Abstract
Chromatin domain organization and the compartmentalized distribution of chromosomal regions are essential for packaging of deoxyribonucleic acid (DNA) in the eukaryotic nucleus as well as regulated gene expression. Nucleoli are the most prominent morphological structures of cell nuclei and nucleolar organization is coupled to cell growth. It has been shown that nuclear scaffold/matrix attachment regions often define the base of looped chromosomal domains in vivo and that they are thereby critical for correct chromosome architecture and gene expression. Here, we show regulated organization of mammalian ribosomal ribonucleic acid genes into distinct chromatin loops by tethering to nucleolar matrix via the non-transcribed inter-genic spacer region of the ribosomal DNA (rDNA). The rDNA gene loop structures are induced specifically upon growth stimulation and are dependent on the activity of the c-Myc protein. Matrix-attached rDNA genes are hypomethylated at the promoter and are thus available for transcriptional activation. rDNA genes silenced by methylation are not recruited to the matrix. c-Myc, which has been shown to induce rDNA transcription directly, is physically associated with rDNA gene looping structures and the intergenic spacer sequence in growing cells. Such a role of Myc proteins in gene activation has not been reported previously.
INTRODUCTION
In eukaryotic organisms, deoxyribonucleic acid (DNA) is packaged into chromatin within the nucleus. The architecture and dynamics of chromatin play a crucial role in establishing and maintaining appropriate gene expression levels (1). Compelling evidence has shown that the equilibrium between the ‘open’ (accessible) and ‘closed’ (inaccessible) states of genes can be attributed to their physical location in an appropriate nuclear ‘compartment’ or ‘domain’ (2). Moreover, epigenetic modifications are important for establishing the functional compartmentalization into chromatin domains within nuclei as well as the regulation of nuclear functions (3). It has been proposed that nuclear compartmentalization could facilitate the efficiency of gene expression by bringing together appropriate genes and transcription factors, involved in ribonucleic acid (RNA) synthesis, processing and modifications, in time and space (4,5).
Nucleoli are the most prominent nuclear domains. The best characterized nucleolar function is ribosomal RNA (rRNA) synthesis, processing and assembly into ribosomal sub-units (5). Nucleolar assembly and ribosome biogenesis are essentially coupled to the architectural and functional compartmentalization of the nucleus, cell growth and proliferation (6). The 400 copies of the rRNA gene are clustered in tandem arrays separated by intergenic spacers (IGS) and are found on human chromosomes 13, 14, 15, 21 and 22. The nucleolus to which these chromosomal regions contribute is a dynamic nuclear structure with a size that is correlated to the level of ribosomal DNA (rDNA) transcription and which is broken down and reformed at every cell division. As for the rest of cellular DNA, the long length of extended rDNA repeats (7) in relation to the size of the nucleolus requires considerable compaction of the rDNA. Using the chromosome conformation capture (3C) technique, we previously showed dynamic changes in the higher order of rDNA chromatin structure that reflects cell growth status (8,9).
Evidence from three-dimensional electron tomography suggests that the rDNA genes are compacted in the form of loop structures within the nucleolus (10). This is reminiscent of the multi-loop sub-compartment model proposed for non-nucleolar genomic regions, by which folding and looping of chromatin domains is mediated by the anchorage to the nuclear matrix via scaffold attachment regions or matrix association regions (S/MARs) (2,11). The S/MARs are involved in the modulation of transcription by promoting chromatin accessibility and histone acetylation (12). Furthermore, MAR-mediated chromatin loop formation has been shown to be important for expression of the human β-globin gene cluster (13) and the cytokine gene cluster (14), as well as the IGF2 (15) and Dlx5 loci (16). Interestingly, these studies may be relevant for nucleolar genes because attachment of rDNA to nuclear matrix has been described previously (17–20).
c-Myc is a key inducer of cell growth in response to growth factors. c-Myc induces functions required for protein synthesis such as the protein subunits of ribosomes, and we and others have shown that c-Myc plays a direct nucleolar role in producing the RNA components of ribosomes (21,22). We further showed that c-Myc plays a key role in the induction of higher-order rDNA chromatin structure changes that accompany cell growth induction. (8,9). However, the mechanism by which c-Myc changes rDNA chromatin structure remained elusive. In this work, we demonstrate that c-Myc is associated with the rDNA IGS region where it regulates association of epigenetically non-silenced rDNA genes to the nucleolar matrix in a growth-dependent fashion.
MATERIALS AND METHODS
Cell lines, culture conditions and chemical treatments
Human embryonic kidney 293 (HEK293) cells, HeLa cells and rat fibroblasts (TGR-1, Rat1MycER and HO15.19) were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) and 0.1% (v/v) gentamycin at 37°C in a 5% CO2 atmosphere. In starvation/re-feeding experiments, cells were grown to confluence and starved in DMEM without FBS for 24–48 h for rat fibroblasts or 72 h for HeLa cells, and then DMEM containing 10% FBS was added for 4 h. The starved Rat1MycER (myc +/+, MycER) cells were untreated or treated with 4-hydroxytamoxifen (4-HT; final concentration, 200 nM) (Sigma-Aldrich) and were added to culture medium for 4 h. For treatments with the c-Myc inhibitor 10058-F4 (Calbiochem), a final concentration of 80 μM was added to culture medium 1 h before serum or 4-HT addition and incubated overnight during serum stimulation or for a further 2 h for 4-HT treatment.
Pre-rRNA transcription analysis
Cells were harvested and total RNA was extracted, using a RNeasy Midi Kit (Qiagen) according to manufacturer instructions, and quantified spectrophotometrically. Following the manufacturer's protocol, 1 ug of whole cell RNA from each sample was reverse transcribed using the RevertAid H Minus First Strand cDNA synthesis Kit (Thermo Scientific). Amounts of pre-rRNA were measured in relation to Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) by real-time polymerase chain reaction (PCR) performed on a MyiQ™ single-color real-time PCR detection system (Bio-Rad) using the 2−ΔΔCT method (23), where ΔΔCT = (CT, Pre-rRNA − CT, GAPDH)experimental group − (CT, Pre-rRNA − CT, GAPDH)control group. The primers used to amplify human and rat pre-rRNA are shown in Supplementary Tables S1 and S2. The forward primer: 5′-tggcattgctctcaatgacaac-3′ and the reverse primer: 5′-tgggatggaattgtgagggagatg-3′ were used to amplify rat GAPDH mRNA (GenBank number X02231.1). The forward primer: 5′-tcgacagtcagccgcatcttc-3′ and the reverse primer: 5′-acgtactcagcgccagcatcg-3′ were used to amplify human GAPDH mRNA (GenBank number AF261085.1). The real-time PCR program used is the same as described under quantitative PCR (qPCR) assays.
Isolation of nucleolar MARs
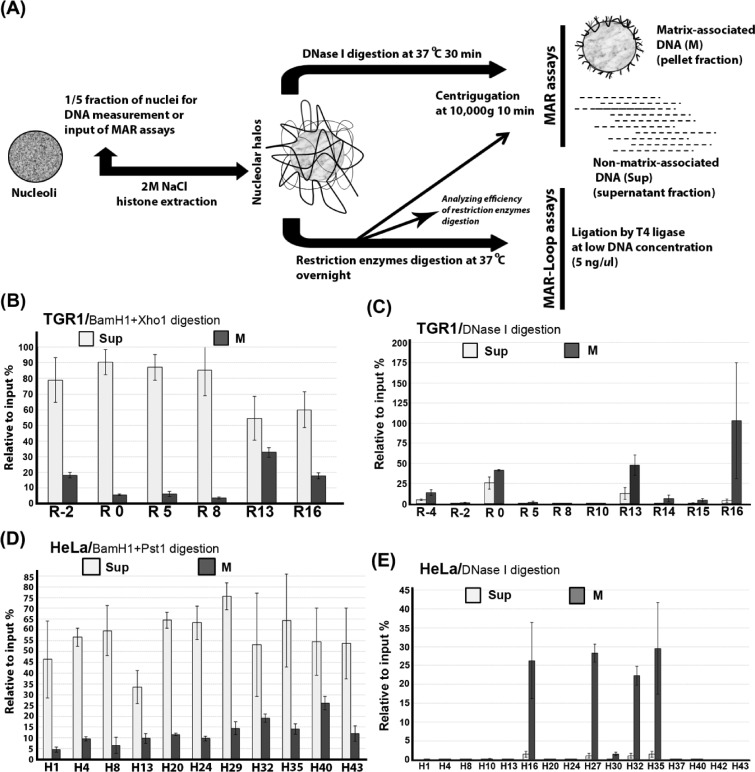
Nucleoli from HeLa, TGR-1, HO15.19 and RatMycER cells were prepared using sucrose gradients (24). Nucleolar matrix with looped DNA domains (halos) was prepared by extraction of histones and other proteins from nuclei in high salt buffer essentially as described previously (17); see Figure 2A. Dithiothreitol (DTT; 1 mM/ml) was added into extraction buffer [2-M NaCl, 40-mM tris(hydroxymethyl)aminomethane hydrochloride (Tris HCl) pH 8.5 and 10-mM ethylenediaminetetraacetic acid (EDTA)] in order to isolate nucleolar matrix with a lower protein content (39). Halos were then incubated in digestion buffer containing DTT (1 mM/ml), DNase I (200 ug/ml) (Sigma-Aldrich) and RNase A (200 mg/ml) (Sigma-Aldrich) at 37°C for 30 min, or were digested by indicated restriction enzymes (BamH1+Pst1 in HeLa cells, BamH1+Xho1 in TGR-1/HO15.19/Rat1Myc cells at 10 units/μg DNA) (New England Biolabs) overnight. After digestion with endonuclease or restriction enzymes, the nucleoid samples were separated into a soluble fraction (supernatant, non-matrix-associated DNAs) and an insoluble fraction (matrix-associated DNAs). The cutting efficiencies of restriction enzymes on nucleolar halos were determined as shown in Supplementary Figure S2. The DNAs purified from pools of matrix and loop fractions were used as templates for qPCR analyses with indicated primer pairs. The sequences and locations of primer sets corresponding to human and rat rRNA are available in Supplementary Tables S1 and S2, respectively, and in Figure 1. The enhancement of matrix-associated DNA was calculated in relation to input DNA (without treatment by endonuclease or restriction enzymes).
Figure 2.
Preferential matrix attachment of rDNA genes via the intergenic spacer (IGS) region. (A) Diagram showing the procedure used for isolation of nucleolar halos and the preparation of nucleolar matrix and subsequent MAR and MAR-loop assays. (B) Preferential attachment of rDNA genes to nuclear matrix via IGS sequences in growing TGR-1 cells. The relative association of different regions of the rDNA repeat to nucleolar matrix (M) after digestion with BamH1 and Xho1 as well as the levels of released non-matrix associated regions (Sup) are shown. (C) Fine mapping of nucleolar-matrix-associated rDNA fragments after DNase I treatment of nucleolar matrix from growing TGR-1 cells. (D) Preferential attachment of rDNA genes to nuclear matrix via sequences throughout the IGS region in growing HeLA cells. The supernatant fraction and pellet fraction are separated after restriction enzymes (BamH1+Pst1) digestion. (E) Fine mapping of nucleolar-matrix-associated rDNA fragments after DNase I treatment of nucleolar matrix from growing HeLa cells. The values plotted in (B–E) are the means and standard deviation of results from three independent experiments.
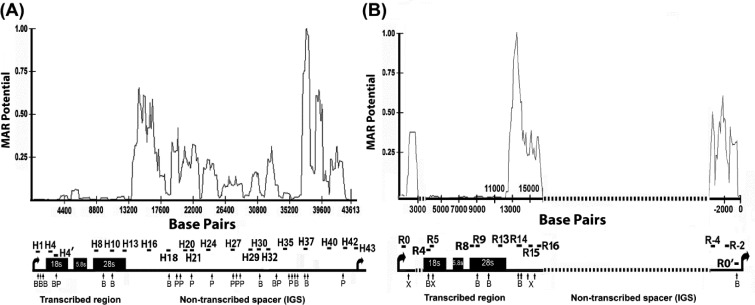
Figure 1.
The non-transcribed intergenic spacer (IGS) region of rRNA genes has an enhanced propensity for nuclear matrix attachment. The line graphs show MAR potential score in relation to nucleotide position for the rDNA repeat sequence from human (A) and rat (B). The lower panels in each figure part show schematic depictions of rRNA gene repeats. Vertical arrows below the rDNA repeat represent the locations of cleavage sites for BamHI (B) and PstI (P) in human (part A) and for BamHI (B) and XhoI (X) in rat (part B). The locations of amplified rDNA regions are shown by horizontal bars. The sequences of primer sets used for amplification of human and rat rDNA are shown in Supplementary Tables S1 and S2. The dotted line in rat rRNA gene represents sequences that are not available from GenBank. Human rDNA sequence, derived from GenBank number U13369, and (B) rat rDNA sequence, derived from accession numbers X04084, X03838, X61110, X00677, X16321, V01270 and X03695, were used. Matrix attachment potential was predicted by the MAR-Wiz software (57) as implemented at http://genomecluster.secs.oakland.edu using a window size of 1000 bp stepped at 100-bp intervals. Scores in excess of 0.6 are generally considered as strong indicators of nuclear matrix binding potential.
DNA methylation assays
The rat rDNA fragments, recovered separately from total DNA or non-matrix-associated DNA and matrix-associated DNA after digestion of restriction enzymes (BamH1+Xho1), were subjected to digestion with HpaII or MspI enzyme (New England Biolabs) prior to qPCR analysis. HpaII/MspI restriction enzyme sensitivity assays were used to measure the methylation status of a promoter CpG site at position −145 (relative to the transcription initiation site) (40). A primer set, the forward primer: 5′-agcatggacttctgaggccgag-3′ and the reverse primer: 5′-cataaagctgccccagagag-3′, was used to amplify a fragment of 450 bp containing an HpaII/MspI restriction site that includes the −145-bp CpG site. CpG methylation at this site inhibits cleavage by HpaII and thus the level of resistance to cleavage can be used as a measure of CpG methylation. The R0 primer pair (Supplementary Table S2) amplifies a 200-bp fragment downstream of the transcription initiation site that was used for normalization between samples.
3C assay and MAR-loop assay
The procedure for 3C has been described in detail previously (8). The MAR-loop assay is a development of the 3C method (25). The obligatory cross-linking step with formaldehyde was omitted because the nucleolar matrix is sufficient to maintain higher-order DNA structures. The formaldehyde cross-linked chromatin (3C) or nucleolar halos (MAR-loop) were digested by the indicated restriction enzymes as shown above at a concentration of 10 units/μg DNA, and then a low concentration of chromatin or nucleolar halo, equivalent to 5 ng DNA/ul, was ligated overnight using T4 ligase (150 U) (Fermentas) at 16°C. DNA was recovered from the ligation mixtures by proteinase K digestion followed by phenol-chloroform extraction and ethanol precipitation prior to use as template for PCR amplification. The sequences of primers for amplifying size marker templates and re-ligated DNA fragments are shown in Supplementary Tables S3–S6. The size marker DNA fragments were amplified, gel purified and sequenced.
Chromatin immunoprecipitation assays
Preparation of nuclei from 0.25% formaldehyde-fixed HEK293 or HeLa cells and sonication of nuclear chromatin was carried out as described previously (8). The average length of DNA fragments in preparations of sonicated nucleolar chromatin ranged from 500 bp to 1 kb. Four micrograms of antibodies against Myc (sc-764) and upstream binding factor UBF (SC-13125) were used as well as normal rabbit immunoglobulin G (IgG) as a negative control (Santa Cruz Biotechnology). After overnight incubation at 4°C in chromatin immunoprecipitation (ChIP) buffer (0.1% sodium dodecyl sulphate (SDS) in phosphate buffered saline), 30 μl of a 50% slurry of salmon sperm DNA/protein G-coated agarose beads (#16–201, Upstate Millipore) was added, and the mixtures were rotated at 4°C for 30 min. The DNA–protein complexes were eluted twice from the beads with 250 μl of buffer containing 0.1-M NaHCO3 and 1% SDS. After reversal of the cross-links by incubation at 65°C overnight, proteinase K and RNase (Roche) treatment, extraction with pheonol-chloroform and ethanol precipitation, the recovered DNA was re-suspended in 500 ul of Tris HCl (pH 8.5) prior to quantitative real-time PCR analysis. For the 3C-ChIP assay, a combination of the 3C assay and the ChIP assay, the formaldehyde-cross-linked nuclei were digested by the indicated restriction enzymes and re-ligated under dilute condition as for the 3C assay prior to chromatin immunoprecipitation as described above. Samples immunoprecipitated by protein agarose beads only were used as negative controls.
qPCR assays
DNA samples recovered from ChIP assays or matrix-association assays were used as templates in real-time PCR, and samples from individual ChIP assays were analyzed in triplicate. Each PCR mixture contains 0.6 μM of each primer, 5-ul DNA templates in a final volume 40 μl of 1x iQ™ SYBRGreen Supermix (Bio-Rad). The cycling program was set as follows: denature at 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, 63°C for 30 s and 72°C for 40 s, followed by a final extension period at 72°C for 5 min. The cycle threshold (CT) was set within the linear range of all PCR reactions. For each primer pair, the fold enrichment was calculated relative to the CT of amplification between samples from endonuclease or restriction enzymes untreated/treated in matrix association assays, or the CT of amplification of DNAs precipitated from antibody against Myc/UBF and IgG in ChIP assays.
Statistical analysis
P values associated with all comparisons were based on unpaired two-tailed Student's tests. For comparisons between transcribed (amplicons R5 and R8 in rat and amplicons H1, H4, H8, H10 and H13 for human) and IGS regions (amplicons R-2 and R16 in rat and amplicons H16, H20, H24, H27, H30, H32, H35, H40 and H42 for human), data from probes close to the inter-region borders (that detect signals from both regions), like amplicons R0 and R13 in rat and amplicon H43 for human, were omitted. P values < 0.05 were considered as significant. Results are presented as mean values (n > = 3) ± standard error of the mean.
RESULTS AND DISCUSSION
Within the nucleus at large, actively transcribed genes are often in close proximity to each other in association with the nuclear scaffold/matrix at S/MARs while the genomic regions that separate such genes form loops more distant from the matrix (25). We wondered whether the nuclear matrix might be involved in the growth-regulated higher-order chromatin structure of the rDNA (8,9,26). The MAR-Wiz algorithm (27,28,29) predicts that the main potential for matrix attachment within the rDNA is restricted to the non-transcribed IGS in human (Figure 1A) and mouse rDNA (Supplementary Figure S1). A consistent pattern is seen for rat rDNA, although sequence information for most of the IGS is not available (Figure 1B).
Next, we used a matrix association assay (30) to experimentally identify regions of the rDNA repeat that are matrix attached. Figure 2A summarizes the procedures used to study the association of rDNA sequences with the nucleolar matrix. To quantify the level of attachment of different rDNA regions to the nucleolar matrix in growing rat fibroblasts (TGR-1), nucleolar matrix preparations were digested with restriction enzymes (BamH1+Xho1) and MAR assays were performed. Figure 2B shows a significantly (P = 0.03) higher level of matrix attachment for rDNA regions outside the transcribed region (e.g. R-2 and R16) than for transcribed regions (e.g. R5 and R8). Figure 2B also shows that about a third of rDNA genes are tethered to the matrix via IGS sequences under these conditions. An attractive possibility is that the matrix-associated genes may correspond to the subset of rDNA genes that are actively transcribed. The restriction enzymes used showed similar cutting efficiency throughout the rDNA locus and thus the level differences are not due to region-specific differences in cutting efficiency (Supplementary Figure S2A). To more precisely map the extent of the matrix-bound rDNA sequences, the nucleolar matrix from the same cells was digested by DNase I and analyzed by MAR assays. Figure 2C shows clearly that the higher levels of observed rDNA attachment are associated with sequences within the non-transcribed rDNA region. Sequences corresponding to the transcribed region and rDNA genes that are not matrix associated are generally digested by DNase I as expected. A DNaseI-resistant signal is detected for some regions in the supernatant fraction, which could result from protection by non-matrix components, but the biological significance of this protection has not been investigated further. A significantly (P = 1.73E−05) higher tendency for matrix association of IGS regions is also observed for growing HeLa cells (Figure 2D). In this case, probes are available throughout the IGS and the results show that matrix association is elevated in regions spread over a 24-kb region (H16-H40), with the highest association levels at H16, H27, H32 and H35 (Figure 2E). Since HeLa cells are derived from cancer cells with elevated Myc levels, we performed the MAR assay on HEK293 cells. The results showed enhanced matrix-mediated protection of IGS sequences but the level of protection was not as high as in HeLa cells (Supplementary Figure S3A). The experimentally determined region of matrix attachment corresponds in part with the computer prediction in Figure 1, indicating that the MAR-Wiz algorithm works well, as has been reported elsewhere (31,32). The results also extend and confirm at least some of the earlier studies showing interaction between rDNA and matrix (17,18). Interestingly, matrix association is seen closer to the regions of promoter and terminator in the IGS of rat rDNA than is seen in human. This might result from the high degree of sequence divergence in non-transcribed rDNA sequences (Supplementary Figure S4).
Emerging evidence suggests that changes in ribosome biogenesis are associated with alterations of nucleolar architecture (5). Matrix attachment could be a way to organize the higher-order structure of rDNA chromatin and growth-state-dependent changes in matrix attachment could thus account for the growth-dependent changes in higher-order rDNA structure that we have reported previously (8). Figure 3A shows that matrix attachment of several regions throughout the IGS region is strongly induced when serum-starved HeLa cells are induced to grow and rDNA transcription is induced by addition of serum. IGS attachment to nucleolar matrix is very low for most regions of the IGS in starved cells. The exception to this is in the H16, H27, H32 and H35 regions where relatively high matrix association is observed compared to other regions within IGS even during starvation. Likewise, both constitutive and facultative matrix attachment sites that have been described in other contexts (33), our results suggest the existence of sites that show a constitutive level of matrix attachment but which also exhibit a facultative-enhanced matrix associated upon growth stimulation.
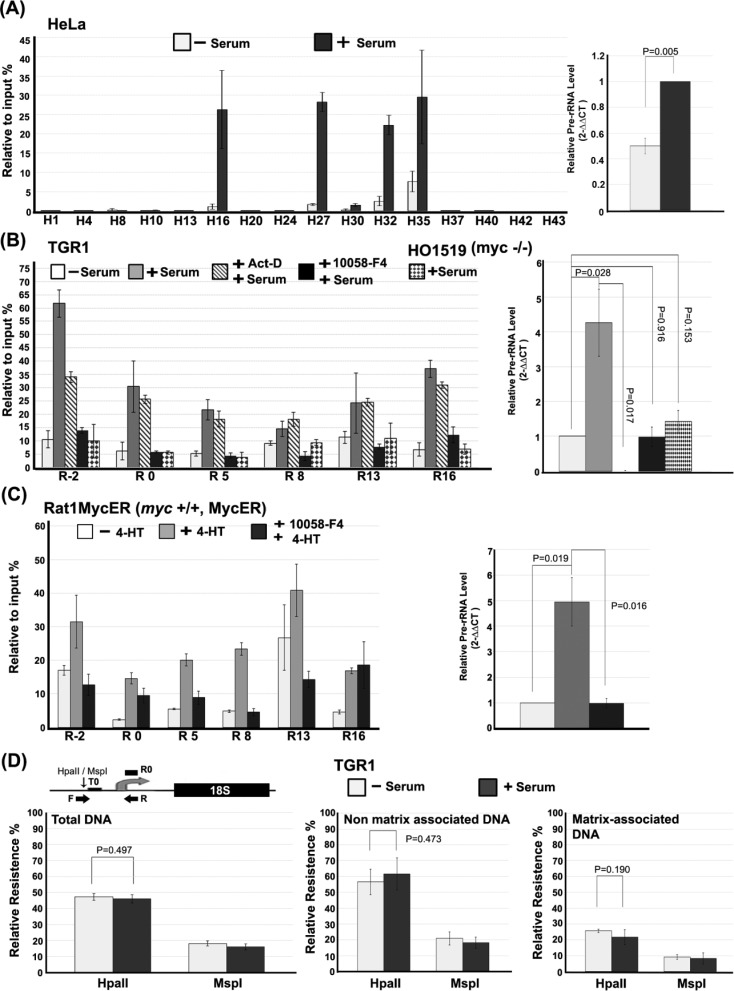
Figure 3.
Growth-dependent and c-Myc-dependent attachment of rDNA to the nucleolar matrix. (A) Matrix attachment of the rDNA IGS is induced upon growth stimulation of HeLa cells. The left panel represents quantitative real-time PCR showing the relative levels of matrix-attached rDNA throughout the rDNA repeat after digestion with DNase I for starved HeLa cells before (−S) or after (+S) re-feeding with serum-containing medium. The right panel shows the corresponding changes in the level of pre-rRNA measured by quantitative real-time PCR. (B) Growth-induced attachment of the rDNA IGS to nuclear matrix requires c-Myc in rat fibroblasts. Levels of matrix attachment after restriction digestion (see Figure 2B) are shown for starved TGR-1 cells re-fed with serum in the absence or presence of actinomycin D (0.1 μg/ml) or the Myc inhibitor, 10058-F4 (80 μM). Growing HO1519 (myc−/−) cells were treated and analyzed in parallel. Pre-rRNA levels corresponding to the different cells and treatments are shown (right panel). (C) Activation of Myc in serum-starved cells is sufficient to induce rDNA IGS matrix attachment. The relative levels of matrix-attached rDNA after restriction digestion (see Figure 2B) at the indicated rDNA regions in Rat1MycER cells, which express a Myc-ER fusion protein, before (−4-HT) and after (+4-HT) activation of Myc-ER by addition of 4-hydroxytamoxifen in the absence or presence of c-Myc inhibitor 10058-F4. Pre-rRNA levels corresponding to the different treatments are shown (right panel). The cutting efficiencies of restriction enzymes on samples in (B) and (C) are shown in Supplementary Figure S2D and E. (D) rRNA genes that associate with nucleolar matrix are hypomethylated in the promoter region. A diagram of the rat rDNA promoter region shows the primer set (forward primer, F; reverse primer, R), described in the Materials and Methods section, used to detect the methylation status of the CpG residue at −145 bp from the transcription start site, while the primer set R0 is for normalization between samples (upper panel; see the Materials and Methods section). The quantitative real-time PCR signal for genomic DNA from serum-starved (−Serum) and growing (+ Serum) TGR-1 cells after cleavage with HpaII or MspI, prior to (left panel) or after separation of non-matrix-associated DNA (middle panel) and matrix-associated DNA (right panel). The values in (A–D) are the means and standard deviations of results from three independent experiments.
To determine whether serum-induced changes in rDNA matrix attachment are dependent on on-going or induced rDNA transcription or Myc activity, we used Rat1 fibroblast-derived cell lines that are appropriate for studying Myc activity because they show a good serum starvation/re-feeding response and derivative cell lines are available that either lack Myc or allow its pharmacological activation, via a Myc-Estrogen Receptor fusion protein (34). First, we investigated the level of matrix binding of transcribed and non-transcribed regions of rat rDNA in the absence or presence of serum (Figure 3B). All measured regions show increased matrix association after serum addition to serum-starved cells but the induced matrix association is significantly greater in the IGS region, compared to the transcribed region (P = 0.003). To determine whether rDNA transcription was required for the serum-induced changes in matrix association, TGR-1 cells were pre-treated with a low concentration of actinomycin D (0.1 ug/ml) prior to serum addition. Figure 3B shows that a significant increase in serum-induced matrix association of rDNA is seen in both transcribed (P = 0.026) and IGS (P = 1.60E−5) regions even in the presence of actinomycin D and that the levels of serum-induced matrix association are generally similar to those seen in the absence of actinomycin D. Serum-induced matrix attachment does not therefore depend on rDNA transcription, which was undetectable in the actinomycin D-treated cells (right panel). Previously, it has been shown that the c-Myc protein is a direct regulator of rDNA transcription (21,22) and that nucleolar c-Myc plays a key role in modulating growth-dependent changes in the higher-order organization of rDNA chromatin (8,9). To determine whether c-Myc is required for serum-induced attachment of rDNA to nucleolar matrix, we compared matrix attachment of rat rRNA gene repeats in cells pre-treated with the small-molecule c-Myc inhibitor, 10058-F4, prior to serum addition in relation to matrix attachment levels in untreated serum-starved cells. Figure 3B shows that there is no significant induction of matrix attachment in cells pre-treated with 10058-F4 (P = 0.941) and that induction of rRNA synthesis is strongly inhibited by 10058-F4. A similar result was seen for serum-stimulated HO15.19 rat cells that lack the c-myc gene (P = 0.716) (Figure 3B). Thus Myc activity is required for both enhanced matrix attachment of the rDNA IGS and induced rDNA transcription during serum-dependent growth stimulation of starved cells. Similarly, the Myc inhibitor reduces serum induction of pre-rRNA levels as well as the level of matrix attachment in growing HeLa cells (Supplementary Figure S3B).
To verify and further characterize the involvement of c-Myc in the regulation of growth-dependent matrix attachment of the rDNA IGS, we used cell lines derived from Rat1 fibroblasts, which express an inducible Myc/Estrogen Receptor (Myc-ER) fusion protein. The Myc-ER can be activated by addition of 4-HT allowing activation of Myc in starved cells in the absence of normal growth-related activating signals. Figure 3C shows that activation of Myc-ER in starved cells was sufficient to cause increased overall association of rDNA sequences with the matrix (P = 1.163E−4) as well as increased levels of rRNA synthesis, even though the Myc-ER fusion protein does not exactly reciprocate the pattern seen by endogenous c-Myc, which could be due to the leakiness of the Myc-ER system (35), a difference in the functionality of the Myc-ER protein, its over-expression or a requirement for other factors in addition to Myc. The Myc-specificity of the Myc-ER effects is further emphasized by the significant overall inhibition of Myc-ER-induced matrix attachment and rRNA transcription by prior treatment of the cells with the 10058-F4 Myc inhibitor (P = 1.533E−5; Figure 3C). At R16, where pre-treatment with 10058-F4 does not reduce Myc-ER stimulates matrix attachment, we cannot exclude that Myc functions in a fashion independent of Max and E-box binding. Such mechanisms of Myc action have been reported (36). Taken together our results are consistent with a dual role of Myc in the nucleolus. The first, rRNA transcription independent, role of Myc in reorganizing matrix attachment of rDNA genes in growth-stimulated cells is likely to overlap with the role of Myc in reorganizing higher-order rDNA structure that was reported previously (8). This first role appears to be separable from and probably precedes a second role that is more directly coupled to the induction of rDNA transcription. This second role is thought to involve Myc-mediated recruitment of the SL1 transcription factor to the RNA PolI promoter (22) perhaps through direct interaction of Myc with the TATA-binding protein subunit of SL1, which has been shown previously (37).
The population of rDNA genes can be divided into at least two, and perhaps more, functionally significant categories (38). A large group of genes is epigenetically silenced by mechanisms involving DNA methylation as well as histone modification. Methylated rDNA genes are thought to be refractory to transcriptional activation and recombination in part due to their dense compaction and peripheral nucleolar localization (39,40). Non-silenced genes are available for transcriptional activation and are transcribed at a level that is regulated in accordance with cellular growth requirements (41). Since c-Myc is an important regulator of growth-coupled rDNA transcription and matrix attachment we might expect the matrix-attached rDNA genes in growing cells to be enriched in non-methylated genes that are competent for transcriptional activation. To test this, we monitored the methylation status of a CpG site in the promoter of rat rDNA (Figure 3D, top panel) in TGR-1 cells using resistance to cleavage by HpaII as a measure of CpG methylation level. Figure 3D (left panel) shows that around 45% of rDNA genes are resistant to HpaII cleavage while over 80% can be cleaved by MspI, an isoschizomer that is not sensitive to DNA methylation. This is consistent with previous reports (42). To test the distribution of silenced and non-silenced genes in relation to their matrix attachment, nucleolar halos were prepared from growing TGR-1 cells and non-matrix-associated DNA was released by cleavage with BamHI and XhoI restriction enzymes. Figure 3D (right panel) shows that the matrix-attached rDNA was cleaved well (≤ 20% uncleaved genes) by both HpaII and MspI, while the non-matrix-attached rDNA showed higher levels of resistance to cleavage by HpaII (≥ 60% uncleaved genes). We conclude that that the non-matrix-associated rDNA genes are more hypermethylated while genes associated with the matrix are enriched in hypomethylated, non-silenced gene copies.
To determine whether changes in methylation status at the CpG upstream of the rat rDNA promoter could be important for the increased levels of matrix association seen in growing cells compared to starved cells, the HpaII cleavage data described above were compared to analogous data from serum-starved cells. As shown in Figure 3D, we were not able to detect a significant difference in the levels of methylated rDNA genes in the matrix-associated (right panel) and non-matrix-associated (center panel) fractions between starved and growing cells. Thus the data do not support a role for rDNA gene methylation status in the observed growth-dependent changes in matrix attachment (Figure 3D). Our data are fully consistent with previously reported results showing that growth-dependent changes in the expression of rDNA genes occur by activating new rDNA genes within the subset of non-CpG methylated genes (38).
Differential, matrix attachment could be the mechanism for the growth- and Myc-dependent changes in higher-order rDNA chromatin structure that we reported previously based on chromosome conformation capture (3C) studies (8,9). Analogous to the 3C assay, the MAR-loop assay has been used to detect DNA sequences that are held in close proximity to each other as a result of interaction with the nuclear matrix (30). Unlike the 3C assay, it is not necessary to use formaldehyde cross-linking to maintain integrity of the samples in the MAR-loop assay. Figure 4A (right panel) shows that rat rDNA IGS sequences can be ligated together even when they are separated from each other by 15 kb or more of DNA sequence, consistent with attachment to the matrix in close proximity to each other. Ligation of fragment combinations involving sequences in the transcribed region of the rDNA is much weaker or undetectable, consistent with loss of transcribed sequences from the matrix upon restriction enzyme cleavage. The position and orientation of primers as well as the primer combinations used are illustrated schematically in Figure 4A (left panel). To study whether the MAR-loop assay detects the same higher-order structure relationships as the 3C assays performed previously, we performed a more extensive side-by-side analysis across the human rDNA repeat unit using both assays in parallel. Figure 4B (right panel) shows that the results of the two assays are very similar such that sequences within the IGS region are found in close proximity to each other by both assays while neither assay detects proximity of sequences in the transcribed region to each other or to IGS sequences. The position and orientation of primers as well as the primer combinations used are illustrated schematically in Figure 4B (left panel). The similarity of the results from these assays provides strong evidence that matrix attachment is important for the higher-order chromatin structures found in the rDNA of growing cells.
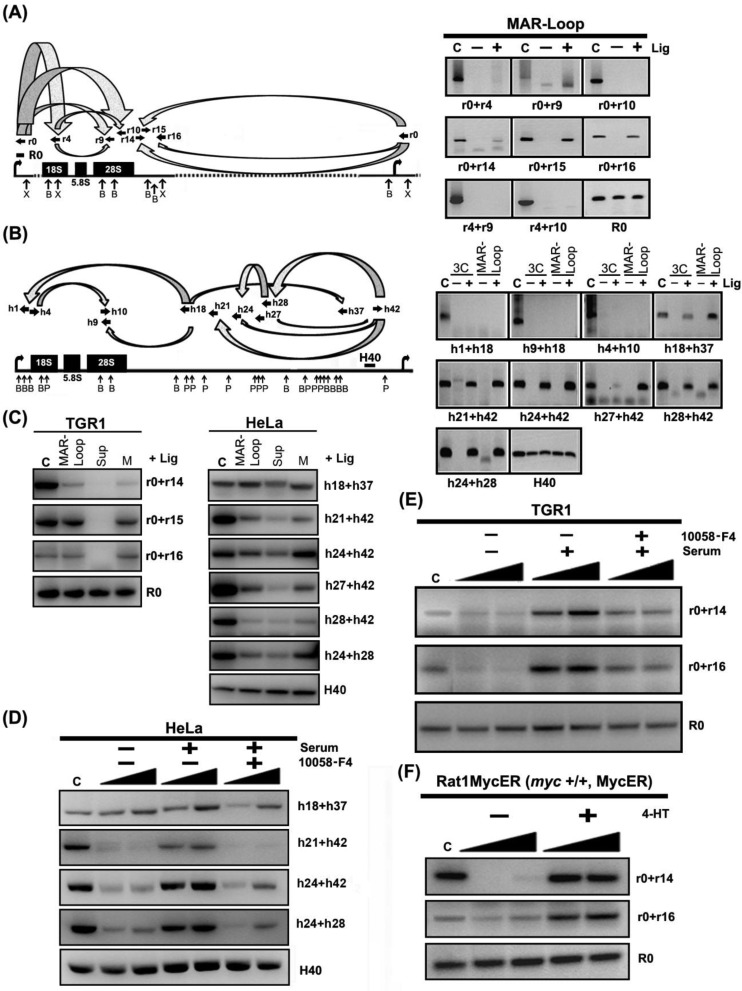
Figure 4.
rDNA IGS matrix attachment can account for growth- and Myc-dependent changes in higher-order rDNA structure. (A) Distant regions within IGS are bound to matrix in close proximity to each other. The left panel shows the combinations of primer sets used and their locations and orientations along rat rDNA repeat (sequences of primer sets are shown in Supplementary Table S4). The right panels show results for different primer pairs from an MAR-loop assay of growing TGR-1 cells, in which DNA fragments, which are held in close proximity to each other after cleavage with XhoI (X) and BamHI (B) by matrix attachment, can be ligated (+ Lig) to create novel DNA fragments. The panels also show the migration of positive control (C) fragments, which indicate the expected size of potential ligation products, and that no novel DNA fragments are detected in the absence of added ligase (− Lig). The last panel (R0) is a loading control (see Figure 1B). (B) The MAR-loop assay and 3C assay identify the involvement of an equivalent set of rDNA IGS regions in the formation of rDNA gene loop structures in growing HeLa cells. Annotations are the same as for part (A) (sequences of primer pairs are shown in Supplementary Table S6) except that the H40 region is amplified in all samples as a loading control. (C) Positive proximity results from the MAR-loop assay are predominately associated with the matrix fraction when the assay is performed on isolated matrix-associated (M) and matrix-non-associated (Sup) fractions. Other annotations are shown as (A) and (B). Growth stimulation of matrix-associated gene loop structures is dependent on the activity of c-Myc in (D) HeLa cells and (E) TGR-1 cells. MAR-ligation assay results for starved HeLa and TGR-1 cells before (−serum) or after (+serum) addition of medium containing serum in the absence or presence of c-Myc inhibitor, 10058-F4. Other annotations are as for part (A) and part (B). The filled ramps indicate that PCR amplifications were performed at increasing substrate concentrations, since product formation is easily saturated at higher product concentrations. (F) Myc-ER activation is sufficient to induce matrix-associated gene looping in cells lacking endogenous c-Myc. MAR-ligation assay results before (−4-HT) or after (+4-HT) treatment of Rat1MycER cells. The cutting efficiencies of restriction enzymes on all the indicated samples are shown in Supplementary Figure S2C–E.
To further verify the relationship between matrix association of the IGS and the formation of higher-order structures in the rDNA, we performed the MAR-loop assay in non-matrix-associated and matrix-associated fractions separately (Figure 4C). The results show that the novel ligation products occur mainly in the matrix-associated fraction, indicating IGS bound to matrix constitutes the base of higher-order rDNA loops in growing cells. To investigate the role of matrix attachment in the formation of growth-induced higher-order rDNA structure, we investigated whether signals in the MAR-loop assay were regulated by growth status and Myc. Figure 4D and E shows that MAR-loop signals involving IGS sequences increase when serum-starved HeLa and TGR-1 cells are stimulated to grow by serum addition. Furthermore, serum induction of the MAR-ligation signal requires Myc activity since the induction is reduced when serum induction is performed in the presence of the small molecule Myc inhibitor, 10058-F4 (Figure 4D and E). Finally, induction of Myc-ER by 4-HT is sufficient to strongly induce a matrix-attached loop structure in starved Rat1MycER cells (Figure 4F). We thus conclude that growth-associated attachment of different rDNA IGS regions in close proximity to one another is involved in mediating the changes in higher-order rDNA looping structures that accompany growth stimulation of cells.
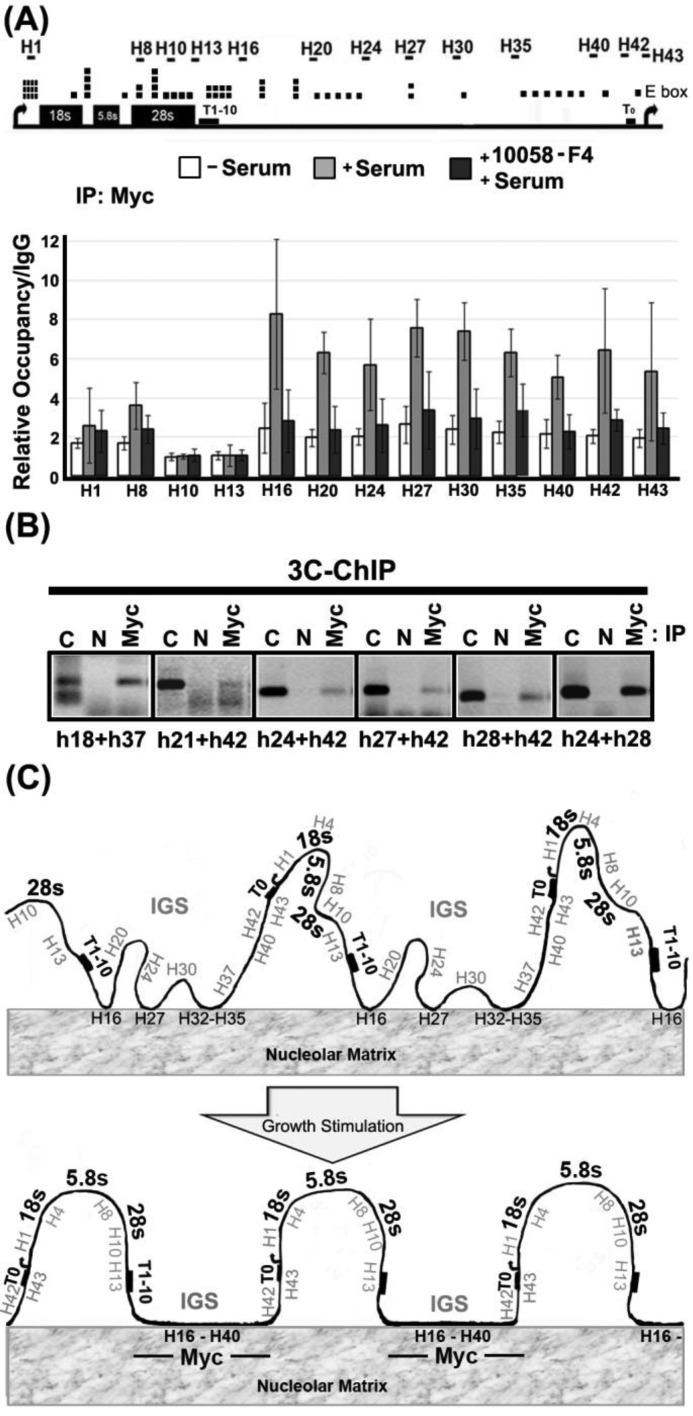
The simplest mechanisms for Myc to induce matrix attachment would involve physical interaction of Myc with the matrix:IGS assembly. To test whether Myc can bind to the IGS, we performed ChIP across the human rDNA repeat. Figure 5A shows that there is little or no binding of Myc to the human rDNA repeat in starved HeLa cells. However, serum-induced growth stimulation causes a significantly higher increase in binding of Myc to the IGS compared to the transcribed region (P = 1.07E−05) and the IGS-enhanced binding is more strongly reduced, compared to transcribed-region binding, in the presence of the Myc inhibitor, 10058-F4 (P = 6.75E−05). Myc binding to the rDNA also shows a significant tendency to be higher in the IGS than in the transcribed region in HEK293 cells (P = 0.012) (Supplementary Figure S5A). Our results confirm and extend previous results showing Myc binding to the IGS (22), although we do not observe the higher relative levels of Myc binding that were measured at the promoter and terminator sites in that study. The differences between our results and the previous study could result from the use of different cell systems. However, serum-induced binding of Myc to the IGS in this study is not seen in cells that were serum-stimulated in the presence of the 10058-F4 Myc inhibitor. The inhibitor inhibits interaction of Myc with its hetero-dimerization partner, Max, suggesting the possibility that Myc/Max could bind to E-box sites (43) that are found throughout the IGS sequence (Figure 5A, top panel). Since only a subset of rDNA genes associates with the matrix upon growth stimulation it was necessary to determine whether the observed Myc binding coincided with the set of genes involved in the formation of higher-order rDNA loops. Figure 5B shows that several fragments across the IGS region that can be ligated to one another in a 3C assay can be specifically precipitated by Myc antibodies (Myc) but not by an irrelevant control antibody (N). Thus IGS-bound Myc is bound to rDNA genes that form growth-associated higher-order rDNA looping structures.
Figure 5.
c-Myc associates with the rDNA IGS region in a growth-dependent fashion and is physically associated with the gene looping structures that are found in growing cells. (A) Quantitative real-time PCR ChIP assays to detect association of c-Myc with rDNA in starved HeLa cells before (−serum) and after addition of serum (+serum) or serum and 10058-F4 (+serum, +10058-F4). The upper panel shows a human rDNA repeat marked with locations of E boxes (canonical and non-canonical) and primer pairs for qPCR. Binding values are expressed relative to levels detected after parallel ChIP reactions with IgG from non-immunized rabbit. Error bars show standard deviation about the mean (n ≥ 4). (B) c-Myc is physically associated with rDNA-IGS-mediated gene looping structures in growing HeLa cells. The re-ligated rDNA chromatin, immunoprecipitated by a specific antibody (Myc) or agarose beads as a negative control (N), is amplified by using primer pairs throughout the rDNA IGS region (sequences of primer sets are shown in Supplementary Table S6). The expected migration of the ligation product is shown by the positive control lane (C). (C) A schematic diagram describing the main conclusions of the work. Hypomethylated (potentially active) rDNA genes associate with nucleolar matrix via the IGS in a growth- and Myc-dependent manner. Matrix association appears to be the cause of the growth-related gene looping structures that characterize the rDNA in growing cells. Myc is associated with the growth-stimulated gene looping structures via interaction with the rDNA IGS.
Figure 5C shows a schematic diagram summarizing the major findings of this work. Namely, that (i) the rDNA IGS is induced to interact with nuclear matrix upon growth stimulation; (ii) Myc is required for matrix attachment; (iii) Myc-dependent attachment of the IGS region to the matrix is a likely mechanism for the growth-stimulated changes in higher-order rDNA structure which have been reported previously (8,9). Myc binds to the rDNA IGS during growth stimulation and is associated with the IGS of genes that form growth-associated rDNA gene loop structures. The results suggest that Myc-associated rDNA gene loop structures form by association of the IGS sequences with nucleolar matrix but it should be noted that we do not yet have direct evidence for ternary complexes containing Myc, rDNA gene loop structures and nucleolar matrix. Our model also suggests that the Myc-dependent changes in rDNA structure involve mainly rDNA genes that are not silenced by methylation and which are therefore available for growth-induced transcriptional activation. Together with previous data (8), our results suggest that the mechanism described may precede induction of rRNA transcription upon growth stimulation and that it may be a part of the mechanism that leads to transcriptional activation of rDNA genes.
Accumulating evidence suggests that nuclear matrix proteins and matrix-attached DNA sequences reflect components of nuclear architecture that are important for regulation of nuclear functions in vivo. For example, recent results suggest that the MAR elements regulate epigenetic switching that is strongly correlated to gene expression (44). Further, the nuclear matrix has been suggested to play a significant role during tumorigenesis (45). Several genome-wide studies have shown preferential localization of Myc at promoter regions in mammalian cells (46), consistent with Myc's function as a transcription factor (47). However, our results showing a role of Myc-regulated nuclear matrix attachment in modulation of higher-order chromatin structure of clustered rDNA genes and their transcription are in line with reports from other nuclear contexts. For example, Yang et al. have observed association of Myc with insulators at non-promoter regions in which Myc is thought to mediate chromatin organization into higher-order topological domains that contribute to the maintenance of chromosome architecture during the cell cycle (48). The purpose of growth-regulated attachment of rDNA to the matrix may be to create so-called chromatin hubs or transcription factories, with associated gene looping structures, like those described for other nuclear contexts (49). Further studies will be required to determine whether the mechanisms we describe here are related to other mechanisms that have been shown to modulate higher-order genomic organization, such as those mediated by the CTCF protein (50).
The key role of the rDNA IGS in growth-induced matrix attachment is interesting in relation to other observations implicating IGS sequences in maintaining the integrity of rDNA clusters and in regulating rDNA transcription (51,52). Consistently, enrichment of active/repressive histone modifications has been shown to be broadly distributed along the IGS of rDNA repeats (53). Furthermore, recent work shows that non-coding RNAs transcribed from the rDNA IGS influence the intra-nuclear distribution of key proteins in response to stress (54). Since stress responses are often associated with cell growth changes it will be interesting to investigate possible linkage between IGS-encoded transcripts, nuclear matrix attachment, stress and growth. The linkage of the IGS region to stress and growth is particularly interesting in the context of the broader realization that the nucleolus is an important sensor of cellular stress, including oncogenic stress, in addition to its classical role in protein synthesis (55). Together with conclusions from others, an important conclusion of this work is that the rDNA IGS is not just an unimportant spacer DNA that separates rDNA genes from each other.
c-Myc has been shown to be a direct activator of rDNA transcription and of associated changes in higher-order structure (8,9,21,22). The growth-induced association of Myc with the IGS, and its association with juxtaposed regions of the IGS found specifically in growth-induced gene looping structures, is consistent with a direct role of Myc in tethering the IGS to matrix in growing cells. The inhibitory effect of 10058-F4 that prevents conversion of Myc to its DNA-binding form (heterodimer with Max) suggests that Myc might bind to binding sites within the rDNA IGS (Figure 5A). It is tempting to speculate that IGS-bound Myc could on principle tether the IGS to matrix by binding to one or more matrix proteins in the nucleolus but further studies will be required to test this.
Other proteins known to be involved in regulating the activity of rDNA could in principle collaborate with c-Myc. One candidate is UBF, which has been shown to be an important factor that determines the active form of rDNA genes (38). However, the level of UBF bound to the IGS is relatively low and this level does not change when rDNA transcription is activated (56). Consistently, we have also seen significantly lower levels of UBF bound to the IGS compared to the transcribed region (P = 0.003) (Supplementary Figure S5B). Taken together with previous reports, this suggests that UBF is not recruited to the IGS during transcriptional activation in a way that parallels the changes in c-Myc binding. During preparation of this manuscript, Zillner et al. reported that Tip5 (TTF1-interacting protein 5), a subunit of nucleolar remodeling complex (NoRC), functions in rRNA gene repression by re-organizing rDNA chromatin and its association with nuclear matrix (57). TTF1 is a factor associated with both transcriptional repression and activation of rRNA genes and has been reported previously to play a role in organizing the higher-order chromatin structure of rDNA genes in mouse cell lines (26). Indeed c-Myc has been shown to be able to enhance association of TTF1 to rDNA (8). Given the difference in matrix association in the TTF1 binding regions (promoter and R13/H13) of rDNA between rat and human (Figure 2), there may be species differences in the mechanisms regulating the higher-order structure of the rDNA and further studies will be required to elucidate the nature of the interplay between c-Myc and TTF1 in the organization of rRNA genes.
FUNDING
Swedish Cancer Society, the Swedish Research Council, the Baltic Sea Foundation and the Center for Biosciences Karolinska Institutet [A.W.]; Department of Laboratory Medicine, Karolinska Institutet [A.N-M].
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors wish to thank other members of the group for their support and suggestions throughout the project.
REFERENCES
- 1.Felsenfeld G. Chromatin unfolds. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 2.Brown K.E. Chromatin folding and gene expression: new tools to reveal the spatial organization of genes. Chromosome Res. 2003;11:423–433. doi: 10.1023/a:1024966424909. [DOI] [PubMed] [Google Scholar]
- 3.Mehta I.S., Figgitt M., Clements C.S., Kill I.R., Bridger J.M. Alterations to nuclear architecture and genome behavior in senescent cells. Ann. N.Y. Acad. Sci. 2007;1100:250–263. doi: 10.1196/annals.1395.027. [DOI] [PubMed] [Google Scholar]
- 4.Prieto J.L., McStay B. Nucleolar biogenesis: the first small steps. Biochem. Soc. Trans. 2005;33:1441–1443. doi: 10.1042/BST0331441. [DOI] [PubMed] [Google Scholar]
- 5.Raska I., Shaw P.J., Cmarko D. New insights into nucleolar architecture and activity. Int. Rev. Cytol. 2006;255:177–235. doi: 10.1016/S0074-7696(06)55004-1. [DOI] [PubMed] [Google Scholar]
- 6.Raska I., Koberna K., Malinsky J., Fidlerova H., Masata M. The nucleolus and transcription of ribosomal genes. Biol. Cell. 2004;96:579–594. doi: 10.1016/j.biolcel.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 7.Miller O.L., Jr, Beatty B.R. Visualization of nucleolar genes. Science. 1969;164:955–957. doi: 10.1126/science.164.3882.955. [DOI] [PubMed] [Google Scholar]
- 8.Shiue C.N., Berkson R.G., Wright A.P. c-Myc induces changes in higher order rDNA structure on stimulation of quiescent cells. Oncogene. 2009;28:1833–1842. doi: 10.1038/onc.2009.21. [DOI] [PubMed] [Google Scholar]
- 9.Shiue C.N., Arabi A., Wright A.P. Nucleolar organization, growth control and cancer. Epigenetics. 2010;5:200–205. doi: 10.4161/epi.5.3.11376. [DOI] [PubMed] [Google Scholar]
- 10.Cheutin T., O’Donohue M.F., Beorchia A., Vandelaer M., Kaplan H., Defever B., Ploton D., Thiry M. Three-dimensional organization of active rRNA genes within the nucleolus. J. Cell Sci. 2002;115:3297–3307. doi: 10.1242/jcs.115.16.3297. [DOI] [PubMed] [Google Scholar]
- 11.Linnemann A.K., Platts A.E., Krawetz S.A. Differential nuclear scaffold/matrix attachment marks expressed genes. Hum. Mol. Genet. 2009;18:645–654. doi: 10.1093/hmg/ddn394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forrester W.C., Fernandez L.A., Grosschedl R. Nuclear matrix attachment regions antagonize methylation-dependent repression of long-range enhancer-promoter interactions. Genes Dev. 1999;13:3003–3014. doi: 10.1101/gad.13.22.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostermeier G.C., Liu Z., Martins R.P., Bharadwaj R.R., Ellis J., Draghici S., Krawetz S.A. Nuclear matrix association of the human beta-globin locus utilizing a novel approach to quantitative real-time PCR. Nucleic Acids Res. 2003;31:3257–3266. doi: 10.1093/nar/gkg424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spilianakis C.G., Flavell R.A. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 15.Murrell A., Heeson S., Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 16.Horike S., Cai S., Miyano M., Cheng J.F., Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 17.Stephanova E., Stancheva R., Avramova Z. Binding of sequences from the 5′- and 3′-nontranscribed spacers of the rat rDNA locus to the nucleolar matrix. Chromosoma. 1993;102:287–295. doi: 10.1007/BF00352403. [DOI] [PubMed] [Google Scholar]
- 18.Smith H.C., Rothblum L.I. Ribosomal DNA sequences attached to the nuclear matrix. Biochem. Genet. 1987;25:863–879. doi: 10.1007/BF00502606. [DOI] [PubMed] [Google Scholar]
- 19.Keppel F. Transcribed human ribosomal RNA genes are attached to the nuclear matrix. J. Mol. Biol. 1986;187:15–21. doi: 10.1016/0022-2836(86)90402-x. [DOI] [PubMed] [Google Scholar]
- 20.Bolla R.I., Braaten D.C., Shiomi Y., Hebert M.B., Schlessinger D. Localization of specific rDNA spacer sequences to the mouse L-cell nucleolar matrix. Mol. Cell Biol. 1985;5:1287–1294. doi: 10.1128/mcb.5.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arabi A., Wu S., Ridderstrale K., Bierhoff H., Shiue C., Fatyol K., Fahlen S., Hydbring P., Soderberg O., Grummt I., et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- 22.Grandori C., Gomez-Roman N., Felton-Edkins Z.A., Ngouenet C., Galloway D.A., Eisenman R.N., White R.J. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 23.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Andersen J.S., Lyon C.E., Fox A.H., Leung A.K., Lam Y.W., Steen H., Mann M., Lamond A.I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 25.Bode J., Benham C., Knopp A., Mielke C. Transcriptional augmentation: modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements) Crit. Rev. Eukaryot. Gene. Expr. 2000;10:73–90. [PubMed] [Google Scholar]
- 26.Nemeth A., Guibert S., Tiwari V.K., Ohlsson R., Langst G. Epigenetic regulation of TTF-I-mediated promoter-terminator interactions of rRNA genes. EMBO J. 2008;27:1255–1265. doi: 10.1038/emboj.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Namciu S.J., Friedman R.D., Marsden M.D., Sarausad L.M., Jasoni C.L., Fournier R.E. Sequence organization and matrix attachment regions of the human serine protease inhibitor gene cluster at 14q32.1. Mamm. Genome. 2004;15:162–178. doi: 10.1007/s00335-003-2311-y. [DOI] [PubMed] [Google Scholar]
- 28.Weber M., Hagege H., Murrell A., Brunel C., Reik W., Cathala G., Forne T. Genomic imprinting controls matrix attachment regions in the Igf2 gene. Mol. Cell. Biol. 2003;23:8953–8959. doi: 10.1128/MCB.23.24.8953-8959.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh G.B., Kramer J.A., Krawetz S.A. Mathematical model to predict regions of chromatin attachment to the nuclear matrix. Nucleic Acids Res. 1997;25:1419–1425. doi: 10.1093/nar/25.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar P.P., Bischof O., Purbey P.K., Notani D., Urlaub H., Dejean A., Galande S. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat. Cell Biol. 2007;9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- 31.Rogozin I.B., Glazko G.V., Glazkov M.V. Computer prediction of sites associated with various elements of the nuclear matrix. Brief. Bioinform. 2000;1:33–44. doi: 10.1093/bib/1.1.33. [DOI] [PubMed] [Google Scholar]
- 32.Evans K., Ott S., Hansen A., Koentges G., Wernisch L. A comparative study of S/MAR prediction tools. BMC Bioinformatics. 2007;8:71–99. doi: 10.1186/1471-2105-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hair A., Vassetzky Y. Determination of the chromatin domain structure in arrayed repeat regions: organization of the somatic 5S RNA domain during embryogenesis in Xenopus laevis. J. Cell. Biochem. 2007;102:1140–1148. doi: 10.1002/jcb.21413. [DOI] [PubMed] [Google Scholar]
- 34.Mateyak M.K., Obaya A.J., Adachi S., Sedivy J.M. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 35.Littlewood T.D., Hancock D.C., Danielian P.S., Parker M.G., Evan G.I. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uribesalgo I., Buschbeck M., Gutierrez A., Teichmann S., Demajo S., Kuebler B., Nomdedeu J.F., Martin-Caballero J., Roma G., Benitah S.A., et al. E-box-independent regulation of transcription and differentiation by MYC. Nat. Cell Biol. 2011;13:1443–1449. doi: 10.1038/ncb2355. [DOI] [PubMed] [Google Scholar]
- 37.McEwan I.J., Dahlman-Wright K., Ford J., Wright A.P. Functional interaction of the c-Myc transactivation domain with the TATA binding protein: evidence for an induced fit model of transactivation domain folding. Biochemistry. 1996;35:9584–9593. doi: 10.1021/bi960793v. [DOI] [PubMed] [Google Scholar]
- 38.Sanij E., Poortinga G., Sharkey K., Hung S., Holloway T.P., Quin J., Robb E., Wong L.H., Thomas W.G., Stefanovsky V., et al. UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 2008;183:1259–1274. doi: 10.1083/jcb.200805146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Derenzini M., Pasquinelli G., O’Donohue M.F., Ploton D., Thiry M. Structural and functional organization of ribosomal genes within the mammalian cell nucleolus. J. Histochem. Cytochem. 2006;54:131–145. doi: 10.1369/jhc.5R6780.2005. [DOI] [PubMed] [Google Scholar]
- 40.McStay B., Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 41.Grummt I., Ladurner A.G. A metabolic throttle regulates the epigenetic state of rDNA. Cell. 2008;133:577–580. doi: 10.1016/j.cell.2008.04.026. [DOI] [PubMed] [Google Scholar]
- 42.Stancheva I., Lucchini R., Koller T., Sogo J.M. Chromatin structure and methylation of rat rRNA genes studied by formaldehyde fixation and psoralen cross-linking. Nucleic Acids Res. 1997;25:1727–1735. doi: 10.1093/nar/25.9.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grandori C., Mac J., Siebelt F., Ayer D.E., Eisenman R.N. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- 44.Galbete J.L., Buceta M., Mermod N. MAR elements regulate the probability of epigenetic switching between active and inactive gene expression. Mol. Biosyst. 2009;5:143–150. doi: 10.1039/b813657b. [DOI] [PubMed] [Google Scholar]
- 45.Sjakste N., Sjakste T., Vikmanis U. Role of the nuclear matrix proteins in malignant transformation and cancer diagnosis. Exp. Oncol. 2004;26:170–178. [PubMed] [Google Scholar]
- 46.Kidder B.L., Yang J., Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS One. 2008;3:e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 48.Yang J., Sung E., Donlin-Asp P.G., Corces V.G. A subset of Drosophila Myc sites remain associated with mitotic chromosomes colocalized with insulator proteins. Nat. Commun. 2013;4:1464–1472. doi: 10.1038/ncomms2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eivazova E.R., Gavrilov A., Pirozhkova I., Petrov A., Iarovaia O.V., Razin S.V., Lipinski M., Vassetzky Y.S. Interaction in vivo between the two matrix attachment regions flanking a single chromatin loop. J. Mol. Biol. 2009;386:929–937. doi: 10.1016/j.jmb.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 50.Phillips J.E., Corces V.G. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Sullivan A.C., Sullivan G.J., McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 2002;22:657–668. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murano K., Okuwaki M., Hisaoka M., Nagata K. Transcription regulation of the rRNA gene by a multifunctional nucleolar protein, B23/nucleophosmin, through its histone chaperone activity. Mol. Cell. Biol. 2008;28:3114–3126. doi: 10.1128/MCB.02078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zentner G.E., Saiakhova A., Manaenkov P., Adams M.D., Scacheri P.C. Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res. 2011;39:4949–4960. doi: 10.1093/nar/gkq1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Audas T.E., Jacob M.D., Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol. Cell. 2012;45:147–157. doi: 10.1016/j.molcel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 55.Boulon S., Westman B.J., Hutten S., Boisvert F.M., Lamond A.I. The nucleolus under stress. Mol. Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poortinga G., Wall M., Sanij E., Siwicki K., Ellul J., Brown D., Holloway T.P., Hannan R.D., McArthur G.A. c-MYC coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation. Nucleic Acids Res. 2011;39:3267–3281. doi: 10.1093/nar/gkq1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zillner K., Filarsky M., Rachow K., Weinberger M., Langst G., Nemeth A. Large-scale organization of ribosomal DNA chromatin is regulated by Tip5. Nucleic Acids Res. 2013;41:5251–5262. doi: 10.1093/nar/gkt218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.