Abstract
During the process of prokaryotic CRISPR adaptation, a copy of a segment of foreign deoxyribonucleic acid referred to as protospacer is added to the CRISPR cassette and becomes a spacer. When a protospacer contains a neighboring target interference motif, the specific small CRISPR ribonucleic acid (crRNA) transcribed from expanded CRISPR cassette can protect a prokaryotic cell from virus infection or plasmid transformation and conjugation. We show that in Escherichia coli, a vast majority of plasmid protospacers generate spacers integrated in CRISPR cassette in two opposing orientations, leading to frequent appearance of complementary spacer pairs in a population of cells that underwent CRISPR adaptation. When a protospacer contains a spacer acquisition motif AAG, spacer orientation that generates functional protective crRNA is strongly preferred. All other protospacers give rise to spacers oriented in both ways at comparable frequencies. This phenomenon increases the repertoire of available spacers and should make it more likely that a protective crRNA is formed as a result of CRISPR adaptation.
INTRODUCTION
CRISPR-Cas are diverse small RNA-based adaptive immunity systems of prokaryotes. A typical CRISPR-Cas system consists of a CRISPR (Clusters of Regularly Interspaced Short Palindromic Repeats) cassette, a deoxyribonucleic acid (DNA) locus consisting of number of identical repeated sequences separated by variable spacers (1–3) and several cas genes (4,5). The non-coding CRISPR cassette transcript (pre-crRNA) is processed by introduction of endonucleolytic cuts inside the repeat sequences (6,7). As a result, a set of crRNAs is produced each containing identical flanking sequences originating from repeats and variable internal sequences corresponding to CRISPR spacers (6,8). Individual crRNAs associate with Cas proteins and guide them to double-stranded DNA (and in some cases, RNA (9,10)) targets matching the spacer sequence (6,8,11). Such sequences are referred to as protospacers (12). When a DNA protospacer fully matches a crRNA spacer and contains an additional short motif (13,14) referred to as target interference motif (TIM) (15), a R-loop at the site of recognition is formed and the target DNA is destroyed (16,17). Thus, when CRISPR spacers match functional TIM-associated sequences in mobile genetic elements such as bacteriophages or plasmids, the cell is able to purge such elements, protecting itself from bacteriophage infection or plasmid conjugation/transformation (3,16,18–22). This protective function is referred to as CRISPR interference (19).
A set of spacers present in a CRISPR cassette determines the ability of a CRISPR-Cas system to recognize and protect the host prokaryote from mobile genetic elements (12,14,18). Spacers are acquired in a pseudo-Lamarckian process from bacteriophage or plasmid DNA. This poorly understood process is referred to as CRISPR adaptation (3,16,18–21). The two most conserved Cas proteins, Cas1 and Cas2, are dispensable for CRISPR interference (6) but essential for CRISPR adaptation (23). For the adaptation process to lead to subsequent interference, the adaptation machinery must insert into the CRISPR cassette of the host foreign DNA fragments associated with TIM sequences that can promote subsequent interference. In addition, differentiation between self and non-self DNA must be somehow accomplished, since acquisition of spacer from self-DNA would lead to an autoimmune response (24–26).
Escherichia coli Cas1 and Cas2 alone, when overexpressed in the absence of other Cas proteins or crRNA, are capable of causing new spacer acquisition and analysis of protospacers shows that many are preceded with an AWG motif (23) referred to as spacer acquisition motif (SAM) (15). Four functional TIMs, ATG, AAG, AGG and GAG are recognized by the E. coli Cse1 protein, a part of a large interfering Cascade-crRNA complex, during CRISPR interference (17). Thus, the AAG and ATG SAMs are also functional TIMs (17), which ensures that many adaptation events lead to interference-capable spacers. In addition to SAM, an ‘acquisition affecting motif’ AAM has been reported within some protospacers which are highly preferred substrates of acquisition (27).
In addition to the Cas1/Cas2 dependent, Cascade-crRNA-independent adaptation process (referred to as ‘unprimed’), a much more efficient ‘primed adaptation’ has been described (28–30). This process requires not just Cas1 and Cas2 but also Cascade and is activated by the presence of target DNA that contains point substitutions in the TIM or the protospacer that render crRNA recognition and CRISPR interference inefficient (28). Under these conditions, the residual ‘priming’ interaction of Cascade-crRNA complex with mutated target strongly stimulates acquisition of spacers from protospacers located in cis with respect to the priming protospacer. Primed spacer acquisition is characterized by a very strong preference for protospacers with an AAG SAM (28,31) and is obviously highly adaptive, as it allows to specifically target foreign DNA that had ‘escaped’ the previous line of CRISPR-Cas defense by acquiring point mutations in the protospacer or TIM.
The enzymology of CRISPR adaptation is currently unknown. Yet, it is clear that Cas1, Cas2 (23,28) or both of these proteins (32), possibly along with some yet-to-be identified host factors (18), must recognize a donor DNA fragment and then initiate a sequence of events that leads either to copying or excision and physical transfer of this fragment into recipient CRISPR cassette. As a result, the cassette is expanded by the addition of a new spacer and an extra repeat copy, which must arise through replication of a preexisting repeat (23,28,29,33). Here, we analyze multiple spacer acquisition events during primed and unprimed CRISPR adaptation in E. coli. We observe that for a given protospacer, acquisition events lead to both possible orientations of resulting spacers. The frequency of both orientations is similar when a protospacer from which complementary spacers are produced does not contain an AAG SAM, thus increasing a chance that a crRNA recognizing a protospacer with a functional TIM is produced. In contrast, for protospacers with an AAG SAM (which is also a functional TIM), a single spacer orientation—the one that leads to functional, protective crRNA—is strongly favored.
MATERIALS AND METHODS
E. coli strains, plasmids and the primed adaptation experiment
The E. coli KD263 strain is a derivative of BW40119 strain described earlier (28). It contains the cas3 gene under the control of the lacUV5 promoter and the casABCDE12 operon under the araBp8 promoter control. The KD263 strains harbors a single genetically modified CRISPR cassette with two repeats and a single g8 spacer described earlier (14). KD263 was transformed with a pG8_C1T plasmid, a derivative of the pT7blue cloning vector harboring a 209-bp fragment of the M13 bacteriophage DNA containing the g8 protospacer (28). The protospacer sequence harbors a C to T change at the position of +1 that renders CRISPR interference by the g8 spacer containing crRNA ineffective (14). KD263 cells transformed with pG8_C1T were grown overnight at 37°C in Luria-Bertani (LB) broth supplemented with 100 μg/ml ampicillin. Aliquots of the culture were diluted 200-fold into six individual tubes with fresh LB broth without ampicillin and supplemented with IPTG (isopropyl β-D-1 thiogalactopyranoside) and arabinose to the final concentration 1 mM each. The cultures were grown at 37°C overnight. The six individual cultures were mixed and genomic DNA was isolated from the pooled cultures. The cells were lyzed by 2-min incubation with 1 mg/ml lysozyme and DNA was purified by phenol, phenol/chloroform, chloroform extractions followed by ethanol precipitation. CRISPR expansion was monitored by polymerase chain reaction (PCR) in 20 μl reactions containing 20–50 ng genomic DNA with primers matched the CRISPR leader sequence and g8 spacer: Ec_LDR-F (5′-AAGGTTGGTGGGTTGTTTTTATGG-3′) and M13g8 (5′-GGATCGTCACCCTCAGCAGCG-3′) using Phusion High-Fidelity DNA Polymerase (New England Biolabs). Six independent amplification reactions were pooled, PCR products corresponding to expanded CRISPR cassette were gel purified using QIAquick Gel Extraction Kit (QIAGEN) and sequenced with MySeq Illumina System at Moscow State University Genomics facility as described (31).
Data processing
Raw sequencing data were analyzed using ShortRead and BioStrings (34) packages. Illumina-sequencing reads were filtered for quality scores of ≥20 and reads containing two repeats (with up to two mismatches) were selected. Reads that contained 33-bp sequences between two CRISPR repeats were next selected. The 33-bp segments were considered spacers. Spacers were next mapped on the pG8_C1T plasmid with no mismatches allowed. R scripts and their package ggplot2 (35) were used for spacers statistics and graphical representation. Logo construction was done with http://weblogo.berkeley.edu (36).
RESULTS
Experimental primed adaptation set up
The E. coli strain KD263 with cas genes fused to inducible promoters and a CRISPR cassette containing two repeats and an M13-phage-derived g8 spacer was transformed with a pT7blue-based plasmid containing a fragment of the M13 phage with g8 protospacer. The protospacer contained a point mutation that introduced a single mismatch between the target DNA and g8 crRNA at the +1 position (see below). Elsewhere, we show that such a mismatch renders CRISPR interference inactive, however, strongly stimulates primed adaptation from DNA located in cis to the protospacer (28). Expression of cas genes was next induced and cells growth was continued in the absence of antibiotics, thus allowing cells that lost the plasmid to survive. PCR analysis with appropriate primers revealed that more than 50% of cells had expanded their CRISPR cassettes after overnight growth without antibiotics. The growth kinetics of plasmid-bearing KD263 was not affected by induction of cas genes expression (data not shown). At the conditions of our experiment there is no strong selective pressure for the cells to acquire an interference-capable spacer (and no penalty for acquiring a non-functional spacer). While cells that acquired a functional spacer may be able to lose the plasmid faster and thus get a small selective advantage during growth in the absence of antibiotic, cells that acquired a non-functional spacer also survive and propagate. Thus, the distribution of spacers (and protospacer choice) in our experiment should largely reflect the preferences of the acquisition machinery only.
A DNA band corresponding to CRISPR cassette expanded by a single repeat-spacer unit was purified and subjected to deep-coverage high-throughput sequencing and analysis. A flow chart describing the general outcome of this analysis is presented in Figure 1. A total of 1 934 605 sequences of newly acquired spacers (defined here as 33-bp DNA fragments separating CRISPR repeat sequences in Illumina reads) were obtained. 80.4% (1 555 829) of spacers were mapped with no mismatches to sequences (protospacers) from a plasmid used to transform the cells. Ninety seven percent of protospacers had an AAG SAM (Figure 1). 93.3% of spacers were mapped to protospacers located in the plasmid DNA strand that is non-targeted by the g8 crRNA. The strong preference for an AAG SAM and a bias toward the non-targeted strand are both the hallmarks of primed adaptation (28,31).
Figure 1.
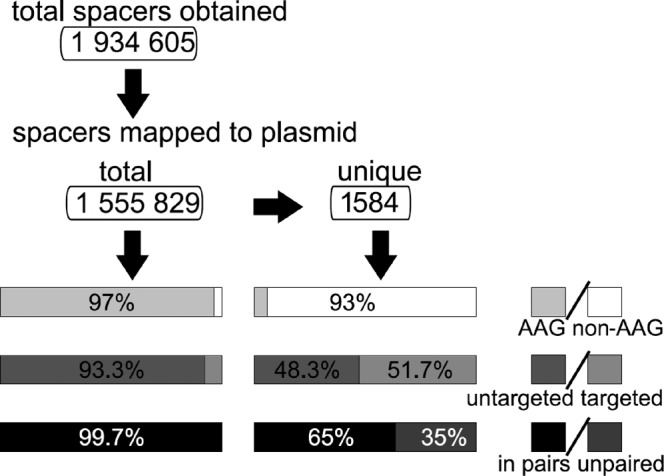
A workflow and statistics of analysis of high-throughput sequencing data on acquisition of plasmid-derived spacers into Escherichia coli CRISPR cassette. See text for details. Results for all spacers (left) and unique (right) spacers are summarized.
The 1 555 829 spacers mapped to just 1584 unique protospacers (Figure 1). Among the unique protospacers just 7% contained an AAG SAM. Further, the unique protospacers were equally distributed between both strands of plasmid DNA (51.7% unique protospacers located in the targeted strand, Figure 1).
Identification of conserved sequence motifs in protospacers used during primed adaptation
Analysis depicted in Figure 1 demonstrates that trends revealed when unique protospacers are analyzed and different from those revealed when the frequency of resulting spacers is considered. This occurs because some protospacers contribute a disproportionately large amount of spacers. At our coverage, spacers from protospacers with an AAG SAM were selected, on an average, 13 483 times (the actual spread of this value was very wide, ranging from 20 to 217 955 times, median = 2352). The different frequencies of protospacer choice are unlikely to be an artifact of PCR amplification and library construction since a good correlation in relative spacer frequencies is observed in independent experiments ((31) and data not shown). Therefore, the frequency of spacer occurrence during sequencing is a good measure of protospacer choice efficiency. Spacers from protospacers without an AAG SAM were selected on average less often (from 1 to 5639 times, median = 3). Despite the fact that spacers acquired from protospacers with AAG SAM were much more frequent, such protospacers constituted only 7% of unique protospacers in our data set.
We constructed a LOGO for all 1584 unique protospacers and adjacent five upstream and downstream nucleotides. The ‘upstream’ and ‘downstream’ directions were set by the orientation of the corresponding spacer in the CRISPR cassette. In E. coli, 33-bp DNA fragments are inserted in CRISPR cassette during spacer acquisition (33). When considering a target DNA sequence that serves as a donor of a spacer, we number SAM/TIM residues from −2 to 0 (i.e. A−2, A−1 and G0 for an AAG SAM). G0 is also the first residue of the protospacer (28,29,33). Subsequent residues of the protospacer are numbered consecutively up to position +32. The numbering then continues (+33, +34, etc.) downstream of the protospacer. The results of our LOGO analysis are presented in Figure 2A. Though during LOGO construction the frequency of protospacer use is not taken into account, the upstream AAG motif was observed, due to the 7% of unique protospacers with AAG SAM (above). A guanine at position 0 was most strongly conserved (recall that in LOGO representation, the overall height of the stack indicates the sequence conservation at position being investigated, while the height of symbols within the stack indicates the relative frequency of each amino or nucleic acid at this position (36)). No preferences for bases within the individual positions throughout the protospacer positions +1 to +31 were observed. Surprisingly, a weak, but clearly detectable preference for a C at position +32 was detected.
Figure 2.
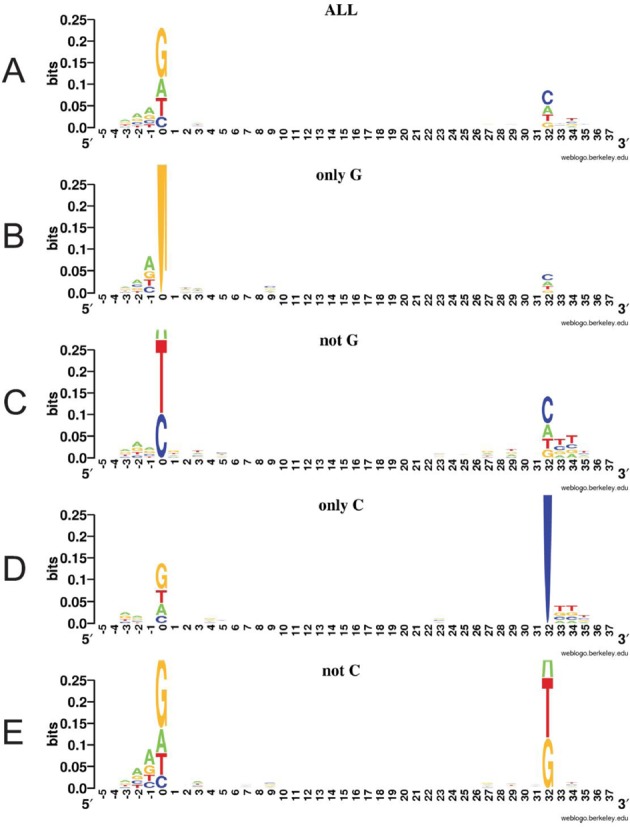
Local context of protospacers used during primed adaptation. Protospacer region LOGOs built for all protospacer in the data set (A), for protospacers with a G at position 0 (B), for protospacers lacking a G at this position (C), for protospacers with a C at position +32 (D) and for protospacers lacking a C at this position (E).
In principle, a preference for a C at position +32 may signal a requirement for a sequence that functions together with the upstream SAM, or, alternatively, may be indicative of a SAM-independent protospacer choice signal. To select between these possibilities we constructed a LOGO only for those protospacers that contained a G at position 0. As can be seen from Figure 2B, the preference for a C at position +32 was decreased in this group of protospacers. A LOGO for a reciprocal set of protospacers, i.e. the ones that do not contain a G at 0, showed an increased preference for a C at position +32 (Figure 2C). Moreover, a weak preference for T at positions +33 and +34 became evident. Complementary analysis considered protospacers with or without a C at position +32. The former group of spacers had a very weak preference for a G at position 0 and no preference for positions −1 and −2 (Figure 2D). In the later group of protospacers, a preference for A−2A−1G0 became stronger than in the total protospacer set (compare Figure 2A and E). The results thus show that (i) cytosine is preferentially found as the last residue of protospacers (position +32) and (ii) the presence of a G originating from a consensus SAM sequence at position 0 decreases the likelihood of finding a C at position +32. The opposite is also true.
Most spacers acquired during primed adaptation are inserted in both orientations
The numerical predominance of non-AAG protospacers over AAG protospacers in our unique protospacer set may be partially explained by interdependencies in spacer choice. For example, up to four conjugated protospacers linked to non-AAG motifs can arise when spacers are selected ‘incorrectly’, one or two nucleotides downstream or upstream from the predominant ‘correct’ site determined by the location of consensus AAG SAM (31). To reveal interdependencies of protospacer choice, overlapping or neighboring protospacers that started in the region extending from position −20 to position +49 of every unique protospacer present in our data set were identified on both strands. Every time a 0 position of an overlapping or neighboring protospacer mapped to a position within the −20/+49 interval of protospacer being considered, a +1 score was added to this position. In Figure 3A, a histogram showing total scores for each position surveyed for all unique protospacers from our collection is shown. For obvious reasons our procedure results in a score of 1584, which equals a total number of unique protospacers, for position 0. For most other positions, the score is uniform and averages about 400. The only clear exception is position +32, where a clear excess of overlapping opposite-strand protospacers starting at this position is observed. A closer inspection also reveals that for both 0 and +32 positions, the immediately adjacent positions (−1, −2 and +1, +2 and +30, +31 and +33, +34, correspondingly) at the same strand tend to be used more often as overlapping protospacer start points, a likely indication of imprecise protospacer selection or ‘slippage’ following the initial recognition of AAG SAM sequence mentioned above.
Figure 3.
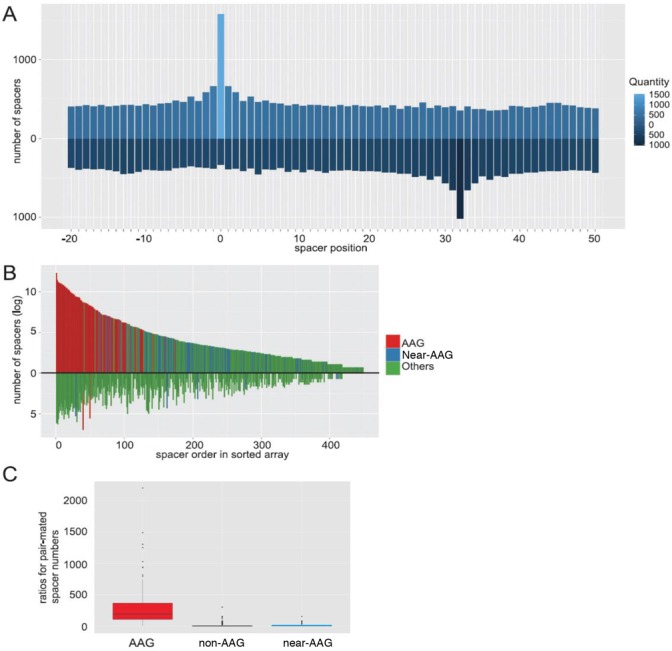
Paired spacer selection during primed CRISPR adaptation. (A) The interdependence of protospacer choice. A summary representation of locations of positions +1 of overlapping or neighboring protospacers relative to the +1 position of every protospacer in the data set. (B) Self-complementary spacer pairs were ranked according to the quantity of the more frequent spacer in the pair (plotted above the horizontal axis). The corresponding less common spacers for each pair are plotted below the axis. Spacers from protospacers with an AAG SAM are highlighted in red. Spacers from non-AAG protospacers are shown either in green or in blue. Blue color indicates the ‘near-AAG’ group of spacers from protospacers that had an AAG-conjugated sequence at the place of SAM (i.e. an AAG sequence was located 1–2 nucleotides upstream or downstream of the protospacer boundary on the SAM side). Green color indicates spacers from all other non-AAG protospacers. (С) A box plot of observed ratios for pair-mated spacers originating from AAG, all non-AAG (including ‘near-AAG’), and ‘near-AAG’ only protospacers.
The clear overrepresentation of overlapping protospacers beginning at position +32 suggests that self-complementary spacers exist in our collection. This notion is also supported by the results of the LOGO analysis, since a G at position 0 and a C and position +32 tend to be present in different subpopulations of protospacers (the same is also true for protospacers with AAG SAM and the complementary downstream CTT motif, Figure 2A and C). In other words, the CTT motif characteristic of some protospacers (Figure 2C) may arise during the acquisition of a spacer derived from a protospacer with an AAG SAM in an inverted orientation. Indeed, a direct search revealed that 65% of unique protospacers in our data set formed self-complementary pairs. The remaining 35% of unique protospacers for which no pairs could be found constituted just 0.3% of the total number of spacers, i.e. were very rare and presumably the corresponding protospacers were poor spacer donors. The median of quantities of spacers with and without a complementary mate was 6 and 1, correspondingly. In principle, both protospacers within a pair in our data set could have been selected independently. If true, this would make the observed pairs essentially random. However, this notion does not match the fact that only 1584 out of 6190 possible unique plasmid-derived spacers (note that the donor plasmid is 3095-bp long) are observed in our data set. Therefore, the appearance of paired spacers originating from the same protospacer reflects an inherent property of the acquisition mechanism.
To further investigate spacer pairs present in our data set, the more common spacer was identified for each pair. The more common spacers were next sorted according to the number of times they occurred in our data set. A plot showing the distribution of observed numbers of spacers in the sorted array is shown in Figure 3B (in the figure, the more common spacers from each pair are presented above the horizontal axis). The corresponding less common pair-mated spacers were not arrayed. Instead, their numbers were plotted below the horizontal axis under the corresponding more common pair mate. As expected (31), spacers corresponding to protospacers with an AAG SAM (indicated in red) were the predominant ones—both overall and within a pair. These spacers should generate functional crRNA capable of interfering with target DNA through recognition of an AAG TIM. The more rare complementary spacers could only result in functional crRNA when a downstream interference-capable TIM was present, presumably an event determined by chance. For pairs of spacers derived from AAG protospacers, the median of ratios between 'correct', functional, and ‘incorrect' and, therefore likely non-functional complementary spacers equaled 194 (Figure 3C). For non-AAG protospacers (indicated in blue and green in Figure 3B), the median value of ratios between more and less common spacers within a pair equaled 4. These values are different with a P-value of less than 2.2e−16 according to Wilcoxon test. Spacers that were selected by imprecise choice/slippage following AAG SAM recognition (indicated in blue in Figure 3B) were analyzed as a separate group. The median of ratios between more and less common spacers within pair from this ‘near-AAG’ group equaled 9 (Figure 3C), which is significantly different from the value obtained for AAG-protospacers (P-value 2e−16).
The Qimron lab has recently reported the results of analysis of high-throughput sequencing data of acquired E. coli spacers that suggested the presence of a downstream spacer ‘AAM’ motif within frequently used protospacers (27). This motif could not be revealed by LOGO analysis but was identified as a difference of nucleotide frequency at each protospacer position among frequently and rarely acquired spacers groups. It was therefore proposed that an AAG SAM and the downstream motif jointly affect the efficiency of spacer acquisition. No downstream motif could be identified in highly used protospacers present in our data set. Thus, the preferential usage of protospacers in our experimental system does not appear to require any additional signals other than an AAG PAM. It should be noted, however, that the downstream spacer motif was observed only among host-derived spacers but not from plasmid-derived spacers. Cells that acquired host-derived spacers should be purged from the population at our conditions due to auto-immune response (26) and the corresponding spacers should thus evade detection.
Spacer choice during unprimed adaptation
Overall, our analysis suggests that cells containing complementary spacers that must have originated from the same plasmid protospacer can oftentimes be found in a population that underwent primed adaptation. The presence of an AAG SAM strongly increases the likelihood that a spacer is inserted in an orientation that generates interference-capable crRNA. This effect appears to be unrelated to the actual frequency of protospacer use, since increased usage of ‘near-AAG’ protospacers does not lead to preferential orientation of spacers derived from them. The above trends have been revealed when analyzing spacers acquired during primed adaptation, which, in addition to Cas1/Cas2 proteins that appear to constitute the adaptation machinery, requires a partial match between crRNA and a priming protospacer, intact Cascade complex, and the Cas3 endonuclease/helicase (28). Recently, a large data set of plasmid-derived spacers acquired in the course of unprimed E. coli adaptation caused by Cas1/Cas2 overproduction was obtained (27). To determine if complementary spacers are generated during unprimed adaptation, we analyzed the available high-throughput sequencing data of Yosef et al. (27). This data set includes 5336 unique spacers from a total of 9422 possible plasmid-derived spacers. Only 122 protospacers (2.3% of the total) contained an AAG SAM. Together, spacers originating from AAG-protospacers constituted 36.7% of all spacers sequenced, reflecting the previously noticed decreased preference for AAG during unprimed adaptation (recall that spacers originating from AAG-protospacers constituted 97% of all spacers sequenced during the primed adaptation experiment, above). We found that 94% of spacers from unprimed adaptation data set have а complementary counterpart. Together, these paired spacers account for 86% of donor protospacers. As in the case of primed adaptation, pairs arising from AAG-protospacers were the most frequent and within such pairs spacers with functional orientation were more common (Figure 3A). The median value of ratios between the more and less common complementary spacers in AAG-protospacer-derived pairs was 128 (Figure 4B). For spacer pairs derived from protospacers without AAG, this value was 2.8 (Figure 4B). Both values are similar to those obtained for corresponding groups of spacers from the primed adaptation experiment (Figure 3C). We therefore conclude that generation of complementary spacers from the same protospacer is a common feature of both primed and non-primed adaptation.
Figure 4.
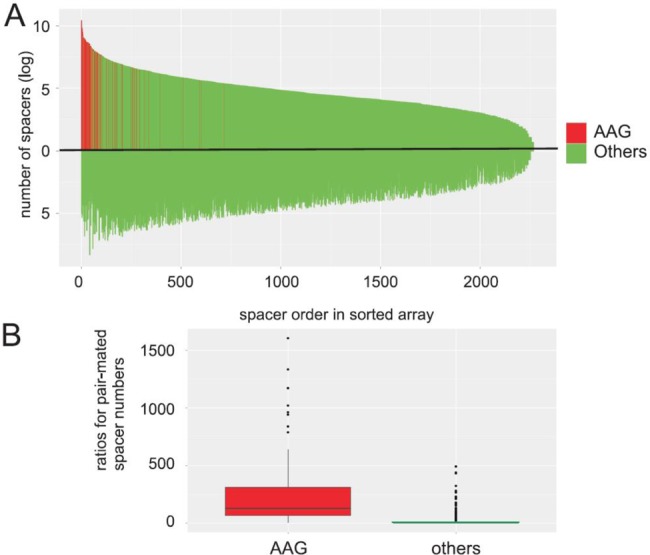
The interdependence of protospacers choice during unprimed CRISPR adaptation. (A) Self-complementary spacer pairs were ranked according to the quantity of the more frequent spacer in the pair (plotted above the horizontal axis). The corresponding less common spacers for each pair are plotted below the axis. Spacers from protospacers with an AAG SAM are highlighted in red. Spacers from protospacers with a non-AAG SAM are shown in green. (B) A box plot of observed ratios for pair-mated spacers originating from AAG and non-AAG protospacers.
DISCUSSION
Our results reveal that when an E. coli culture undergoes CRISPR adaptation, the population of cells that results invariably contains clones with complementary CRISPR spacers that are generated from the same protospacer. Simple statistical analysis shows that pair-mated spacers that account for most newly acquired spacers are not generated through independent recognition of the same protospacer sequence on both strands of plasmid DNA but are rather a result of two possible outcomes of insertion of material from once recognized protospacer into a CRISPR cassette. This protospacer ‘flippage’ is a characteristic feature of both primed and unprimed adaptation. In the absence of consensus AAG SAM, the ratio between the two complementary spacers that originate from the same protospacer is about 1:4 or less (Figure 3C and Figure 4B). This bias is unrelated to strand polarity during primed spacer acquisition (data not shown) and is likely determined by sequence preferences of Cas1, Cas2 and their putative partners encoded by the bacterial genome to either CRISPR repeat or protospacer intermediate en route to insertion in the cassette (or both). When a protospacer is preceded by an AAG, which is both a preferred SAM during selection of protospacers for spacer acquisition and a functional TIM for interference, the preference for a pair-mated spacer that results in crRNAs capable of target interference is increased 30-fold or more. This bias is observed during both primed and unprimed adaptation. It therefore follows that preferential ‘correct’ orientation of AAG-protospacer-derived spacers is determined by Cas1/Cas2 and their putative non-Cas partners, but not by other Cas proteins. This, however, creates an interesting conundrum, since only the protospacer sequence and the last G of SAM become inserted into CRISPR cassette. In other words, if a hypothetical intermediate of the adaptation reaction corresponds to material that is actually inserted in CRISPR cassette, there does not seem to be a way to distinguish between intermediates derived from AAG- and XXG-protospacers. Yet the two types of protospacers are clearly different with respect to their ability to be preferentially inserted in only one orientation. This apparent paradox may be explained by assuming that spacer acquisition machinery assumes a particular conformation when adopting spacers from AAG-protospacers. For example, the adaptation machinery may be able to effectively recognize a protospacer with an AAG SAM only when approaching it from one direction and this directionality is somehow maintained during the generation of new spacer. The situation may be formally similar to that described for some site-specific recombination systems where directionality is maintained over long distances and even for DNA sequences located in trans (37). One difficulty with this scenario, however, is that for numerous spacers that arise upon ‘slippage’ of the adaptation machinery after the recognition of the AAG SAM, the ability to maintain the preferred orientation of spacers is lost (Figure 3C). An alternative and more radical mechanism that can account for the maintenance of spacer orientation bias for AAG protospacers is shown in Figure 5. According to this model, an intermediate of spacer insertion reaction contains the entire AAG SAM, with the two adenine residues contributing to spacer orientation but being removed at some later stages of the process (Figure 5).
Figure 5.
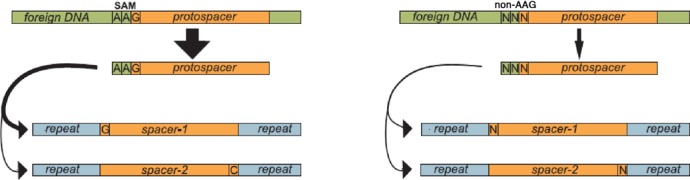
A possible model of CRISPR-Cas adaptation. The models schematically present the likely sequence of events during adaptation of spacers from AAG (left) and non-AAG protospacers (right). Both kinds of protospacers are schematically shown at the top of the figure as part of foreign DNA (olive green). A fragment that gets inserted in the cassette is shown in orange. AAG protospacers are much more efficient spacer donors compared to non-AAG protospacers (indicated by the width of black downward arrows). During the adaptation, an intermediate of the spacer acquisition reaction, here depicted as a double-stranded DNA fragment, is formed. In principle, this intermediate may also be single-stranded and/or copied from target DNA. During adaptation from AAG protospacers, an orientation of intermediate insertion in the cassette that results in interference-capable crRNA is strongly favored. In the course of adaptation from non-AAG protospacers both orientations are likely.
The reciprocal orientations of CRISPR spacers has been reported for Streptococcus agalactiae (38). Mick et al. (39) analyzed CRISPR spacers from an intestinal metagenome. Among the 4171 unique spacers present in their data set, there are eight complementary pairs. While the source of these spacers cannot be determined one spacer pair matches a spacer from a CRISPR cassette from Bifidobacterium longum. Similarly, acquisition of complementary spacers was reported in Sulfolobus (40). Thus, it is possible that generation of two oppositely oriented spacers from the same protospacer is a common feature of the CRISPR adaptation process rather than a specific feature of the E. coli system. Such a dual mode of spacer insertion may have an adaptive value. The two possible outcomes increase 2-fold the number of unique spacers and thus increase the likelihood that at least one of crRNAs resulting from spacers derived from non-AAG protospacers will support interference. The effect may not be significant for primed adaptation, where most spacers are selected from protospacers with AAG SAM but can become important in the case of unprimed adaptation, which is not only ∼50 times less frequent than primed adaptation (28), but is also less specific (63% of newly acquired spacers derived from non-AAG protospacers and thus likely leading to non-functional crRNA (27)). Conversely, increased bias towards ‘correct’ orientation for spacers derived from AAG protospacers should also be biologically relevant since this orientation is guaranteed to result in crRNA capable of interference.
ACKNOWLEDGMENT
We wish to thank F. Mojica, S. Brouns, and A. Hynes for critical reading of the manuscript and M. Terns for fruitful discussions.
Footnotes
These authors contributed equally to this work.
The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors
FUNDING
National Institutes of Health (NIH) [GM10407 to K.S.]; Russian Foundation for Basic Research Foundation [11-04-01373-a to K.S. and 14-04-00916 to E.S.]; Ministry of Education and Science of Russian Federation project 14.B25.31.0004 [to K.S.]. Source of open access funding: Ministry of Education and Science of Russion Federation project 14.B25.31.0004.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mojica F.J., Diez-Villasenor C., Soria E., Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 2000;36:244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- 2.Jansen R., van Embden J.D., Gaastra W., Schouls L.M. Identification of a novel family of sequence repeats among prokaryotes. OMICS. 2002;6:23–33. doi: 10.1089/15362310252780816. [DOI] [PubMed] [Google Scholar]
- 3.Bhaya D., Davison M., Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 4.Jansen R., Embden J.D., Gaastra W., Schouls L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 5.Makarova K.S., Haft D.H., Barrangou R., Brouns S.J., Charpentier E., Horvath P., Moineau S., Mojica F.J., Wolf Y.I., Yakunin A.F., et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brouns S.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J., Snijders A.P., Dickman M.J., Makarova K.S., Koonin E.V., van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pougach K., Semenova E., Bogdanova E., Datsenko K.A., Djordjevic M., Wanner B.L., Severinov K. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol. Microbiol. 2010;77:1367–1379. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jore M.M., Lundgren M., van Duijn E., Bultema J.B., Westra E.R., Waghmare S.P., Wiedenheft B., Pul U., Wurm R., Wagner R., et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 9.Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J., Rouillon C., Kerou M., Reeks J., Brugger K., Graham S., Reimann J., Cannone G., Liu H., Albers S.V., et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiedenheft B., Lander G.C., Zhou K., Jore M.M., Brouns S.J., van der Oost J., Doudna J.A., Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deveau H., Barrangou R., Garneau J.E., Labonte J., Fremaux C., Boyaval P., Romero D.A., Horvath P., Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mojica F.J., Diez-Villasenor C., Garcia-Martinez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 14.Semenova E., Jore M.M., Datsenko K.A., Semenova A., Westra E.R., Wanner B., van der Oost J., Brouns S.J., Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah S.A., Erdmann S., Mojica F.J., Garrett R.A. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garneau J.E., Dupuis M.E., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadan A.H., Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 17.Westra E.R., van Erp P.B., Kunne T., Wong S.P., Staals R.H., Seegers C.L., Bollen S., Jore M.M., Semenova E., Severinov K., et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 19.van der Oost J., Jore M.M., Westra E.R., Lundgren M., Brouns S.J. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Horvath P., Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 21.Deveau H., Garneau J.E., Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 22.Marraffini L.A., Sontheimer E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yosef I., Goren M.G., Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar R., Qimron U. The Escherichia coli CRISPR system protects from lambda lysogenization, lysogens, and prophage induction. J. Bacteriol. 2010;192:6291–6294. doi: 10.1128/JB.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stern A., Keren L., Wurtzel O., Amitai G., Sorek R. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 2010;26:335–340. doi: 10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vercoe R.B., Chang J.T., Dy R.L., Taylor C., Gristwood T., Clulow J.S., Richter C., Przybilski R., Pitman A.R., Fineran P.C. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 2013;9:e1003454. doi: 10.1371/journal.pgen.1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yosef I., Shitrit D., Goren M.G., Burstein D., Pupko T., Qimron U. DNA motifs determining the efficiency of adaptation into the Escherichia coli CRISPR array. Proc. Natl. Acad. Sci. U.S.A. 2013;110:14396–14401. doi: 10.1073/pnas.1300108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Datsenko K.A., Pougach K., Tikhonov A., Wanner B.L., Severinov K., Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 29.Swarts D.C., Mosterd C., van Passel M.W., Brouns S.J. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M., Wang R., Zhao D., Xiang H. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. 2013;42:2483–2492. doi: 10.1093/nar/gkt1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savitskaya E., Semenova E., Dedkov V., Metlitskaya A., Severinov K. High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli. RNA Biol. 2013;10:716–725. doi: 10.4161/rna.24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plagens A., Tjaden B., Hagemann A., Randau L., Hensel R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J. Bacteriol. 2012;194:2491–2500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goren M.G., Yosef I., Auster O., Qimron U. Experimental definition of a clustered regularly interspaced short palindromic duplicon in Escherichia coli. J. Mol. Biol. 2012;423:14–16. doi: 10.1016/j.jmb.2012.06.037. [DOI] [PubMed] [Google Scholar]
- 34.Pages H., Aboyoun P., Gentleman R., DebRoy S. 2012. R package version 2.24.1.
- 35.Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer; 2009. [Google Scholar]
- 36.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Craigie R., Mizuuchi K. Role of DNA topology in Mu transposition: mechanism of sensing the relative orientation of two DNA segments. Cell. 1986;45:793–800. doi: 10.1016/0092-8674(86)90554-4. [DOI] [PubMed] [Google Scholar]
- 38.Lopez-Sanchez M.J., Sauvage E., Da Cunha V., Clermont D., Ratsima Hariniaina E., Gonzalez-Zorn B., Poyart C., Rosinski-Chupin I., Glaser P. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol. Microbiol. 2012;85:1057–1071. doi: 10.1111/j.1365-2958.2012.08172.x. [DOI] [PubMed] [Google Scholar]
- 39.Mick E., Stern A., Sorek R. Holding a grudge: persisting anti-phage CRISPR immunity in multiple human gut microbiomes. RNA Biol. 2013;10:900–906. doi: 10.4161/rna.23929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erdmann S., Garrett R.A. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol. Microbiol. 2012;85:1044–1056. doi: 10.1111/j.1365-2958.2012.08171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


