Abstract
Ribonucleic acid (RNA)-binding proteins are key players of gene expression control. We have shown that Gemin5 interacts with internal ribosome entry site (IRES) elements and modulates initiation of translation. However, little is known about the RNA-binding sites of this protein. Here we show that the C-terminal region of Gemin5 bears two non-canonical bipartite RNA-binding sites, encompassing amino acids 1297–1412 (RBS1) and 1383–1508 (RBS2). While RBS1 exhibits greater affinity for RNA than RBS2, it does not affect IRES-dependent translation in G5-depleted cells. In solution, the RBS1 three-dimensional structure behaves as an ensemble of flexible conformations rather than having a defined tertiary structure. However, expression of the polypeptide G51383–1508, bearing the low RNA-binding affinity RBS2, repressed IRES-dependent translation. A comparison of the RNA-binding capacity and translation control properties of constructs expressed in mammalian cells to that of the Gemin5 proteolysis products observed in infected cells reveals that non-repressive products accumulated during infection while the repressor polypeptide is not stable. Taken together, our results define the low affinity RNA-binding site as the minimal element of the protein being able to repress internal initiation of translation.
INTRODUCTION
Ribonucleic acid (RNA)-binding proteins play a pivotal role in the regulation of gene expression. Due to their capacity to interact with different targets, either proteins or RNAs, this diverse group of proteins plays multifunctional roles in multiple RNA-dependent processes (1–4). In earlier studies, we identified Gemin5 as one of the proteins interacting directly with two different viral internal ribosome entry site (IRES) elements, foot-and-mouth disease virus (FMDV) and hepatitis C virus (5). Subsequently, we found that it mediates translation repression (6). Gemin5 is a peripheral protein of the survival of motor neuron complex (7), a multiprotein complex found in metazoan cells (8). This complex, which is composed of 7 Sm proteins and the Gemins 2–8, plays a critical role on the biogenesis of spliceosomal small nuclear ribonucleoproteins (snRNPs) (9). In turn one of the Gemins, Gemin5, has been reported to be responsible for the interaction with the Sm site of snRNAs (10). Thereby, the finding that Gemin5 down-regulated translation (6) revealed a new role for this protein on translation control beyond its role in snRNP biogenesis (11). However, little is known about how this protein recognizes distinct RNA targets.
Initiation of translation in eukaryotic messenger RNAs (mRNAs) involves a set of initiation factors (eIFs) that recruit the small ribosome subunit to the m7GTP residue (or cap) located at the 5′end of most mRNAs (12). In addition, cap-independent mechanisms have been exploited by mRNAs translated under conditions that compromise the general cap-dependent translation, as it has been illustrated in virus infected cells (13–15). To evade cap-dependent inhibition and, at the same time, benefit from the translation shut down of the vast majority of cellular mRNAs, some viral RNAs have evolved a mechanism based on IRES elements. Interestingly, cellular mRNAs translated under strong cap-dependent inhibition also make use of IRES elements to start translation (16). In the case of picornaviruses and other positive-strand viral RNAs, these elements are long, heavily structured regions that recruit the ribosomal subunits internally, promoting translation initiation at internal start codons independent of the 5′end of the mRNA (17–20). IRES-dependent translation of viral and cellular mRNAs depends on a large variety of RNA-binding proteins, in addition to a subset of eIFs (21–23). However, how distinct RNA-binding proteins regulate the activity of IRES elements remains to be elucidated.
We have recently shown that Gemin5 recognizes the IRES element via its interaction with a stem loop placed at the 3′end (24). This was achieved by preparing immunoprecipitated photocrosslinked complexes with soluble cell extracts or by using RNA-binding assays with recombinant proteins. To gain a higher resolution understanding of this interaction, selective 2′hydroxyl acylation analysed by primer extension RNA reactivity was performed with the FMDV IRES and purified proteins. This experiment demonstrated that incubation of Gemin5 with the IRES induced the specific protection of residues within domain 5, out-competing the IRES stimulator protein PTB (24). This effect is consistent with its down-regulatory role in IRES-dependent translation.
Interestingly, Gemin5 is proteolyzed in FMDV infected cells by the action of the L protease (25). This feature is shared with other host factors involved in gene expression control, such as eIF4G, eIF5B, poly(A)-binding protein (PABP), polypyrimidine tract-binding protein (PTB), Gemin3 and cleavage stimulation factor, together with other proteins controlling various steps of RNA biology (26–31).
The three-dimensional structure of full length Gemin5 is yet unknown. Notably, we have found that a polypeptide encompassing the C-terminal region of the protein was able to interact directly with the IRES to a similar extent than the full-length protein (24). Furthermore, in experiments carried out in parallel the N-terminal region of the protein had no IRES-binding capacity, suggesting that the C-terminal region harbors all the necessary features to promote an interaction with the IRES element. Thus, the RNA-binding domain recognising the IRES element differs from the region responsible for the interaction with snRNAs, which is located on the fifth tryptohan-aspartic motif (WD) repeat within the N-terminal region of Gemin5 (10). These results led us to investigate the possibility that different regions of the protein could be involved in the recognition of RNA targets with different sequences, structural organisations and RNA functions.
An in silico analysis of the C-terminal region of the protein did not reveal any apparent sequence similarities to other well-known RNA recognition motifs present in canonical RNA-binding proteins. This lack of similarity to other RNA recognition motifs prompted us to study the molecular basis of the RNA-binding properties of this fragment and also its capacity to repress translation. Here we show that the region of Gemin5 encompassing amino acids 1383–1508 is coincident with a proteolysis product resulting from viral L protease activity. In addition, it bears an RNA-binding site (RBS2). Expression of this polypeptide in G5-depleted cells represses IRES-dependent translation initiation. Moreover, the RNA-binding capacity of Gemin5 is not confined to this region but it is further extended upstream of the 1383–1508 motif. Indeed, an overexpressed protein, encompassing amino acids 1297–1412, exhibits the highest affinity for RNA in both gel-shift and UV-crosslink assays. This region (RBS1), however, does no affect translation initiation in G5-depleted cells. Interestingly, the RBS1 fragment in solution does not contain a unique tertiary structure. Indeed, the 1H–15N nuclear magnetic resonance (NMR) dispersion is poor in agreement with the presence of an ensemble of flexible conformations, which probably accounts for the functional versatility of the protein.
MATERIALS AND METHODS
Constructs
Plasmids expressing domain 5 of the FMDV IRES element (32), CMVpBIC (expressing the bicistronic RNA CAT–FMDV IRES–luciferase) (6), pcDNA3-G5FLAG and pRSETBG5 were described (24). To express proteins in mammalian cells, we generated the vector pcDNA3/Xpress that allowed the expression of amino terminal HIS-Xpress tagged fusion proteins. For this, the HindIII/EcoRI fragment from pcDNA3.1-HIS6CDaxx (25) was inserted into pcDNA3.1/ZEO (Invitrogen), similarly digested.
pcDNA3/XpressG5845–1508 was created in two steps. First, the polymerase chain reaction (PCR) product obtained with the pair of primers G5–1s/G5–10as (see Supplementary Table S1 for primers) and template pcDNA3G5FLAG was inserted into pcDNA3/Xpress via BamHI-EcoRI. This construct was used to insert the PCR product resulting with primers G5–11s/G5–2as via EcoR1-Not1. Construct pcDNA3/XpressG5845–1436 was similarly generated using primers G5–11s/G5–12as in the second PCR step. To create pcDNA3/XpressG51287–1508, the PCR product obtained with primers G5–5s/G5–6as using as template pET-G51287–1508 was inserted via BamHI-NotI into pcDNA3/Xpress. Construct pcDNA3/XpressG51383–1508 was generated using primers G5–3s/G5–4as and inserted via KpnI-BamHI into pcDNA3/Xpress. pcDNAXpress-G51297–1412 was generated by PCR with primers G5–7s/G5–8as and pcDNAG5Flag template, then inserted via KpnI-NotI into pcDNAXpress.
To purify proteins expressed in bacteria, the construct pET-G5845–1436 (expressing His-G5845–1436) was generated in two steps. First, the PCR product obtained using primers G5–9s/G5–10as with pCDNA3G5Flag as template was inserted via EcoRI-NdeI into pET28a+. The later construct was used to insert via EcoRI-SalI the PCR product obtained with primers G5–11s/G5–12as on template pcDNA3/XpressG5845–1436. The construct pET-G51287–1508 expressing His-G51287–1508 was generated using primers G5–13s/G5–14as with pcDNA3G5Flag as template and inserted into the EcoRI-XhoI sites of pET28a+. Construct His-G51383–1508 was generated by insertion of the EcoRI-HindIII fragment of pcDNA3/XpressG51383–1508 into pET28a+. Construct pRSETB-G5CtΔ1365–1394 was generated by substitution of the Sph-HindIII fragment by a PCR product obtained with the pair of primers G5–15s/G5–16as using as template pcDNA3G5-V5.
The plasmid pYES2-G5845–1508 to express Gemin5 in yeast was generated via PCR with primers G5–17s/G5–18as using as template pcDNA3.1-XpressG5845–1508, then digested with Kpn1-Not1 and inserted into pYES2. The nucleotide sequence of all constructs was verified by sequencing (Macrogen).
RNA synthesis
In vitro transcription was performed using T7 RNA polymerase with linearized DNA, 40 mM Tris-HCl, 50 mM DTT, 0.5 mM rNTPs, as described (33). To obtain RNAs expressing G51217–1508, G51287–1508, G51217–1436 or G51383–1508, the corresponding plasmids were linearized with NheI, SalI or HindIII. Bicistronic RNAs [cloramphenicol acetyl transferase (CAT)–IRES–luciferase (LUC)] were produced as described (34). IRES transcripts were uniformly labelled using α32P-CTP (500 Ci/mmol) (35). RNA was extracted with phenol-chloroform, ethanol precipitated and resuspended in Tris10 mM, pH 8, EDTA 1mM (TE) to a concentration of 0.04 pmol/μl. RNA integrity was examined in 6% acrylamide 7 M urea denaturing gel electrophoresis (36).
Expression and purification of proteins
Escherichia coli BL21 transformed with plasmids pET-G5845–1436, pET-G51287–1508, pET-G51383–1508, pET-G5Δ1365–1394 grown at 37°C were induced with Isopropylβ-D-1-thiogalactopyranoside (IPTG) 0.5 M during 2 h. Bacterial cell lysates were prepared in binding buffer (20 mM NaH2PO4, 500 mM NaCl, 20 mM Imidazole) using a French press, and cell debris was eliminated by centrifugation at 16000 g 30 min at 4°C twice. The lysate was loaded in His-GraviTrap columns (HealthCare) and the recombinant protein was eluted using Imidazole 500 mM. Proteins were dialysed against phosphate buffer pH 6.8, 1 mM DTT, and stored at −20°C in 50% glycerol.
Saccharomyces cerevisiae W303, grown in rich yeastextract peptone dextrose (YEPD) medium, was transformed with plasmid pYES2-G5845–1508 using lithium acetate. Transformants were grown in syntheticcomplete (SC) selection medium lacking uracil. Yeast S30 fraction was prepared as described (37) and subsequently used to purify G5845–1508 in His-GraviTrap columns performing extensive washings with 20 mM Imidazole buffer prior to protein elution.
RNA-protein photocrosslinking
Uniformly radiolabelled RNA (IRES domain 5, 0.04 pmol, ∼5 × 105 cpm) was incubated with purified proteins (50–200 ng) and UV-irradiated as described (6). Following extensive RNase A treatment samples were subjected to 10–15% sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and 32P-labelled proteins were visualized by autoradiography (38).
RNA-protein binding assay
Ni-NTA/HIS-Gemin5 complexes were assembled by incubating Ni-NTA agarose resin (25 μl) (Qiagen), pre-washed four times with cold BBH(binding buffer Hepes) buffer (10 mM HEPES; pH 7.4, 100 mM NaCl, 2.5 mM MgCl2, 0.01% NP-40), with the polypeptide of interest (0.7 pmol) in 100 μl of BBH buffer during 4 h at 4°C in a rotating wheel. Unbound proteins were removed by washing three times with cold BBH buffer. 32P-labelled domain 5 RNA (80 fmol) was added to each protein-beads complex and incubated in cold BBH buffer in the presence of cytoplasmic RNA (300-fold excess), during 2 h at 4°C in a rotating wheel. Unbound RNA was removed by washing five times with cold BBH buffer. Bound RNAs were extracted with phenol-chloroform, ethanol precipitated, and fractionated by 6% acrylamide 7 M urea denaturing gel electrophoresis, and visualized by autoradiography.
RNA electrophoretic mobility shift assay
RNA-binding reactions were carried out for 20 min at 4°C in 10 μl of binding buffer [40 mM Tris (pH 7.5), 250 mM NaCl, 0.1% (w/v) βME]. Increasing amounts of protein were incubated with a fixed concentration of 32P-labelled RNA (∼2 nM). Electrophoresis was performed in non-denaturing 8.0% (29:1) polyacrylamide gels. The gels were run for 4 h in 1X TBE (Tris-borate-EDTA) buffer (90 mM Tris, 64.6 mM boric acid, 2.5 mM EDTA, pH 8.4) at 100 V at 4°C. The gels were dried and exposed to a PhosphorImager screen overnight. The screen was scanned on a Molecular Dynamics Storm 840 PhosphorImager.
Immunodetection
Gemin5 was immunodetected by western blot (WB) using anti-Gemin5 (Novus) or anti-Xpress (Invitrogen) antibodies. Immunodetection of tubulin (Sigma) was used as loading control. Secondary antibodies (Thermo Scientific) were used according to the manufacturer instructions.
Gemin5 siRNA interference and Gemin5 polypeptides expression in mammalian cells
siRNAs targeting Gemin5 mRNA (siG5.1 GCAUAGUGGUGAUAAUUGAUU, siG5.2 CAAUGAAGAUGGAUCAAUAUU, siG5.3 CCUUAAUCAAGAAGAGAAAUU) or a control sequence (control siRNA AUGUAUUGGCCUGUAUUAGUU) were purchased from Dharmacon. HEK293 cells grown to 70% confluent were treated with 100 nM siRNA using lipofectamine 2000 (Invitrogen) according to the manufacturer instructions, 30 h prior to transfection with plasmids expressing the desired G5 polypeptides with, or without, of CMVpBIC construct, using lipofectamine (Invitrogen). The total amount of DNA in the transfection mixture was balanced with empty vector, pcDNA3.1. Cell lysates were prepared 24, 48 or 56 h later in 100 μl of lysis buffer (50 mM Tris-HCl pH 7.8, 100 mM NaCl, 0.5% NP40) and the protein concentration in the lysate was determined by Bradford assay. LUC and CAT activity was determined in each lysate. Values represent the mean ± SD of three independent assays. Equal amounts of protein were loaded in SDS-PAGE to determine the efficiency of interference as well as the efficiency of translation of the transfected Gemin5 polypeptides.
IRES activity assays
IRES activity was quantified as the expression of LUC normalized to the amount of protein in the lysate, while CAT activity reported cap-dependent translation from bicistronic mRNAs in transfected HEK293 cell monolayers (6). Each experiment was carried out in duplicate wells and repeated independently at least three times. Values represent the mean ± SD. Differences in IRES-dependent translation (LUC activity) induced by the coexpression of Gemin5 polypeptides were analysed by a paired two-sided Student t-test; differences were considered significant when P < 0.01.
NMR spectroscopy
All experiments were recorded on a Bruker Avance III 600-MHz spectrometer equipped with a z pulse field gradient unit and either a triple (1H, 13C, 15N) or quadruple (plus 31P) resonance probe heads. Double labelled (15N and 13C) samples were prepared to obtain sequence-specific assignments using HNCACB/HN(CO)CACB or CBCA(CO)NH/CBCANH experiments. The samples were prepared in 50 mM Phosphate buffer with 150 mM NaCl, at pH 6.7 All spectra were processed with the NMRPipe/NMRDraw (39) software and were analysed with Computer Aided Resonance Assignment software (CARA) (40).
NMR titration experiments with RNA fragments
For the 15N-HSQC experiments, 15N-labelled protein domains were prepared at 0.25 mM concentration, in the same buffer as described above, and unlabelled ligand was added to the sample up to a final molar ratio 3:1. Measurements were performed at 285 K or 295 K.
RESULTS
Direct interaction of Gemin5 with the IRES element involves two sites within the C-terminal region
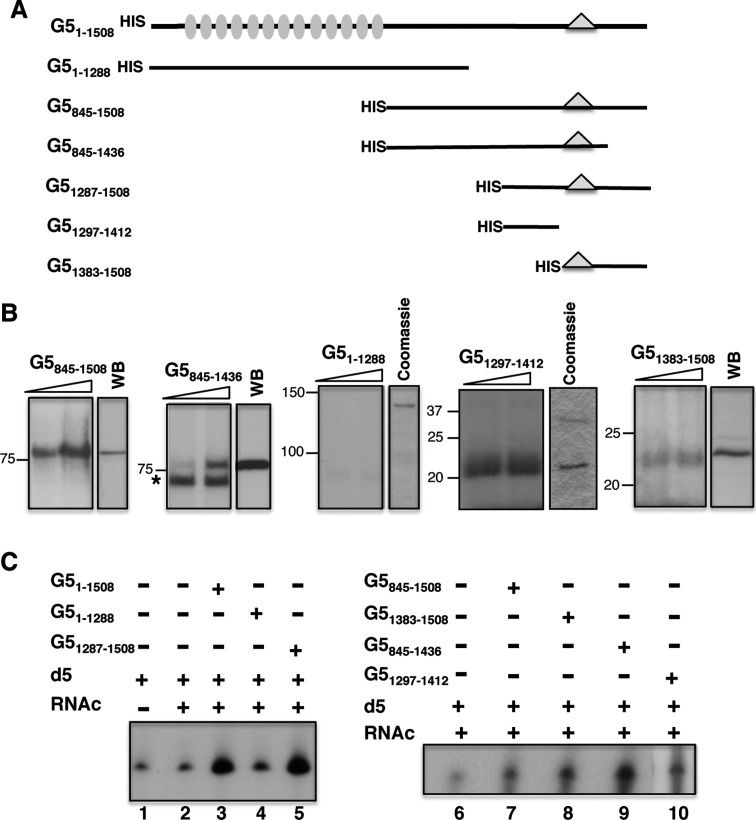
We have shown that Gemin5 has the ability to bind directly to domain 5 of the FMDV IRES element (24). Therefore, to identify the specific region of Gemin5 involved in the interaction with the IRES element, we conducted UV-crosslink assays using increasing amounts of various overexpressed polypeptide fragments corresponding to the C-terminal region of Gemin5 (Figure 1A) using radiolabelled domain 5 as probe. The N-terminal region (residues 1–1288) of the protein, encompassing the WD repeats, was used as a control. Incubation of polypeptide G5845–1508 with 32P-labelled domain 5 followed of UV irradiation resulted in the detection of a 85 kDa product that migrated with a polypeptide of similar size detected by WB (Figure 1B). Similarly, a band of about 75 kDa was observed using the G5845–1436 purified polypeptide in UV-crosslinking coincident with the protein observed by WB. An asterisk in this panel depicts a non-specific product. No detectable UV-crosslinking product was observed with G51–1288.
Figure 1.
Identification of the IRES-binding site in Gemin5. (A) Schematic of HIS-tagged polypeptides used in RNA-binding assays. Numbers indicate amino acid residues referred to the full-length protein. Grey ovals depict WD motifs located within the N-terminal region of the protein. A triangle within the C-terminal region of the protein depicts the position of the epitope recognized by Gemin5 antibody. (B) UV-crosslinking (UV-XL) assay conducted with increasing amounts of purified HIS-tagged Gemin5 polypeptides depicted at the top and radiolabelled domain 5, fractionated in SDS-PAGE and visualized by autoradiography. In each case, the mobility of the protein detected by WB using anti-Gemin5 or Coomassie Blue staining of the purified protein is shown on the right. Mobility of Mw markers is indicated at the left. (C) Gemin5-RNA binding assay. Autoradiograph of denaturing 6% acrylamide gels, 7 M Urea loaded with RNAs isolated from Ni-agarose beads coupled to the indicated proteins (0.07 pmol). D5 and RNAc are used for radiolabelled domain 5 and total cytoplasmic RNA, respectively.
Since the C-terminal region G51287–1508 was shown to interact directly with IRES RNA (24), we divided this region in two products aimed to map precisely the RNA-binding domain of Gemin5. Interestingly, a prominent interaction was observed with purified G51297–1412, which had the same mobility as the peptide in a Coomassie Blue stained gel (Figure 1B). Similarly, and although more weakly, a band of about 22 kDa was observed in UV-crosslinking carried out with purified G51383–1508, which was also coincident with the protein observed by WB (Figure 1B). Therefore, we conclude that a bipartite RNA-binding site, comprising residues 1297–1412 (RBS1) and 1383–1508 (RBS2), within the C-terminal region of Gemin5 enables a direct interaction with the IRES element with RBS1 being more intense than RBS2 in UV-crosslinking experiments.
To further support these findings, we used overexpressed proteins in RNA-pull down assays. Three constructs including an N-terminal HIS-tag to facilitate the protein purification were prepared, full-length Gemin5 (amino acids 1–1508), and the two halves, G51287–1508 and G51–1288 (Figure 1C). Then, 32P-labelled domain 5 was incubated with the HIS-tagged purified proteins immobilized on pre-washed Ni-agarose beads. Following extensive washing, bound RNAs were isolated and analysed by denaturing urea polyacrylamide gel electrophoresis. The mobility of the isolated RNAs was similar to that of the free probe (input RNA, lane 1). A negative control containing RNA incubated with Ni-agarose beads free from protein was used as a measure of the background signal (lane 2). The RNA-binding capacity of the C-terminal region (G51287–1508, lane 5) was similar to that of the full-length protein (G51–1508, lane 3) and above the signal observed with the N-terminal fragment G51–1288 (lane 4). Notably, polypeptides G51383–1508 and G51297–1412 (Figure 1C, lanes 8 and 10, respectively) exhibited a positive RNA-binding capacity relative to the background signal (lane 6), which was also observed with the longer overlapping polypeptides G5845–1508 and G5845–1436 (lanes 7 and 9). This result was in full agreement with data obtained in UV-crosslinking assays, reinforcing our conclusion that a bipartite site spanning amino acids 1297–1412 (RBS1) and 1383–1508 (RBS2) within the most C-terminal region of Gemin5 is necessary and sufficient to interact with the FMDV IRES.
N-terminal deletions of the RNA-binding site 1 severely decrease binding capacity
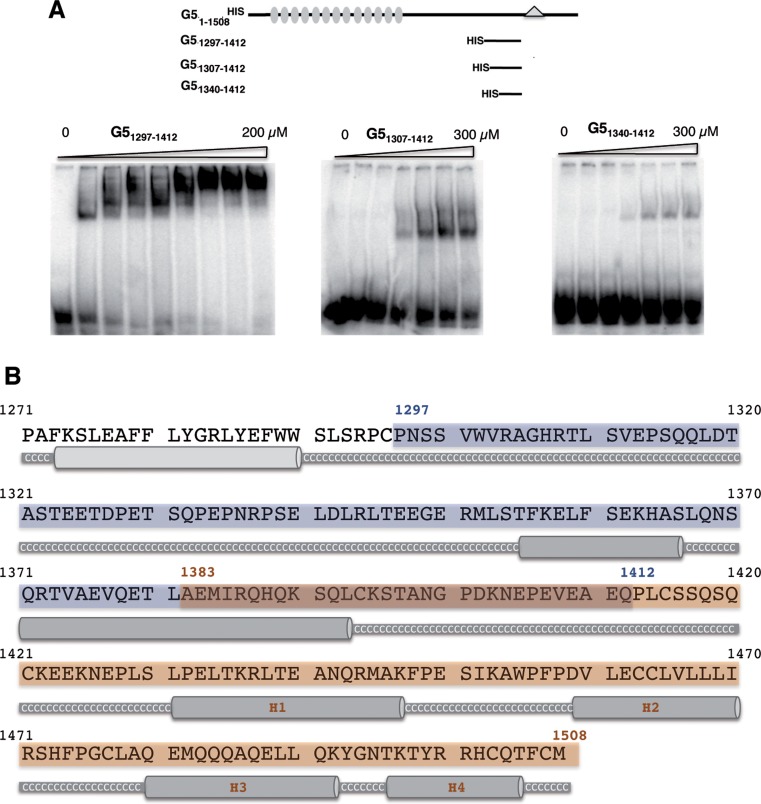
The RNA-binding capacity of G51297–1412 observed in photo-crosslinking assays (Figure 1B) prompted us to define in further detail the boundaries of RBS1 using gel-shift assays with radiolabelled domain 5 and purified HIS-tagged proteins. As shown in Figure 2A, the polypeptide G51297–1412 exhibited the highest RNA-binding capacity, a Kd of approximately 20 μM, since two constructs harbouring N-terminal deletions of 10 and 43 residues (G51307–1412, G51340–1412, respectively) showed a dramatic decrease in RNA binding, a Kd in excess of 350 μM. These results suggest that the entire RBS1 (blue shaded region in Figure 2B), which includes two serine residues at its coil-coiled N-terminus, is important for RNA binding. On the other hand, according to secondary prediction models of the Gemin5 C-terminal region, the RBS2 (brown shaded region in Figure 2B) comprises four helices, of which the second (H2) and third (H3) ones are enriched in leucine and glutamine residues, respectively.
Figure 2.
Determination of the RNA-binding capacity of Gemin5 truncated polypeptides. (A) Gel-shift assay conducted with increasing amounts of purified His-tagged Gemin5 polypeptides depicted at the top and radiolabelled domain 5 RNA, fractionated in native gels and visualized by autoradiography. (B) Gemin5 C-terminal region structure prediction. Jpred3 program (Jnet version: 2.2, www.compbio.dundee.ac.uk) was used for the secondary structure prediction analysis. Helical prediction scores below 7 are shown in light grey and scores between 8 and 9 in dark grey. High scores represent higher probability of adopting a helical conformation. Coil-coiled regions are displayed as a grey thick line and labelled in white Cs. Blue and brown boxes represent the boundaries of the two regions analyzed; dark brown box depicts the overlapping sequence.
Next, we attempted to generate a structural model of polypeptide 1297–1412 which showed the highest RNA-binding capacity. All experimental conditions we assayed yielded samples that provide NMR data in agreement with the absence of tertiary structure (Supplementary Figure S1). Only ∼80% of the signals corresponding to the RBS1 domain were defined in the spectrum, precluding the unambiguous assignment of all resonances. Still, we have assigned some fragments of the sequence using standard backbone triple resonance experiments. Based on these assignments and on the dispersion of the NMR signals shown in the spectrum, we conclude that the RBS1 construct has a short helical conformation surrounded by unstructured regions, yielding the ensemble of flexible conformations observed in 2D and 3D NOESY (Nuclear Overhauser effect spectroscopy) experiments.
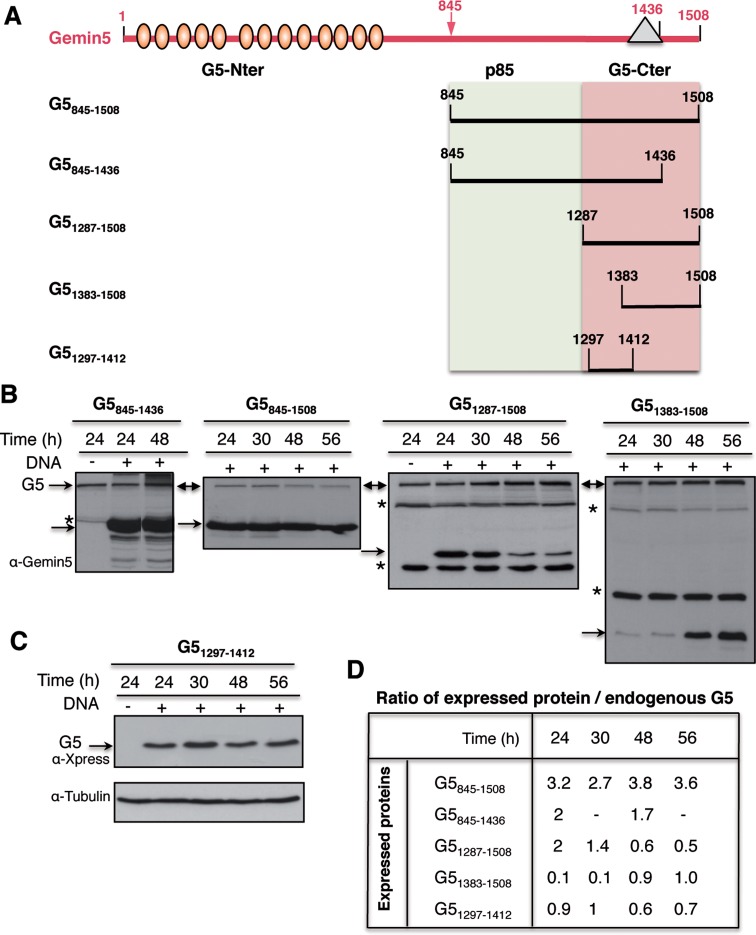
Expression of truncated Gemin5 proteins in mammalian cells reveals differences in protein accumulation
In order to assign the translational regulatory capacity to specific regions of the Gemin5 protein, we generated a set of constructs that allowed the expression of partially overlapping polypeptides in mammalian cells (Figure 3A). Immunodetection with anti-Gemin5 or anti-Xpress antibodies monitored polypeptide expression levels at different post-transfection times. Protein loading levels were verified using anti-tubulin. The results obtained indicated that each of these proteins was differentially accumulated in the cell, requiring different expression times to detect an equivalent amount of each polypeptide. For instance, accumulation of G5845–1508 and G5845–1436 in HEK293 cells was very efficient at 24 h post-transfection time (hpt), remaining stable up to 48 hpt (Figure 3B). Under the same conditions, expression of G51287–1508 was efficient up to 30 hpt, declining by 48–56 hpt. Surprisingly, when G51287–1508 was split into two fragments, the G51383–1508 showed a clear detection only 48–56 hpt (Figure 3B) while G51297–1412 exhibited a uniform expression over a period from 24 to 56 hpt (Figure 3C). Accordingly, the ratio of the ectopically expressed protein to the endogenous Gemin5 (immunodetected with the same antibody in the same sample) was about 2-fold for G5845–1508, G5845–1436 and G51287–1508 24 hpt. In contrast, the G51297–1412 ratio reached maximum values (around 1) 30 hpt while G51383–1508 needed 48 hpt to detect similar expression levels (Figure 3D). Therefore, the post-transfection time necessary to accumulate similar levels of ectopically expressed protein varies in accordance with the region of Gemin5 expressed in mammalian cells.
Figure 3.
(A) Schematic of Gemin5 polypeptides expressed in transfected cells. Numbers indicate amino acid residues referred to in the text. Ovals depict WD motifs located within the N-terminal region of the protein. A triangle within the C-terminal region of the protein depicts the position of the epitope recognized by Gemin5 antibody. (B) Analysis of the optimal time of expression of Gemin5 polypeptides in HEK293 cells. Plasmids expressing the indicated proteins were transfected in HEK293 cells; transfected (+) and mock-transfected (−) cells were harvested at the indicated time (h) in lysis buffer. Equal amount of total protein was loaded in SDS-PAGE (8% G5845–1436, and G5845–1508, 15% G51287–1508, G51383–1508) and immunoblotted with anti-Gemin5 antibody. Arrows depict the position of the full-length Gemin5 (G5) as well as the expressed proteins. Asterisks depict unspecific products. (C) Expression of G51297–1412 determined by anti-Xpress WB; loading control was assessed by anti-tubulin. (D) Ratio of the level of expression of Gemin5 polypeptides relative to the amounts of intracellular Gemin5 protein, detected by WB on the same membrane by anti-Gemin5 antibody. For G51297–1412 (which does not contain the anti-G5 epitope) the intensity of bands detected with anti-Xpress was normalized to the intensity observed with the same antibody in a parallel experiment with G51287–1508, blotted on the same membrane.
Expression of Gemin5 polypeptides reveals that RNA-binding site 2 acts as a translation repressor
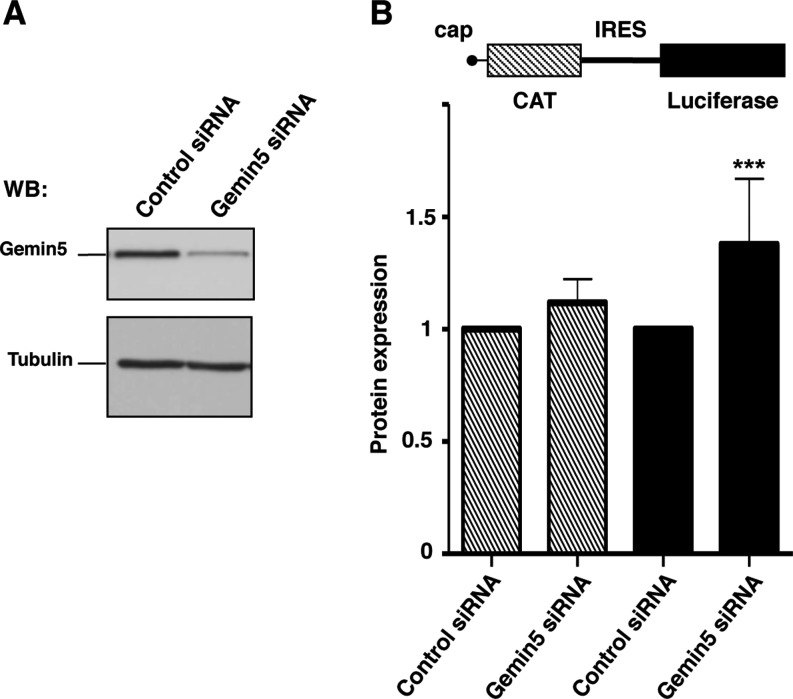
With the aim to determine the influence of Gemin5 polypeptides on translation control, we designed siRNAs to deplete Gemin5 in HEK293 cells (Supplementary Figure S2), of which siG5.3 was the most efficient 48 hpt, remaining active up to 72 hpt. In comparison to a control siRNA, the amount of Gemin5 protein detected by WB was reduced by 90% with siG5.3 (Figure 4A). To monitor the effect of Gemin5 depletion on translational efficiency, we used the construct CMVpBIC that allows the synthesis of a single bicistronic transcriptional unit (Figure 4B) in which the expression of LUC is monitored to assay IRES-dependent translation while the levels of CAT are monitored to assay 5′end-dependent translation initiation. The efficiency of LUC expression in Gemin5-depleted cells normalized to the activity observed in cells transfected with a control siRNA indicated that depletion of Gemin5 induced an increase of IRES-dependent translation reported by LUC activity, as indicated by the P values (P < 0.005) obtained in the t-test (Figure 4B). This result is in agreement with previous data using shRNAs targeting a different sequence of Gemin5 mRNA (6).
Figure 4.
Effect of Gemin5 depletion on protein synthesis. (A) WB analysis of HEK293 cell extracts transfected with siRNA G5.3 targeted to Gemin5 or a control siRNA with no target sequence in mammalian mRNAs. Tubulin was used as loading control. (B) Gemin5-depleted cells were used to monitor IRES- and cap-dependent translation upon transfection with equal amounts of the bicistronic plasmid (diagram at the top). Each experiment was repeated at least three times in duplicate wells. Effect on protein synthesis was calculated as the% of CAT (striped bars) and LUC (black bars) values observed in the control siRNA. Values represent the mean ± SD. (***P < 0.005).
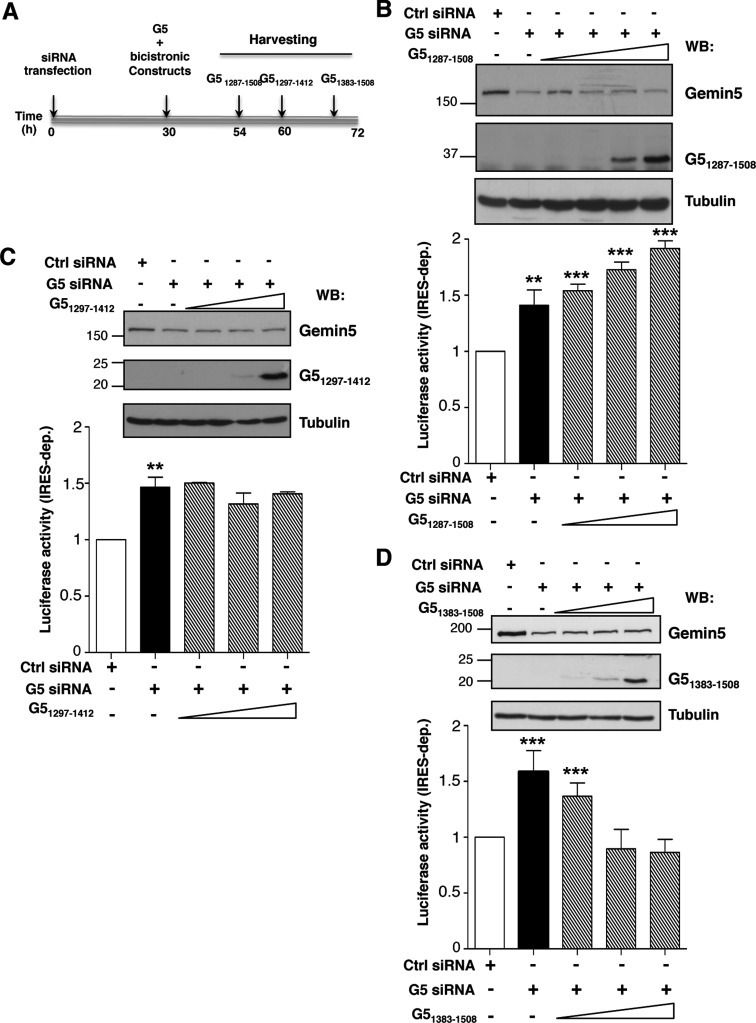
Expression of Gemin5 polypeptides in G5-depleted cells was used to assess the effect of different C-terminal regions of the protein in translation control (Figure 5A). The efficiency of depletion was monitored by immunodetection on the same membrane as that used to monitor the expression of the desired Gemin5 polypeptide (Figure 5B–D) using Gemin5 antibody, with the exception of G51297–1412 that was detected with anti-Xpress (Figure 5C). Loading control was verified by anti-tubulin immunoblot (Figure 5B–D). The effect on translation efficiency was measured by the expression of CAT (cap-dependent) and LUC (IRES-dependent) from the co-transfected plasmid CMVpBIC. In all cases, IRES-dependent translation was normalized to the levels of LUC observed in cells treated with the control siRNA (set to 100%) (empty bars in Figure 5B–D). Gemin5 depletion induced an increase of about 1.5-fold in IRES-dependent translation initiation in every experimental setting (black bars in Figure 5B–D), in agreement with earlier data (6). Expression of G51287–1508 affected IRES-dependent translation in a dose-dependent manner behaving as a stimulator of IRES-dependent translation (P < 0.005) (Figure 5B). However, expression of G51297–1412, corresponding to RBS1, did not alter the levels of IRES-dependent expression (Figure 5C) despite similar levels of the truncated Gemin5 polypeptide being detected. Therefore, none of these polypeptides mimic the repressor effect observed with the full-length protein (Figure 4A).
Figure 5.
Differential effect of Gemin5 C-terminal truncated polypeptides on IRES activity. (A) Diagram of the silencing and co-transfection assay with indication of harvesting time (h), depending on the expressed Gemin5 construct. Effect of expression of G51287–1508 (B), G51297–1412 (C) and G51383–1508 (D) on IRES-dependent translation (striped bars) monitor by LUC activity, relative to the values observed in control siRNA treated cells (empty bars). Black bars depict the effect of Gemin5 silencing on IRES-dependent translation. Values represent the mean ± SD (***P < 0.005; **P < 0.01). A Gemin5 WB (top panel) shows the silencing (Gemin5, p170 band) and the expression of the indicated G5 construct separated on 15% SDS-PAGE. Tubulin is used as loading control. Mobility of MW markers is indicated at the left of each WB.
Next, using the same experimental procedure, we observed that the most C-terminal region of the protein containing RBS2, G51383–1508, induced a dose-dependent inhibition of IRES-dependent translation, reaching levels similar to those observed in the control siRNA-depleted cells (Figure 5D). In this case, longer post-transfection times were required to detect a similarly intense polypeptide, consistent with expression data shown in Figure 3B. No significant changes in cap-dependent translation monitored by CAT expression were observed by expression of these polypeptides (Supplementary Figure S3). The IRES-dependent inhibitory capacity of G51383–1508 was also observed in in vitro translation assays when its mRNA was expressed immediately prior to bicistronic RNA (Supplementary Figure S4).
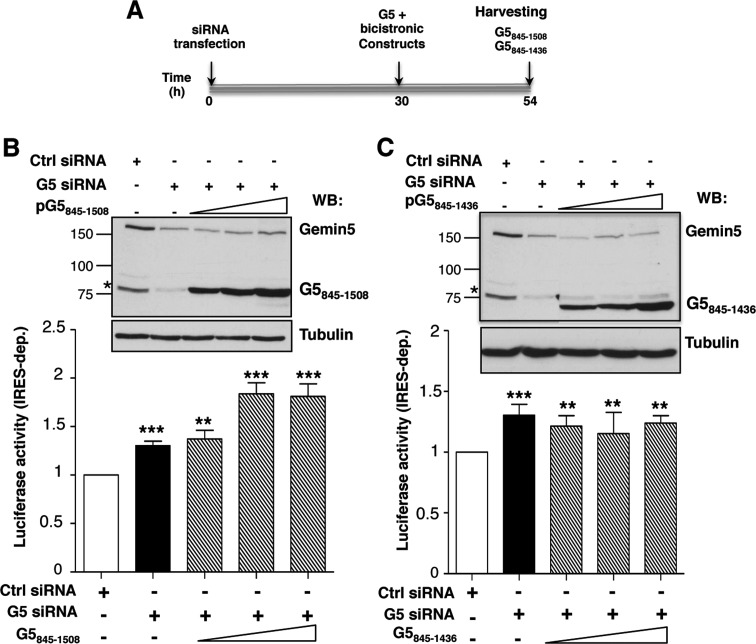
Using a similar experimental procedure (Figure 6A), we analysed the effect of proteins G5845–1508 and G5845–1436, adjusting the expression time of each protein to the optimal conditions shown in Figure 3B. Analysis of these proteins was of interest due to the fact that these polypeptides correspond to Gemin5 proteolysis fragments detected in infected cells, p85 and p57, respectively (25). While the p85 product appeared as a weak band in infected cells, p57 was accumulated during infection. Expression of G5845–1508 in G5-depleted cells showed a stimulation of IRES-dependent translation (P < 0.005) (Figure 6B), consistent with the G51287–1508 data (Figure 5B). In contrast, expression of G5845–1436, lacking the most C-terminal region of G5 (Figure 2B), had no effect on internal initiation (Figure 6C).
Figure 6.
Effect of G5845–1508 and G5845–1436 polypeptides on IRES activity. (A) Diagram of the silencing and co-transfection assay with indication of harvesting time (h). Effect of expression of G5845–1508 (B) and G5845–1436 (C) on IRES-dependent translation (striped bars) monitor by LUC activity, relative to the values observed in control siRNA treated cells (empty bars). Black bars depict the effect of Gemin5 silencing on IRES-dependent translation. Values represent the mean ± SD (***P < 0.005; **P < 0.01). A Gemin5 WB (top panel) shows the silencing (Gemin5, p170 band) and the expression of the indicated G5 construct separated on 8% SDS-PAGE. Tubulin is used as loading control. Mobility of MW markers is indicated at the left of each WB.
Collectively, these results led us to conclude that the IRES-dependent translational down-regulation of Gemin5 observed in tissue culture cells specifically resides in RBS2, within amino acids 1383–1508 corresponding to the most C-terminal region of the protein.
Relationship between the capacity to control translation initiation in mammalian cells and the proteolysis products found in infected cells
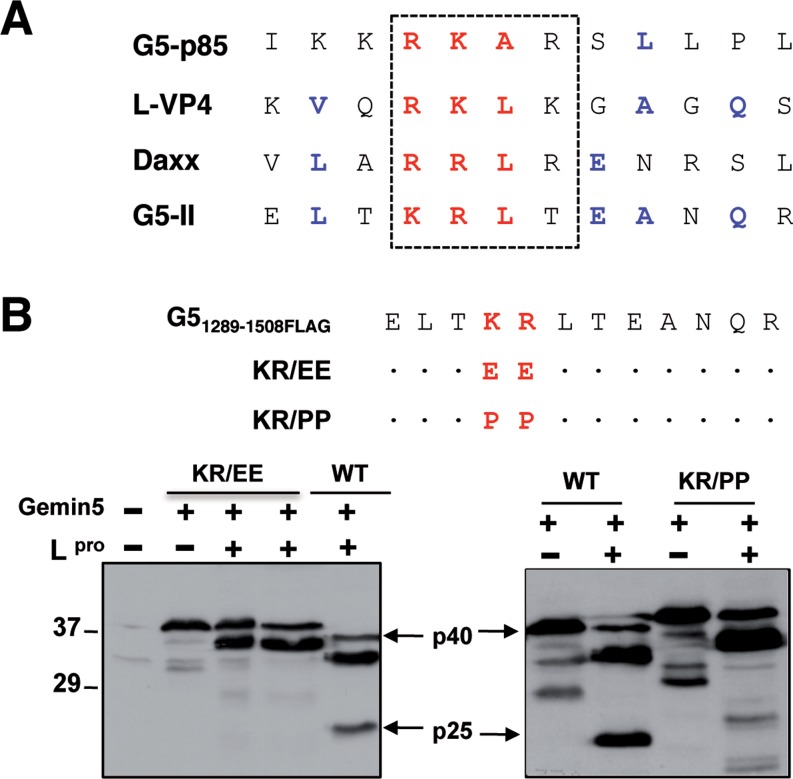
As mentioned above, Gemin5 is proteolyzed in FMDV infected cells by the action of the L protease, rendering at least two products p85 and p57 (25). The recognition motif for the L protease rendering p85 included a stretch of basic amino acids (arginine-lysine-alanine-arginine) RKAR (position 845) (Figure 7A), partially coincident with the cleavage site within the viral polyprotein (L-VP4) and Daxx. The aforementioned data suggested that a cleavage site near the C-terminal end could be responsible for the p57 product. However, no products <20 kDa were detected by WB with the available antibodies (either anti-Gemin5 or anti-FLAG using FLAG-tagged overexpressed proteins) in infected cells. In addition, the protease recognition motif was difficult to predict due to the large diversity between Lpro cleavage sites in host factors eIF4GI or eIF4GII (27,41). Furthermore, whether or not these products play a role on translation control remained to be elucidated.
Figure 7.
Mutational analysis of L protease cleavage site on Gemin5. (A) Amino acid sequence alignment of candidate L protease recognition motif near the C-terminal sequence of Gemin5 (G5-II) with known Lpro substrates (G5-p85, L-VP4 FMDV polyprotein and Daxx). (B) Diagram of the double amino acid substitutions introduced in the wild-type Gemin5. A dot in the sequence alignment is used to indicate no change with respect to the wild-type sequence. Plasmids expressing G51289–1508FLAG (WT) (p40) or the substitution mutants (carrying substitutions indicated at the top) in IRES-dependent manner were co-transfected with increasing amounts of a plasmid expressing the FMDV L protease in BHK-21 cells. Cell extracts were analysed by 12% SDS-PAGE and WB using Gemin5 antibody. A slight decrease in the mobility of the mutated proteins was observed. Arrows indicate the position of L-induced cleavage product (p25), which was detected in the WT sequence loaded in parallel.
To gain information about the L protease cleavage site near the C-terminal end of Gemin5, we used the information available for Gemin5 p85, L-VP4 and Daxx (Figure 7A) searching for similarities in amino acid sequence. The motif (threonine-lysine-arginine-leucine) TKRL located in G51383–1508 downstream of the epitope recognized by the Gemin5 antibody was a promising candidate. To determine whether this sequence provided an authentic Lpro recognition motif, we used the transfection assay successfully used to identify the RKAR motif (25). Mutational analysis of the truncated FLAG-tagged Gemin5 sequence (p40) indicated that substitution of basic amino acids by glutamic or proline (KR (lysine-arginine) to EE (glutamic-glutamic) or PP (proline-proline)) fully abrogated the cleavage by L protease in transfected cells (Figure 7B). This result indicated that cleavage around the TKRL motif of Gemin5 generates a small peptide (p25), corresponding to the most C-terminal fragment of the protein.
The identified Lpro cleavage products (Figure 7) and (25) are closely coincident with the G5 polypeptides G5845–1508, G5845–1436, G51383–1508, analysed in Figures 5 and 6. Therefore, given their differential capacity to mediate IRES-dependent translation in transfected cells it is tempting to suggest that the proteolysis products accumulated in infected cells may exert similar effects on translation initiation than the Gemin5 polypeptides analysed in this study.
DISCUSSION
The focus of this work was to decipher the critical regions of Gemin5 responsible for both an interaction with the IRES element and to mediate translation regulation. Here we show that two minimal regions of Gemin5 encompassing amino acids 1297–1412 (RBS1) and 1383–1508 (RBS2) possess RNA-binding capacity, being responsible for the interaction with the 3′end domain of a picornavirus IRES element. By expressing various fragments of the protein in Gemin5-depleted cells, we show that the polypeptide 1383–1508 specifically down-regulates IRES-dependent translation in mammalian cells. In addition, this polypeptide retains the capacity to interact directly with the IRES RNA in vitro in the absence of other factors.
By using UV-crosslinking, RNA-binding pull down and gel-shift assays, we show that two short regions of Gemin5, RBS1 and RBS2, are sufficient to bind directly to domain 5 of the FMDV IRES, with RBS1 having a higher affinity than RBS2. Longer polypeptides including these RNA-binding sites also interact with the IRES. Furthermore, deletion mutant G5Δ1365–1394 removing a long region of RBS1 (see Figure 2B) retains RNA-binding capacity in UV-crosslinking (Supplementary Figure S5), demonstrating that the C-terminal region of RBS1 is dispensable for RNA binding. In contrast, no RNA-binding capacity above the background signal was found for an N-terminal fragment corresponding to two thirds of the protein.
No apparent similarity has been detected between the Gemin5 C-terminal region and known RNA-binding motifs. Therefore, the RNA-binding capacity of the C-terminal region of Gemin5 was unpredictable using bioinformatics’ approaches. To get some insights into the molecular basis of this interaction, we analysed by NMR the polypeptide carrying RBS1 that showed the highest RNA-binding capacity. However, only ∼80% of the signals were defined in the spectrum (Supplementary Figure S1), precluding the assignment of all resonances. Based on the assignments of some fragments of the sequence using standard backbone triple resonance experiments and on the dispersion of the NMR signals shown in the spectrum, we conclude that the RBS1 region has a short helical conformation surrounded by unstructured regions. The presence of these elements of secondary structure is in agreement with the obtained secondary structure prediction (Figure 2B).
Intrinsically unstructured proteins (IUPs) are generally flexible in order to perform their functions while others fold only in complex with target structures (42). IUPs play roles in cell signalling and cell cycle regulation, DNA recognition molecules, protein-RNA recognition, among others (43–45). Compared to sequences of ordered proteins, disordered protein sequences are depleted in I, L, V, W, F, Y and C, and enriched in E, K, R, G, Q, S, P and A. As a consequence of the low representation of hydrophobic amino acids in a given protein sequence, the abundant hydrophobic interactions present in folded structures are nearly absent and display physicochemical characteristics resembling those of random coils. When analysed by NMR, disordered samples display poor signal dispersion, as we observed in the NMR data acquired with the Gemin5 construct studied. This flexibility probably represents a great advantage for a scaffold protein since the protein can easily select the best conformation among the ensemble to recognize many ligands. However, in our specific case it hampers the identification of the residues involved in RNA recognition, since both bound and free protein chemical shifts are in fast exchange.
According to secondary prediction models of the Gemin5 C-terminal region, RBS2 comprises four helices (Figure 2B). Interestingly, the sequence of the second one (H2) is enriched in leucine (LECCLVLLLI) while the third predicted helix (H3) contains a glutamine-rich (QEMQQQAQELLQ) sequence. Despite none of these sequences are canonical RNA-binding motifs, leucine-rich and particularly, glutamine-rich motifs are known to mediate the interaction of proteins with RNA, as reported in the case of LRPRPC protein (46) and Tia 1 (47), giving support to the RNA-binding capacity of the G51383–1508 shown in this study.
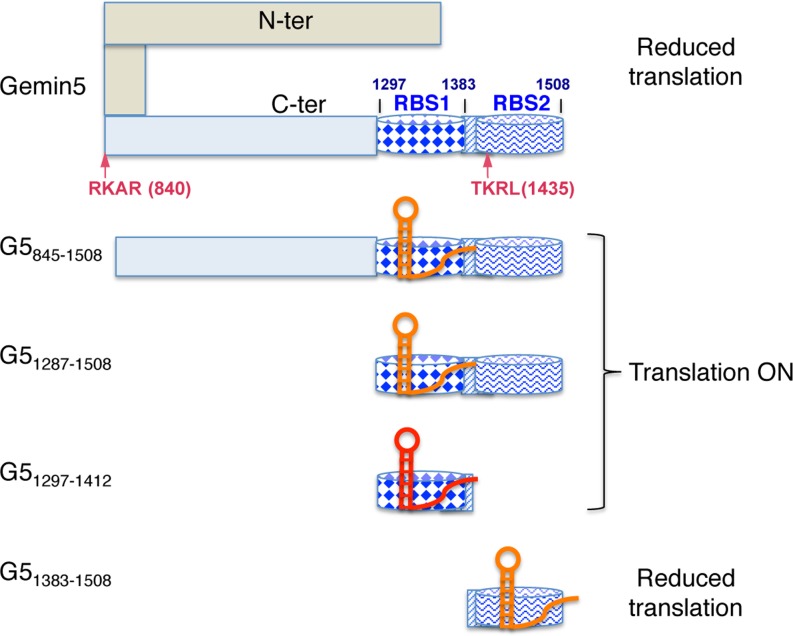
Functional analysis involving expression of various Gemin5 polypeptides in mammalian cells with reduced amounts of the endogenous protein provided insights into the mechanism mediating translation control. The repressor effect of Gemin5 on IRES-dependent translation precisely maps to residues 1383–1508, corresponding to RBS2. In contrast, overexpression of the G51297–1412 or G5845–1436 fragments, containing RBS1, did not appreciably affect translation efficiency, whereas expression of G5845–1508 and G51287–1508 stimulated translation slightly. These differential effects on translation are summarized in a model (Figure 8) that illustrates the mechanism deciphered in this study. Gemin5 is proteolyzed during FMDV infection generating at least two fragments, p85 and p57 (25), corresponding to G5845–1508 and G5845–1436, respectively. Proteolysis was observed at similar times as PABP or PTB, and later than eIF4G, presumably contributing to inhibit cellular gene expression by eIF4G-independent mechanisms. Given the fact that the amounts of p85 and p57 in infected cells were differentially accumulated (25), we investigated the potential role of each of these products in translation control. Remarkably, while the full-length protein down-regulates translation, G845–1436 corresponding to the product accumulated in infected cells does not impair IRES-dependent translation. Instead, the short peptide G51383–1508 that was undetected in infected cells, consistent with the observation that its expression declined over time (Figure 3), represses translation. In contrast, its counter part G51297–1412 does not affect translation efficiency, as it also happens with G5845–1436. Thus, we would like to conclude that the repressor polypeptide G51435–1508 produced in infected cells is not stable while the stable products (p85, p57) do not interfere IRES-dependent translation. Indeed, G5845–1508 slightly stimulates translation. The close correlation between the effect on IRES-dependent translation reported in this study and the amounts of proteolysis products accumulated in infected cells is consistent with the hypothesis that conformational changes of the protein induced by the differential RNA-binding capacity of RBS1 and RBS2 and/or their capacity to interact with other factors are at the basis of the role of Gemin5 on translation control (Figure 8). Conversely, changes in RNA structure induced by the binding of the IRES to RBS1 or RBS2 depending upon the Gemin5 polypeptides could also be compatible with the differential effect on translation. Further research to clarify the conformations adopted by these domains might contribute to illuminate the crosstalk between the elements in the bipartite RNA-binding site of Gemin5.
Figure 8.
Hypothesized model for the differential effect of Gemin5 truncated polypeptides on translation control. The C-terminal region of Gemin5 starting at residue 845 is shown in pale blue. Within this region, dark blue barrels depict the RNA-binding sites RBS1 (diamonds) and RBS2 (wavy line), respectively. The striped rectangle located between these sites depicts the overlapping region of the polypeptides analysed in this study, which was partially deleted in construct G5Δ1365–1394. Arrows depict the RKAR (840) and TKRL (1435) recognition sites of the FMDV L protease. Domain 5 of the FMDV IRES is shown as a red or orange hairpin, indicating the higher or lower efficiency of binding to the Gemin5 polypeptides based on gel-shift or UV-crosslinking assays. In this model, we hypothesize that the IRES RNA binds to RBS2 on the most C-terminal fragment G51383–1508 and the full-length Gemin5 protein reducing translation efficiency, while binding of the IRES RNA to RBS1 in proteins G5845–1508, G51287–1508 and G51297–1412 does not adversely affect internal initiation of translation. Details are provided in the text.
In addition to these properties, Gemin5 was described as a scaffolding protein with the capacity to interact with eIF4E (48) as well as m7GTP-resin (49), two features that link Gemin5 with translation events. Gemin5 that is found in the cell cytoplasm (50) was recently included in the mRNA interactome catalogue (51), congruent with a multifunctional role of this protein. It is likely that Gemin5 may recruit (or interfere with) other factors that also have mRNA-binding capacity and thus regulate translation. Importantly, we have discovered that Gemin5 directly binds to the IRES through a specific stem loop allowing a large degree of RNA sequence flexibility (24). The RNA-binding capacity of this region of Gemin5 differs from that reported to recognize the sm site of snRNAs (10). This difference suggests the existence of several RNA targets recognized by specialized domains likely assembled in distinct functional complexes. Further studies will be necessary to identify novel targets of Gemin5 and understand the possible role of this protein on the expression of other mRNAs within the cell.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are indebted to E. Pedroso for providing us with chemically synthesized RNA and C. Gutierrez for valuable comments on the manuscript. M.J.M. is an ICREA Programme Investigator.
FUNDING
Ministeriode Economia y Competitividad (MINECO) (CSD2009-00080, BFU2011-25437); Fundación Ramón Areces. Funding for open access charge: MINECO, BFU2011-25437.
Conflict of interest. None declared.
REFERENCES
- 1.Muller-McNicoll M., Neugebauer K.M. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 2013;14:275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 2.Sanford J.R., Gray N.K., Beckmann K., Caceres J.F. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durie D., Lewis S.M., Liwak U., Kisilewicz M., Gorospe M., Holcik M. RNA-binding protein HuR mediates cytoprotection through stimulation of XIAP translation. Oncogene. 2011;30:1460–1469. doi: 10.1038/onc.2010.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castello A., Fischer B., Hentze M.W., Preiss T. RNA-binding proteins in Mendelian disease. Trends Genet. 2013;29:318–327. doi: 10.1016/j.tig.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Pacheco A., Reigadas S., Martinez-Salas E. Riboproteomic analysis of polypeptides interacting with the internal ribosome-entry site element of foot-and-mouth disease viral RNA. Proteomics. 2008;8:4782–4790. doi: 10.1002/pmic.200800338. [DOI] [PubMed] [Google Scholar]
- 6.Pacheco A., Lopez de Quinto S., Ramajo J., Fernandez N., Martinez-Salas E. A novel role for Gemin5 in mRNA translation. Nucleic Acids Res. 2009;37:582–590. doi: 10.1093/nar/gkn979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otter S., Grimmler M., Neuenkirchen N., Chari A., Sickmann A., Fischer U. A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J. Biol. Chem. 2007;282:5825–5833. doi: 10.1074/jbc.M608528200. [DOI] [PubMed] [Google Scholar]
- 8.Borg R., Cauchi R.J. The gemin associates of survival motor neuron are required for motor function in Drosophila. PloS One. 2013;8:e83878. doi: 10.1371/journal.pone.0083878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Battle D.J., Kasim M., Yong J., Lotti F., Lau C.K., Mouaikel J., Zhang Z., Han K., Wan L., Dreyfuss G. The SMN complex: an assembly machine for RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:313–320. doi: 10.1101/sqb.2006.71.001. [DOI] [PubMed] [Google Scholar]
- 10.Battle D.J., Lau C.K., Wan L., Deng H., Lotti F., Dreyfuss G. The Gemin5 protein of the SMN complex identifies snRNAs. Mol. Cell. 2006;23:273–279. doi: 10.1016/j.molcel.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 11.Yong J., Kasim M., Bachorik J.L., Wan L., Dreyfuss G. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol. Cell. 2010;38:551–562. doi: 10.1016/j.molcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonenberg N., Hinnebusch A.G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsh D., Mohr I. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 2011;9:860–875. doi: 10.1038/nrmicro2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez-Salas E., Pacheco A., Serrano P., Fernandez N. New insights into internal ribosome entry site elements relevant for viral gene expression. J. Gen. Virol. 2008;89:611–626. doi: 10.1099/vir.0.83426-0. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald K.D., Semler B.L. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim. Biophys. Acta. 2009;1789:518–528. doi: 10.1016/j.bbagrm.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spriggs K.A., Bushell M., Willis A.E. Translational regulation of gene expression during conditions of cell stress. Mol. Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 17.Martinez-Salas E. The impact of RNA structure on picornavirus IRES activity. Trends Microbiol. 2008;16:230–237. doi: 10.1016/j.tim.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez N., Fernandez-Miragall O., Ramajo J., Garcia-Sacristan A., Bellora N., Eyras E., Briones C., Martinez-Salas E. Structural basis for the biological relevance of the invariant apical stem in IRES-mediated translation. Nucleic Acids Res. 2011;39:8572–8585. doi: 10.1093/nar/gkr560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filbin M.E., Kieft J.S. Toward a structural understanding of IRES RNA function. Curr. Opin. Struct. Biol. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraser C.S., Hershey J.W., Doudna J.A. The pathway of hepatitis C virus mRNA recruitment to the human ribosome. Nat. Struct. Mol. Biol. 2009;16:397–404. doi: 10.1038/nsmb.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komar A.A., Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–240. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pacheco A., Martinez-Salas E. Insights into the biology of IRES elements through riboproteomic approaches. J. Biomed. Biotechnol. 2010;2010:458927. doi: 10.1155/2010/458927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreev D.E., Fernandez-Miragall O., Ramajo J., Dmitriev S.E., Terenin I.M., Martinez-Salas E., Shatsky I.N. Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot-and-mouth disease virus RNA. RNA. 2007;13:1366–1374. doi: 10.1261/rna.469707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pineiro D., Fernandez N., Ramajo J., Martinez-Salas E. Gemin5 promotes IRES interaction and translation control through its C-terminal region. Nucleic Acids Res. 2013;41:1017–1028. doi: 10.1093/nar/gks1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pineiro D., Ramajo J., Bradrick S.S., Martinez-Salas E. Gemin5 proteolysis reveals a novel motif to identify L protease targets. Nucleic Acids Res. 2012;40:4942–4953. doi: 10.1093/nar/gks172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Almstead L.L., Sarnow P. Inhibition of U snRNP assembly by a virus-encoded proteinase. Genes Dev. 2007;21:1086–1097. doi: 10.1101/gad.1535607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gradi A., Foeger N., Strong R., Svitkin Y.V., Sonenberg N., Skern T., Belsham G.J. Cleavage of eukaryotic translation initiation factor 4GII within foot-and-mouth disease virus-infected cells: identification of the L-protease cleavage site in vitro. J. Virol. 2004;78:3271–3278. doi: 10.1128/JVI.78.7.3271-3278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez Pulido M., Serrano P., Saiz M., Martinez-Salas E. Foot-and-mouth disease virus infection induces proteolytic cleavage of PTB, eIF3a,b, and PABP RNA-binding proteins. Virology. 2007;364:466–474. doi: 10.1016/j.virol.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Bonderoff J.M., Larey J.L., Lloyd R.E. Cleavage of poly(A)-binding protein by poliovirus 3C proteinase inhibits viral internal ribosome entry site-mediated translation. J. Virol. 2008;82:9389–9399. doi: 10.1128/JVI.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Breyne S., Bonderoff J.M., Chumakov K.M., Lloyd R.E., Hellen C.U. Cleavage of eukaryotic initiation factor eIF5B by enterovirus 3C proteases. Virology. 2008;378:118–122. doi: 10.1016/j.virol.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weng K.F., Li M.L., Hung C.T., Shih S.R. Enterovirus 71 3C protease cleaves a novel target CstF-64 and inhibits cellular polyadenylation. PLoS Pathog. 2009;5:e1000593. doi: 10.1371/journal.ppat.1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez de Quinto S., Lafuente E., Martinez-Salas E. IRES interaction with translation initiation factors: functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA. 2001;7:1213–1226. doi: 10.1017/s1355838201010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez N., Garcia-Sacristan A., Ramajo J., Briones C., Martinez-Salas E. Structural analysis provides insights into the modular organization of picornavirus IRES. Virology. 2011;409:251–261. doi: 10.1016/j.virol.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 34.Lopez de Quinto S., Martinez-Salas E. Involvement of the aphthovirus RNA region located between the two functional AUGs in start codon selection. Virology. 1999;255:324–336. doi: 10.1006/viro.1999.9598. [DOI] [PubMed] [Google Scholar]
- 35.Fernandez-Miragall O., Martinez-Salas E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA. 2003;9:1333–1344. doi: 10.1261/rna.5950603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernandez-Miragall O., Ramos R., Ramajo J., Martinez-Salas E. Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA. 2006;12:223–234. doi: 10.1261/rna.2153206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez-Gabriel M.A., Remacha M., Ballesta J.P. Phosphorylation of ribosomal protein P0 is not essential for ribosome function but can affect translation. Biochemistry. 1998;37:16620–16626. doi: 10.1021/bi981396i. [DOI] [PubMed] [Google Scholar]
- 38.Lopez de Quinto S., Martinez-Salas E. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA. 2000;6:1380–1392. doi: 10.1017/s1355838200000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 40.Bartels C., Xia T.H., Billeter M., Guntert P., Wuthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 41.Kirchweger R., Ziegler E., Lamphear B.J., Waters D., Liebig H.D., Sommergruber W., Sobrino F., Hohenadl C., Blaas D., Rhoads R.E., et al. Foot-and-mouth disease virus leader proteinase: purification of the Lb form and determination of its cleavage site on eIF-4 gamma. J. Virol. 1994;68:5677–5684. doi: 10.1128/jvi.68.9.5677-5684.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 43.Weiss M.A., Ellenberger T., Wobbe C.R., Lee J.P., Harrison S.C., Struhl K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature. 1990;347:575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]
- 44.Romero P., Obradovic Z., Li X., Garner E.C., Brown C.J., Dunker A.K. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 45.Markus M.A., Hinck A.P., Huang S., Draper D.E., Torchia D.A. High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA. Nat. Struct. Biol. 1997;4:70–77. doi: 10.1038/nsb0197-70. [DOI] [PubMed] [Google Scholar]
- 46.Mili S., Pinol-Roma S. LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol. Cell. Biol. 2003;23:4972–4982. doi: 10.1128/MCB.23.14.4972-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dember L.M., Kim N.D., Liu K.Q., Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- 48.Fierro-Monti I., Mohammed S., Matthiesen R., Santoro R., Burns J.S., Williams D.J., Proud C.G., Kassem M., Jensen O.N., Roepstorff P. Quantitative proteomics identifies Gemin5, a scaffolding protein involved in ribonucleoprotein assembly, as a novel partner for eukaryotic initiation factor 4E. J. Proteome Res. 2006;5:1367–1378. doi: 10.1021/pr0504539. [DOI] [PubMed] [Google Scholar]
- 49.Bradrick S.S., Gromeier M. Identification of gemin5 as a novel 7-methylguanosine cap-binding protein. PLoS One. 2009;4:e7030. doi: 10.1371/journal.pone.0007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hao le T., Fuller H.R., Lam le T., Le T.T., Burghes A.H., Morris G.E. Absence of gemin5 from SMN complexes in nuclear Cajal bodies. BMC Cell Biol. 2007;8:28–44. doi: 10.1186/1471-2121-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M., et al. Insights into RNA Biology from an Atlas of Mammalian mRNA-Binding Proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.