Abstract
A variety of human tumors employ alternative and recombination-mediated lengthening for telomere maintenance (ALT). Human RecQ helicases, such as BLM and WRN, can efficiently unwind alternate/secondary structures during telomere replication and/or recombination. Here, we report a novel role for RECQL1, the most abundant human RecQ helicase but functionally least studied, in telomere maintenance. RECQL1 associates with telomeres in ALT cells and actively resolves telomeric D-loops and Holliday junction substrates. RECQL1 physically and functionally interacts with telomere repeat-binding factor 2 that in turn regulates its helicase activity on telomeric substrates. The telomeric single-stranded binding protein, protection of telomeres 1 efficiently stimulates RECQL1 on telomeric substrates containing thymine glycol, a replicative blocking lesion. Loss of RECQL1 results in dysfunctional telomeres, telomere loss and telomere shortening, elevation of telomere sister-chromatid exchanges and increased aphidicolin-induced telomere fragility, indicating a role for RECQL1 in telomere maintenance. Further, our results indicate that RECQL1 may participate in the same pathway as WRN, probably in telomere replication.
INTRODUCTION
Telomeres are protein–DNA complexes at the ends of linear chromosomes that provide chromosomal stability and ensure faithful segregation of genetic material. These structures not only protect the terminal chromosomal DNA from end degradation, but are also involved in its replication (1). Mammalian telomeres are composed of several kilobases of TTAGGG repeats and a complex of shelterin proteins that assist in both chromosome end protection and telomere replication (2). The telomeric repeat-binding factors TRF1, TRF2 and telomeric single-stranded (ss) binding protein, protection of telomeres 1 (POT1) directly recognize TTAGGG repeats and are further interconnected by three additional proteins TIN2, TPP1 and RAP1 to form the shelterin complex that safeguards the human telomeres (2). The ends of linear chromosomes shorten with each round of DNA replication unless actively maintained (3), and telomere attrition is associated with genomic instability, cell cycle arrest and eventually senescence or apoptosis.
Most human stem cells and reproductive cells utilize telomerase for maintaining telomere length (4), while some immortalized mammalian cell lines and a variety of human tumor cells maintain telomere length in the absence of telomerase activity by one or more mechanisms referred to as alternative lengthening of telomeres (ALT) (5). While the exact mechanism is still unclear, ALT cells exhibit both intra- and inter-telomeric homologous recombination (HR) mediated replication to maintain telomere length and involve several recombination intermediates, such as telomeric D-loops and Holliday junctions (HJs) (5,6).
Telomeres need to avoid being recognized as DNA double-strand breaks (DSBs) in order to prevent fusion with each other during normal DNA repair mechanisms. Mammalian telomeres form a higher order structure by sequestering the ss-terminus into the double-stranded (ds) telomere DNA—thus forming a T-loop and protecting the chromosome terminus (7). Further, the highly repetitive G-rich sequences on the lagging strand can adopt unusual secondary structures, such as G-quadruplexes (G4-DNA) (8). Such alternate or secondary structures must be resolved prior to DNA replication. Emerging evidence in the past decade has established roles for the human RecQ helicases in dissociating such alternate structures or intermediates during DNA replication and recombination at the telomeres (9). In particular, BLM, WRN and RECQL4 all localize to telomeres in ALT cells during S-phase and interact with the major shelterin proteins, such as TRF1, TRF2 and POT1, which all regulate the activity of these RecQ helicases and enable resolution of several secondary structures (10–17). It is not known if RECQL1 or RECQL5 are also involved in telomere maintenance; however, it has been reported that RECQL5 does not efficiently unwind telomeric D-loops (18). Recent proteomics analysis of human telomere chromatin indicates a possible association of RECQL1 with the telomeres in ALT cells (19), but no functional involvement has been reported. Here, we investigate a novel role for human RECQL1 in telomere maintenance.
RECQL1 was the first discovered and is the smallest of the five human RecQ helicases. It is the most abundant member and is expressed throughout the cell cycle (20). The crystal structure and oligomeric nature of RECQL1 has been solved recently (21,22), but its functional roles are still poorly understood. RECQL1 is not yet associated with any human disease. It binds to the origins of replication sites at the onset of S-phase and depletion of RECQL1 results in shorter replication fiber tracts, suggesting a possible role for RECQL1 in replication fork progression (23). We have recently shown that RECQL1 promotes strand exchange between homologous sequences on synthetic stalled replication forks in vitro and that loss of RECQL1 leads to activation of CHK1 and hyper-phosphorylation of replication protein A (RPA), indicating signs of replicative stress (24). RECQL1 was also proposed to play a critical role in the restart of stalled replication forks following replicative stress with camptothecin, a topoisomerase I inhibitor (25). RECQL1 may also be involved in controlling hyper-recombination events, perhaps at stalled replication forks, because loss of RECQL1 leads to elevation of sister-chromatid exchanges (SCEs) and ultimately genomic instability (26). Here, we propose a novel role for RECQL1 in telomere maintenance. RECQL1 associates with telomeres in ALT cells and can efficiently unwind telomeric D-loops. RECQL1 physically and functionally interacts with the major shelterin proteins that also regulate its helicase activity on telomeric substrates. Loss of RECQL1 induces telomere dysfunction-induced foci, leads to telomere loss and aphidicolin-induced telomere fragility, indicating a possible role for RECQL1 in telomere replication. RECQL1 also efficiently resolves telomeric HJs and loss of RECQL1 elevates telomeric sister-chromatid exchanges (T-SCEs). Further, our results indicate that RECQL1 may act in the same pathway as WRN and that loss of RECQL1 or WRN results in telomere loss specifically during S-phase, further strengthening their involvement in telomere replication.
MATERIALS AND METHODS
Recombinant proteins
Recombinant His-tagged RECQL1 protein was over-expressed and purified using a baculovirus/Sf9 insect system as previously described (26). Dual-tagged WRN protein with N-terminal 6xHis and C-terminal InteinCBD tags was expressed and purified from High FiveTM insect cells as previously described (27). Recombinant glutathione s-transferase (GST)-tagged human POT1 and histidine-tagged human TRF1 and TRF2 proteins were purified using a baculovirus expression as described previously (14,15,28). TPP1-POT1 heterodimer was a kind gift from Dr Lei Ming, University of Michigan.
Preparation and characterization of telomeric D-loops and HJ substrates
All of the unmodified oligonucleotides were from Integrated DNA Technologies (Coralville, IA) and were PAGE-purified by the manufacturer. The modified oligonucleotides (containing either 8-oxo-2-deoxyguanosine or thymine glycol) were synthesized and purified by The Midland Certified Reagent Co. (Midland, TX). All the oligonucleotides used in the study were listed in Supplementary Table S1. The telomeric repeat sequences are underlined. Both the telomeric and non-telomeric D-loops were prepared and characterized as described previously (18,29). Telomeric D-loop DL1 was constructed using the SS1 as the invading strand, and BB and BT to form the parental duplex. Similarly damaged telomeric D-loops (DL2–DL6) were generated using SS2–SS6 as the invading strands, respectively, along with BB and BT. Non-telomeric D-loop was constructed using SSmix, BBmix and BT. Static telomeric HJ substrate is prepared by using the four oligonucleotides Tel HJ B1–B4. Telomeric fork duplex consisting of 33 bp duplex and 15 nt overhang was generated using Tel G and Tel C. Thymine glycol containing fork duplex is generated by using Tel G Tg and Tel C Tg oligonucleotides.
Helicase assays
All the helicase assays were carried out (in a 10 μl reaction volume) in the buffer containing 20 mM Tris-HCl, pH 7.5, 8 mM DTT, 5 mM MgCl2, 10 mM KCl, 10% glycerol and 80 μg/ml BSA as described previously (30). Indicated amounts of POT1, TRF1, TRF2 or WRN were incubated with RECQL1 in the helicase buffer followed by the addition of the 32P-labeled substrate and incubated for 30 min at 37°C. The reactions were then stopped by the addition of equal volumes of stop buffer (35 mM EDTA, 0.9% SDS, 25% glycerol, 0.04% bromophenol blue, 0.04% xylene cyanol) and reaction products were separated by non-denaturing PAGE.
Telomeric ChIP
To investigate the presence of RECQL1 at telomeres, telomeric chromatin immunoprecipitation (ChIP) analysis was carried out as described previously (31). In brief, either the U2OS or HeLa cells were digested with trypsin, washed with phosphate buffered saline (PBS), fixed in 1% formaldehyde in PBS for 60 min at room temperature and lysed in 1% SDS, 50 mM Tris-HCl, pH 8.0, 10 mM EDTA at a density of 107 cells/ml. Lysates were sonicated to obtain chromatin fragments and further diluted in the dilution buffer. Indicated antibodies (either RECQL1 or TRF1 at 5 μg) were added to the reaction mixture, incubated at 4°C overnight on a rotator followed by the addition of protein G agarose beads. After extensive washes, bound chromatin was separated from the protein G beads with 200 μl of elution buffer (1% SDS, 0.1 M Na2CO3) twice. After addition of 16 μl of 5 M NaCl, cross-links were reversed by incubation of the reaction mixture at 65°C for 4 h followed by treatment with 20 μg DNase-free RNase A. Samples were then digested with 40 μg proteinase K at 37°C for 60 min and phenol extracted. The precipitated DNA was denatured at 95°C for 5 min, and dot blotted onto Hybond membranes. Membranes were treated with 1.5 M NaCl–0.5 N NaOH for 15 min and with 1 M NaCl–0.5 M Tris-HCl (pH 7.0) for 15 min. Hybridization was performed with radiolabeled (TTAGGG)5 or Alu probes. Membranes were washed four times in 2× saline-sodium citrate buffer (SSC) with 0.1% SDS, and signals were visualized by Phosphor Imager. The percentage of precipitated DNA was quantitated by Image Quant software (Molecular Dynamics). For experiments with TRF2-depletion, two independent siRNA against TRF2 from Qiagen (cat no. SI00742637 and SI04261159, respectively) were employed. Negative control siRNA was also purchased from Qiagen (cat no. 1027310).
Cell culture and stable RECQL1 knockdown cells
U2OS, HeLa and HeLa 1.2.11 cells were maintained in high glucose Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Sigma) and 1% penicillin-streptomycin at 37°C in 5% CO2. Stable RECQL1 knockdown (KD) cells were generated using the lentiviral shRNA against RECQL1 (TRCN0000289591) from Sigma Aldrich as described previously (24). Similarly, stable WRN KD cells were also generated by using the shRNA against WRN (TRCN0000004899) from Sigma Aldrich. Transient depletion of RECQL1 was performed by using the target-specific siRNA against RECQL1(5′-GCAAGGAGAUUUACUCGAA-3′) from Dharmacon as described previously (26). RECQL1 KD cells were cultured for at least 8–10 passages before subjected to quantitative fluorescence in situ hybridization (Q-FISH) or flow cytometry-cell sorting fluorescence in situ hybridization (flow-FISH) analysis. For the double depletion experiments, siRNA-RECQL1 was used to transiently knockdown RECQL1 in lentiviral stable WRN KD cells.
Fluorescence in situ hybridization (Q-FISH)
Telomeric Q-FISH was performed using peptide nucleic acid probes as described previously (32). Both the scrambled and RECQL1 KD cells (U2OS or HeLa 1.2.11cells) were treated with 200 μl of 0.5% colcemid for 3 h. Cells were then harvested and immediately incubated in 75 mM KCl for 20 min at 37°C followed by fixation in ice-cold (3:1) methanol and glacial acetic acid. Metaphase spreads were then treated with pepsin (1 mg/ml, Sigma) and fixed with 4% formaldehyde. The slides containing the spreads were then dehydrated by ethanol wash (70, 90 and 100%, respectively) and hybridized with a Cy3-labeled p-nitroanilide (CCCTAA)3 C-telomere probe (0.3 mg/ml, Panagene) in hybridization buffer (70% formamide, 2.5% blocking protein, 10 mm Tris, pH 7.4 and 2.5 mm MgCl2). Then the slides were washed with washing solution I (70% formamide, 1% BSA and 10 mm Tris, pH 7.4) and washing solution II (1× TBS and 0.7% Tween 20). After dehydration, the spreads were counterstained with 4',6-diamidino-2-phenylindole (DAPI) and mounted with ProLong Gold anti-fade reagent (Invitrogen). Images were captured using CytoVisionTM software (Applied Imaging Corp.) on a fluorescence microscope (Axio2, Carl Zeiss, Germany) at ×100 magnification. Q-FISH was analyzed for telomere length analysis by measuring the average fluorescence of telomere spots and analyzed by TFL-TELO software (32). At least 20 metaphases were analyzed from three different sets of RECQL1 KD and scrambled U2OS and HeLa 1.2.11 cells. The metaphase spreads were also scored manually for any telomeric abnormalities (i.e. presence of signal free ends (SFEs) and telomere doublets).
Chromosome Orientation FISH (CO-FISH)
Chromosome orientation FISH (CO-FISH) was used to measure the frequency of T-SCE as described previously (33). Cells were treated with 5-bromo-2-deoxyuridine (BrdU) and 5-bromo-2-deoxycytidine (BrdC) (Sigma) (3:1) at a final concentration of 1 × 10−5 m for 16 h, followed by incubation with 0.1 mg/ml colcemid (Invitrogen) for 4 h. The cells were then suspended in prewarmed 75 mM KCl hypotonic solution for 20 min at 37ºC followed by 3:1 methanol/acetic acid fixation. Metaphase spreads were prepared by dropping the fixed suspension cells onto glass slides and then air-drying the slides. The slides were rehydrated in PBS for 15 min followed by 0.5 mg/ml RNAase treatment at 37ºC for 10 min and subsequent 0.5 µg/ml Hoechst 33258 (Sigma) staining in 2× SSC for 15 min at room temperature (RT). Slides were then rinsed with 2× SSC and exposed to 365 nm UV light (Stratalinker 1800 UV irradiator) for 30 min, and then digested the DNA with 10 U/µl exonuclease III (Promega) for 10 min at RT. Slides were then washed with PBS and series of ethanol dehydration processes, 70%, 90% and 100%, 5 min each. TelC-cyc3 (Panagene) hybridization mixture A (70% formamide (Sigma), 0.25% blocking reagent (Roche), 5% MgCl2 in 10 mM Tris (pH 7.2)) was added to the slides and incubated 2 h in the dark, humid chamber. Slides were washed twice (each 15 min) in wash buffer 1 (70% formamide, 0.1% BSB, 10 mM Tris (pH 7.2)) and then three times in wash buffer 2 (0.1M Tris, 0.15M NaCl and 0.08% Tween-20) 5 min each. Slides were mounted with vectashield (Vector) containing DAPI and stored protected from light. The images were taken as described above under ‘fluorescence in situ hybridization (Q-FISH).’ A chromosome with more than two telomeric DNA signals by Cy3-labeled C-telomere probe was scored as T-SCE-positive. The frequencies of T-SCEs were obtained from at least 50 metaphases from three different RecQ KD and scrambled U2OS cells.
Flow-FISH
Cells (1X106) were trypsinized and suspended in PBS for each sample. All samples were spun down at max speed for 30 s and then resuspended in TelG-FITC probe (Panagene) containing hybridization solution (70% formamide, 1% BSA, 20 mM Tris (pH 7.4)). Cells were denatured at 86ºC for 10 min and then hybridized for 2 h in the dark at RT. Cells were washed twice with wash solution 1 (70% formamide, 1% BSA, 20 mM Tris (pH 7.4), 0.1% Tween 20)) followed by a one time wash with solution 2 (1× PBS, 0.1% BSA, 0.1% Tween 20). Cells were resuspended in staining solution (1× PBS, 0.1% BSA, 10 U/ml RNAse A, 0.6 µg/ml 7AAD (Sigma)) for 2 h at RT. Samples were stored at 4ºC and protected from light. Samples were analyzed by BD FACSCantoII. For flow-FISH experiments, a t-test was performed on triplicates for each cell type from three independent experiments using sigma-plot and the p-value was obtained as indicated.
Co-immunoprecipitation (co-IP)
For in vivo co-IP, U2OS cells were lysed in cell lysis buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 1 mM PMSF and 1% Triton X-100 and 1× protease inhibitors (Roche Applied Science)) and co-IP was performed as described elsewhere (34) using an anti-RECQL1 antibody (anti-rabbit, Santa Cruz Inc.) and probing for anti-TRF2 antibody (anti-mouse, Imgenex). Lysis was performed in the presence of ethidium bromide (50 μg/ml) by rotating end over end for 30 min at 4ºC and centrifuged at 14 000 g for 20 min. The supernatants or whole cell extracts were collected and subjected for preclearing with Protein A/G beads (Thermo Fisher Scientific, Waltham, MA, USA). The cell extracts were incubated overnight at 4ºC with anti-RECQL1 antibody or with rabbit IgG (Santa Cruz Inc., Santa Cruz, CA, USA). Immune complexes were subsequently incubated with Protein A/G agarose beads for 5 h at 4ºC. The beads were then washed 4–5 times with 50 mM Tris pH 7.4, 150 mM NaCl and 0.2% Triton X-100, and finally resuspended in 20 μl of 2× SDS loading buffer and denatured at 95°C for 5 min to release bound proteins. The immunoprecipitates were analyzed by western blotting and probed with custom-made mouse monoclonal antibody for TRF2. Transient depletion of TRF2 was performed in U2OS cells using the target-specific siRNA (purchased from Dharmacon) against TRF2: 5′-AGGCUGGAGUGCAGAAAUAdTdT-3′. Myc-POT1 was transiently over-expressed in U2OS cells by using pLPC myc hPOT1 plasmid from Addgene (Plasmid 12387) and myc pull down was performed using the mouse monoclonal c-Myc antibody (9E10) from Santa Cruz (sc-40) as described above. Rabbit anti-Myc tag antibody from Abcam (ab9106) was used for the western blotting.
In vitro immunoprecipitation
Both RECQL1 (10 pmol) and TRF2 (20 pmol) were preincubated with either rabbit IgG or anti-RECQL1 antibody in 200 μl of helicase buffer with 5 μg/ml BSA and incubated at 4°C for 90 min. Protein A/G beads were subsequently added to the protein mix and incubated at 4°C for 2 h. The beads were then isolated, washed five times in 500 μl of helicase buffer containing 150 mM NaCl and 0.1% BSA, and processed as described above.
Immunofluorescence
U2OS or HeLa cells (both scrambled and RECQL1 KD cells, as indicated) were plated on Lab-Tek II chambered glass slides (Thermo-Fisher Scientific) at a density of 20 000 cells/chamber grown overnight. Cells were treated with 2 mM hydroxyurea (HU; Sigma) for 16 h, as indicated. Cells were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature, washed with PBS, and permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature. Following a final wash with PBS, cells were blocked for 1 h at room temperature with 5% FBS in PBS. Then the cells were treated with primary antibodies overnight at 4°C, followed by washes in PBS (three times, 5 min each) and finally incubated with secondary antibodies for 1 h at room temperature. Then the cells were washed again with PBS (five times for 3 min) and treated with Vectashield mounting medium containing DAPI (Vector Laboratories). The following primary antibodies were used: anti-rabbit 53BP1 (Novus Biologicals, 1:100), anti-mouse TRF1 (Abcam TRF-78, 1:50), anti-mouse TRF2 (Imgenex, 1:50), anti-phospho-RPA32 (S4/S8) antibody (Bethyl Laboratories, 1:200), anti-rabbit RPA32 (Genetex, 1:100) and anti-rabbit RECQL1 antibody (Santa Cruz Inc., 1:200). Secondary antibodies used were: donkey anti-rabbit Alexa Fluor 488 (1:1000) and donkey anti-mouse Alexa Fluor 647 (1:1000) (Invitrogen), diluted in blocking solution (5% FBS). Images were captured on a Nikon Eclipse TE2000 confocal microscope (×40 magnification) with a Hamamatsu C9100–13 camera. Data acquisition and analysis were performed using Volocity 5.5 software (PerkinElmer Life Sciences). Images of individual representative cells were cropped from the original images and are shown in Figures 5 and 9 and Supplementary Figure S1. The Pearson coefficients of these individual foci were calculated using Volocity software. For a better representation, co-localization channels (positive products of the differences of the means for the two channels) were generated.
Figure 5.
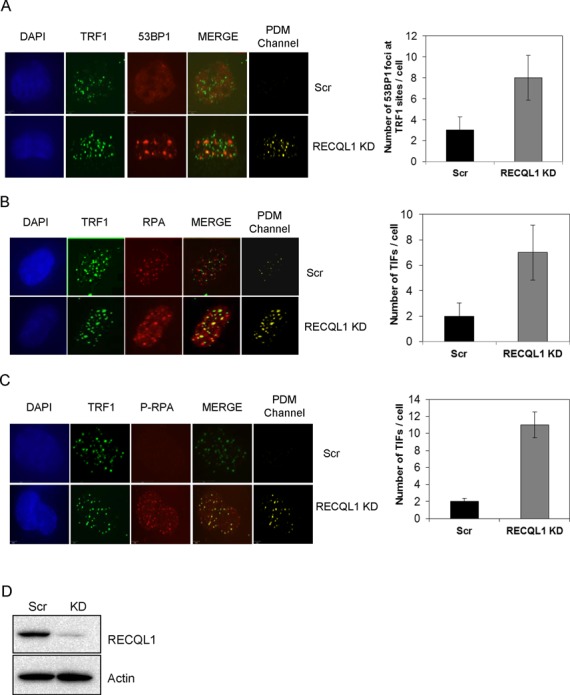
Stable depletion of RECQL1 accumulates telomere dysfunction-induced foci. Stable depletion of RECQL1 was performed in U2OS cells using lentiviral shRNA. Both RECQL1 KD and scrambled cells were analyzed for accumulation of 53BP1 foci (A), RPA32 (B) and phospho-RPA (C) at telomere regions (TRF1 sites) by immunofluorescence. Images were shown to the left with the corresponding co-localization channels, generated by Volocity 5.0 (Improvision, PerkinElmer), and the representative plots were shown to the right. (D) Western blot showing knockdown efficiency of RECQL1 in U2OS cells.
Figure 9.
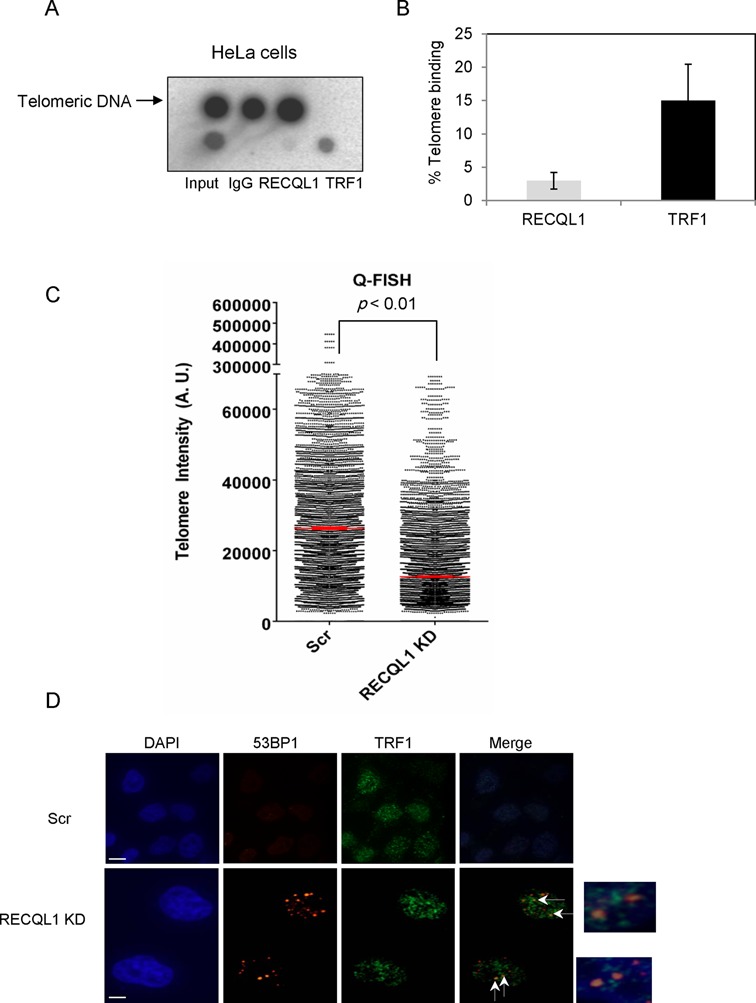
Association of RECQL1 to telomeres in telomerase-positive (HeLa) cells. Telomeric ChIP was performed in HeLa cells using anti-RECQL1 antibody with the anti-TRF1 antibody used as a positive control. (A) Dot blot showing the precipitated DNA and hybridized with the telomeric probe. (B) The graph showing the signal/input ratio from the RECQL1 IPs as a function of the quantity of chromatin (input) used in the assay. The results were visualized by Phosphor Imager and quantitated with Image Quant software (Molecular Dynamics) and are expressed in % binding to the telomeres. (C) Q-FISH analysis was performed in RECQL1-depleted HeLa 1.2.11 cells and showing the median telomere length in both scrambled and RECQL1 KD cells. (D) Immunofluorescence showing dysfunctional telomeres or accumulation of 53BP1 foci at TRF1 sites in RECQL1-depleted HeLa 1.2.11 cells.
RESULTS
Human RECQL1 associates with telomeres in ALT cells
Recent proteomics analysis of telomeric binding proteins indicates possible association of human RECQL1 at telomeres in ALT cells (19). To test this association, telomeric ChIP analysis was performed in U2OS (ALT) cells, in which RECQL1-associated telomeric DNA was detected by hybridization using α-32P-labeled telomere-specific probe and visualized by dot blot. A small but significant fraction of telomeric DNA was detected in RECQL1 immunoprecipitations (IPs) from U2OS cells, and a control IP carried out using the same quantity of non-immune IgG yielded no telomeric DNA (Figure 1A). Quantification of these results indicates that the signal for RECQL1 is about 8% of the input DNA. An increase (to about 14% of the input DNA) in the signal was obtained after treatment with HU (Figure 1C). A control IP with TRF1 antibody was used as a positive control for the presence of telomeric DNA and as much as 20–25% of the input signal was detected (Figure 1C). These ChIP samples were also confirmed by western blotting for the presence of RECQL1 and TRF1 (Figure 1B). To control for the co-precipitation of nonspecific DNA, a probe for the Alu repeat sequence was used as a negative control. The signal/input ratio for the RECQL1 IP using the Alu probe was less than 0.1% of that using the telomere probe (Figure 1A), indicating that RECQL1's association with telomeric DNA is not a nonspecific finding. These observations indicate that RECQL1 associates with telomeres in ALT cells.
Figure 1.
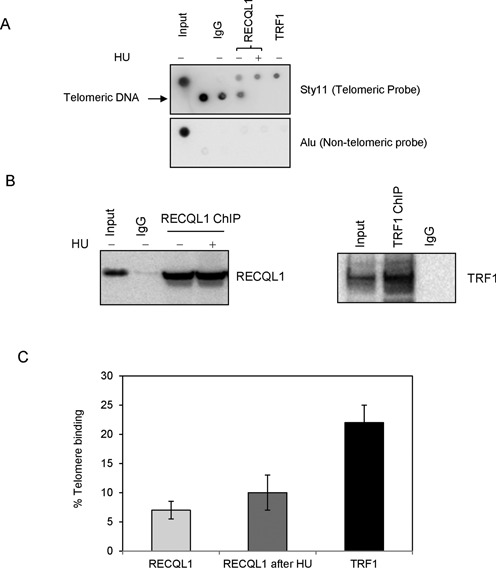
RECQL1 associates to telomeres in ALT cells. (A) Telomeric ChIP was performed in U2OS cells using anti-RECQL1 antibody in the presence and absence of HU (treated with 5 mM for 18 h). The precipitated DNA was hybridized with the sty-11 telomeric probe (top) or an Alu probe (bottom). Dot blots were performed using anti-RECQL1 antibody, normal IgG and anti-TRF1 antibody that is used as a positive control. (B) Telomeric ChIP samples were confirmed by western blotting for the presence of RECQL1 and TRF1. (C) The graph showing the signal/input ratio from the RECQL1 IPs as a function of the quantity of chromatin (input). The results were visualized by Phosphor Imager and quantitated with Image Quant software (Molecular Dynamics) and are expressed in % binding to the telomeres. The error bars represent the mean and standard deviation from three independent ChIP assays.
RECQL1 actively unwinds both damaged and undamaged telomeric D-loops in vitro
Telomeres are protected in the form of T-loops and these structures must be unwound in order to resolve topological stress and to allow for efficient and complete replication of telomeric DNA. To investigate if RECQL1 can unwind telomeric D-loops, both telomeric and non-telomeric D-loops were generated. RECQL1 efficiently disrupted a telomeric D-loop (DL1) and the activity was significantly higher than for a non-telomeric D-loop (DLmix) (Figure 2A). The unwinding activity of RECQL1 was less than 10% at a concentration of 20 nM and only about 30% at 50 nM on DLmix compared to about ∼50% of unwinding activity observed at 20 nM of RECQL1 and almost complete unwinding (> 90%) at 50 nM on DL1 (Figure 2B). To explore the possibility that the observed preference of RECQL1 for unwinding telomeric substrates stems from enhanced binding of RECQL1 to telomeric DNA, electrophoretic mobility shift assays (EMSA) were performed. Our results indicate that RECQL1 has a higher binding affinity toward telomeric D-loops than non-telomeric D-loops (Supplementary Figure S1A). The binding dissociation constant (Kd) was ∼10 nM for DL1 and a Kd > 50 nM for DLmix (Supplementary Figure S1A).
Figure 2.
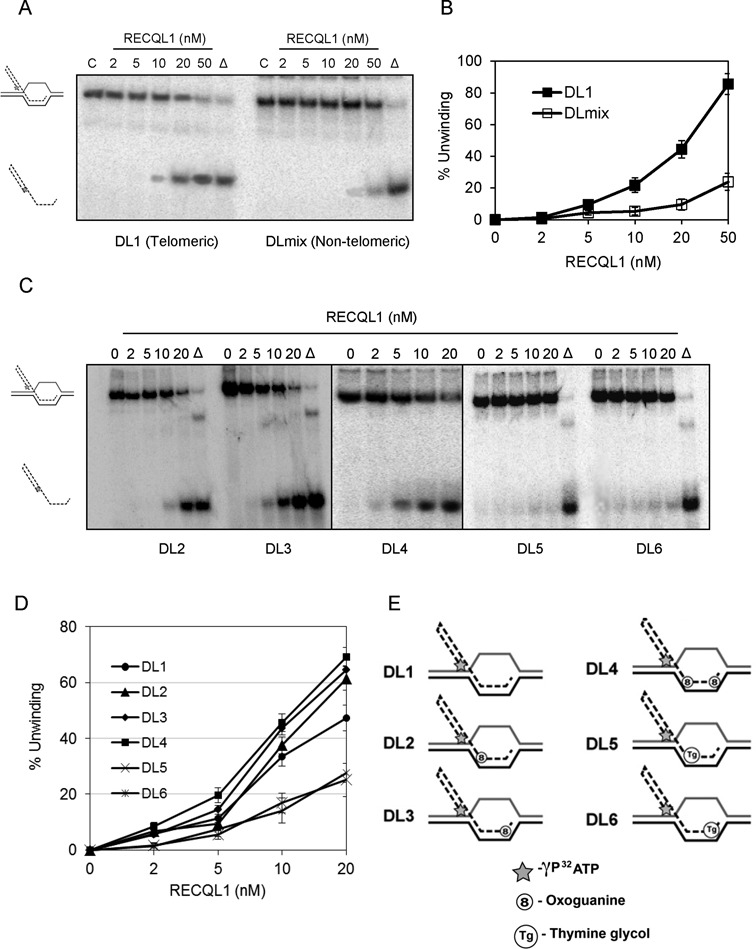
RECQL1 actively unwinds both damaged and undamaged telomeric D-loops. (A) A representative gel showing RECQL1 helicase activity on both telomeric (DL1) and non-telomeric (DLmix) D-loops. (B) Plot showing unwinding efficiency of RECQL1 on both DL1 and DLmix. (C) Representative gels showing RECQL1 helicase activity on telomeric D-loops containing 8-oxoguanine located toward either 3′-end (DL2), 5′-end (DL3) or both the ends (DL4) of the invading strand and thymine glycol located toward either 3′-end (DL5) or 5′-end (DL6) of the invading strand. (D) Plot comparing the unwinding efficiency of RECQL1 on both undamaged and damaged telomeric D-loops (DL1-DL6). (E) Schematic representation of both undamaged (DL1) and damaged telomeric D-loops (DL2–DL6) containing either an 8-oxoguanine (8) or a thymine glycol (Tg) located toward 5′- or 3′- end of the invading strand. The asterisk represents the γP32 ATP labeling at 5′-end of the invading strand.
Telomere integrity is affected by either oxidative DNA damage or a replication block. Therefore, we wanted to further investigate if RECQL1 could disrupt damaged telomeric D-loops containing an oxidized base, 8-oxogunanine, or by a thymine glycol lesion that blocks DNA replication (35). Our previous observations indicate that both BLM and WRN can efficiently disrupt telomeric D-loops containing either of these lesions (18,29), while RECQL4 shows a slight preference for thymine glycol containing D-loops (29). Our present results show that RECQL1 more efficiently disrupts 8-oxoguanine than thymine glycol containing telomeric D-loops (Figure 2C). RecQ helicases have 3′–5′ polarity and we found that RECQL1 efficiently unwound 8-oxoguanine containing D-loops (∼60–70% of activity at a concentration of 20 nM) irrespective of whether the lesion was placed toward 5′-end (DL2), or 3′-end (DL3) or located at both the positions (DL4) of the invading strand (Figure 2D). A schematic representation of these structures is shown in Figure 2E. On the other hand, RECQL1 showed poor unwinding activity of < 25% at 20 nM with thymine glycol placed either toward 5′-end (DL5) or 3′-end (DL6) of the invading strand (Figure 2D). Nevertheless, RECQL1 can unwind thymine glycol containing D-loops (both DL5 and DL6) and shows efficient unwinding activity at higher concentrations of about 100–200 nM (Supplementary Figure S1B). Collectively, these observations indicate novel substrate specificity for RECQL1 and that RECQL1 can efficiently unwind telomeric D-loops.
RECQL1 physically and functionally interacts with TRF2
Both BLM and WRN localize to the telomeres in ALT cells and interact with TRF1 and TRF2 (15,17). In particular, BLM co-localizes with TRF2 during late S- and G2-phases of the cell cycle and overlaps with BrdU incorporation suggesting a role for BLM in telomeric DNA synthesis. To test the telomeric localization of RECQL1 in ALT cells, we examined its possible interaction with shelterin proteins. We initially tested if RECQL1 is enriched at TRF2 sites in response to replicative stress. However, our immunofluorescence experiments performed in U2OS cells indicate that RECQL1 does not form clear foci but rather shows a pan nuclear staining (Supplementary Figure S1C), probably due to its abundant expression. To our knowledge, RECQL1 has not been shown to form punctate foci even in response to DNA damage, unlike BLM or WRN. This presents some difficulty in assessing its localization in ALT cells. Nevertheless, our co-IP experiments indicate that RECQL1 specifically complexes with TRF2 (Figure 3A), but not with TRF1 (data not shown). The interaction was unchanged following replicative stress with HU (Figure 3A). The reverse co-IP was also performed using the TRF2 antibody and a consistent signal for RECQL1 was found in the TRF2 IP (Figure 3B). To further confirm this interaction, transient depletion of TRF2 was performed using siRNA against TRF2 and the interaction was lost in TRF2-depleted cells (Figure 3C), confirming the physical interaction between RECQL1 and TRF2. Our in vitro immunoprecipitation experiments also indicate a direct physical interaction between the two purified proteins (Figure 3D). Collectively, these observations indicate a physical and a direct interaction of RECQL1 with TRF2. Further, to investigate if TRF2 is required for the recruitment of RECQL1 to the telomeres, a telomeric ChIP analysis was performed in TRF2-deleted cells using two independent siRNAs (Supplementary Figure S1D and E). The results indicate that there was no significant change in the association of RECQL1 to the telomeres, suggesting that the telomeric binding of RECQL1 may be independent of TRF2.
Figure 3.
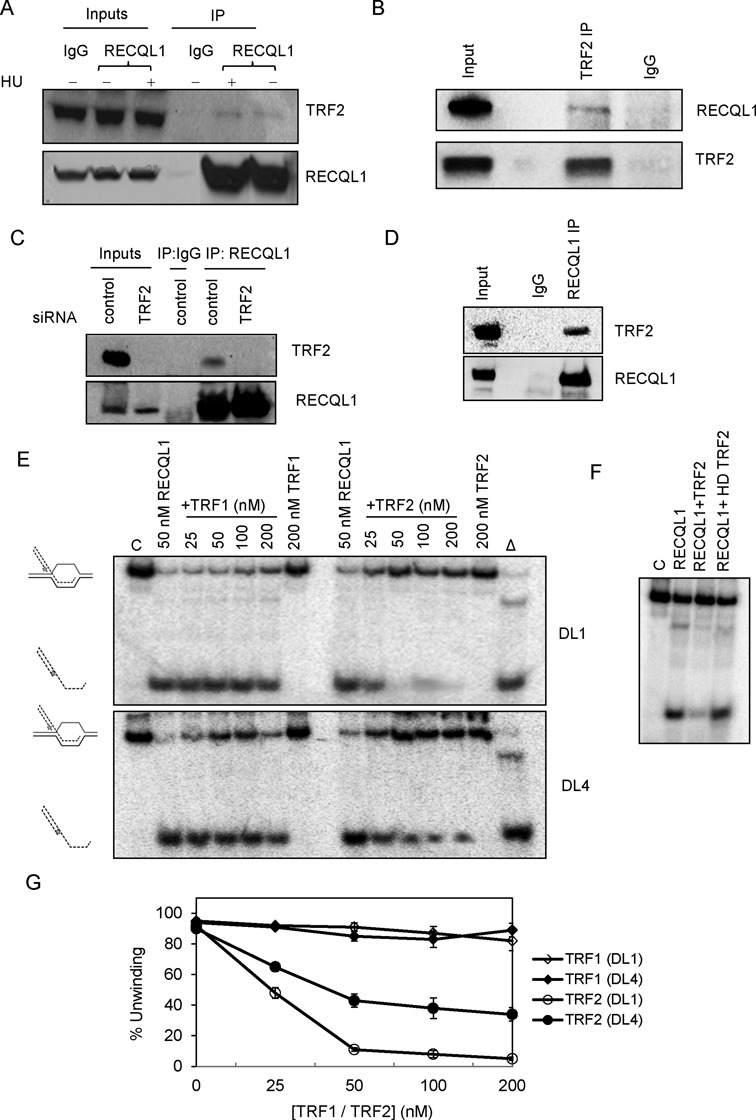
RECQL1 physically and functionally interacts with TRF2. (A) Co-immunoprecipitation (co-IP) of RECQL1 and TRF2 from whole cell extracts of U2OS cells, performed in the presence of ethidium bromide, both in the presence and absence of HU (treated with 5 mM for 18 h). (B) Reverse co-IP was performed with the antibody against TRF2 and blotted for RECQL1. (C) Co-IP of RECQL1 and TRF2 from whole cell extracts of U2OS cells transiently depleted with siRNA against TRF2. All the co-IP assays were performed in the presence of ethidium bromide to ensure that the interaction is not DNA mediated. (D) In vitro co-IP performed using both the purified proteins TRF2 and RECQL1. (E) Helicase activity of RECQL1 (50 nM) performed in the presence of increasing concentrations of either TRF1 (25–200 nM) or TRF2 (25–200 nM) using an undamaged telomeric D-loop (DL1) or a damaged telomeric D-loop containing 8-oxoguanine (DL4). (F) Effect of heat denatured (HD) TRF2 on the helicase activity of RECQL1 using the telomeric D-loop. (G) Plot showing the effect of increasing concentrations of both TRF1 and TRF2 on the helicase activity of RECQL1.
To test if there is any functional interaction between the two proteins, unwinding activity of RECQL1 was analyzed in the presence of increasing concentrations of TRF2. Our results indicate that TRF2 inhibits the helicase activity of RECQL1 on both telomeric D-loops (Figure 3E) and fork duplexes (Supplementary Figure S2A). At a molar ratio of 1:0.5 (RECQL1:TRF2), more than 50% inhibition of the activity of RECQL1 was observed on undamaged D-loop, DL1, and molar ratios of equal to or greater than 1:1 (RECQL1:TRF2) resulted in complete inhibition of RECQL1 (Figure 3E). Heat denatured TRF2 did not inhibit RECQL1 under similar buffer conditions (Figure 3F). To determine if the inhibition was due to the competition in DNA binding, we studied the DNA binding efficiency of both the proteins and analyzed the binding coefficients (Kd values) toward the telomeric D-loops. We have previously reported EMSA or binding patterns of both TRF1 and TRF2 with these telomeric D-loops (29). As observed by us and others, the DNA binding ability of TRF2 is technically challenging to study because of lack of detectable products at lower concentrations (12,14). Our present results similarly indicate that TRF2 binds the telomeric D-loops weakly, with a Kd ∼ 100 nM, while RECQL1 binds very efficiently with Kd ∼ 10 nM under similar buffer conditions (Supplementary Table S2). On the other hand, TRF1, which showed higher binding affinity than TRF2 with a Kd ∼ 50 nM (Supplementary Table S2), did not affect the helicase activity of RECQL1, indicating that the inhibition is specific to TRF2 (Figure 3E). Hence, we speculate that the inhibition is due to a functional interaction with TRF2 and not merely a competition for DNA binding. The inhibition by TRF2 is partially overcome on damaged D-loops containing an 8-oxogunanine, DL4 (Figure 3E). Even at a molar ratio of 1:4 (RECQL1:TRF2), RECQL1 showed an activity of ∼40% on DL4 compared to the complete inhibition observed on DL1 (Figure 3G). Similar results were obtained using thymine glycol containing D-loops (data not shown). On the other hand, neither TRF2 nor TRF1 affected the helicase activity of RECQL1 on non-telomeric D-loops (Supplementary Figure S2B). Collectively, these observations indicate that RECQL1 physically interacts with TRF2 and that TRF2 in turn actively modulates the helicase activity of RECQL1 on telomeric substrates.
RECQL1 physically interacts with POT1 and POT1 efficiently stimulates RECQL1 on telomeric substrates containing thymine glycol
Another major interaction partner for the human RecQ helicases (apart from TRF2) is POT1, the telomeric ss-binding protein, and it was shown that POT1 stimulates both BLM and WRN on telomeric substrates (16,18). To investigate if RECQL1 also interacts with POT1, Myc-POT1 was over-expressed in U2OS cells and a myc pull down was performed. Our results indicate a possible interaction between RECQL1 and POT1 (Figure 4A). To investigate any functional interaction, the effect of POT1 on RECQL1 helicase activity was analyzed. The results indicate that POT1 stimulates RECQL1 on telomeric D-loops and that the stimulation is higher on thymine glycol containing D-loops (compare Figure 4B and C) on which RECQL1 alone shows the least preference. At a molar ratio of 1:2 (RECQL1:POT1), we observed that the helicase activity of RECQL1 was increased ∼3-fold (Figure 4B, lane 5) and at a molar ratio of 1:5 (RECQL1:POT1) the helicase activity of RECQL1 was increased 4-fold (lane 6) on an undamaged telomeric D-loop (DL1) (Figure 4B and D). Similar modest stimulation of RECQL1 (about 2–3-fold at a molar ratio of 1:5, RECQL1:POT1) was observed using the telomeric D-loop containing 8-oxoguanine, DL4 (Figure 4B, lanes 10–14). Whereas using thymine glycol containing D-loops (DL5 and DL6), we observed significantly higher stimulation (Figure 4C). A 2.5-fold increase in the helicase activity of RECQL1 (Figure 4C, lanes 4 and 11) was observed at a molar ratio of 1:1 (RECQL1:POT1) and at molar ratios of 1:2.5 and 1:5, the activity was further increased to 5-fold (lanes 5 and 12) and almost 7-fold (lanes 6 and 13), respectively (also refer Figure 4D). Similar results were obtained on telomeric fork substrates and the stimulation is higher on thymine glycol containing fork duplex (Figure 4E and F). For example, at a molar ratio of 1:1 (RECQL1:POT1) the unwinding activity of RECQL1 was increased by 3-fold using an undamaged fork substrate when compared to a more than 5-fold increase on a thymine glycol containing fork duplex (Figure 4E, compare lanes 4 and 11). Thymine glycol is a potential blocker for DNA replication (35) and collectively our observations indicate that POT1 might help RECQL1 overcome thymine glycol lesions and enable replication fork unwinding at telomeres.
Figure 4.
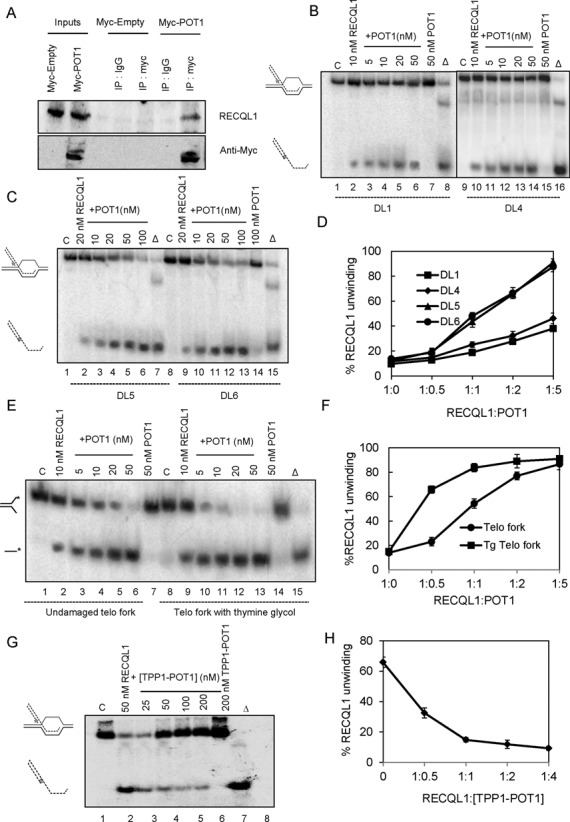
Functional interaction of RECQL1 and POT1 to unwind D-Loops containing a thymine glycol lesion. (A) Myc-POT1 was over-expressed in U2OS cells and a myc pull down was performed and blotted for endogenous RECQL1. (B) Helicase activity of RECQL1 performed in the presence of indicated and increasing concentrations of POT1 using an undamaged telomeric D-loop (DL1) and damaged telomeric D-loop containing 8-oxo guanine (DL4). (C) Helicase activity of RECQL1 performed in the presence of indicated and increasing concentrations of POT1 using telomeric D-loop containing thymine glycol toward either 3′-end (DL5) or 5′-end (DL6) of the invading strand. (D) Plot showing the % increase in RECQL1 unwinding of telomeric D-loops with increasing concentrations of POT1. (E) Helicase activity of RECQL1 (10 nM) on a telomeric fork duplex with and without thymine glycol lesion in the presence of increasing concentrations of POT1 (5–50 nM). (F) Plot showing % increase in RECQL1 unwinding of telomeric fork substrates. (G) Helicase activity of RECQL1 (50 nM) performed in the presence of increasing concentrations of TPP1-POT1 heterodimer (25–200 nM) using an undamaged telomeric D-loop (DL1). (H) Plot showing the effect of TPP1-POT1 on RECQL1 unwinding of an undamaged telomeric D-loop.
Studies indicate that POT1 can independently bind to telomeric substrates in vitro (18), and DNA binding by human POT1 is enhanced by the N-terminus of TPP1, despite the fact that TPP1 has no discernible DNA-binding activity by itself (36). POT1 plays an important role in the protection of telomeres and is essential for repression of replication stress-induced ATR signaling (37), but this function requires the TPP1-POT1 heterodimer complex (38). The TPP1-POT1 heterodimer complex functions as a unit to protect human telomeres by regulating telomerase access to telomere DNA and also by inhibiting DNA damage response at telomeres (36,38). Hence, we wanted to investigate the effect of TPP1-POT1 heterodimer on the unwinding activity of RECQL1. In contrast to the POT1 findings, the TPP1-POT1 heterodimer inhibits RECQL1 on telomeric D-loops (Figure 4G and H).
Stable depletion of RECQL1 induces accumulation of telomere dysfunction-induced foci
Next, we determined if there were any telomere defects in the absence of RECQL1. We have extensively characterized stable RECQL1 KD cells (24) and shown that loss of RECQL1 leads to a replication-stress-like phenotype. Here, we tested if loss of RECQL1 would lead to further accumulation of DNA damage at telomeres. RECQL1 KD cells accumulated over 2–3-fold more 53BP1 foci per cell at TRF1 sites than the scrambled cells (Figure 5A), indicating that stable depletion of RECQL1 induces accumulation of DNA DSB foci (53BP1) at telomeres in U2OS cells. Further, we also observed accumulation of RPA (Figure 5B) and hyper-phosphorylation of RPA (Figure 5C) at telomeric sites in RECQL1 KD cells. A positive co-localization channel generated using the Volocity 5.0 was also shown. The telomeric RPA foci represent accumulation of recombinogenic ssDNA that leads to replicative stress and activation of DNA damage signaling at telomeres (39,40). Greater than 4-fold accumulation of telomeric RPA foci were observed in RECQL1 KD cells compared to the scrambled cells (Figure 5B and C). The knockdown efficiency of RECQL1 is shown in Figure 5D and is near complete.
Loss of RECQL1 leads to telomere loss and aphidicolin-induced telomere fragility
Defects in telomere replication, improper resolution of T-loops and hyper-recombination events all affect telomere length maintenance in ALT cells (41). Consistently, loss of BLM leads to telomere shortening in ALT cells (42). Loss of WRN and TERC, the RNA component of telomerase, in mTerc−/− Wrn−/− cell lines also show telomere shortening and engagement of the ALT pathway (43). We observed a similar phenotype of telomere shortening in RECQL1 depleted ALT cells by telomere Q-FISH (Figure 6A) and by flow-FISH (Figure 6B). The median telomere length was significantly shorter in RECQL1 KD cells compared to the scrambled cells using both methods of analysis (Figure 6A and B). The metaphase spreads from the Q-FISH analysis were shown in Supplementary Figure S3A and the near complete knockdown efficiency of RECQL1 was shown in Supplementary Figure S3B. Short telomeres may eventually fail to give rise to an observable signal at the ends, resulting in an increased frequency of signal-free ends or telomere loss. Our Telomere-FISH analysis revealed an increase in telomere loss (i.e. loss of detectable telomere signal at one or more ends) and also in telomere doublets (i.e. the presence of an extra telomeric signal at one chromatid end) in RECQL1 KD cells (Figure 6C). A higher number of telomere loss was observed when compared to telomere doublets (Figure 6E). Both telomere loss and telomere doublet formation may be due to incomplete replication or may result from accumulation of T-loops (44,45). Telomere doublets are often described as fragile telomeres and observed in cells treated with low doses of aphidicolin (a specific inhibitor of DNA polymerases) or after the loss of TRF1 (46). Indeed, a higher frequency of telomere doublets was observed in RECQL1-depleted cells than in control cells after treatment with aphidicolin (Figure 6D). In particular, RECQL1 KD cells showed over 3–4-fold higher accumulation of fragile telomeres than the scrambled cells after treatment with aphidicolin (Figure 6F). These observations collectively indicate a potential role for RECQL1 in telomere replication.
Figure 6.
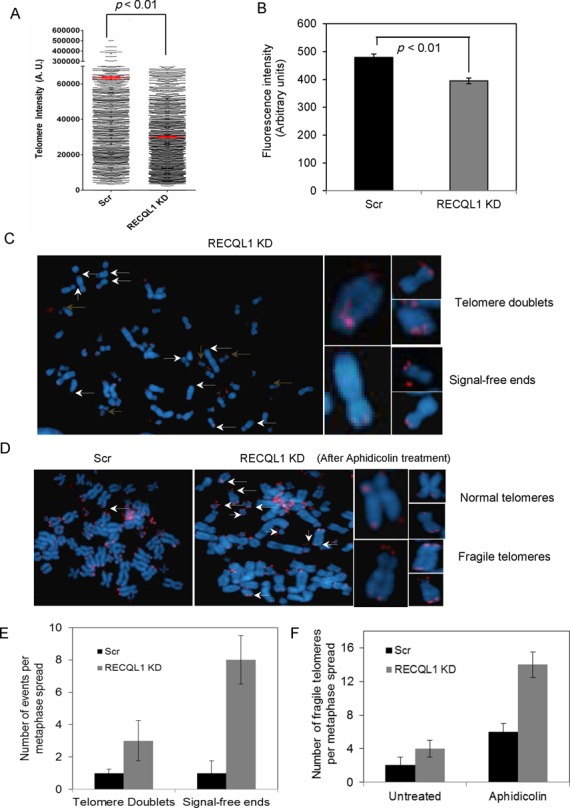
Loss of RECQL1 leads to telomere shortening and increases aphidicolin-induced telomere fragility in ALT cells. (A) Q-FISH analysis was performed on RECQL1 KD cells (U2OS) and the quantification of Q-FISH analysis showing median telomere length in red in both scrambled and RECQL1 KD cells. (B) Flow-FISH analysis was performed in RECQL1 KD cells (U2OS). The significance of the results was calculated by performing Student's t-test and the observed p-value is indicated. (C) Left: metaphase spreads of RECQL1 KD cells indicating signal-free ends (indicated in white arrows) and telomere doublets (indicated in brown arrows); representative images of single chromosomes with the respective defects were shown on right. Q-FISH analysis was performed on RECQL1 KD cells (U2OS) followed by treatment with low doses aphidicolin (0.3 μM for 18 h). (D) Metaphase spreads of scrambled and RECQL1 KD cells after treatment with aphidicolin shown on the left and images of single chromosomes showing normal and fragile telomeres shown on the right. (E) Plot showing average number of telomere doublets and SFEs per metaphase spread in scrambled and RECQL1 KD cells. (F) Plot showing average number of fragile telomeres per metaphase spread followed by treatment with aphidicolin. At least 25 metaphase spreads were analyzed for each cell type from two independent experiments.
Loss of RECQL1 results in accumulation of T-SCEs
It has been previously shown that RECQL1 can resolve HJ substrates (30). HJs are important intermediates of the recombination events that take place at stalled replication forks and are also reported to form preferentially at telomeres during telomere replication (47). Conversely, telomere shortening may also lead to defective resolution of crossovers and promote hyper-recombination events in ALT cells (48). Hence, we evaluated if RECQL1 could unwind telomeric HJs and if loss of RECQL1 elevated recombination events at telomeres. RECQL1 can disrupt a static telomeric HJ substrate (Figure 7A). 20 nM of RECQL1 caused about 25% HJ disruption and formation of a duplex intermediate, while increasing concentrations to 50 and 100 nM of RECQL1 resulted in further efficient disruption of the substrate and unwinding to ssDNA (Figure 7A). Further, CO-FISH indicated an accumulation over 3-fold more T-SCEs in RECQL1 KD cells compared to the scrambled cells (Figure 7B and C). Collectively, these observations indicate elevated hyper-recombination events at telomeres in the absence of RECQL1 and that RECQL1 may be involved in disrupting telomeric recombination intermediates.
Figure 7.
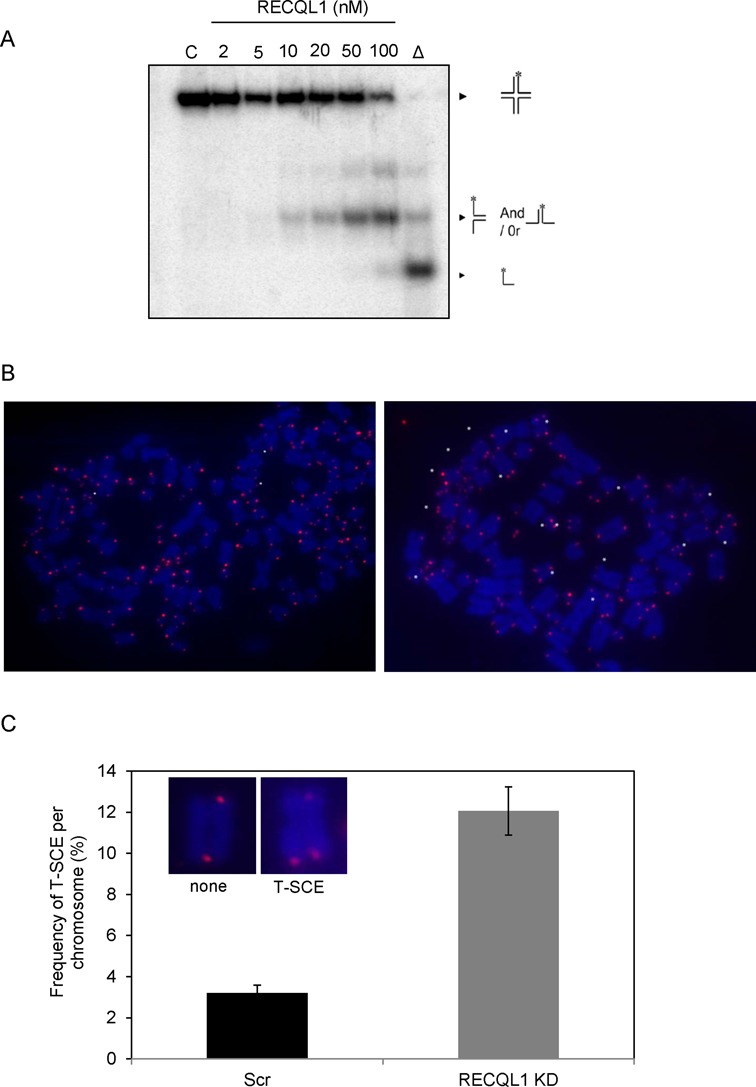
Loss of RECQL1 leads to telomere sister-chromatid exchanges (T-SCEs). (A) Helicase activity of RECQL1 (2–100 nM) on static telomeric HJ substrate. (B) Co-FISH analysis was performed on RECQL1 KD cells (U2OS) and representative images of the metaphase spreads of RECQL1 KD and scrambled cells showing the T-SCE events shown in white asterisks. (C) Plot showing % T-SCEs in scrambled and RECQL1 KD cells. Representative images of a single chromosome showing T-SCEs were shown in the inset.
Both RECQL1 and WRN may be involved in telomere replication
WRN can also efficiently unwind telomeric HJs (49) and loss of WRN has also been reported to elevate T-SCEs in ALT cells (43). WRN plays a role in telomere replication and loss of WRN also leads to telomere loss, eventually eliciting formation of dysfunctional telomeres, telomere shortening and telomere loss (44,50). Hence, we evaluated if RECQL1 and WRN participate in the same pathway or act in different pathways. Double depletion of RECQL1 and WRN was performed in U2OS cells and subsequently cells were quantified for telomere lengths by flow-FISH. Cells with double knockdowns of RECQL1 and WRN exhibited similar levels of telomere shortening compared to the singly depleted cells (Figure 8A), suggesting that RECQL1 and WRN may act in the same pathway, probably in telomere replication. It is important to note that transient depletion of RECQL1 using siRNA (Figure 8A) also resulted in telomere shortening similar to that observed in stable RECQL1 KD cells, further strengthening our observations. The knockdown efficiency of both proteins was high and is shown in Figure 8B. Further, flow-FISH coupled with cell cycle analysis showed that loss of either RECQL1 or WRN led to telomere shortening especially during S-phase, further indicating their involvement in telomere replication (Figure 8C). Although WRN has previously been implicated in telomere replication, this is the first report, to our knowledge, indicating telomere shortening specifically during S-phase in the absence of WRN.
Figure 8.
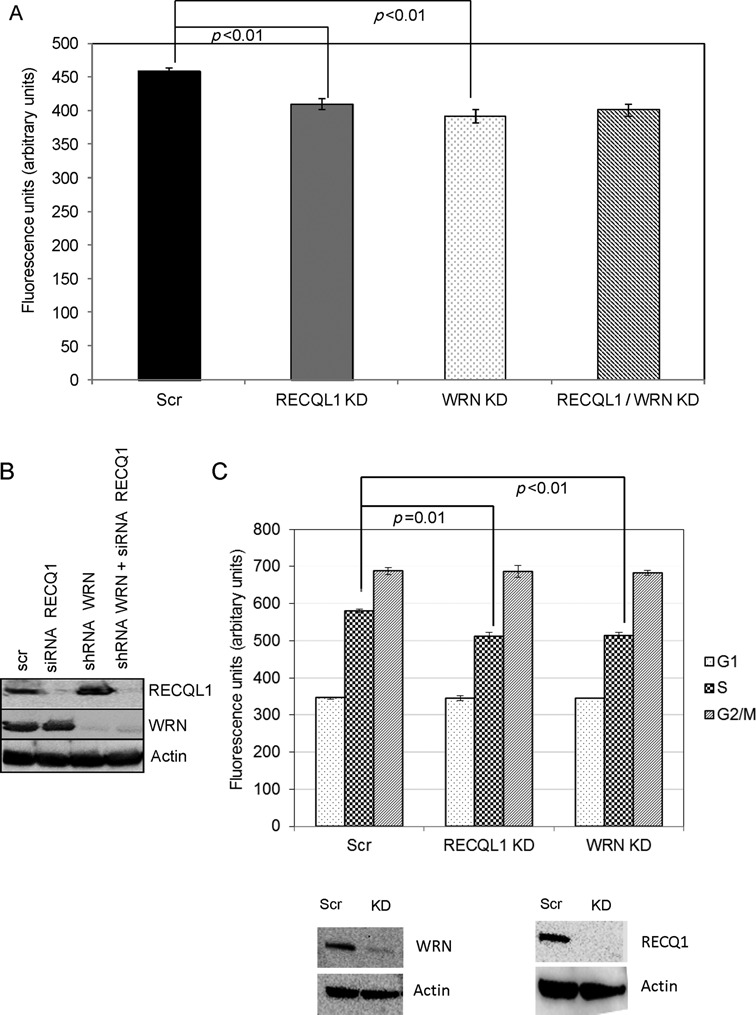
RECQL1 and WRN may act in the same pathway. (A) Double depletion of both RECQL1 and WRN was performed in U2OS cells and analyzed for telomere lengths by using flow-FISH analysis. (B) Western blot showing double depletion of RECQL1 (by using siRNA) and WRN (using shRNA). (C) Telomere length analysis was further studied by performing flow-FISH coupled with cell cycle analysis on stable KD cells of RECQL1 or WRN, and the western blots indicating the knockdown efficiency were shown below.
Telomeric association of RECQL1 in telomerase-positive cells
Our above results indicate a potential involvement of RECQL1 in telomere maintenance in ALT cells. Next, we evaluated the involvement of RECQL1 in telomerase-positive cells. Telomeric ChIP experiment was performed in HeLa cells (Figure 9A). The quantification of these results indicates that only about 3% of the input signal for RECQL1 was present at the telomere, while more than 15% of the signal was obtained for TRF1 (Figure 9B). However, this quantification may not be directly comparable with the one in U2OS cells as both cell types may have different telomere lengths. Conversely, RECQL1 may be less abundant at the telomeres in the HeLa cells than the U2OS cells. Recent studies also indicate a possible involvement of both BLM and WRN in telomere maintenance in normal telomerase-positive human fibroblasts (52) but their involvement is well characterized in ALT cells (15,17,42). To characterize the role of RECQL1 at telomeres in telomerase-positive cells, stable depletion of RECQL1 was performed in telomerase-positive HeLa 1.2.11 cells and subsequently analyzed for telomere defects. Similar to U2OS cells, RECQL1 depletion caused telomere shortening (Figure 9C) and an accumulation of DNA DSB foci (53BP1) at TRF1 sites (Figure 9D). These results indicate that RECQL1 may bind to telomeres in telomerase-positive cells, thereby regulating telomere integrity.
DISCUSSION
Maintenance of telomeres is essential for the conservation of genomic integrity. Telomerase is involved in synthesis of telomeres, however, some human cancers activate the ALT pathway (5). Dissociation of telomeric D-loop structures is not only required for progression of the ALT-based DNA elongation events but is also essential for complete repair of any DNA damage (5). T-loops may also contribute to telomere length maintenance in ALT cells, by copy template, involving a mechanism of rolling circle replication, in which the lagging strand synthesis may be templated using the invading 3′-ss overhang by branch migration (5). Within the last decade, much research has implicated potential roles for human RecQ helicases in telomere maintenance, particularly in ALT cells (10,15,17,44,52). Here, we identify a novel role for RECQL1 in telomere maintenance in ALT cells. RECQL1 can efficiently unwind telomeric D-loops and HJs and this resolution would not only benefit telomere replication but might also help regulating telomere length in ALT cells. Telomeric ss-binding protein, POT1 stimulates RECQL1 and this functional interaction may enable RECQL1 to overcome a replicative blocking lesion such as a thymine glycol. Consequently, loss of RECQL1 induces accumulation of TIFs, S-phase-associated telomere shortening, and increased telomere fragility, indicating a potential role for RECQL1 in telomere replication. Collectively, RECQL1 may be involved in disrupting alternative or secondary structures at stalled replication forks, combating hyper-recombination and ensuring efficient fork progression at telomeres. Consistently, loss of RECQL1 leads to hyper-recombination events and elevation of T-SCEs.
TRF2 binds to the duplex region of the telomeric DNA and is directly involved in inhibition of DNA damage signaling at telomeres and protection of T-loops (37,51,53). RECQL1 interacts with TRF2 as does other human RecQ helicases, BLM, WRN and RECQL4 (10,11,14). TRF2 inhibits the helicase activity of RECQL1 on telomeric substrates, while it has been previously shown that TRF2 promotes the helicase activity of BLM, WRN and RECQL4 (10,11,14). RECQL1 is the most abundant human RecQ helicase, expressed throughout the cell cycle, while BLM, WRN and RECQL4 are less abundant than RECQL1 and associate with telomeres specifically during S-phase (10,11,15,17,44) and play an important role in the resolution of telomeric D-loops (54,55). Although RECQL1 binds to the telomeres, independently of TRF2, TRF2 regulates the helicase activity of RECQL1 through its physical interaction. TRF2 may be required to modulate the activity of RECQL1, but not other human RecQ helicases, in order to maintain the T-loop structural confirmation, and this inhibition by TRF2 might be overcome after DNA damage or under conditions required for D-loop unwinding (probably during S-phase). We find that the inhibition is partially overcome on damaged telomeric D-loops, indicating that RECQL1 might be involved in resolution of telomeric D-loops following DNA damage. The TPP1-POT1 heterodimer complex, also involved in repression of DNA damage signaling at telomeres and maintenance of T-loop structures (38), inhibits RECQL1, similarly to TRF2. Interestingly, the inhibition of RECQL1 by TRF2 is overcome in the presence of WRN that localizes to the telomeres specifically during S-phase (15,44) (Supplementary Figure S4A). These observations support our notion that TRF2 might regulate RECQL1 unwinding of the T-loops in cell cycle phases other than S-phase. TRF2 stimulates WRN (compare lanes 5 and 6) but the resultant activity is lower than the reaction containing RECQL1 in addition to TRF2 and WRN (compare lane 4 with lane 6), indicating that the additional activity may be due to RECQL1. Similar modulation of RECQL1 by Poly [ADP-ribose] polymerase 1 (PARP-1) was shown to be important in regulating its role at stalled replication forks (25).
Incomplete replication or replication fork stalling and/or collapse at telomeric ends can lead to telomere loss or aberrations. Loss of RECQL1 leads to telomere shortening, increased signal-free ends and accumulation of fragile telomeres (or telomere doublet formation). Fragile telomeres may result from defective processing of stalled replication forks and may often represent accumulation of ssDNA at telomeres (46). Loss of RECQL1 leads to accumulation of RPA and phospho-RPA at telomeres, indicating replication stress and accumulation of recombinogenic ssDNA at the telomeres. The accumulation of fragile telomeres is more significant after treatment with aphidicolin, indicating that RECQL1 may be involved in protecting telomere fragile sites upon replicative block. Consistently, RECQL1 has been shown to accumulate at common fragile sites in response to replicative block with aphidicolin and that RECQL1-depleted cells are hypersensitive to aphidicolin (56).
Telomere shortening activates DNA damage signaling and triggers permanent cell cycle arrest or apoptosis. Recent evidence indicates that telomere shortening induces growth arrest at the G1 phase of the cell cycle and activates ATM signaling (41). Incidentally, we and others have shown that loss of RECQL1 activates ATM signaling (24,57) and leads to accumulation of cells in G0/G1 phase with lower number of cells in S-phase (probably due to replication defects) (23,24). Short telomeres may also initiate increased level of telomere recombination events (48) and consistently, we observe that loss of RECQL1 leads to accumulation of T-SCEs. SCEs may occur during DNA replication (S-phase of the cell cycle) when lesions that stall and/or collapse the replication fork are encountered (58). T-SCEs were specifically deemed a marker of ALT cells that may reflect recombination-based telomere maintenance (33,59). Additionally, oxidative stress may enhance telomere shortening (60). It has been shown that 8-oxoguanine in telomeric DNA can inhibit DNA binding of shelterin proteins (61) and can stimulate HR at telomeres (62). Emerging studies also provide evidence for a unique role of RECQL1 in repair of oxidative DNA damage and that RECQL1-deficient cells are sensitive to oxidative DNA damage (63). Here, we show that RECQL1 can efficiently unwind 8-oxoguanine containing telomeric D-loops.
WRN plays an important role in telomere replication, and loss of WRN leads to a similar phenotype of accumulation of RPA, telomere loss and elevation of T-SCEs (43,44,50). Our present results indicate that RECQL1 may act in the same pathway as WRN, as double depletion of both proteins did not significantly affect telomere length when compared with the singly depleted cells. Loss of WRN or loss of RECQL1 results in telomere shortening specifically during S-phase. We wanted to further investigate if RECQL1 and WRN might cooperate during telomere replication. We have previously shown that WRN and RECQL4 can cooperate with each other in the resolution of telomeric D-loops (10). However, we did not observe such cooperation between RECQL1 and WRN using damaged telomeric D-loops, DL2 and DL5. They do not either compete with each other even at a molar ratio of 1:5 (WRN:RECQL1) and the observed effect was limited to the combined activity of the two proteins (Supplementary Figure S4B). The human RecQ helicases have long been proposed to play both cooperative and overlapping, but yet non-redundant roles with each other. We have previously shown that WRN cooperates with RECQL5 during general genomic replication (34) and with RECQL4 during telomere replication (10). Here, we speculate that RECQL1 plays an overlapping, yet non-redundant role with WRN in telomere replication. For instance, WRN can unwind G4-DNA to ensure proper telomere replication (44,64). RECQL1 cannot unwind G4-DNA (30), but may be involved in disruption of other secondary structures and aid in the restart of stalled replication forks at telomeres that would otherwise result in telomere fragility and/or telomere loss. Incidentally, it has been shown that RECQL1, but not BLM or WRN, plays a unique role during replication origin firing and nascent DNA synthesis (23) and is also critical for replication fork restart (25). This division of labor among the RecQ helicases may relate to their non-redundant functions and distinct substrate specificity.
CONCLUSION
In summary, telomere shortening in ALT cells may be due to hyper-recombination events coupled with defects in DNA replication. In general, telomeric DNA is difficult to replicate and replication forks within telomeres often stall (65). This may be more likely in ALT cells where the telomeres are much longer and prone to replication fork stalling. RECQL1 may function at the stalled replication forks in addition to regulating ALT-specific recombination events. Here, we propose a novel role for RECQL1 in telomerase-negative ALT cells, and possibly to a lesser extent, in telomerase-positive cells (HeLa cells). Future studies would be required to investigate role of RECQL1 in telomere replication in telomerase-positive cells. Previous studies indicate that RecQL1 knockout mice did not show any differences in telomere length abnormalities (66), but it is important to note that mice have longer telomeres than humans and similarly Wrn knockout mice were shown to exhibit the characteristic aging symptoms only in the presence of shorter telomeres, with the loss of telomerase RNA template Terc (67).
Telomere shortening can be a result of incomplete replication and telomere attrition and telomere shortening has been part of tumor-suppressor pathways (68). A large number of human tumors do not activate telomerase and may rely on the ALT mechanisms to lengthen telomeres (5,6). Both RECQL1 and WRN are highly expressed in various tumors and are proposed to be ideal therapeutic targets for tumor suppression (69,70). Additionally, current data indicate that specific depletion of RECQL1 leads to mitotic catastrophe in cancer cells but does not affect normal cells (57). We speculate that telomere attrition might be an important consequence after loss of RECQL1 in cancer cells, as was previously proposed for WRN (71). It may be possible that loss of RECQL1 leads to a rare human, yet undiscovered disease characterized by aging symptoms and cancer predisposition.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGMENTS
We thank Drs Huiming Lu and Raghavendra Shamanna for critical reading of the manuscript. We would like to thank Jade Scheers and Dr Robert Wersto for their technical and analytical help with the flow-FISH experiments. We thank Drs Robert Brosh Jr and Joshua A. Sommers for the purification of recombinant RECQL1. We thank Dr Tomasz Kulikowicz for the purification of recombinant dual-tagged WRN.
FUNDING
Intramural Research Program of the National Institute on Aging (NIH) [AG000726–20]. Funding for open access charge: NIH Intramural Program.
Conflict of interest statement None declared.
REFERENCES
- 1.O'Sullivan R.J., Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de L.T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 3.Harley C.B., Futcher A.B., Greider C.W. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 4.Autexier C., Lue N.F. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 5.Henson J.D., Neumann A.A., Yeager T.R., Reddel R.R. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 6.Muntoni A., Neumann A.A., Hills M., Reddel R.R. Telomere elongation involves intra-molecular DNA replication in cells utilizing alternative lengthening of telomeres. Hum. Mol. Genet. 2009;18:1017–1027. doi: 10.1093/hmg/ddn436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greider C.W. Telomeres do D-loop-T-loop. Cell. 1999;97:419–422. doi: 10.1016/s0092-8674(00)80750-3. [DOI] [PubMed] [Google Scholar]
- 8.Verdun R.E., Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 9.Singh D.K., Ghosh A.K., Croteau D.L., Bohr V.A. RecQ helicases in DNA double strand break repair and telomere maintenance. Mutat. Res. 2012;736:15–24. doi: 10.1016/j.mrfmmm.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh A.K., Rossi M.L., Singh D.K., Dunn C., Ramamoorthy M., Croteau D.L., Liu Y., Bohr V.A. RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. J. Biol. Chem. 2012;287:196–209. doi: 10.1074/jbc.M111.295063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lillard-Wetherell K., Machwe A., Langland G.T., Combs K.A., Behbehani G.K., Schonberg S.A., German J., Turchi J.J., Orren D.K., Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- 12.Machwe A., Xiao L., Orren D.K. TRF2 recruits the Werner syndrome (WRN) exonuclease for processing of telomeric DNA. Oncogene. 2004;23:149–156. doi: 10.1038/sj.onc.1206906. [DOI] [PubMed] [Google Scholar]
- 13.Mendez-Bermudez A., Hidalgo-Bravo A., Cotton V.E., Gravani A., Jeyapalan J.N., Royle N.J. The roles of WRN and BLM RecQ helicases in the alternative lengthening of telomeres. Nucleic Acids Res. 2012;40:10809–10820. doi: 10.1093/nar/gks862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Opresko P.L., von K.C., Laine J.P., Harrigan J., Hickson I.D., Bohr V.A. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 15.Opresko P.L., Otterlei M., Graakjaer J., Bruheim P., Dawut L., Kolvraa S., May A., Seidman M.M., Bohr V.A. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 16.Opresko P.L., Mason P.A., Podell E.R., Lei M., Hickson I.D., Cech T.R., Bohr V.A. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J. Biol. Chem. 2005;280:32069–32080. doi: 10.1074/jbc.M505211200. [DOI] [PubMed] [Google Scholar]
- 17.Stavropoulos D.J., Bradshaw P.S., Li X., Pasic I., Truong K., Ikura M., Ungrin M., Meyn M.S. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum. Mol. Genet. 2002;11:3135–3144. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh A., Rossi M.L., Aulds J., Croteau D., Bohr V.A. Telomeric D-loops containing 8-oxo-2’-deoxyguanosine are preferred substrates for Werner and Bloom syndrome helicases and are bound by POT1. J. Biol. Chem. 2009;284:31074–31084. doi: 10.1074/jbc.M109.027532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dejardin J., Kingston R.E. Purification of proteins associated with specific genomic loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawabe T., Tsuyama N., Kitao S., Nishikawa K., Shimamoto A., Shiratori M., Matsumoto T., Anno K., Sato T., Mitsui Y., et al. Differential regulation of human RecQ family helicases in cell transformation and cell cycle. Oncogene. 2000;19:4764–4772. doi: 10.1038/sj.onc.1203841. [DOI] [PubMed] [Google Scholar]
- 21.Muzzolini L., Beuron F., Patwardhan A., Popuri V., Cui S., Niccolini B., Rappas M., Freemont P.S., Vindigni A. Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS Biol. 2007;5:e20. doi: 10.1371/journal.pbio.0050020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pike A.C., Shrestha B., Popuri V., Burgess-Brown N., Muzzolini L., Costantini S., Vindigni A., Gileadi O. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc. Natl. Acad. Sci. U.S.A. 2009;106:1039–1044. doi: 10.1073/pnas.0806908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thangavel S., Mendoza-Maldonado R., Tissino E., Sidorova J.M., Yin J., Wang W., Monnat R.J., Jr, Falaschi A., Vindigni A. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol. Cell Biol. 2010;30:1382–1396. doi: 10.1128/MCB.01290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popuri V., Croteau D.L., Brosh R.M., Jr, Bohr V.A. RECQ1 is required for cellular resistance to replication stress and catalyzes strand exchange on stalled replication fork structures. Cell Cycle. 2012;11:4252–4265. doi: 10.4161/cc.22581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berti M., Chaudhuri A.R., Thangavel S., Gomathinayagam S., Kenig S., Vujanovic M., Odreman F., Glatter T., Graziano S., Mendoza-Maldonado R., et al. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol. 20:347–354. doi: 10.1038/nsmb.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma S., Brosh R.M., Jr Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges. PLoS ONE. 2007;2:e1297. doi: 10.1371/journal.pone.0001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tadokoro T., Kulikowicz T., Dawut L., Croteau D.L., Bohr V.A. DNA binding residues in the RQC domain of Werner protein are critical for its catalytic activities. Aging (Albany. NY) 2012;4:417–429. doi: 10.18632/aging.100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lei M., Podell E.R., Cech T.R. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 29.Ferrarelli L.K., Popuri V., Ghosh A.K., Tadokoro T., Canugovi C., Hsu J.K., Croteau D.L., Bohr V.A. The RECQL4 protein, deficient in Rothmund-Thomson syndrome is active on telomeric D-loops containing DNA metabolism blocking lesions. DNA Repair (Amst.) 2013;12:518–528. doi: 10.1016/j.dnarep.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popuri V., Bachrati C.Z., Muzzolini L., Mosedale G., Costantini S., Giacomini E., Hickson I.D., Vindigni A. The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J. Biol. Chem. 2008;283:17766–17776. doi: 10.1074/jbc.M709749200. [DOI] [PubMed] [Google Scholar]
- 31.Loayza D., de L.T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 32.Lansdorp P.M., Verwoerd N.P., van de Rijke F.M., Dragowska V., Little M.T., Dirks R.W., Raap A.K., Tanke H.J. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 33.Bailey S.M., Brenneman M.A., Goodwin E.H. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res. 2004;32:3743–3751. doi: 10.1093/nar/gkh691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Popuri V., Huang J., Ramamoorthy M., Tadokoro T., Croteau D.L., Bohr V.A. RECQL5 plays co-operative and complementary roles with WRN syndrome helicase. Nucleic Acids Res. 2013;41:881–899. doi: 10.1093/nar/gks1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aller P., Rould M.A., Hogg M., Wallace S.S., Doublie S. A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc. Natl. Acad. Sci. U.S.A. 2007;104:814–818. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xin H., Liu D., Wan M., Safari A., Kim H., Sun W., O’Connor M.S., Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 37.Denchi E.L., de L.T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 38.Hockemeyer D., Palm W., Else T., Daniels J.P., Takai K.K., Ye J.Z., Keegan C.E., de L.T., Hammer G.D. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat. Struct. Mol. Biol. 2007;14:754–761. doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- 39.Martinez P., Flores J.M., Blasco M.A. 53BP1 deficiency combined with telomere dysfunction activates ATR-dependent DNA damage response. J. Cell Biol. 2012;197:283–300. doi: 10.1083/jcb.201110124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takai K.K., Kibe T., Donigian J.R., Frescas D., de L.T. Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol. Cell. 2011;44:647–659. doi: 10.1016/j.molcel.2011.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cesare A.J., Reddel R.R. Telomere uncapping and alternative lengthening of telomeres. Mech. Ageing Dev. 2008;129:99–108. doi: 10.1016/j.mad.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 42.Bhattacharyya S., Sandy A., Groden J. Unwinding protein complexes in ALTernative telomere maintenance. J. Cell Biochem. 2010;109:7–15. doi: 10.1002/jcb.22388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laud P.R., Multani A.S., Bailey S.M., Wu L., Ma J., Kingsley C., Lebel M., Pathak S., DePinho R.A., Chang S. Elevated telomere-telomere recombination in WRN-deficient, telomere dysfunctional cells promotes escape from senescence and engagement of the ALT pathway. Genes Dev. 2005;19:2560–2570. doi: 10.1101/gad.1321305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crabbe L., Verdun R.E., Haggblom C.I., Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 45.Wang R.C., Smogorzewska A., de L.T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 46.Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., de L.T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fouche N., Ozgur S., Roy D., Griffith J.D. Replication fork regression in repetitive DNAs. Nucleic Acids Res. 2006;34:6044–6050. doi: 10.1093/nar/gkl757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morrish T.A., Greider C.W. Short telomeres initiate telomere recombination in primary and tumor cells. PLoS Genet. 2009;5:e1000357. doi: 10.1371/journal.pgen.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nora G.J., Buncher N.A., Opresko P.L. Telomeric protein TRF2 protects Holliday junctions with telomeric arms from displacement by the Werner syndrome helicase. Nucleic Acids Res. 2010;38:3984–3998. doi: 10.1093/nar/gkq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arnoult N., Saintome C., Ourliac-Garnier I., Riou J.F., Londono-Vallejo A. Human POT1 is required for efficient telomere C-rich strand replication in the absence of WRN. Genes Dev. 2009;23:2915–2924. doi: 10.1101/gad.544009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van S.B., Smogorzewska A., de L.T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 52.Barefield C., Karlseder J. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res. 2012;40:7358–7367. doi: 10.1093/nar/gks407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karlseder J., Hoke K., Mirzoeva O.K., Bakkenist C., Kastan M.B., Petrini J.H., de L.T. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bachrati C.Z., Borts R.H., Hickson I.D. Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Opresko P.L., Sowd G., Wang H. The Werner syndrome helicase/exonuclease processes mobile D-loops through branch migration and degradation. PLoS ONE. 2009;4:e4825. doi: 10.1371/journal.pone.0004825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu X., Parvathaneni S., Hara T., Lal A., Sharma S. Replication stress induces specific enrichment of RECQ1 at common fragile sites FRA3B and FRA16D. Mol. Cancer. 2013;12:29. doi: 10.1186/1476-4598-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Futami K., Kumagai E., Makino H., Goto H., Takagi M., Shimamoto A., Furuichi Y. Induction of mitotic cell death in cancer cells by small interference RNA suppressing the expression of RecQL1 helicase. Cancer Sci. 2008;99:71–80. doi: 10.1111/j.1349-7006.2007.00647.x. [DOI] [PubMed] [Google Scholar]
- 58.Wilson D.M., III, Thompson L.H. Molecular mechanisms of sister-chromatid exchange. Mutat. Res. 2007;616:11–23. doi: 10.1016/j.mrfmmm.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 59.Londono-Vallejo J.A., Der-Sarkissian H., Cazes L., Bacchetti S., Reddel R.R. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 2004;64:2324–2327. doi: 10.1158/0008-5472.can-03-4035. [DOI] [PubMed] [Google Scholar]
- 60.von Z.T., Petrie J., Kirkwood T.B. Telomere-driven replicative senescence is a stress response. Nat. Biotechnol. 2003;21:229–230. doi: 10.1038/nbt0303-229b. [DOI] [PubMed] [Google Scholar]
- 61.Opresko P.L., Fan J., Danzy S., Wilson D.M., III, Bohr V.A. Oxidative damage in telomeric DNA disrupts recognition by TRF1 and TRF2. Nucleic Acids Res. 2005;33:1230–1239. doi: 10.1093/nar/gki273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu X.D., Niedernhofer L., Kuster B., Mann M., Hoeijmakers J.H., de L.T. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- 63.Sharma S., Phatak P., Stortchevoi A., Jasin M., Larocque J.R. RECQ1 plays a distinct role in cellular response to oxidative DNA damage. DNA Repair (Amst.) 2012;11:537–549. doi: 10.1016/j.dnarep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mohaghegh P., Karow J.K., Brosh R.M., Jr, Bohr V.A., Hickson I.D. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohki R., Ishikawa F. Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats. Nucleic Acids Res. 2004;32:1627–1637. doi: 10.1093/nar/gkh309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma S., Stumpo D.J., Balajee A.S., Bock C.B., Lansdorp P.M., Brosh R.M., Jr, Blackshear P.J. RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Mol. Cell Biol. 2007;27:1784–1794. doi: 10.1128/MCB.01620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang S., Multani A.S., Cabrera N.G., Naylor M.L., Laud P., Lombard D., Pathak S., Guarente L., DePinho R.A. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat. Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 68.Lansdorp P.M. Telomeres and disease. EMBO J. 2009;28:2532–2540. doi: 10.1038/emboj.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mendoza-Maldonado R., Faoro V., Bajpai S., Berti M., Odreman F., Vindigni M., Ius T., Ghasemian A., Bonin S., Skrap M., et al. The human RECQ1 helicase is highly expressed in glioblastoma and plays an important role in tumor cell proliferation. Mol. Cancer. 2011;10:83. doi: 10.1186/1476-4598-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arai A., Chano T., Futami K., Furuichi Y., Ikebuchi K., Inui T., Tameno H., Ochi Y., Shimada T., Hisa Y., et al. RECQL1 and WRN proteins are potential therapeutic targets in head and neck squamous cell carcinoma. Cancer Res. 2011;71:4598–4607. doi: 10.1158/0008-5472.CAN-11-0320. [DOI] [PubMed] [Google Scholar]
- 71.Bai Y., Murnane J.P. Telomere instability in a human tumor cell line expressing a dominant-negative WRN protein. Hum. Genet. 2003;113:337–347. doi: 10.1007/s00439-003-0972-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


