STREs, previously defined as Msn2 binding sites. We conclude that most Msn2 binding is targeted to specified Msn2 binding motifs in promoter regions of genes.
Comparing our data with those previously obtained for Msn2 binding following peroxide treatment revealed a common core of Msn2-bound genes as well as condition-specific binding. Since we examined binding at only one time point following nutrient shift, we may not have captured all the nutrient-dependent binding sites. Nonetheless, the binding pattern mirrors the transcriptional changes associated with different stresses, in which a core stress response is augmented by activation/repression of genes targeted to specific stresses (20,21). Consistent with this interpretation, those genes bound by Msn2 uniquely following peroxide treatment are enriched for those associated with remediation of oxidative stress while those uniquely bound following glucose downshift are involved in carbohydrate metabolism. These results suggest that the stress-specific transcriptional response is mediated at least in part by Msn2.
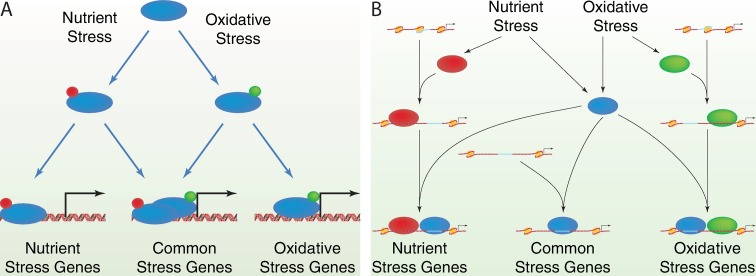
How might Msn2 binding be responsive to specific stimuli? One possibility is that post-translational modification of Msn2 dictated by different stress signaling pathways modifies the binding specificity of the protein (Figure 8A). In this context, different stresses yield different patterns of Msn2 nuclear entry and exit, patterns that may well be perceived by different promoters in different ways (34–36). We found in this study that Msn2 responsive genes had quite divergent delay times in response to Msn2 activation when Msn2 was largely cytoplasmic and exhibited only random bursts of nuclear occupancy. This difference was essentially eliminated when Msn2 resided predominately in the nucleus. This divergence in response times may reflect the distinction recently described by Hansen and O'Shea (36), who noted that some genes respond to both sustained and pulsatile Msn2 nuclear localization (fast promoters) while others responded only to sustained Msn2 localization (slow promoters). In fact, two of the three slow-responding genes defined by Hansen and O'Shea, ALD3 and TKL2, were among the slowest responding genes to wild-type Msn2 induction in our study, while all four of the fast-responding genes from Hansen and O'Shea, DDR2, DCS2, RTN2 and HXK1, initiated mRNA accumulation significantly earlier, along with the majority of induced genes (Figure 3A–D). The third slow-responding gene characterized by Hansen and O'Shea was not captured adequately for analysis in our microarray study. We could not identify a correlation between any aspect of nucleosome remodeling with response delay times: both fast- and slow-responding genes exhibited Msn2/4-dependent clearance of nucleosomes from their NDRs, for instance. However, further analysis of Msn2 binding, response kinetics and nucleosome remodeling may prove informative.
Figure 8.
Possible mechanisms for stress-specific binding of Msn2 to different sets of genes. (A) Different stresses could result in distinct modifications (red versus green dots) of Msn2 (blue oval), which could alter the binding specificity or the nuclear occupancy dynamics and which would lead to interaction with different but overlapping sets of stress-responsive genes. (B) Different stresses could activate Msn2 (blue oval) as well as a stress-specific transcription factor (red oval for nutrient stress and green oval for oxidative stress). Those genes with STREs lying in nucleosome-free domains would bind Msn2 under either condition. However, binding of the stress-specific transcription factor could partially unwrap adjacent nucleosomes (beige ovals) to reveal additional, previously inaccessible STREs to which Msn2 could bind.
Another possible scenario to explain stress-specific Msn2 promoter binding is that transcription factors responsive to specific stress signals, e.g. Yap1 for oxidative stress or the Hap1–4 complex for glucose downshift (56–59), bind to promoters in a stress-specific manner and stimulate chromatin clearance at those promoters, allowing access of Msn2 to STRE sites that would otherwise be occluded (Figure 8B). This may represent an example of cooperative binding of transcription factors by sequential unwrapping of DNA from nucleosomes, such that binding of a transcription factor to its cognate site near the periphery of a positioned nucleosome provides access to a binding site for a second transcription factor that would be otherwise buried under the interior of the nucleosome (60,61). Consistent with this model, we note a significant overlap (P < 10−24) between Msn2 binding and promoters at which Floer et al. mapped RSC-associated and partially unwrapped nucleosomes (62). Moreover, we find that more than 70% of the promoters to which Msn2 binds and activates transcription undergo nucleosome remodeling and for 40% of those the remodeling is independent of Msn2. Thus, other transcription factors may well clear the space to allow Msn2 binding and that clearance may well be stress specific.
Msn2 promotes both transcriptional activation and transcriptional repression
We find that Msn2 binding stimulates both transcriptional activation and transcriptional repression. The capacity of Msn2 to promote transcriptional activation is well documented and consistent with the structural features of the protein (26,63). The activity as a repressor is less well documented. Our data demonstrate that repression is not an indirect effect, as might result from transcriptional activation of a repressor protein or inhibition of growth. Rather, Msn2 binds to promoters of repressed genes and in some cases is responsible for recruitment of nucleosomes into the NDR. How Msn2 binding results in activation in some cases and repression in others is certainly not clear but may involve the type of combinatorial interaction with transcriptional modulators as mentioned above. Moreover, in a companion paper (Elfving et al. 2014, submitted), we show that Msn2 recruits mediator complex, most often to promote recruitment and activation of Pol II but occasionally to reposition Mediator to a non-productive position in the promoter. Thus, the same basic activity can function both in activation and repression. Finally, since stress is associated with at least a transient cessation of growth (58,64,65), we were interested in understanding whether Msn2 might repress genes whose expression is necessary for growth. In fact, a significant component of the ESR consists of repression of genes that promote growth, such as ribosomal protein and ribosome biogenesis genes (21). We do find that many of the genes repressed upon activation of Msn2 are highly enriched for those involved in ribosome biogenesis. However, few of these genes are bound by Msn2, at least under conditions of nutrient downshift. Rather, we observed that Msn2 activates transcription of DOT1, which encodes a repressor of ribosome biogenesis genes (see Supplementary Tables S1 and S2). Moreover, we find that Msn2 binds to and activates transcription of XBP1, which encodes a repressor of a number of genes required for cell cycle progression (66). Accordingly, Msn2, while a primary purveyor of the ESR, may indirectly repress the growth-associated genes encompassed in the ESR.
A complex interplay between Msn2 binding and nucleosome occupancy
More than 10,000 canonical Msn2 recognition sites reside in the yeast genome and yet only a small fraction of these serve as binding sites for Msn2 in vivo. Comparing nucleosome occupancy to subsequent Msn2 binding, we see that those STREs that fail to serve as binding sites generally lie in regions of well-ordered nucleosomes. Moreover, those STREs lying under the core of a well-positioned nucleosome show diminished binding of Msn2, relative to sites outside nucleosomes or at the edges of positioned nucleosomes. Thus, positioned nucleosomes serve to restrict Msn2 binding. Moreover, the gradient of Msn2 binding as a function of the distance of an STRE from the edge of a nucleosome is consistent with partial unwrapping of DNA from a nucleosome in vivo. Finally, we note that Msn2 fails to bind to STREs lying in NDRs of limited width. This may suggest that such NDRs may be stably obstructed by factors other than nucleosomes or by nucleosomal subparticles of non-canonical length. In short, well-positioned nucleosomes, as well as other unidentified chromatin features, occlude STRE binding of Msn2 and thereby dictate which of the many elements are available for promoting regulatory changes in response to stress-induced Msn2 entry into the nucleus.
While nucleosomes generally restrict access of Msn2 to its cognate sites on the genome, Msn2 can serve to alter nucleosome positioning. Shivaswamy et al. (11) showed that nucleosomes over some STREs were lost following heat shock and suggested that such loss allowed binding of Msn2 and subsequent transcriptional activation, arguing for a passive role for Msn2 in nucleosome remodeling. However, by examining nucleosome positioning in both a wild-type and an msn2 msn4 strain, we have shown that in many cases nucleosome repositioning is dependent on Msn function and thus that Msn2, Msn4 or both play an active role in nucleosome remodeling. Our results are consistent with those of Huebert et al. (41) who found that Msn2 binding to STREs occluded by nucleosome preceded nucleosome removal following peroxide treatment. We find that Msn2 does not bind to STREs located under the central core of nucleosomes but rather can bind to sites located near the periphery of the nucleosomes. This suggests a model in which partial unwrapping of nucleosomes in vivo allows initial binding of Msn2 to STREs at the edges of nucleosomes. Subsequent recruitment of chromatin modifying and remodeling proteins by Msn2 would lead to reposition or eviction of the occluding nucleosome. In a similar way, recruitment of Msn2 to STREs in NDRs could result in restructuring the local chromatin environment. Thus, while nucleosomes obstruct Msn2 binding, Msn2 binding can evict nucleosomes from some STREs and can alter the nucleosome positioning in its vicinity.
Global measurement versus individual responses
The studies described in this report measure Msn2 binding and nucleosome positioning over the entire population of cells in a culture and thus provide a measure only of the average behavior over all cells. However, microfluidics-based studies demonstrate that stress elicits a variable Msn2 response, with some cells exhibiting substantial and sustained Msn2 nuclear occupancy while others showing no accumulation (34,35). Moreover, Msn2 shows bursting behavior in which the cellular cohort of Msn2 makes random transient forays into the nucleus. Thus, the pattern of binding we observe by ChIP-Seq likely does not reflect the pattern of Msn2 genome occupancy in any individual cell, particularly since the number of Msn2 molecules is apparently less than the number of bound sites identified in our and other studies. This raises the possibility that the cohort of genes induced byMsn2 may differ significantly among different cells, either in a completely random fashion or in distinct subsets of responses. Such diversity in transcriptional output may allow genetically identical cells in a population to respond individually and in quite distinct ways to a common stress, enhancing the number of survival strategies available to the population of cells as a whole.
FUNDING
Swedish Cancer Society, the Swedish Research Council and the Kempe Foundation [S.B.]; National Institutes of Health [R01 HG004708 to A.V.M.]; Alfred P. Sloan Research Fellowship [A.V.M.]; National Institutes of Health [GM076562 to J.R.B.]; Center for Quantitative Biology/National Institutes of Health [P50 GM071508 to J.R.B]. Funding for open access charge: NIH GM076562.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors would like to thank Scott McIsaac for providing strains and sharing unpublished data, Mark Rutledge for assistance with analysis of microarray data and John Matese for management of microarray data archiving.
REFERENCES
- 1.Segal E., Fondufe-Mittendorf Y., Chen L., Thastrom A., Field Y., Moore I.K., Wang J.P., Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tolkunov D., Zawadzki K.A., Singer C., Elfving N., Morozov A.V., Broach J.R. Chromatin remodelers clear nucleosomes from intrinsically unfavorable sites to establish nucleosome-depleted regions at promoters. Mol. Biol. Cell. 2011;22:2106–2118. doi: 10.1091/mbc.E10-10-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zawadzki K.A., Morozov A.V., Broach J.R. Chromatin-dependent transcription factor accessibility rather than nucleosome remodeling predominates during global transcriptional restructuring in Saccharomyces cerevisiae. Mol. Biol. Cell. 2009;20:3503–3513. doi: 10.1091/mbc.E09-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang C., Pugh B.F. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009;10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee W., Tillo D., Bray N., Morse R.H., Davis R.W., Hughes T.R., Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 6.Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 7.Morse R.H. Transcription factor access to promoter elements. J. Cell. Biochem. 2007;102:560–570. doi: 10.1002/jcb.21493. [DOI] [PubMed] [Google Scholar]
- 8.Workman J.L. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 9.Hogan G.J., Lee C.K., Lieb J.D. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet. 2006;2:e158. doi: 10.1371/journal.pgen.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee C.K., Shibata Y., Rao B., Strahl B.D., Lieb J.D. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 11.Shivaswamy S., Bhinge A., Zhao Y., Jones S., Hirst M., Iyer V.R. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L., Ma H., Pugh B.F. Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res. 2011;21:875–884. doi: 10.1101/gr.117465.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geng F., Laurent B.C. Roles of SWI/SNF and HATs throughout the dynamic transcription of a yeast glucose-repressible gene. EMBO J. 2004;23:127–137. doi: 10.1038/sj.emboj.7600035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martens J.A., Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 2003;13:136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 15.Mellor J., Morillon A. ISWI complexes in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2004;1677:100–112. doi: 10.1016/j.bbaexp.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 16.Saha A., Wittmeyer J., Cairns B.R. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 17.Sudarsanam P., Winston F. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 2000;16:345–351. doi: 10.1016/s0168-9525(00)02060-6. [DOI] [PubMed] [Google Scholar]
- 18.Whitehouse I., Tsukiyama T. Antagonistic forces that position nucleosomes in vivo. Nat. Struct. Mol. Biol. 2006;13:633–640. doi: 10.1038/nsmb1111. [DOI] [PubMed] [Google Scholar]
- 19.Lorch Y., Griesenbeck J., Boeger H., Maier-Davis B., Kornberg R.D. Selective removal of promoter nucleosomes by the RSC chromatin-remodeling complex. Nat. Struct. Mol. Biol. 2011;18:881–885. doi: 10.1038/nsmb.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Causton H.C., Ren B., Koh S.S., Harbison C.T., Kanin E., Jennings E.G., Lee T.I., True H.L., Lander E.S., Young R.A. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berry D.B., Gasch A.P. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol. Biol. Cell. 2008;19:4580–4587. doi: 10.1091/mbc.E07-07-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berry D.B., Guan Q., Hose J., Haroon S., Gebbia M., Heisler L.E., Nislow C., Giaever G., Gasch A.P. Multiple means to the same end: the genetic basis of acquired stress resistance in yeast. PLoS Genet. 2011;7:e1002353. doi: 10.1371/journal.pgen.1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 25.Klosinska M.M., Crutchfield C.A., Bradley P.H., Rabinowitz J.D., Broach J.R. Yeast cells can access distinct quiescent states. Genes Dev. 2011;25:336–349. doi: 10.1101/gad.2011311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Pastor M.T., Marchler G., Schuller C., Marchler-Bauer A., Ruis H., Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 27.De Wever V., Reiter W., Ballarini A., Ammerer G., Brocard C. A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation. EMBO J. 2005;24:4115–4123. doi: 10.1038/sj.emboj.7600871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorner W., Durchschlag E., Martinez-Pastor M.T., Estruch F., Ammerer G., Hamilton B., Ruis H., Schuller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorner W., Durchschlag E., Wolf J., Brown E.L., Ammerer G., Ruis H., Schuller C. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 2002;21:135–144. doi: 10.1093/emboj/21.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santhanam A., Hartley A., Duvel K., Broach J.R., Garrett S. PP2A phosphatase activity is required for stress and Tor kinase regulation of yeast stress response factor Msn2p. Eukaryot. Cell. 2004;3:1261–1271. doi: 10.1128/EC.3.5.1261-1271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garmendia-Torres C., Goldbeter A., Jacquet M. Nucleocytoplasmic oscillations of the yeast transcription factor Msn2: evidence for periodic PKA activation. Curr. Biol. 2007;17:1044–1049. doi: 10.1016/j.cub.2007.05.032. [DOI] [PubMed] [Google Scholar]
- 32.Gonze D., Jacquet M., Goldbeter A. Stochastic modelling of nucleocytoplasmic oscillations of the transcription factor Msn2 in yeast. J. R. Soc. Interface. 2008;5(Suppl. 1):S95–S109. doi: 10.1098/rsif.2008.0141.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao N., Budnik B.A., Gunawardena J., O'Shea E.K. Tunable signal processing through modular control of transcription factor translocation. Science. 2013;339:460–464. doi: 10.1126/science.1227299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hao N., O'Shea E.K. Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat. Struct. Mol. Biol. 2012;19:31–39. doi: 10.1038/nsmb.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrenko N., Chereji R.V., McClean M.N., Morozov A.V., Broach J.R. Noise and interlocking signaling pathways promote distinct transcription factor dynamics in response to different stresses. Mol. Biol. Cell. 2013;24:2045–2057. doi: 10.1091/mbc.E12-12-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen A.S., O'Shea E.K. Promoter decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression. Mol. Syst. Biol. 2013;9:704. doi: 10.1038/msb.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchler G., Schuller C., Adam G., Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., Ye S., Erkine A.M. Analysis of Saccharomyces cerevisiae genome for the distributions of stress-response elements potentially affecting gene expression by transcriptional interference. In Silico Biol. 2009;9:379–389. doi: 10.3233/ISB-2009-0412. [DOI] [PubMed] [Google Scholar]
- 39.Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 40.Venters B.J., Wachi S., Mavrich T.N., Andersen B.E., Jena P., Sinnamon A.J., Jain P., Rolleri N.S., Jiang C., Hemeryck-Walsh C., et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huebert D.J., Kuan P.F., Keles S., Gasch A.P. Dynamic changes in nucleosome occupancy are not predictive of gene expression dynamics but are linked to transcription and chromatin regulators. Mol. Cell. Biol. 2012;32:1645–1653. doi: 10.1128/MCB.06170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amberg D.C., Burke D.J., Strathern J.N. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, 2005 Edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- 43.Sabourin M., Tuzon C.T., Fisher T.S., Zakian V.A. A flexible protein linker improves the function of epitope-tagged proteins in Saccharomyces cerevisiae. Yeast. 2007;24:39–45. doi: 10.1002/yea.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winston F., Dollard C., Ricupero-Hovasse S.L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 45.McIsaac R.S., Oakes B.L., Wang X., Dummit K.A., Botstein D., Noyes M.B. Synthetic gene expression perturbation systems with rapid, tunable, single-gene specificity in yeast. Nucleic Acids Res. 2013;41:e57. doi: 10.1093/nar/gks1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaman S., Lippman S.I., Schneper L., Slonim N., Broach J.R. Glucose regulates transcription in yeast through a network of signaling pathways. Mol. Syst. Biol. 2009;5:245. doi: 10.1038/msb.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fisher T.S., Taggart A.K., Zakian V.A. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 2004;11:1198–1205. doi: 10.1038/nsmb854. [DOI] [PubMed] [Google Scholar]
- 48.Teytelman L., Thurtle D.M., Rine J., van Oudenaarden A. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. 2013;110:18602–18607. doi: 10.1073/pnas.1316064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Badis G., Chan E.T., van Bakel H., Pena-Castillo L., Tillo D., Tsui K., Carlson C.D., Gossett A.J., Hasinoff M.J., Warren C.L., et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fordyce P.M., Gerber D., Tran D., Zheng J., Li H., DeRisi J.L., Quake S.R. De novo identification and biophysical characterization of transcription-factor binding sites with microfluidic affinity analysis. Nat. Biotechnol. 2010;28:970–975. doi: 10.1038/nbt.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu C., Byers K.J., McCord R.P., Shi Z., Berger M.F., Newburger D.E., Saulrieta K., Smith Z., Shah M.V., Radhakrishnan M., et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chenais B., Caruso A., Hiard S., Casse N. The impact of transposable elements on eukaryotic genomes: from genome size increase to genetic adaptation to stressful environments. Gene. 2012;509:7–15. doi: 10.1016/j.gene.2012.07.042. [DOI] [PubMed] [Google Scholar]
- 53.Servant G., Pinson B., Tchalikian-Cosson A., Coulpier F., Lemoine S., Pennetier C., Bridier-Nahmias A., Todeschini A.L., Fayol H., Daignan-Fornier B., et al. Tye7 regulates yeast Ty1 retrotransposon sense and antisense transcription in response to adenylic nucleotides stress. Nucleic Acids Res. 2012;40:5271–5282. doi: 10.1093/nar/gks166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Azvolinsky A., Giresi P.G., Lieb J.D., Zakian V.A. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol. Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shor E., Warren C.L., Tietjen J., Hou Z., Muller U., Alborelli I., Gohard F.H., Yemm A.I., Borisov L., Broach J.R., et al. The origin recognition complex interacts with a subset of metabolic genes tightly linked to origins of replication. PLoS Genet. 2009;5:e1000755. doi: 10.1371/journal.pgen.1000755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Temple M.D., Perrone G.G., Dawes I.W. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 2005;15:319–326. doi: 10.1016/j.tcb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Toone W.M., Jones N. AP-1 transcription factors in yeast. Curr. Opin. Genet. Dev. 1999;9:55–61. doi: 10.1016/s0959-437x(99)80008-2. [DOI] [PubMed] [Google Scholar]
- 58.Broach J.R. Nutritional control of growth and development in yeast. Genetics. 2012;192:73–105. doi: 10.1534/genetics.111.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zaman S., Lippman S.I., Zhao X., Broach J.R. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- 60.Moyle-Heyrman G., Tims H.S., Widom J. Structural constraints in collaborative competition of transcription factors against the nucleosome. J. Mol. Biol. 2011;412:634–646. doi: 10.1016/j.jmb.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tims H.S., Gurunathan K., Levitus M., Widom J. Dynamics of nucleosome invasion by DNA binding proteins. J. Mol. Biol. 2011;411:430–448. doi: 10.1016/j.jmb.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Floer M., Wang X., Prabhu V., Berrozpe G., Narayan S., Spagna D., Alvarez D., Kendall J., Krasnitz A., Stepansky A., et al. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141:407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmitt A.P., McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5777–5782. doi: 10.1073/pnas.93.12.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Airoldi E.M., Huttenhower C., Gresham D., Lu C., Caudy A.A., Dunham M.J., Broach J.R., Botstein D., Troyanskaya O.G. Predicting cellular growth from gene expression signatures. PLoS Comput. Biol. 2009;5:e1000257. doi: 10.1371/journal.pcbi.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brauer M.J., Huttenhower C., Airoldi E.M., Rosenstein R., Matese J.C., Gresham D., Boer V.M., Troyanskaya O.G., Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol. Biol. Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miles S., Li L., Davison J., Breeden L.L. Xbp1 directs global repression of budding yeast transcription during the transition to quiescence and is important for the longevity and reversibility of the quiescent state. PLoS Genet. 2013;9:e1003854. doi: 10.1371/journal.pgen.1003854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.