Abstract
In a chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) experiment, an important consideration in experimental design is the minimum number of sequenced reads required to obtain statistically significant results. We present an extensive evaluation of the impact of sequencing depth on identification of enriched regions for key histone modifications (H3K4me3, H3K36me3, H3K27me3 and H3K9me2/me3) using deep-sequenced datasets in human and fly. We propose to define sufficient sequencing depth as the number of reads at which detected enrichment regions increase <1% for an additional million reads. Although the required depth depends on the nature of the mark and the state of the cell in each experiment, we observe that sufficient depth is often reached at <20 million reads for fly. For human, there are no clear saturation points for the examined datasets, but our analysis suggests 40–50 million reads as a practical minimum for most marks. We also devise a mathematical model to estimate the sufficient depth and total genomic coverage of a mark. Lastly, we find that the five algorithms tested do not agree well for broad enrichment profiles, especially at lower depths. Our findings suggest that sufficient sequencing depth and an appropriate peak-calling algorithm are essential for ensuring robustness of conclusions derived from ChIP-seq data.
INTRODUCTION
Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) has become a standard technique for profiling transcription factors, chromosomal proteins and histone modifications (1–3). Identification of binding sites for transcription factors is relatively easy, as they are ‘point-source’ factors that produce localized, sharp peaks; histone marks, on the other hand, range from point-source (e.g. H3K4me3) to ‘broad-source’ factors that produce large enrichment domains (e.g. H3K27me3), Here, we focus on histone modifications, as these present the most challenging case. Among the key considerations in the design of ChIP-seq experiments are the following: (i) quality of the antibody, as large-scale validation efforts of the modENCODE and ENCODE consortia have found that nearly ∼1/4 of the tested histone antibodies failed specificity criteria by dot blot or western blot (4); (ii) which set of histone modifications are sufficient to capture the interested aspects of chromatin organization; (iii) appropriate controls, either an ‘input’ chromatin without an immunoprecipitation step or the use of a mock antibody and (iv) number of replicates necessary to capture biological variability. Recent guidelines by the modENCODE and ENCODE consortia deal with some of these topics (5).
In this work, we address another critical question: how many reads should we sequence to obtain reliable results in a cost-effective manner? In the early days, the cost of sequencing was the determining factor in deciding on the depth of sequencing, and some of the initial papers had only 3–6 million reads for DNA-binding factors in human (1,2). With the rapidly decreasing cost of sequencing, the average depth of sequencing per experiment has substantially increased. With the relative ease of multiplexing (combining multiple samples with barcoding on one unit of sequencing), an experimentalist now has a much greater control over the number of reads obtained in an experiment. For instance, a ‘lane’ on the Illumina HiSeq 2000 currently generates up to 200 million reads, and the experimentalist can choose to sequence, for instance, 20 ChIP libraries for 10 million reads per library, or 4 ChIP libraries for 50 million reads per library. In any given genome, ChIP-seq enrichment profiles are expected to saturate in terms of enrichment regions if the library is sequenced to sufficient depth. However, how many reads constitute a sufficient depth remains unclear, especially for profiling broad histone modifications.
Evaluating the influence of sequencing depth on the result of a ChIP-seq experiment is not simple. Even for transcription factors, the number of peaks increases without saturation as more reads are sequenced if only statistical significance is used, since even very small peaks become statistically significant when the number of reads at those peaks gets larger. Thus, an additional criterion (e.g. 2-fold enrichment in ChIP over background) is needed to reach saturation. This means that the exact criteria used in a specific peak-calling algorithm have a significant impact on the number of peaks detected. The problem is exacerbated for broad histone modifications, where the enrichment ratios are lower and it is more difficult to define a biologically meaningful enrichment ratio.
The diminishing return for additional reads beyond some minimal number is clear. In fly, Chen and colleagues analyzed deep-sequenced ChIP-seq data for two factors (Su(HW) and H3K36me3) and found that more than half of the Su(HW) peaks (a point-source insulator protein) detected at 120 million reads were recaptured at 2.7–5.4 million reads (6). However, this effect is much less dramatic for broad-source factors. How the saturation point for the enriched regions in each of the broad histone modifications scales with the number of reads is largely unexplored (e.g. the same analysis as Su(HW) was not done for H3K36me3 in (6)) and is the subject of this study. We also study how genome size impacts the saturation point. The human genome is ∼18 times larger than the fly genome, but the saturation point depends heavily on the type of histone mark, and the required increase in the read number is typically much less than 18-fold, depending on the mark distribution. For example, H3K36me3 should scale with the size of expressed exons, while H3K9me3 should scale with the size of the heterochromatic regions.
Numerous algorithms have been developed to detect enriched regions in ChIP-seq data, mainly for transcription factor (TF) binding proteins (7–9). Some have been designed or modified to identify broad enrichment regions (10–12). Several studies have reported comparisons of the performance of peak callers. However, whether and how their performance depends on sequencing depth has not been studied previously. The present study utilizes multiple peak callers to ensure that the main conclusions do not depend on specific features of a single peak caller.
In this study, we generated deep-sequenced fly and human ChIP-seq datasets for select histone modifications (H3K4me3, H3K36me3, H3K27me3 and H3K9me2/H3K9me3), which are representative of marks associated with promoters, transcriptional elongation, Polycomb-regulated repression and heterochromatin, respectively. Using these data, we explore several features of ChIP signals as a function of sequencing depth, including tag density profiles, genomic coverage, size and number of ChIP enriched regions. We also compare the performance of five popular peak-calling algorithms at different sequencing depths on these datasets, as well as on public mouse datasets that include ChIP-qPCR validation data (13,14). Furthermore, we predict genomic coverage at full saturation and sufficient sequencing depths for >20 marks in the fly.
MATERIAL AND METHODS
ChIP-seq datasets
The procedures for ChIP sample preparation and sequencing were previously described for human (15) and fly (16). Additional details including the information on cell lines/tissue types can be found at http://www.modENCODE.org (fly) and http://encodeproject.org/ENCODE (human). Datasets generated and analyzed in this study, including accession numbers, are summarized in Supplementary Table S1. To ensure high quality and consistency of ChIP profiles, we performed the cross-correlation analysis and compared enriched regions between replicates, as described in (5).
Alignments
For deep-sequenced datasets in fly and human, reads that passed default parameters of the Illumina quality filter were aligned using Bowtie (17). To build reference indices, Flybase reference r5.22 and hg19 were used for fly and human, respectively, using the Bowtie-build function with default parameters. The parameters of alignments were -n 2 -l 28 -e 70 -m 1 for unique mapping.
Correlation analysis in tag density profiles
To examine whether ChIP profiles reached saturation, we calculated the genome-wide Pearson correlation coefficients between full and subsampled data. The tag density profiles were generated using get.smoothed.tag.density() with the parameters of bandwidth = 100 bp and step size = 50 bp in the SPP R package (11).
Detecting enriched regions
To identify significantly enriched regions, we used the same sequencing depth for ChIP and input data in human and fly, with an assumption that in practice a similar number of reads for ChIPs and inputs are likely to be sequenced. It has also been suggested that equal numbers of ChIP and input reads result in best performance of peak callers (6). For Figures 1–3, we detected ChIP-enriched regions by comparing scaled ChIP and input tag counts to see if their ratio exceeded that expected from a Poisson process, using get.broad.enrichment.cluster() in the SPP R package (11) with a sliding window of 1 kb (default) (find.binding.positions() is typically used instead for point-source peaks). The clusters of significant windows with Z-score > 3 (default) were determined as enriched regions. The same parameters were used throughout this study. In Figure 5, to detect enriched regions, we used SPP, MACS2, PeakSeq, Scripture and ZINBA (7,8,10–12) with default parameters, except for MACS2 and ZINBA. The parameters for model estimation were turned off for MACS2 and ZINBA. MACS2, which is a newer version of MACS, was modified to perform peak calling in a distinctive mode for the broad peak detection, by adding the parameter ‘-broad’. Additional details can be found at https://github.com/taoliu/MACS/. For Figure 5, the results from human chromosome 1 were used.
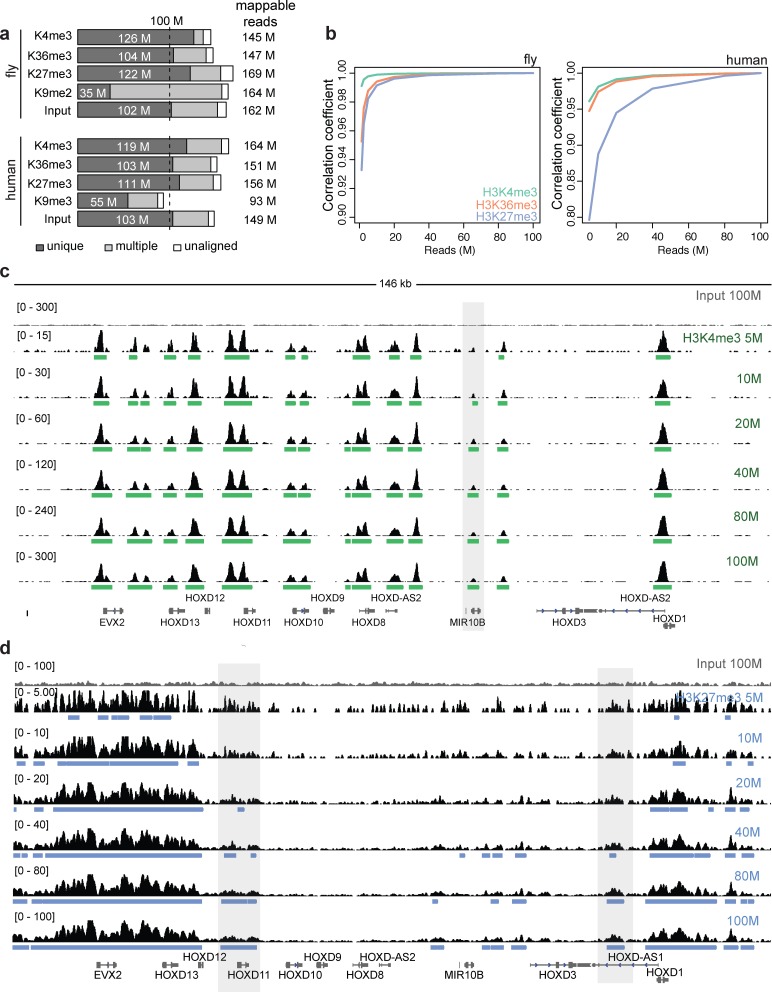
Figure 1.
Deep-sequenced ChIP-seq profiles for key histone modifications in human and fly. (a) Data overview. Bar graphs indicate the number of uniquely aligned reads (dark gray), multiply aligned reads (light gray) and unaligned reads (white). See also Supplementary Table S1 for full datasets analyzed in this study. Fly data are from late embryos and human data are from the A549 cell line. (b) Genome-wide Pearson correlation coefficients between tag density profiles from the 100 million reads and those from subsampled data in fly (left) and human (right). (c) ChIP tag density profiles at the HOXD loci for human H3K4me3 at different sequencing depths. Numbers on the y-axis denote the tag density ranges with a Gaussian kernel smoothing (σ = 100 bp). An input profile is on the top row for comparison. The green boxes below the ChIP profiles correspond to the significantly enriched regions based on the broad peak detection method of SPP (11). The enriched region highlighted is not detected at 5 million reads for this mark. (d) Same as (c) but for H3K27me3. The enriched regions at HOXD11 and HOXD-AS1 loci highlighted are not detected at low depths for this mark.
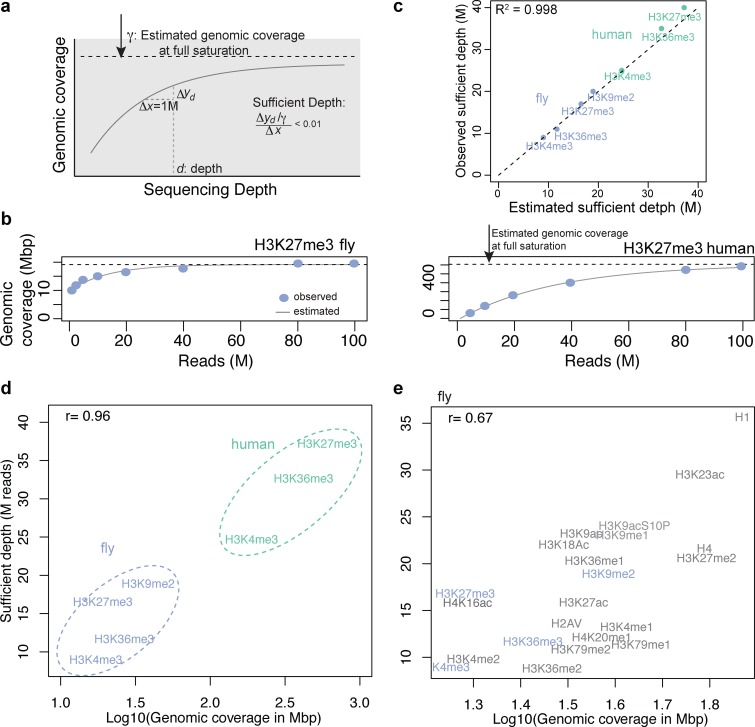
Figure 3.
Estimation of fully saturated genomic coverage and sufficient depth. (a) Diagram of how fully saturated genomic coverage and sufficient depth were estimated. (b) Example of observed and estimated genomic coverage for H3K27me3 in fly (left) and human (right). (c) Comparison between the estimated sufficient depth using the model and the observed sufficient depth calculated from the full data in Figure 2b. (d) Estimated genomic coverage at full saturation and sufficient depth. Human H3K9me3 was not included because 55 million reads were insufficient for this estimation. (e) Predicted genomic coverage at full saturation and sufficient depths for 22 fly marks in late embryos. Marks in blue indicate results from the deep-sequenced data, as in panel (c).
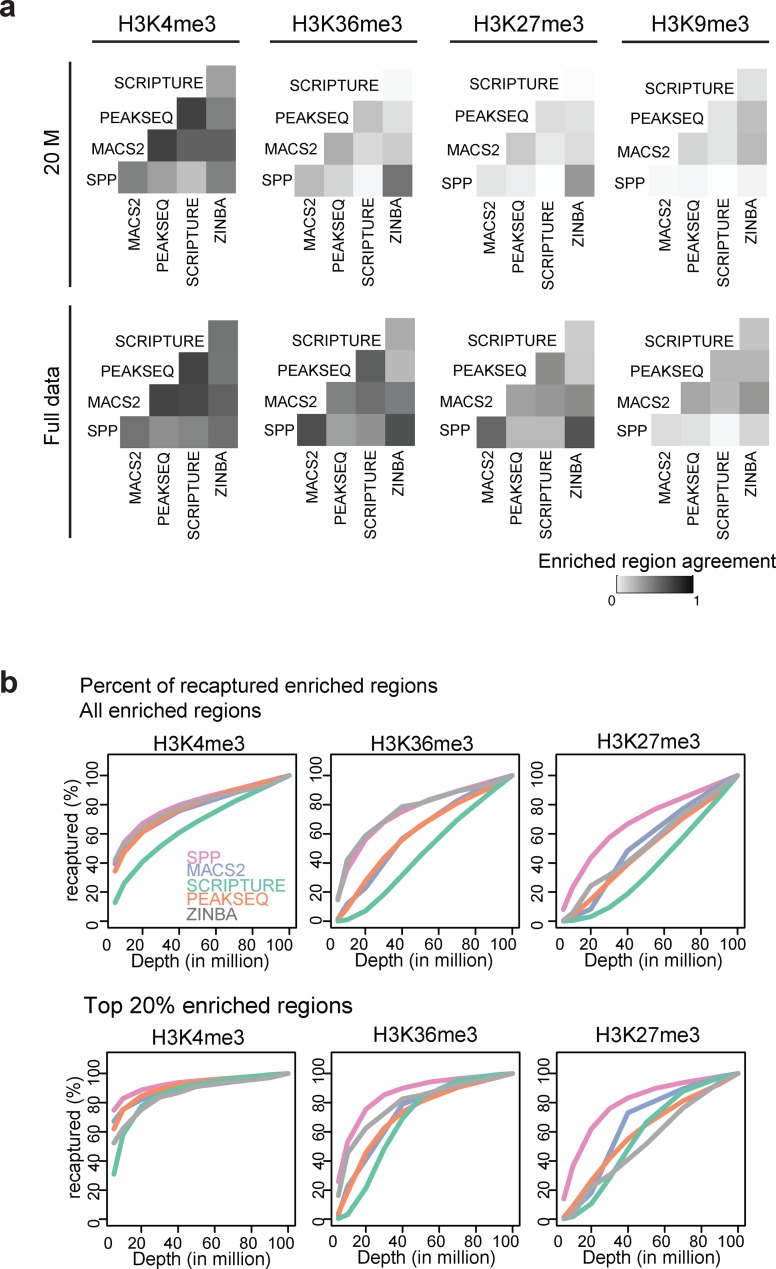
Figure 5.
Comparison of enriched regions detected by various algorithms. (a) Each cell in the heatmaps shows the Jaccard similarity coefficient between the enriched regions (in bp) by each pair of methods at 20 million and 100 million reads for H3K4me3, H3K36me3 and H3K27me3, and at 20 million and 55 million reads for H3K9me3. (b) Percentage of the enriched regions recaptured at different sequencing depths for all regions (upper panels) and percentage of top 20% enriched regions recaptured at different sequencing depths (lower).
In Figure 2c and d, the genomic coverage was defined as  , where si was each enriched region identified and N is the total number of the enriched regions. The mean size of enriched regions was
, where si was each enriched region identified and N is the total number of the enriched regions. The mean size of enriched regions was  .
.
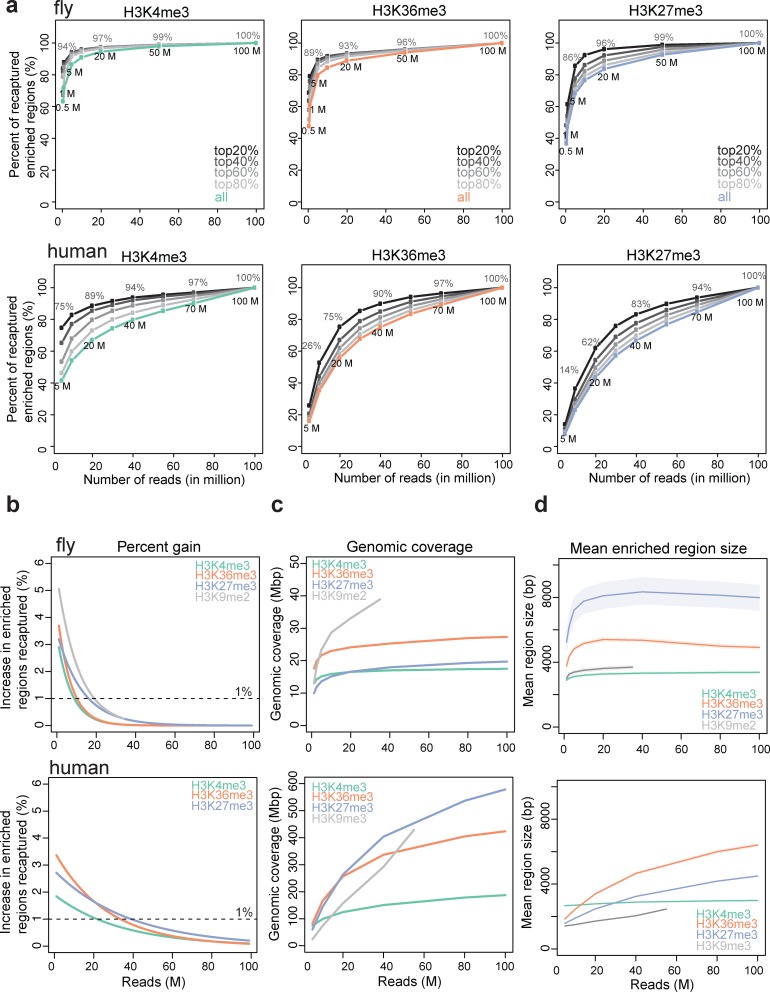
Figure 2.
Enriched regions with variable sequencing depths. (a) Percentage of significantly enriched regions from the full data recovered in each subsample for H3K4me3, H3K36me3 and H3K27me3 in fly (upper) and human (lower). The five lines correspond to the top 20%, 40%, 60%, 80% and all enriched regions from the full data. (b) Percentage of increase in enriched regions recaptured when an additional 1 million reads were sequenced for fly H3K4me3, H3K36me3, H3K27me3 and H3K9me2 (upper) and human H3K4me3, H3K36me3 and H3K27me3 (lower). (c) Genomic coverage of significantly enriched regions for fly H3K4me3, H3K36me3, H3K27me3 and H3K9me2 (upper) and human H3K4me3, H3K36me3, H3K27me3 and H3K9me3 (lower). (d) Mean enriched region size in fly (upper) and human (lower). The shaded lines indicate 95% confidence intervals.
Derivation of a model for genomic coverage as a function of sequencing depth
We assume that the observed tag distribution along a genome follows a Poisson distribution. If we assume a perfect ChIP, where tag distributions are only constrained within protein-binding sites, the Poisson rate λ would be approximately proportional to d/γ, where d is the total number of mappable reads and γ is total genomic coverage of the true protein-binding sites. Then the probability that we observe a read count c at a particular genomic location is
 |
where c is the number of reads at each bin (or bp), and λ is the mean coverage at each bin. The probability that we observe c > 0 becomes
 |
where θ1 is a factor that determines how steep the curve is, including the sizes of each enriched region. θ2 incorporates background noise, such as antibody efficiency. Although the Poisson model tends to underestimate the variance and we introduced additional simplifications such as using p(C > 0), this model explains the exponential behavior and results in reasonable estimates, in agreement with our observed values. From this, we modeled the observed genomic coverage (GC) as a function of sequencing depth d:
 |
where γ is genomic coverage when d is infinite. The values of α, β and γ could be estimated from the observed genomic coverage at each sequencing depth d (i.e. the number of bases found in peaks called at depth d) using a nonlinear regression. The sufficient depth is d (in million reads), beyond which  falls below 0.01.
falls below 0.01.
Measurement of variability in the detected regions by various algorithms
To assess agreement in the identified regions between the methods in Figure 5a, we calculated the Jaccard similarity coefficient between the enriched regions detected by each pair as J(Sa, Sb) = |Sa ∩ Sb| / |Sa ∪ Sb|, where Sa and Sb are the enriched regions in base pairs detected by the pair of algorithms denoted as a and b and | • | refers to the size of the set.
Analysis of mouse data
We obtained the datasets for H3K27me3 and H3K36me3 ChIP-seq profiles in mouse myoblasts and myotubes from Asp et al. (13). The uniquely aligned reads were downloaded from Gene Expression Omnibus (accession no. GSE25308). We used the ChIP-qPCR validated sites available in Asp et al. and Micsinai et al. (13,14). In (14), the number of validated sites in myoblasts were 197 and 94 for H3K27me3 and H3K36me3, respectively. Since the data in (14) were mainly designed to maximize the differences in detected peaks between algorithms, we used data in (13) as a primary dataset for our analysis. The sensitivity and specificity were calculated by comparing enriched regions in bp identified by SPP, MACS2, PeakSeq, Scripture and Hotspot (7,8,11,12,18) for positively and negatively validated sites. For mouse datasets, because the library size for input data was smaller than for ChIP data, the full dataset for the input was not subsampled.
RESULTS
Generation of deep-sequenced ChIP-seq data in human and fly
To investigate the effect of sequencing depths in histone modification experiments, we generated ∼150 million reads for some key histone modifications in fly embryos (50 bp reads) and human A549 (lung adenocarcinoma epithelial cell line, 35 bp reads), as part of the fly modENCODE and human ENCODE consortia (Figure 1a). Our approach is to compare these datasets with their read-subsampled datasets, considering the full data as an approximation of the true profile. As we will discuss later, most of the deep-sequenced data reached saturation based on several measures. The percentage of uniquely mappable reads is the highest for H3K4me3, followed by H3K27me3, H3K36me3 and H3K9me2 or H3K9me3, in both fly and human data (Figure 1a). Since reads that originate from the repeat-enriched regions are often not uniquely mappable, it is not surprising that the percentage of uniquely mappable reads in H3K9me2/H3K9me3 marks is substantially lower than other marks (Figure 1a). Although including multiply mappable reads for marks involving predominantly repetitive regions may increase the overall mappability, this operation would introduce additional uncertainty in genomic mapping and therefore was not considered in this study. In subsequent analyses, ‘sequencing depth’ refers to the number of uniquely mappable reads; this number divided by the mapping rate is the total number of reads to be sequenced.
To perform a fair comparison across multiple marks with different numbers of uniquely mapped reads, we first randomly sampled 100 million uniquely mapping reads for H3K4me3, H3K36me3, H3K27me3 and input. A different threshold was used for H3K9me3 (55 million) or H3K9me2 (35 million) because they have smaller numbers of uniquely mapped reads. We considered the 100 (or 35 or 55) million reads as the full data, and always performed subsampling from these reads.
Tag profiles with variable sequencing depth
To assess variations in tag density profiles with respect to sequencing depth, we calculated genome-wide Pearson correlation coefficients between the tag profiles from the 100 million-read full data and the subsamples. We observed that the correlation increases at greater sequencing depth as expected, reaching a plateau after a rapid initial increase (Figure 1b). The saturation points differ depending on the histone modification and the genome under study. H3K4me3, a point-source modification, is saturated at a lower depth, while H3K27me3, a broad-source modification, requires more reads for saturation. We found that human profiles tend to have a higher saturation point than fly profiles: for the three marks (H3K4me3, H3K36me3 and H3K27me3), the human profiles reach a plateau at around 40–50 million reads, whereas the fly profiles reach a plateau by ∼20 million reads. These patterns are evident when viewed in a genome browser (Figure 1c and d for human H3K4me3 and H3K27me3; see Supplementary Figures S1–S3 for other marks in human and fly). For most fly histone modifications, the tag density profiles become highly similar to that of the full data at >20 million reads (Supplementary Figures S2–S3). In human, for H3K4me3, the profiles at different sequencing depth are largely identical, and most enriched regions are identified at <20 million reads (Figure 1c). In contrast, for H3K27me3, the profile at 40 million reads was different from those at greater depths; some regions are not detected at low depths for this mark. In the HOXD clusters that are often targeted by Polycomb group proteins (19), the enriched regions of H3K27me3 at the HOXD11 and HOXD-AS1 loci are not consistently identified until the read count is >40 million.
Enriched regions with variable sequencing depths
We examined significantly enriched genomic regions in subsampled and full datasets, using SPP as the base method (11). This method (developed in the senior author's laboratory) features options for sharp and broad profiles and has been used to process ChIP-seq data for ENCODE along with MACS (12). With the broad peak detection option, enriched regions are determined by comparing scaled ChIP and input tag counts to assess if their ratio exceeds that expected from a Poisson process. The clusters of significant windows are determined as enriched regions. We then calculated the percentage of significantly enriched regions from the full data, recaptured in each subsample (Figure 2a; see Supplementary Figure S4 for H3K9me2/me3). This measure is analogous to sensitivity; specificity is not informative since the enriched regions identified from a subsample are almost entirely a subset of the regions identified from the full data. For fly marks, our analysis suggests that 80% of enriched regions in the H3K4me3, H3K36me3 and H3K27me3 data are detected at 3, 6 and 15 million reads, respectively (Figure 2a, upper panels). Human data exhibit a similar trend, although a higher sequencing depth is required to reach saturation: 80% of enriched regions in the full data are called at 50 million reads for both H3K4me3 and H3K36me3, and at 60 million reads for H3K27me3 (Figure 2a, lower panels). Notably, we did not observe saturation for H3K9me3 regions in human at any depth examined (up to 55 million reads) (Supplementary Figure S4).
To explore the relationship between the strength of ChIP signal and sequencing depths, we analyzed peaks with different ChIP signal strength as measured by the ChIP/Input ratio. The top 20% of regions sorted by ChIP signal strength are detected even at low depth of coverage (Figure 2a). In fly, 90% of these regions are identified at depths of 3, 4 and 5 million reads for H3K4me3, H3K36me3 and H3K27me3, respectively (Figure 2a, upper panels). In humans, 90% of the top 20% regions are identified at slightly higher depths: 20, 40 and 55 million reads for H3K4me3, H3K36me3 and H3K27me3, respectively (Figure 2a, lower panels). This suggests that saturation in human requires a large number of reads if the peaks display low ChIP/input ratios.
Defining sufficient sequencing depth
To further quantitate the relationship between sequencing depth and detection sensitivity, as measured by recovery of true enrichment region, we computed the percent increase in enriched regions recaptured when an additional 1 million reads are sequenced. Here we define a ‘sufficient depth’ to be the sequencing depth at which the percent gain per 1 million additional sequence reads falls below 1%—a point at which we deem the gain of additional sequencing to be minimal. The 1% threshold is arbitrary but reasonable in our experience. In fly, the sufficient depth was 9, 11, 17 and 20 million reads for H3K4me3, H3K36me3, H3K27me3 and H3K9me2, respectively (Figure 2b, upper). For the human marks, the sufficient depth was 25, 35 and 40 million reads for H3K4me3, H3K36me3 and H3K27me3, respectively (Figure 2b, lower). H3K9me3 does not fall below 1% gain within the total 55 million reads that we have. These results suggest that there is little gain in identification of enriched regions for fly data beyond 20 million reads, and that 40–50 million reads is a cost-effective choice for most human data.
We also measured genomic coverage and mean size of enrichment regions as a function of sequencing depth. As observed in Figure 2a, the genomic coverage for fly marks saturates at >20 million reads, except for H3K9me2. In human, the genomic coverage continues to increase even at very high depth, although it does slow down. The coverage for H3K9me3 in particular appears to increase linearly with the number of reads (Figure 2c). Examination of the mean enriched region size exhibits a similar trend. In fly, the mean enriched region size becomes stable beyond 10 million reads for most marks. In human, the size of enriched regions still increases with more reads, except for H3K4me3 (Figure 2d).
Mathematical modeling of genomic coverage and sufficient depth
For most datasets, sequencing is not as deep as in these test datasets. Thus, we devised a simple mathematical model for the relationship between sequencing depth and maximum genomic coverage of the enriched region (i.e. when an infinite number of reads are available) (see Methods). This model allows one to estimate genomic coverage and sufficient sequencing depth from less deep datasets (this extrapolation obviously does not work if the coverage of the available data is too low). By fitting the genomic coverage of subsampled reads in a ChIP-seq dataset to the model, we estimated the maximum genomic coverage of a particular enrichment mark, as well as the sufficient sequencing depth (Figure 3 and Supplementary Figures S5–S6). For H3K4me3, H3K36me3 and H3K27me3 in both fly and human, this model predicts that the genomic coverage at 100 million reads captured > 95% of the estimated genomic coverage at full saturation (Figure 3b; see Supplementary Figure S5 for other marks). The estimated genomic coverage was in agreement with the observed genomic coverage (R2 > 0.98; Supplementary Figure S5). The estimated sufficient depth was concordant with those calculated from the full data in Figure 2b (R2 = 0.998; Figure 3c and Supplementary Figure S6) and was also highly correlated with the estimated full genomic coverage on a log scale (Pearson correlation coefficient r = 0.96; Figure 3d). Our method can be applied to TF data as well because the number of peaks detected in a TF binding profile is analogous to the number of basepairs identified in broad marks (Supplementary Figure S6).
We then predicted the maximum genomic coverage and sufficient sequencing depth for 18 additional fly histone modifications generated by the modENCODE consortium, ranging from 17 to 35 million reads. Similar to above, the estimated sufficient depth was positively correlated with the estimated genomic coverage on a log scale (r = 0.67; Figure 3e, also see Table 1). The genomic coverage and sufficient depths were the smallest for marks associated with active promoters, such as H3K4me3 and H3K4me2, followed by transcription-related marks such as H3K36me3 and enhancer-related marks such as H3K4me1 and H3K27ac. Repressive marks such as H3K27me3 and H3K9me2 were located in the middle. Core histones (e.g. H1 and H4) and some ubiquitous histone modifications such as H3K23ac exhibited maximum genomic coverage, requiring greatest sequencing depth. Although we provide the estimated genomic coverage, sufficient depth, and required total library size, it is important to note that these numbers could vary depending on the mapping rate of each sample, which is affected by several factors including the library protocol and the genomic locations where the marks are enriched.
Table 1. Estimated genomic coverage when fully saturated and sufficient depth for 22 fly marks. A factor is the ratio of sufficient depths between the given mark and H3K4me3. To infer the total library size from the sufficient depth, we used the mapping rates from fly late embryos data (estimated total library size = sufficient depth/mapping rate).
| Mark | Genomic coverage (Mbp) | Sufficient depth (million) | Factor | Required total library size (million) |
|---|---|---|---|---|
| H1 | 75 | 36 | 4 | ∼55 |
| H2AV | 32 | 14 | 1.5 | ∼20 |
| H3K18Ac | 31 | 23 | 2.5 | ∼30 |
| H3K23ac | 61 | 30 | 3.3 | ∼35 |
| H3K27ac | 35 | 16 | 1.8 | ∼20 |
| H3K27me2 | 63 | 21 | 2.3 | ∼30 |
| H3K27me3 | 20 | 17 | 1.8 | ∼25 |
| H3K36me1 | 37 | 21 | 2.3 | ∼30 |
| H3K36me2 | 30 | 9 | 1 | ∼10 |
| H3K36me3 | 27 | 12 | 1.3 | ∼15 |
| H3K4me1 | 43 | 14 | 1.5 | ∼15 |
| H3K4me2 | 20 | 10 | 1.1 | ∼10 |
| H3K4me3 | 18 | 9 | 1 | ∼10 |
| H3K79me1 | 45 | 12 | 1.3 | ∼15 |
| H3K79me2 | 34 | 11 | 1.2 | ∼15 |
| H3K9ac | 36 | 24 | 2.6 | ∼30 |
| H3K9acS10P | 44 | 25 | 2.7 | ∼35 |
| H3K9me1 | 41 | 24 | 2.6 | ∼35 |
| H3K9me2 | 39 | 19 | 2.1 | ∼60 |
| H4 | 62 | 22 | 2.3 | ∼30 |
| H4K16ac | 20 | 17 | 1.8 | ∼25 |
| H4K20me1 | 38 | 13 | 1.4 | ∼15 |
Comparison of detected enrichment regions by various algorithms
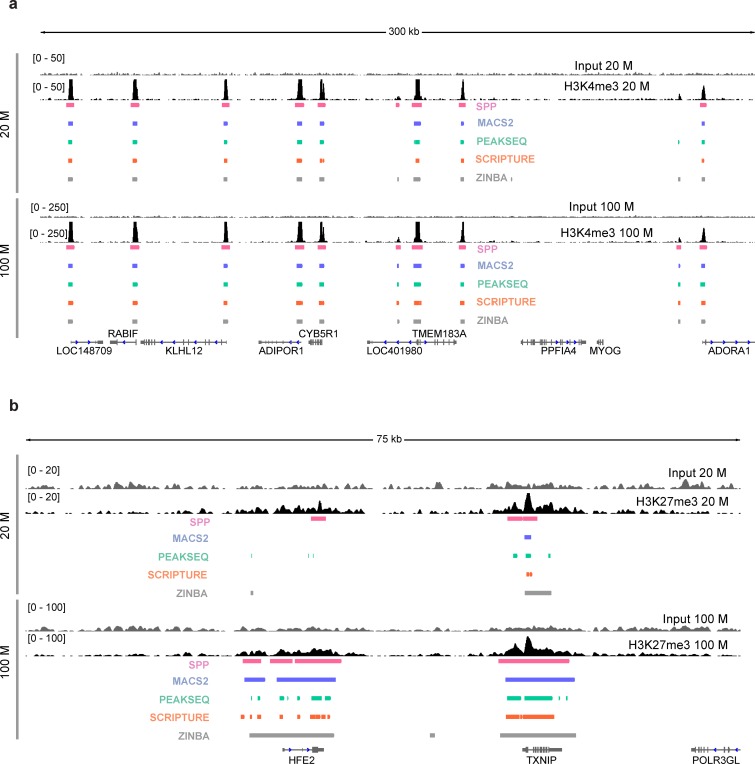
To explore the variability in broad enrichment regions identified by different algorithms at different sequencing depths, we used the human data to compare the results from five widely used peak callers: ZINBA (10), PeakSeq (7), MACS2 (12), Scripture (8) and SPP (11). The examples of tag density profiles along with enrichment regions detected by these algorithms are shown in Figure 4. For human H3K4me3, the regions called by these methods are concordant at both 20 and 100 million reads, with higher consistency at 100 million (Figure 4a). In contrast, the enriched regions detected for H3K27me3 differ drastically, especially at a low sequencing depth (Figure 4b). For example, at the HFE2 locus, all the algorithms identified some part of the region as H3K27me3-enriched at 100 million reads, but with variable levels of fragmentation. At 20 million reads, the differences from the 100 million case and between the methods were dramatic, with only three methods calling tiny fractions of the region. The algorithms designed to detect both broad peaks and sharp peaks (SPP, MACS2 and ZINBA) tend to be more stable at identifying broader enrichment domains across different read depths, compared to those designed primarily for sharp peaks. Examination of the genomic coverage and cluster sizes of the enrichment regions shows that the results vary between the methods, with the largest variation observed for H3K27me3 and H3K9me3 (Supplementary Figures S7–S8). We also observed that the genomic coverage and number of the enrichment regions tend to increase at higher sequencing depths.
Figure 4.
Enriched regions detected by various algorithms. (a) ChIP tag density profiles for human H3K4me3 at a sequencing depth of 20 million (upper panel) and 100 million (lower) reads. An input profile is on the top row for comparison. The boxes below the ChIP profiles indicate the enriched regions identified by SPP, MACS2, PeakSeq, Scripture and ZINBA (7,8,10–12). (b) Same as above but for H3K27me3.
To assess genome-wide agreement in the identified regions between the methods, we calculated the Jaccard similarity coefficient between the enriched regions detected by each pair of methods at 20 and 100 million reads for H3K4me3, H3K36me3 and H3K27me3 (for H3K9me3, the coefficients were obtained at 20 and 55 million reads). Our results show that identified regions are most consistent for H3K4me3 followed by H3K36me3, H3K27me3 and H3K9me3 (Figure 5a). Similarity of identified regions was higher for the full data compared to subsets of 20 million reads. This indicates that enriched regions detected by different algorithms tend to be more variable at a lower depth for broader marks, and that use of multiple algorithms might be beneficial in such cases. Next, we repeated our calculation of the percentage of genomic coverage recovered at each sequencing depth for each of the algorithms (Figure 5b; see Supplementary Figure S9 for H3K9me3). A similar trend was found as before, but there was also a substantial variation among the methods and across marks. The results were more consistent for the top 20% enriched regions (the top regions were ordered based on ChIP fold enrichment values when the algorithm outputs those; otherwise, P-values were used).
Comparison of peak calling algorithms using validated loci
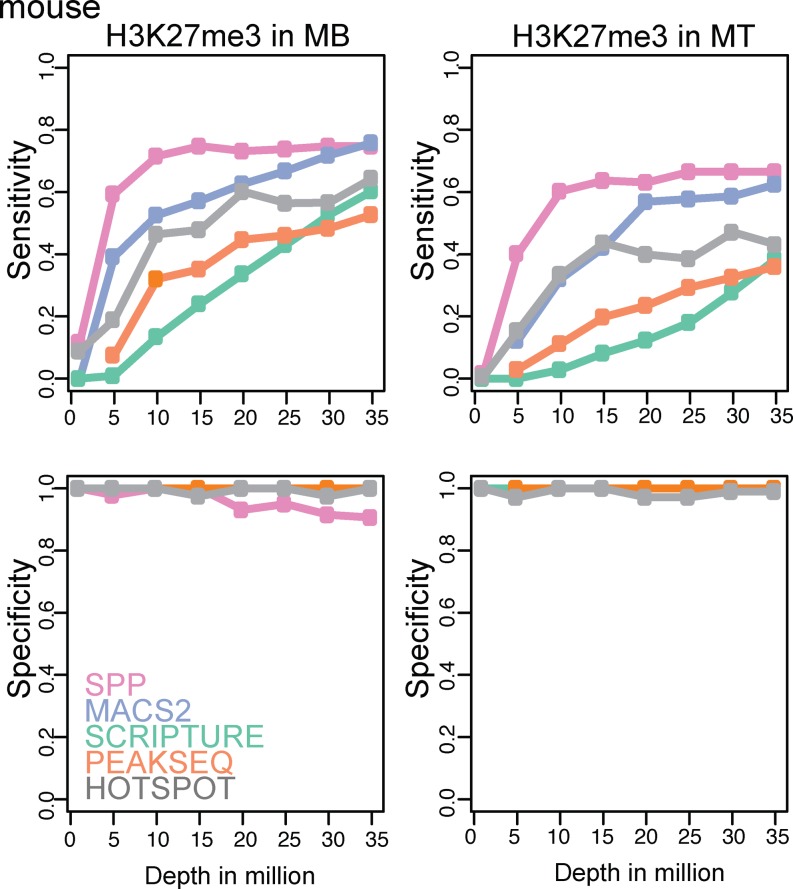
Instead of considering the enriched regions from the full data as the true set, another approach is to use an externally validated set. In a study of epigenetic changes underlying myogenesis, several histone marks were profiled in mouse myoblasts and terminally differentiated myotubes (13), ranging from 25 to 29 million reads for H3K27me3 and H3K36me3. Importantly, they also provided H3K27me3 ChIP-qPCR data for 200 loci. We determined the sensitivity and specificity as a function of sequencing depth for the H3K27me3 profiles from this paper (Figure 6) using the same subsampling analysis as before and using these five algorithms: SPP (11), MACS2 (12), PeakSeq (7), Scripture (8) and Hotspot (18). We also analyzed different ChIP-qPCR datasets for H3K36me3 and H3K27me3 in mouse myoblasts (14) (Supplementary Figure S10). As expected, all algorithms showed increased sensitivity at greater sequencing depths. For most methods, the sensitivity began to saturate at 15–20 million reads, but there was a fair amount of variation between the algorithms (Figure 6 and Supplementary Figure S10).
Figure 6.
Performance of peak calling algorithms using the qPCR-validated loci. Sensitivity (upper) and specificity (lower) for H3K27me3 in mouse muscle cells MB (myoblasts; left) and MT (myotubes; right) (13), considering qPCR-validated sites as the true set. Sensitivity and specificity were calculated based on the overlapped regions in bp.
DISCUSSION
This subsampling analysis shows that most fly histone modification profiles saturate at 10–20 million reads, depending on the mark. For human data, the saturation point is not apparent from the subsampling analysis, except for H3K4me3. We proposed to define sufficient sequencing depth based on the incremental change in the size of the enriched region per one million additional reads, and overall the estimates of sufficient sequencing depth obtained this way is in accordance with results using the full data. This definition suggests that in human ∼20 million reads is likely to be sufficient for H3K4me3 and ∼40 million for H3K36me3 and H3K27me3 (Figure 2b). We also observed that for the strongest 20% of the peaks, 90% of the enriched regions from the full data were reproduced at > 40–50 million reads for H3K36me3 and H3K27me3 (Figure 2a). Based on these results, we suggest that 40–50 million reads is a practical minimum depth for human marks, except for few special cases such as H3K9me3 that cover very large contiguous domains. Although we generated deep-sequenced data for just four marks, the estimated genomic coverage and sufficient depths in Table 1 and the nature of the marks (i.e. how broad they are) can be used to estimate the number of reads needed. For H3K9me3, core histones, and other broadly distributed factors, a future study with much larger datasets will be helpful in determining what depth is required. Here we showed the results using the cell/tissue types for which deep-sequenced dataset was available. The estimated sequencing depth is expected to change slightly for other cell/tissue types.
Although the human genome is ∼18 times larger than the fly genome, the ratio between sufficient fly and human H3K27me3 depth is only ∼2–3, suggesting that the required read count does not increase proportionally with genome size. This might be explained by the relationship between the predicted sequencing depth and genomic coverage we estimated in Figure 3c, where the sufficient depth increased linearly as a function of genomic coverage on the log scale. Thus, although the ratio of genomic coverage between fly and human may be ∼10–20, the resulting ratio on the log scale becomes much smaller (generally 2- to 3-fold, as seen in Figure 3c).
Determining the enriched regions and their boundaries for broad marks is more challenging due to several factors. First, the size of the enriched region differs significantly depending on the modification, and a single algorithm is unlikely to perform optimally at all length scales. Second, the enriched regions are more sensitive to the depth of sequencing compared to TF binding sites, because their ChIP fold enrichment values tend to be lower. Third, larger domains enriched for broad histone marks will have higher variability in genomic coverage and other sequence features, introducing greater fluctuations in profiles that may need to be accounted for through effective normalizations. Our results show high variability in the performance of different algorithms, especially at lower sequencing depth and for very large domains (e.g. H3K27me3). This suggests that researchers should choose proper algorithms that are specifically designed for broad marks in the study using histone modification profiles and that use of multiple algorithms can help reduce false positive regions.
Although this is typically not done in practice, our study makes it clear that it is more cost-effective to employ different depth of sequencing for different factors, using the expected genomic coverage as a guide. Fewer reads are needed for point-source protein factors that bind to a relatively small number of sites in the genome, whereas more reads are needed for broad histone modifications that cover large domains. If target numbers are set for read counts (constrained by the discrete units allowed by the number of barcodes), they can be achieved by appropriate barcoding of the samples. We provided the estimated genomic coverage at full saturation for >20 marks, and those numbers can serve as a valuable guide in inferring the optimal sequencing depth for each mark.
Finally, it is important to note that, although we described ‘sufficient’ depth here, this definition refers to how accurately the true underlying genome-wide features are captured by the data. At a higher level, a ‘sufficient depth’ depends on the purpose of the study. Although the cost of sequencing has decreased dramatically, sequencing multiple marks in multiple time points or cell types can be expensive. Thus, it is important that the investigator makes a judicious choice in experimental design to maximize biological insight given limited resources. In some cases, a ‘sufficient depth’ in a study may be substantially less than what we describe in this paper. For instance, when using transcription factor profiling to discover binding motifs, identifying even 10% of the true peaks may be sufficient. To understand the general characteristics of where enhancers are located genome-wide, a subset of the H3K27ac or H3K4me1 sites may be sufficient to give a reasonable approximation of the overall distribution. In a community mapping projects such as ENCODE or Epigenomics Roadmap, comprehensive mapping is important; in individual projects where a particular mark will be the subject of further studies, it may also be important to have detailed information including gene-specific features. In all cases, it is imperative to understand how many reads are needed to accurately characterize a mark using the framework and the numbers described in this paper and to realize what the limitations of under-sequenced datasets are.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online, including Supplementary Table S1 and Supplementary Figures S1–S10.
ACKNOWLEDGEMENTS
We acknowledge the members of the Broad Peak working group of the modENCODE and ENCODE consortia for helpful discussions.
FUNDING
This work was supported by U01HG004258 to G.K., R01GM101958 to M.K., and R01GM082798 to P.J.P. Funding for open access charge: institutional grant to P.J.P.
REFERENCES
- 1.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Johnson D.S., Mortazavi A., Myers R.M., Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 3.Park P.J. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egelhofer T.A., Minoda A., Klugman S., Lee K., Kolasinska-Zwierz P., Alekseyenko A.A., Cheung M.S., Day D.S., Gadel S., Gorchakov A.A., et al. An assessment of histone-modification antibody quality. Nat. Struct. Mol. Biol. 2011;18:91–93. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P., et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome. Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. .Chen Y., Negre N., Li Q., Mieczkowska J.O., Slattery M., Liu T., Zhang Y., Kim T.-K., He H.H., Zieba J. Systematic evaluation of factors influencing ChiP-seq fidelity. 2012;9:609–614. doi: 10.1038/nmeth.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rozowsky J., Euskirchen G., Auerbach R.K., Zhang Z.D., Gibson T., Bjornson R., Carriero N., Snyder M., Gerstein M.B. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guttman M., Garber M., Levin J.Z., Donaghey J., Robinson J., Adiconis X., Fan L., Koziol M.J., Gnirke A., Nusbaum C., et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jothi R., Cuddapah S., Barski A., Cui K., Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rashid N.U., Giresi P.G., Ibrahim J.G., Sun W., Lieb J.D. ZINBA integrates local covariates with DNA-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome Biol. 2011;12:R67. doi: 10.1186/gb-2011-12-7-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kharchenko P.V., Tolstorukov M.Y., Park P.J. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nature Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asp P., Blum R., Vethantham V., Parisi F., Micsinai M., Cheng J., Bowman C., Kluger Y., Dynlacht B.D. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc. Natl. Acad. Sci. USA. 2011;108:E149–E158. doi: 10.1073/pnas.1102223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Micsinai M., Parisi F., Strino F., Asp P., Dynlacht B.D., Kluger Y. Picking ChIP-seq peak detectors for analyzing chromatin modification experiments. Nucleic Acids Res. 2012;40:e70–e70. doi: 10.1093/nar/gks048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ENCODE Project Consortium. Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C.A., Doyle F., Epstein C.B., Frietze S., Harrow J., et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kharchenko P.V., Alekseyenko A.A., Schwartz Y.B., Minoda A., Riddle N.C., Ernst J., Sabo P.J., Larschan E., Gorchakov A.A., Gu T., et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.John S., Sabo P.J., Thurman R.E., Sung M.-H., Biddie S.C., Johnson T.A., Hager G.L., Stamatoyannopoulos J.A. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo C.J., Kharchenko P.V., Daheron L., Park P.J., Kingston R.E. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.