Abstract
Unlike short interfering RNAs (siRNAs), which are commonly designed to repress a single messenger RNA (mRNA) target through perfect base pairing, microRNAs (miRNAs) are endogenous small RNAs that have evolved to concurrently repress multiple mRNA targets through imperfect complementarity. MicroRNA target recognition is primarily determined by pairing of the miRNA seed sequence (nucleotides 2–8) to complementary match sites in each mRNA target. Whereas siRNA technology is well established for single target knockdown, the design of artificial miRNAs for multi-target repression is largely unexplored. We designed and functionally analysed over 200 artificial miRNAs for simultaneous repression of pyruvate carboxylase and glutaminase by selecting all seed matches shared by their 3′ untranslated regions. Although we identified multiple miRNAs that repressed endogenous protein expression of both genes, seed-based artificial miRNA design was highly inefficient, as the majority of miRNAs with even perfect seed matches did not repress either target. Moreover, commonly used target prediction programs did not substantially discriminate effective artificial miRNAs from ineffective ones, indicating that current algorithms do not fully capture the features important for artificial miRNA targeting and are not yet sufficient for designing artificial miRNAs. Our analysis suggests that additional factors are strong determinants of the efficacy of miRNA-mediated target repression and remain to be discovered.
INTRODUCTION
MicroRNAs (miRNAs) direct the coordinated repression of multiple messenger RNA (mRNA) transcripts, forming complex gene regulatory networks. Target recognition for metazoan miRNAs is based on partial complementarity between the miRNA and the 3′ untranslated region (UTR) of each target mRNA, although targeting is also observed in open reading frames (ORFs) at lower frequency (1). In particular, perfect or near-perfect Watson–Crick base pairing occurs between the miRNA seed region (nucleotides 2–8) and the 3′ UTRs of multiple target transcripts, while complementarity is incomplete across the remaining miRNA sequence. Pairing between the seed region and 3′ UTR is generally necessary, and in some cases sufficient, for repression (2–4), and conservation of seed region base pairing is a key predictor of miRNA targeting (5,6). Further evidence for the importance of the seed region for target recognition was provided by the crystal structure of the miRNA effector protein Argonaute 2, which revealed that the miRNA seed region is held in an A-form helix suitable for pairing with the mRNA (7). Beyond the seed region, 3′ UTR context, secondary structure and accessibility have been suggested as additional contributors to targeting (8–12), while base pairing at the miRNA 3′ end can increase site efficacy (10) or compensate for imperfect seed matches (2). Taken together, the seed region has emerged as a critical determinant of miRNA target recognition, with non-seed factors further shaping the efficacy of seed match sites (1).
Matches between the miRNA seed and an mRNA target are categorized by the extent of base pairing and the presence of an A nucleotide across from miRNA position 1 that enhances recognition. 8mer matches contain perfect Watson–Crick base pairing at miRNA nucleotides 2–8 and an A at target position 1 (5). 7mer-m8 matches are the same as 8mer sites but lack the A at target position 1, while 7mer-A1 sites have the A but have a mismatch at miRNA position 8 (2,5,6). 8mer, 7mer-m8 and 7mer-A1 sites account for the majority of preferentially conserved matches to conserved miRNAs and are referred to as canonical sites (1,13). Rules for a non-canonical seed match have also been described, in which matches to miRNA nucleotides 2–6 nucleate hybridisation to the mRNA target, but the target bulges out across from position 6 to allow further base pairing to miRNA nucleotides 6–8 (14). These non-canonical bulge matches represent an alternative seed targeting mechanism that has not been comprehensively compared in efficacy to canonical seed matches.
One striking characteristic of miRNAs is the ability of an individual miRNA sequence to regulate multiple genes that have corresponding seed match sites. There are an average of 300 conserved targets predicted for each evolutionarily conserved miRNA (13), and the introduction or depletion of a single miRNA can directly alter the abundance of hundreds of proteins (15,16). With nearly 60% of human protein-coding genes having conserved seed match sites in their 3′ UTRs (13), miRNAs are able to form regulatory networks that modulate the expression of many genes in concert (16–18). Moreover, miRNAs can exert strong biological effects, such as controlling cell fate (19,20).
While miRNAs are naturally occurring small RNA repressors of gene expression, artificial short interfering RNAs (siRNAs) have been developed as powerful tools for experimental gene repression. In contrast to the partial complementarity observed with miRNA target interactions, siRNAs are designed to be perfectly complementary to a single mRNA, enabling the repression of individual genes (21). An approach for the experimental repression of multiple genes in concert has been reported but not extensively developed (22). Since miRNAs have a well-studied, naturally evolved seed-based mechanism for targeting multiple genes, they are appealing as the basis for developing a multi-target RNA interference (RNAi) system. However, a framework for the design of artificial miRNAs has not been established.
We sought to systematically design artificial miRNAs and analyse their efficacy for multiple gene repression. Similar to endogenous miRNAs, these artificial miRNAs feature seed matches to multiple target transcripts and have partial base pairing at the 3′ end. Using these artificial miRNAs, we also set out to understand the contributions of the seed and non-seed regions for miRNA targeting. We designed over 200 artificial miRNAs with common seed matches in two non-essential metabolic genes, pyruvate carboxylase (PC) and glutaminase (GLS). We then quantified miRNA activity with luciferase reporter genes and immunoblotting. We found that the artificial miRNAs were effective for simultaneous gene repression, with canonical seed matches supporting stronger repression than bulge matches. However, seed matches were not sufficient for miRNA activity, as the majority of artificial miRNAs failed to repress targets, even among miRNAs with perfect seed complementarity. Although repression was enhanced by base pairing at the miRNA 3′ end, additional non-seed factors (e.g. factors related to the mRNA target) appeared to make a major contribution to miRNA activity. Our study not only establishes the feasibility of artificial miRNAs for repressing multiple genes simultaneously but also demonstrates the importance of non-seed factors for artificial miRNA targeting.
MATERIALS AND METHODS
Cell culture
293T cells were maintained in RPMI 1640 medium (Gibco) with 10% fetal bovine serum (Atlanta Biologicals) under 5% CO2.
mRNA sequence data
Human RefSeq protein-coding transcripts with annotated 5′ UTR, ORF and 3′ UTR were downloaded from the UCSC Genome Browser, build hg19 (http://genome.ucsc.edu) (23–25). For PC and GLS, RefSeq transcript versions NM_000920.3 and NM_014905.4, respectively, were used for designing artificial miRNAs targeting PC and GLS.
Design of non-targeting control artificial miRNAs
We synthesized a set of control artificial miRNAs that lacked seed matches to PC or GLS but were otherwise similar to the miRNAs designed to target PC and GLS. We started by calculating the frequency of each nucleotide appearing at each position within the PC/GLS-targeting miRNAs. From this position frequency matrix, 106 random artificial miRNA sequences were generated. This set of sequences was filtered to remove those with seeds that matched anywhere within the PC or GLS transcripts. To ensure that the seeds of the PC/GLS-targeting miRNAs were similar to the seeds of control sequences, each PC/GLS miRNA seed was matched to a seed present in the remaining control sequences that was most similar based on four measures: (i) whether the seed has matches in the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) 3′ UTR, (ii) whether the seed has matches in the Renilla luciferase ORF, (iii) the number of G or C nucleotides in the seed and (iv) the frequency of seed matches in a set of 4505 genes (Supplementary Table S7) that are highly expressed across multiple cell lines from the Sanger Cell Line Project (http://www.broadinstitute.org/mpr/publications/projects/Integrative_Genomic_Analysis/Sanger_Cell_Line_Project_Affymetrix_QCed_Data_n798.gct). In cases where multiple control seeds matched equally, one was picked at random. The pool of control sequences was then further filtered to contain only those sequences with one of these matched seeds. Negative controls were picked from the remaining sequences at random, with the exception that picking was biased to reproduce the frequency of seeds targeting the control GAPDH 3′ UTR that was present in the PC/GLS-targeting miRNAs. In addition, after each control sequence was picked, any other controls with the same seed were eliminated from the pool to ensure that no two negative controls had the same seed. For each control artificial miRNA sequence picked, a passenger strand was designed as the perfect complement of the miRNA, plus a 2 nt 3′ overhang that was the same as the 3′ overhang of the guide strand.
Synthesis of artificial miRNAs
RNA oligonucleotides representing the guide and passenger strand of each artificial miRNA were synthetized (Integrated DNA Technologies). Oligonucleotide sequences are provided in Supplementary Table S3. To generate artificial miRNA duplexes for transfection, 40 uM guide and passenger strand oligonucleotides were combined and diluted to 4 uM final concentration in annealing buffer (5 mM NaCl, 1 mM Tris-HCl, 1 mM MgCl2 and 0.1 mM DTT, final concentrations). The reaction was denatured at 90°C for one minute then annealed at 37°C for 1 h. Duplexed miRNAs were stored at −80°C until use.
Luciferase reporter assays
Artificial miRNAs were screened for luciferase reporter repression activity in 293T cells in triplicate. Ten picomoles artificial miRNAs were reverse co-transfected into 5 × 103 cells along with 50 ng pLightSwitch_3UTR reporter plasmid (SwitchGear Genomics) and 25 ng pGL3-Control Vector plasmid (Promega, E1741) using 2.5 ul DharmaFECT Duo Transfection Reagent (Thermo Scientific) in 100 ul total volume. The 3′ UTR reporter plasmids used were PC (S803754), GLS (S812820), GAPDH (S801378) and the empty vector (S890005). Twenty-four hours post-transfection, cells were assayed for luciferase activity using the Dual-Glo Luciferase Assay (Promega) according to the manufacturer's protocol, except that 75 ul of each reagent was used. Firefly and Renilla luciferase activity were measured on a GloMax Luminometer (Promega) using a 1 s integration time with an 18 min incubation time following addition of each reagent. Analysis of luciferase reporter data was performed with R and the plyr, qvalue and ggplot2 packages (26–29).
Immunoblotting
To determine protein knockdown in cells transfected with artificial miRNAs, 4 × 105 293T cells were reverse transfected in triplicate with 500 pmol artificial miRNA using 25 ul RNAiMAX (Invitrogen). Five hundred picomoles of Allstars non-targeting control siRNA (1027281; Qiagen) was used as a negative control and 250 pmol each of siPC siRNA (SI05128914; Qiagen) and siGLS siRNA (HSS178458; Invitrogen) combined were used as a positive control. Medium was changed 24 h post-transfection. At 72 h post-transfection, cells were lysed in 1.25% Nonidet P-40, 1.25% SDS, 12.5 mM NaH2PO4 pH 7.2, 2 mM EDTA, 50 mM NaF and protease inhibitor cocktail. Thirty-five microgram of soluble protein was subject to sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblotting. Antibodies used for immunoblotting were PC (sc-271493; Santa Cruz Biotechnology), GLS (ab93434; Abcam), GAPDH (2118S; Cell Signaling Technologies) and β-Actin (A-5441; Sigma). Secondary antibodies used were IRDye 800CW Goat anti-Mouse (926–32210; LI-COR) and IRDye 680CW Goat anti-Rabbit (926–32221; LI-COR). Protein levels were measured on the Odyssey Imaging System (LI-COR) and quantified using ImageJ software (NIH).
Gene expression profiling
2 × 105 293T cells were reverse transfected in triplicate with 250 pmol of artificial miRNA (amiR-104, amiR-143, amiR-175 or amiR-268) or Allstars non-targeting control siRNA (Qiagen) using 12.5 ul RNAiMAX (Invitrogen). Medium was changed 24 h post-transfection. Total RNA was extracted at 48 h post-transfection using the miRNeasy Kit (Qiagen) according to the manufacturer's protocol. Gene expression was assayed using the HumanHT-12 v4 Expression BeadChip Kit (Illumina) on the iScan System (Illumina). Raw data were imported into GenomeStudio (V2011.1, Illumina) for processing with the Gene Expression module (V1.9.0, Illumina). Quantile normalisation was applied, and differential expression for each probe relative to the non-targeting control samples was calculated using the Illumina Custom algorithm. Subsequent analysis was done using R (26). Non-specific filtering was performed to remove probes that did not correspond to RefSeq protein-coding transcripts or that were not detected (P value ≥ 0.01 for detection in all samples). After filtering, transcript-level expression values and differential expression scores were calculated as the mean of probes that detect the same RefSeq transcript. Transcripts were classified as repressed if the differential expression score corresponded to P < 0.01 and expression level was below the non-targeting control reference. Gene expression data are available in the NCBI GEO database (accession number GSE50249). To analyse the relationship between seed matches and gene repression, canonical and bulge matches to the seeds of the four artificial miRNAs were identified throughout each detected transcript. Transcripts with seed matches in the 5′ UTR or ORF were excluded from analysis for the corresponding miRNA.
Prediction of 3′ UTR site accessibility
Sfold V2.2 (12,30,31) was used to predict site accessibility for each artificial miRNA seed match site. Specifically, the mRNA transcripts of the PC and GLS 3′ UTR reporter gene, including the Renilla ORF sequence, were computationally folded with Sfold, which also calculates the probability that each region of four consecutive nucleotides would be single stranded (i.e. all four nucleotides are unpaired in the folded structure). For a given seed match site, the site accessibility was calculated as the maximum single-stranded probability among the set of four-nucleotide regions within the seven-nucleotide seed match site. When an artificial miRNA had multiple seed match sites in a given 3′ UTR reporter, the site with the greatest accessibility score was used.
Prediction of artificial miRNA activity
Minimum free energy for the hybridisation between an artificial miRNA and the PC or GLS 3′ UTRs was calculated using RNAhybrid V2.1 (32,33). For each seed match site, a 42-nucleotide region of the 3′ UTR encompassing the seed match site and the full sequence of the artificial miRNA were used as inputs to RNAhybrid. When an artificial miRNA had multiple seed match sites in a given 3′ UTR, the site with the most negative free energy was used. Probability of Interaction by Target Accessibility (PITA) scores for each artificial miRNA/3′ UTR pair were calculated with the PITA executable V6 (11), and the corresponding ORF sequence was provided as an optional input to ensure more complete thermodynamic analysis. Default values for seed matching parameters were used except that a one-nucleotide loop was allowed within the seed region to account for bulge seed match sites. The PITA programme was run once without considering flanking nucleotides and once with settings to use 3 upstream and 15 downstream flanking nucleotides. TargetScan context+ scores and component scores (local AU context, position effect, site pairing stability, target abundance and miRNA 3′ end pairing) for artificial miRNA seed match sites were calculated using TargetScan V6.1 (5,10,13,34). For each artificial miRNA/3′ UTR pair, the context+ scores and each component score for all seed match sites in the 3′ UTR were added to produce the total context+ score and each total component score. Analysis of minimum free energy, PITA and TargetScan scores for prediction of artificial miRNA activity was performed with R and the plyr, pROC and ggplot2 packages (26,28,29,35).
RESULTS
Design of artificial miRNAs
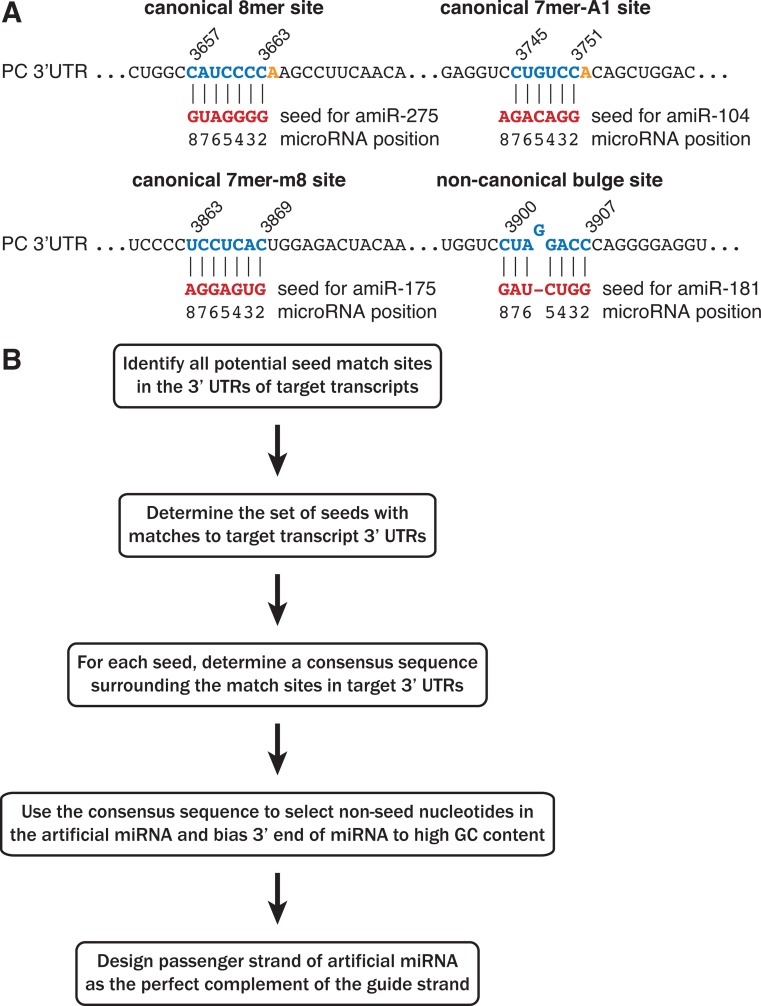
To systematically analyse multiple gene repression by artificial miRNAs, we developed an algorithm to design artificial miRNAs that should recognize a desired set of target transcripts and mimic the features of endogenous miRNAs. Because of the importance of the miRNA seed region in target recognition, we wanted to ensure that any artificial miRNA designed to repress a transcript contained at least one seed match within the 3′ UTR of that transcript. Therefore, our design approach started with identifying all sites in each desired transcript that matched each of the 16 384 theoretically possible seven-nucleotide seed sequences. Transcript sites that corresponded to 7mer-A1, 7mer-m8 and 8mer matches to a seed were referred to collectively as canonical target sites (Figure 1A). To enable the comparison of canonical seed matches to non-canonical bulge matches, we also determined transcript sites that could match each seed according to the mechanism described in (14), and we referred to these as bulge target sites (Figure 1A, bottom right panel). The seed sequences with either canonical or non-canonical match sites present in all of the desired target genes were then identified, and these common seeds served as the basis for designing artificial miRNAs (Figure 1B).
Figure 1.
Design of artificial miRNAs with seed matches to multiple target genes. (A) Artificial miRNAs (amiRs) were designed with seeds that matched canonical or non-canonical sites within each target transcript. Examples of base pairing between a miRNA seed region and the PC 3′ UTR are shown for each seed match type. (B) Schematic of the artificial miRNA design algorithm.
We next sought to design each artificial miRNA with partial complementarity between the non-seed nucleotides and the desired target genes, which is similar to interactions observed for endogenous miRNAs to their targets. For each common seed, a consensus sequence was built from the mRNA sequences adjacent to the match sites in the target genes (Figure 1B). We then designed the miRNA non-seed nucleotides 9–18 as the perfect complement of this consensus sequence, with nucleotides chosen at random when multiple bases were equally likely at a given consensus position. The remaining positions of the artificial miRNA were used to reproduce the thermodynamic asymmetry between the 5′ and 3′ ends that promotes proper strand selection within the RNA-induced silencing complex (RISC) (36–38). Because strong selection for U at position 1 and moderate bias toward G or C at positions 18–21 have been observed in a screen for highly effective shRNAs (36), we designed all of the artificial miRNAs to contain a U at position 1 and G or C at positions 18–20. However, since positions 21 and 22 are not involved in duplex unwinding but may enhance miRNA:target hybridisation, these nucleotides were designed to the consensus sequence of the target genes in the same way as for positions 9–18. Passenger strands for each artificial miRNA were generated as the perfect complement of the guide strand with a two-nucleotide 3′ overhang, similar to siRNAs and shRNAs. The result is an artificial miRNA duplex that can be transfected into cells or cloned into an shRNA vector.
To test our hypothesis that artificial miRNAs could be rationally designed to repress multiple genes of interest, as well as to investigate requirements for effective miRNA targeting, we chose to target two non-essential metabolic genes, PC and GLS. We designed artificial miRNAs targeting the 3′ UTRs of both genes by first identifying the 234 seeds that matched the 3′ UTRs of both PC and GLS through either the canonical or bulge mechanisms (Supplementary Tables S1 and S2). We then applied the algorithm above to generate artificial miRNAs with each of those common seeds (Supplementary Table S3), and each miRNA was chemically synthesized.
Rational design of artificial miRNAs yields sequences that significantly repress two target genes
To determine which artificial miRNAs were functional for repressing PC and GLS, we used 3′ UTR Renilla luciferase reporter assays. Empty vector and a GAPDH 3′ UTR reporter were used as negative control reporters. To obtain a set of negative control miRNAs, we also chose 74 seeds at random that did not match anywhere within the PC or GLS transcripts (Supplementary Tables S4 and S5) and used these to synthesize 74 non-targeting artificial miRNAs (Supplementary Table S3). These non-targeting control miRNAs were designed to have the same nucleotide frequency distribution and to match the PC/GLS targeting miRNAs on four criteria: (1) frequency of seed matches to the control GAPDH 3′ UTR, (2) frequency of seed matches in Renilla luciferase coding sequence, (3) frequency of seed G/C content and (4) abundance of seed match sites in highly expressed transcripts (see Materials and Methods for details).
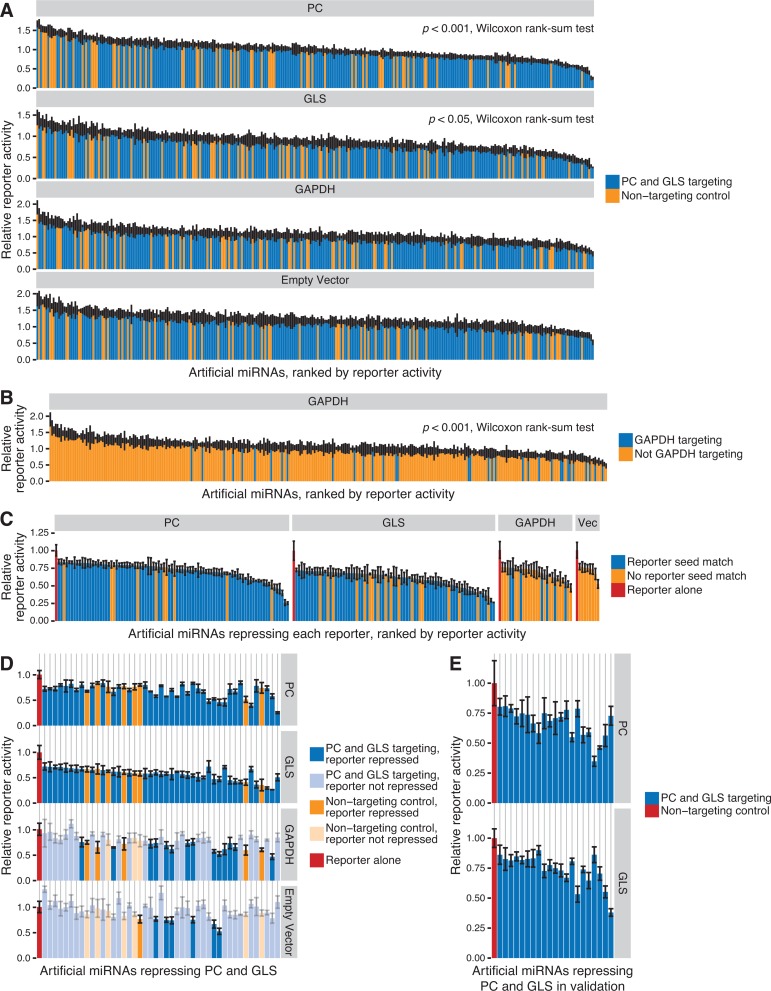
293T cells were transiently co-transfected with each reporter vector and each targeting or non-targeting artificial miRNA. Reporter activity was normalized to the activity observed in the absence of miRNA co-transfection. As we expected given the importance of seed matches for miRNA targeting, we found that PC and GLS reporter activity was significantly lower following co-transfection with artificial miRNAs designed to target PC and GLS compared to non-targeting controls, which lack seed matches (P < 0.05, Wilcoxon rank-sum test) (Figure 2A). In contrast, the empty vector and GAPDH reporters responded similarly to PC/GLS-targeting and non-targeting miRNAs. However, we noted that the GAPDH reporter contained seed match sites for 26 of the PC/GLS-targeting miRNAs and eight of the non-targeting control miRNAs (Supplementary Table S6). Consistent with the hypothesis that seed targeting mediates gene repression, we also observed that GAPDH reporter activity was significantly lower with GAPDH-targeting miRNAs compared to those without predicted seed target sites (Figure 2B).
Figure 2.
Rationally designed artificial miRNAs significantly repress PC and GLS 3′ UTR luciferase reporter genes. (A) Artificial miRNAs with seed matches targeting PC and GLS 3′ UTRs (blue) or non-targeting control miRNAs without seed matches (orange) were screened for repression of PC, GLS and GAPDH 3′ UTR luciferase reporter activity or empty vector. Bars represent the mean ± SD of reporter activity for triplicate transfections relative to the reporter alone control. Wilcoxon rank-sum tests were performed for each 3′ UTR to compare reporter activity with targeting miRNAs to activity with non-targeting controls, and only PC and GLS yielded P < 0.05, as indicated. (B) Relative GAPDH 3′ UTR reporter activity as in (A), but bars for miRNAs with seed matches to the GAPDH 3′ UTR are blue, while miRNAs lacking seed matches are orange. (C) Artificial miRNAs from (A) that repressed relative reporter activity > 2 SD below the mean of the reporter alone control and had a q < 0.05 (t-test versus the reporter alone control). Bars are coloured by whether the miRNA has a seed match to the indicated 3′ UTR reporter (blue) or lacks a seed match (orange). Bars representing the reporter alone control are red. Vec is the empty vector control. (D) Artificial miRNAs from (A) that significantly repressed both PC and GLS reporter activity. In each column, bars represent the activity of the indicated reporter for a single miRNA. Artificial miRNAs along the x-axis are sorted by the minimum relative activity across reporters for each miRNA. Bars are coloured by whether the miRNA was designed to target PC and GLS (blue) or was a non-targeting control (orange). Dark shading indicates the miRNA significantly repressed the corresponding reporter, while light shading indicates the reporter was not repressed. Bars representing the reporter alone control are red. (E) Artificial miRNAs that repressed both PC and GLS in the initial screen were re-assayed, and PC and GLS reporter activity for those miRNAs that validated is shown. Bars represent the mean ± SD of reporter activity for triplicate transfections of each miRNA (blue) relative to a commercially available non-targeting control (red). Bars in a column represent PC and GLS reporter activity for a single artificial miRNA, and miRNAs are sorted by the minimum relative activity across both reporters for each miRNA.
While artificial miRNAs designed to target PC and GLS yielded lower levels of reporter activity, we wanted to determine the set of artificial miRNAs that significantly repressed each reporter gene in order to establish whether designed artificial miRNAs were capable of multi-gene repression and to study the properties of miRNA targeting. Our criteria for reporter repression by an artificial miRNA were (i) reporter activity was greater than 2 SD below the mean activity without miRNA and (ii) the false discovery rate for a t-test of the artificial miRNA compared to the no miRNA control had a q value < 0.05. Artificial miRNAs that passed both criteria are shown in Figure 2C. Because the cloned 3′ UTRs in the reporter vectors differed from the reference sequences used to design the artificial miRNAs, some predicted seed match sites were absent from the reporters, and the corresponding miRNAs were classified as no seed match for that reporter (Supplementary Table S6). Among the 308 designed and negative control artificial miRNAs tested, only eight (2.6%) repressed the empty vector lacking a 3′ UTR, indicating that non-specific repression of the vector was infrequent. For both the PC and GLS reporters, ∼30% of miRNAs with seed matches repressed each reporter, in contrast to ∼15% for miRNAs without seed matches. These differences in reporter repression were significant by Fisher's exact test (P < 0.02) and indicate that artificial miRNAs designed with our algorithm have significantly higher rates of target repression compared to the off-target effects of control miRNAs (Table 1). The importance of seed matches was further supported by the GAPDH reporter, which also showed significant association between artificial miRNAs with seed matches and reporter repression (P < 0.02, Fisher's exact test).
Table 1. Frequency of 3′ UTR reporter repression for artificial miRNAs with and without seed matches in the reporter.
| Reporter | Artificial miRNA seed match in reporter | No artificial miRNA seed match in reporter | P value, Fisher's test | ||||
|---|---|---|---|---|---|---|---|
| Reporter repressed | Reporter not repressed | % Reporter repressed | Reporter repressed | Reporter not repressed | % Reporter repressed | ||
| PC | 78 | 151 | 34.1% | 10 | 69 | 12.7% | 0.00026 |
| GLS | 58 | 142 | 29.0% | 18 | 90 | 16.7% | 0.01849 |
| GAPDH | 7 | 27 | 20.6% | 20 | 254 | 7.3% | 0.01870 |
| EMPTY | 8 | 300 | 2.6% | ||||
For each 3′ UTR reporter indicated, the artificial miRNAs were classified by whether they had a seed match in the reporter and whether the reporter luciferase activity was significantly repressed by the miRNA (activity > 2 SD below the reporter alone mean and q < 0.05). The significance of the association between seed matches and reporter repression for each 3′ UTR reporter was calculated using Fisher's exact test.
Among the 234 artificial miRNAs that were designed to target the PC and GLS reference sequences, only 195 had predicted seed target sites in the sequences of both 3′ UTR reporter constructs that were used (Supplementary Table S6). Thirty-seven of these 195 (19%) repressed both PC and GLS reporters, while only 8 of the 74 non-targeting control miRNAs (11%) did so (Figure 2D). This set of 45 targeting and non-targeting miRNAs was re-assayed to confirm that both PC and GLS reporters were specifically repressed. Of the 37 PC/GLS-targeted miRNAs that repressed both reporters in the initial screen, 32 (86%) validated for repression of the PC reporter and 24 (65%) validated for repression of the GLS reporter. Overall, we verified that 21 of the 37 PC/GLS-targeted miRNAs significantly repressed both PC and GLS compared to a commercially available non-targeting siRNA (q < 0.05, t-test) (Figure 2E and Table 2). In contrast, only two of the eight non-targeted controls that were re-assayed validated for repression of both PC and GLS, confirming that non-specific repression of multiple genes is rare in the absence of seed match sites. Taken together, we found that PC/GLS-designed miRNAs were significantly more likely to repress both PC and GLS than the non-targeted controls (P < 0.05, Fisher's exact test) (Table 2).
Table 2. Frequency of artificial miRNA repression of both PC and GLS 3′ UTR reporters.
| Artificial miRNA | Number of miRNAs | Number of artificial miRNAs that repress | P value, Fisher's test | ||||
|---|---|---|---|---|---|---|---|
| Neither | PC only | GLS only | PC and GLS | Not both | |||
| PC and GLS targeting | 195 | 114 | 38 | 22 | 21 | 174 | 0.048 |
| Non-targeting control | 74 | 54 | 8 | 10 | 2 | 72 | |
Each targeting and non-targeting control artificial miRNA was classified by whether the miRNA significantly repressed luciferase activity from the PC and/or GLS 3′ UTR reporters. The number of miRNAs that failed to repress both reporters is also indicated. The significance of the association between whether a miRNA was targeting and whether it repressed both reports was calculated using Fisher's exact test. The values used for the Fisher's exact test are in italics.
Artificial miRNAs simultaneously repress multiple endogenous genes
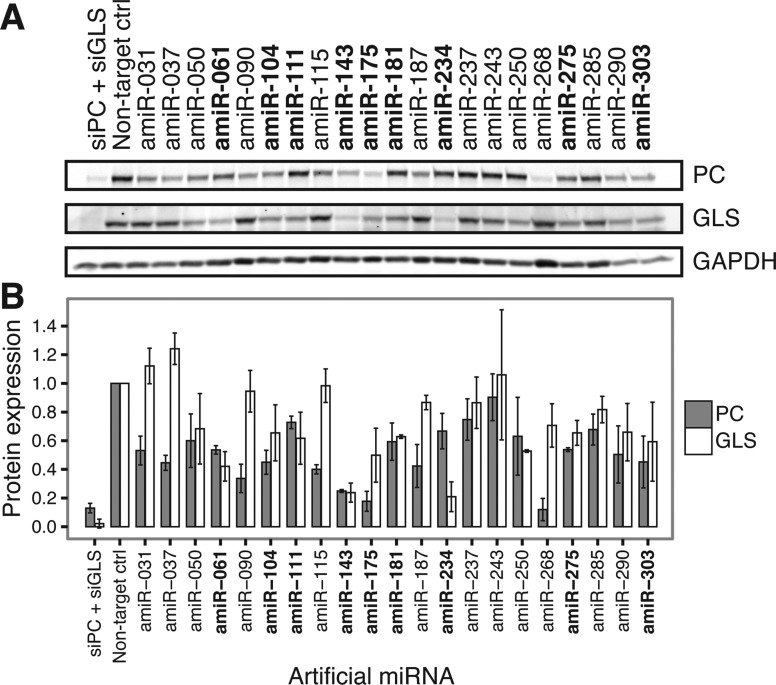
While we had demonstrated that rationally designed artificial miRNAs repress multiple reporter genes that were assayed individually (i.e. in separate transfection experiments), we wanted to confirm that these miRNAs could also concurrently repress multiple endogenous genes. We transfected the 21 validated PC/GLS artificial miRNAs into 293T cells and determined endogenous PC and GLS protein levels by immunoblotting. We observed that nine of the artificial miRNAs reproducibly knocked down both PC and GLS protein expression (Figure 3). Among the 12 miRNAs that failed to validate, three had seeds that matched sites in the Renilla luciferase reporter ORF, suggesting that ORF-based targeting may have been responsible for some non-specific repression in the reporter screen. The most potent artificial miRNAs showed activity comparable to siRNAs, despite being designed to repress multiple targets. Therefore, rationally designed artificial miRNAs can be an effective approach to multi-target gene repression.
Figure 3.
Artificial miRNAs simultaneously repress endogenous PC and GLS protein expression. (A) Following transfection of the indicated artificial miRNAs (amiRs) into 293T cells, endogenous PC and GLS protein were detected by immunoblotting. A non-targeting control siRNA served as a negative control, while highly optimized siRNAs targeting PC and GLS (siPC + siGLS) were co-transfected as a positive control for knockdown. GAPDH served as a loading control for immunoblotting. Artificial miRNAs shown in bold repressed both PC and GLS expression below 80% of negative control levels in duplicate experiments. (B) PC and GLS protein expression relative to the negative control transfection was quantified and normalized to GAPDH. Bars represent the mean ± SD of duplicate experiments. Artificial miRNAs shown in bold are as in (A).
Canonical seed target sites yield more robust gene repression than non-canonical bulge sites
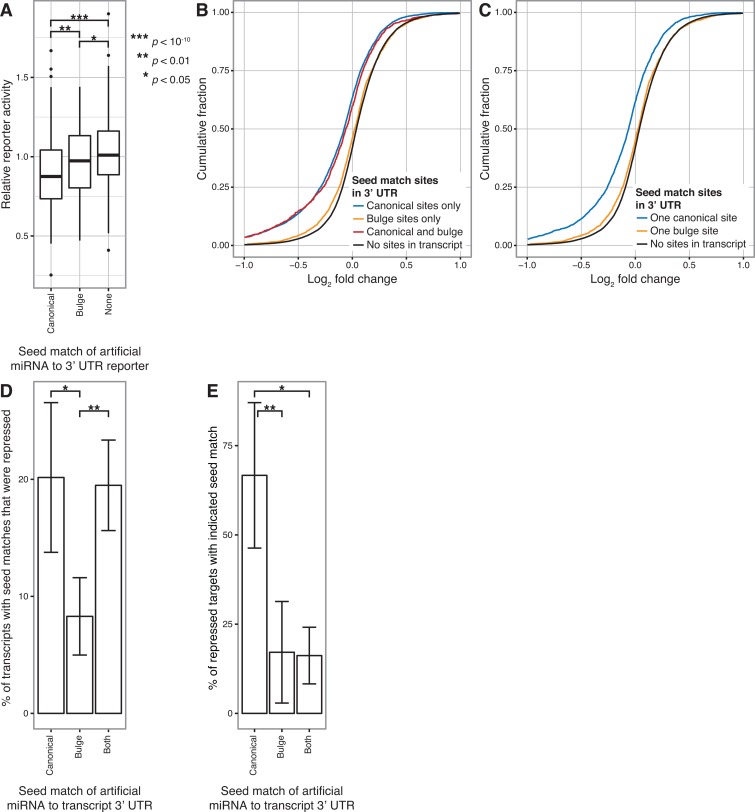
Since the artificial miRNAs were designed to recognize both canonical and non-canonical (bulge) seed target sites, we compared the ability of each site type to direct gene repression. Artificial miRNA/reporter gene pairs were categorized by whether the miRNA seed matched a canonical or bulge site in the 3′ UTR for the PC, GLS and GAPDH reporters (Supplementary Table S6). Artificial miRNAs that could potentially recognize both site types in a reporter were excluded from the analysis for that reporter. In addition, we filtered out miRNAs with seed match sites in the Renilla luciferase ORF (Supplementary Table S6), since these might yield repression independently of the sites in the 3′ UTR. The results for all three reporters were then combined. When we examined the frequency at which canonical or bulge sites lead to reporter repression below our threshold, both types of sites were more effective compared to cases where the miRNA seed did not match the reporter (P < 10−13 for canonical and P < 0.001 for bulge, Fisher's exact test) (Table 3). However, we did not detect a significant difference between the frequencies of reporter repression for the two types of seed matches (34% for canonical versus 24% for bulge, P > 0.05, Fisher's exact test). When we examined the magnitude of reporter repression, we observed that artificial miRNAs with either canonical or bulge seed matches significantly repressed reporter activity compared to no seed matches, as expected (P < 10−10 for canonical and P < 0.05 for non-canonical, t-test) (Figure 4A). In addition, we determined that canonical seed matches overall were significantly more potent than bulge matches (P < 0.01, t-test). These results suggest that canonical sites provide stronger and more effective miRNA targeting than bulge sites, although both site types can mediate gene repression.
Table 3. Frequency of 3′ UTR reporter repression for artificial miRNAs with canonical, bulge or no seed matches.
| Artificial miRNA seed match in reporter | miRNAs with matches (total for all reporters) | Reporter repressed | Reporter not repressed | % Reporter repressed | P values, Fisher's test | ||
|---|---|---|---|---|---|---|---|
| Canonical versus none | Canonical versus bulge | Bulge versus none | |||||
| None | 362 | 32 | 330 | 8.8% | < 10−13 | 0.10 | 0.00031 |
| Canonical | 226 | 76 | 150 | 33.6% | |||
| Bulge | 88 | 21 | 67 | 23.9% | |||
Each pair of artificial miRNA and 3′ UTR reporter (PC, GLS or GAPDH) was classified by whether that miRNA had a canonical, bulge or no seed match in the 3′ UTR of the reporter. For each seed match category, the miRNA / reporter pairs were further classified by whether the miRNA significantly repressed the reporter luciferase activity. The significance of the association between seed match category and reporter repression was calculated using Fisher's exact test for all pairs of seed match categories.
Figure 4.
Canonical seed match sites are more effective than non-canonical bulge sites for mediating gene repression. (A) Relative reporter activity (mean of triplicate transfections) for artificial miRNAs with only canonical seed matches, only bulge seed matches, or no seed matches in a 3′ UTR reporter. Boxplots represent the combined data for artificial miRNAs assayed with the PC, GLS and GAPDH reporters. t-tests with P < 0.05 are indicated. (B) Changes in gene expression after transfection with PC/GLS-targeting artificial miRNAs relative to a non-targeting control transfection were profiled by microarray. Cumulative distributions of fold changes for mRNAs with the indicated type of seed match sites in their 3′ UTR are shown. Data were combined from four artificial miRNAs, each transfected in triplicate. Transcripts with seed matches outside the 3′ UTR were excluded from analysis in this and subsequent panels. A one-sided K-S test that repression of transcripts with 3′ UTR canonical sites was greater than with bulge sites yielded P < 10−51. Similar tests comparing transcripts with canonical sites, bulge sites or both to those with no sites yielded P < 10−189, P < 10−4 and P < 10−42, respectively. (C) Cumulative distributions of fold changes as in (B), except that only mRNAs with single canonical or single bulge seed match sites in the 3′ UTR or no sites were analysed. A one-sided K-S test that transcripts with a single canonical site are more strongly repressed than those with a single bulge site yielded P < 10−31, while similar tests comparing transcripts with single canonical or single bulge sites to those with no sites yielded P < 10−111 and P < 0.002, respectively. (D) Among transcripts with the indicated type of 3′ UTR seed match, the mean percent that was repressed following triplicate transfections was determined for each PC/GLS-targeting artificial miRNA. Bars represent the mean ± SD of the results from four artificial miRNAs. t-tests with P < 0.05 are indicated as in (A). (E) Among repressed transcripts with any 3′ UTR seed match, the mean percent that contained the indicated type of match site was determined from triplicate transfections with each PC/GLS artificial miRNA. Bars represent the mean ± SD of the results from four artificial miRNAs. t-tests with P < 0.05 are indicated as in (A).
To more broadly analyse the difference between canonical and bulge target sites, we used microarray profiling to assess the global impact of artificial miRNAs on gene expression. Four miRNAs that gave a range of PC and GLS repression were transiently transfected into 293T cells, and we measured changes in expression of all detectable RefSeq protein-coding transcripts compared to a non-targeting control transfection. As we observed with reporter assays, genes with canonical or bulge seed match sites in the 3′ UTR were significantly repressed compared to genes lacking sites (P < 10−189 for canonical sites, P < 10−4 for bulge sites and P < 10−42 for both, all compared to no sites, one-sided Kolmogorov–Smirnov (K-S) tests) (Figure 4B). However, repression was significantly greater when the 3′ UTRs contained canonical seed matches compared to bulge sites (P < 10−51, one-sided K-S test). Significant differences in repression were also seen when transcripts contained only a single canonical or bulge site in the 3′ UTR (P < 10−111 for single canonical versus no sites, P < 0.002 for single bulge versus no sites and P < 10−31 for single canonical versus single bulge sites, one-sided K-S tests) (Figure 4C). Across the four artificial miRNAs examined, on average 20% of transcripts with only canonical seed match sites in the 3′ UTR were down-regulated, which was significantly greater than the 8% of transcripts with only bulge sites that were repressed (P < 0.05, t-test) (Figure 4D). Transcripts with both types of sites were down-regulated at a similar frequency (19%) as canonical site transcripts, suggesting that when both sites are present, the canonical sites are the primary determinant of gene repression. Moreover, among the transcripts with seed matches in the 3′ UTR that were repressed by each miRNA, an average of 67% contained canonical sites but only 17% contained non-canonical sites, excluding those that contained both site types (P < 0.01, t-test) (Figure 4E). Taken together, our results demonstrate that canonical target sites are the primary drivers of strong repression.
Seed match sites are not sufficient for miRNA activity
While seed regions are an important determinant of miRNA targeting, several studies have demonstrated that non-seed pairing enhances seed-based target recognition and in some cases enables targeting in the absence of perfect seed matches (2,10,39,40). From our analysis of artificial miRNAs containing 195 different seed sequences that targeted both PC and GLS reporters, we observed substantial variation in miRNA activity, even among those with canonical seed matches (Figure 4A). For each reporter used, the majority of artificial miRNAs with seed matches failed to repress the reporter gene, indicating that seed matches alone are not sufficient to drive target repression (Table 1). Moreover, almost a third of the PC/GLS artificial miRNAs were effective for repressing one reporter gene but ineffective against the other (Table 2). These results demonstrate that miRNA activity is strongly dependent on factors other than the presence of a seed match.
Endogenous miRNA target prediction algorithms are weak predictors of artificial miRNA activity
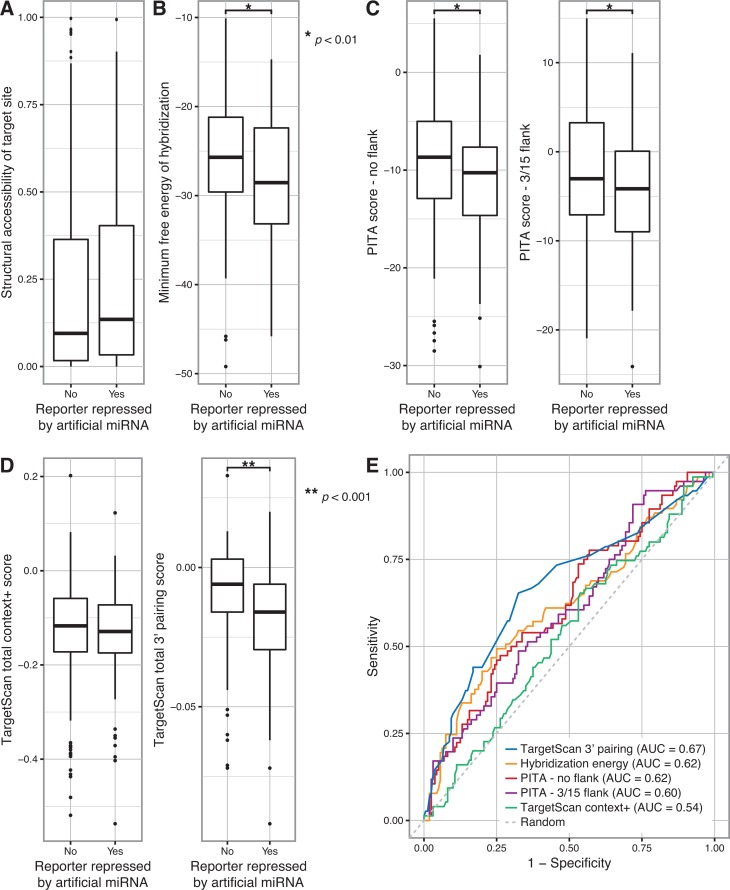
Because many artificial miRNAs showed divergent activity against PC and GLS reporters despite the presence of an identical seed sequence match in both PC and GLS 3′ UTRs, we hypothesized that the differences were due to (1) the local secondary structure of target sites and/or (2) hybridisation between the non-seed miRNA nucleotides and the target transcript. We first used Sfold (30,31) to computationally fold each transcript and predict whether the artificial miRNA target sites were accessible within the secondary structure of the PC and GLS 3′ UTRs. We defined the structural accessibility as the maximum probability that a four nucleotide long single-stranded region exists within the target site, because such a region could serve to nucleate hybridisation (12). We found that there was no significant association between this measure of target site accessibility and reporter repression (Figure 5A). We next calculated the minimum free energy of hybridisation between the artificial miRNAs and their binding sites in the PC and GLS 3′ UTRs using RNAhybrid (32,33), excluding miRNAs that contained seed matches to the Renilla luciferase ORF that might drive non-specific repression. When multiple sites were present for a given miRNA, the site with the lowest free energy was used, under the simple model that the strongest binding site would predominantly drive miRNA activity. We determined that artificial miRNAs that repressed a given target transcript had significantly lower hybridisation energy to that transcript, indicative of stronger binding (P < 0.01, t-test) (Figure 5B). These results suggest that strong binding between the artificial miRNA and transcript, rather than seed recognition alone, is necessary for target recognition and repression.
Figure 5.
Artificial miRNA activity is modestly predicted by base pairing at the miRNA 3′ end. (A) The probability of each artificial miRNA seed match site in the PC and GLS 3′ UTR reporter genes being structurally accessible (i.e. single-stranded) was calculated, and the most accessible site was determined for each miRNA/reporter pair. Boxplots represent the structural accessibility probability values for miRNA/reporter pairs in which the miRNA did or did not repress the reporter. A t-test yielded P > 0.05. (B) The minimum free energy of hybridisation between each artificial miRNA and the PC and GLS 3′ UTRs was calculated, and the hybridisation site with the lowest free energy was determined for each miRNA/gene pair. Boxplots represent the minimum free energy for miRNA/gene pairs in which the miRNA did or did not repress the corresponding 3′ UTR reporter. A t-test with P < 0.01 is indicated by the single asterisk. (C) Target prediction scores were calculated using the PITA algorithm for each artificial miRNA and the PC and GLS 3′ UTRs, using settings that excluded flanking nucleotides (left) or considered 3 upstream and 15 downstream flanking nucleotides (right). Boxplots represent the scores for miRNA/gene pairs in which the miRNA did or did not repress the corresponding 3′ UTR reporter. t-tests with P < 0.01 are indicated by a single asterisk. (D) TargetScan total context+ scores (left) were calculated for each artificial miRNA and the PC and GLS 3′ UTRs. The total sub-score for 3′ end base pairing, calculated as part of the context+ score, was analysed separately (right). Boxplots represent the indicated scores for miRNA/gene pairs in which the miRNA did or did not repress the corresponding 3′ UTR reporter. A t-test of the 3′ pairing score with P < 0.001 is indicated by the double asterisk. A t-test of the context+ score yielded P > 0.05. (E) The ability of the indicated measures of miRNA targeting to predict repression of PC and GLS 3′ UTR reporter genes by each artificial miRNA were compared by plotting sensitivity versus specificity. The area under the curve (AUC) for each prediction method is indicated. A random prediction is shown as a dashed line.
Because seed match sites were not sufficient to mediate gene repression, we sought to determine whether the activity of an artificial miRNA could be predicted using currently available tools for endogenous miRNAs. We examined two published miRNA prediction algorithms, TargetScan (5,10,13,34) and PITA (11), and also tested whether hybridisation energy alone was predictive of activity. Each algorithm was used to calculate a prediction score for each artificial miRNA/gene target pair. In the case of the PITA algorithm, two different settings for the inclusion of flanking nucleotides in the calculations were used (either no flanking nucleotides or 3 upstream and 15 downstream nucleotides) (11). Target sites that could not be scored by the algorithms were excluded from analysis. PITA scores were significantly different between artificial miRNAs that repressed or did not repress the reporters (P < 0.01, t-test), and the difference was similar when PITA scores were calculated with or without taking into account the nucleotides flanking the target site (Figure 5C). TargetScan measures several seed and non-seed factors, including pairing at the miRNA 3′ end, local adenosine-uridine (AU) context, site position effects, seed pairing stability and target site abundance, and combines these into an overall “context+ score.” While the TargetScan 3′ end pairing sub-score, which is based on scoring contiguous Watson–Crick base pairing but ignores the energy changes from that pairing (10), was significantly associated with reporter repression (P < 0.001, t-test), the other scores were not significantly different (Figure 5D and Supplementary Figure S1A). To examine the sensitivity and specificity of the algorithms for predicting artificial miRNA activity, we computed the area under the curve (AUC). 3′ end pairing was modestly predictive of artificial miRNA activity (AUC = 0.67, 95% CI = 0.59–0.75), but none of the algorithms that we tested were strong predictors (Figure 5E and Supplementary Figure S1B). To ensure that the predictions were not limited by our analysis of only two transcripts or our use of reporter assays, we repeated the TargetScan analysis with the microarray profiling data from four artificial miRNAs. Although the context+ scores and several sub-scores differed significantly between repressed and unchanged transcripts, we again observed that none of the scores strongly predicted artificial miRNA activity (Supplementary Figure S2). Taken together, these results suggest that Watson–Crick base pairing at the 3′ end is a partial determinant of artificial miRNA activity and has a bigger impact than target site secondary structure or favourable energy dynamics. However, overall, differences in activity across artificial miRNAs are not driven by factors measured by prediction algorithms derived from endogenous miRNAs.
DISCUSSION
Artificial miRNAs are a new class of short RNAs that combine properties of endogenous miRNAs and artificial siRNAs and, like endogenous miRNAs, have the potential for combinatorial control of gene expression. Our study provides the first systematic analysis of artificial miRNAs and demonstrates that they can repress multiple genes. However, our results showed that rational design of miRNAs based on currently defined rules for miRNA target repression is inefficient at identifying functional multi-targeting artificial miRNAs.
As with endogenous miRNAs, seed matches are clearly important for artificial miRNA activity, and our results show that canonical seed match sites with perfect base pairing are more robust for mediating gene repression than non-canonical bulge sites. Our finding that bulge sites appear to be inferior to canonical sites may also be relevant to endogenous miRNA targeting. In a recent study, bulge sites accounted for at least 15% of Argonaute–miRNA interactions in mouse brain (14), but our analysis suggests that the magnitude of repression from these sites should be expected to be significantly weaker than with canonical sites.
Although studies of endogenous miRNA targeting have emphasized the importance of seed matches, these matches are not sufficient for silencing of all targets, and non-seed factors influence gene repression (8,10,15,16). Consistent with these observations, we found that the majority of the artificial miRNAs did not repress their targets, even among those perfectly matched for the seed. Thus, non-seed factors determine which seed match sites are utilized for gene repression. In previously reported endogenous miRNA transfection experiments, about one-third of predicted seed match targets are altered at the protein level (15), which corresponds well with our result that only 34% of canonical seed match sites result in reporter repression by artificial miRNAs. Therefore, our study further supports the view that target site context is a major determinant of whether a given seed sequence is effective.
In order to understand the non-seed factors contributing to miRNA activity, we utilized published algorithms that have defined non-seed determinants and mRNA context features that predict endogenous miRNA target recognition. While we expected these prediction algorithms to perform similarly with artificial miRNAs, we found that most of the reported measures of target context that we tested were poor predictors of artificial miRNA activity. Several groups have suggested that mRNA secondary structure is a key determinant of miRNA targeting (11,12), but site accessibility due to secondary structure was no different between active and inactive target sites in our system. The PITA algorithm (11), which is based on the miRNA:mRNA hybridisation energy and predicted local mRNA structure, also performed no better than measuring the hybridisation energy alone, again suggesting a minor role for secondary structure in target recognition. The features that contribute to the TargetScan context+ score (10,34) also showed limited evidence of predicting artificial miRNA activity. While differences in the relative importance of targeting determinants have been linked to the methods used to assay miRNA activity (41), we saw similar results between reporter assays and microarray profiling. However, using TargetScan to measure the extent of Watson–Crick base pairing at the 3′ end was the best predictor for artificial miRNA reporter activity and outperformed energy-based predictions.
The failure of endogenous miRNA targeting algorithms to perform robustly with artificial miRNAs raises important questions about the potential for differences between artificial and endogenous miRNAs and the effectiveness of target prediction in general. One possible explanation for our results is that despite their structural similarities, artificial miRNAs do not respond to the same context features that impact endogenous miRNAs, and therefore current prediction algorithms do not fully capture the rules that govern artificial miRNA targeting. Such differences may be due to the fact that endogenous miRNA-target interactions are the product of evolution and have been subject to selective pressures that impact target prediction, whereas artificial miRNAs have not been subject to such evolutionary constraints. At this point, however, this remains a speculative hypothesis and additional studies of artificial miRNAs, utilising a larger variety of 3′ UTR targets, will be necessary to more reliably compare features of targeting by endogenous versus artificial miRNAs. Given their similar frequency for repression of seed matched targets, artificial and endogenous miRNAs most likely adhere to the same general underlying targeting mechanisms. Therefore, our work is consistent with the notion that non-seed factors such as secondary structure, sequence context and base-pairing strength are relatively limited determinants of miRNA targeting and that important additional factors that drive both endogenous and artificial miRNA targeting remain to be discovered.
Although additional non-seed determinants and target context features are strongly implicated in miRNA targeting, their nature is not clear, and they may well be mRNA factors rather than intrinsic factors of the miRNAs themselves. It is possible that RNA-binding proteins play an important role in determining the effectiveness of miRNA targeting. For example, the mRNA-binding protein HuR can antagonize the activity of endogenous miRNAs that bind to the same target mRNA (42–45), although cooperation between HuR and miRNAs has also been observed (46,47). Transcriptome-wide analyses have demonstrated that HuR shares many targets with miRNAs and modulates miRNA activity, with the HuR and miRNA binding sites often being adjacent but not overlapping (48,49). The RNA-binding protein Dnd1 was also shown to regulate miRNA target interactions (50). Thus, specific combinations of proteins along an mRNA, as well as higher-order mRNA structure, may facilitate or block miRNA binding and target repression.
Beyond the implications for miRNA targeting, our study addresses the rational design of artificial miRNAs to repress multiple genes of interest at once for multi-target RNAi. While multi-target RNAi has been demonstrated as a proof of concept (22), a systematic analysis of artificial miRNAs designed to target specific genes has not been previously reported. We identified nine artificial miRNAs with distinct seed regions that repress the protein output of two targeted endogenous genes simultaneously, providing a more extensive demonstration of multi-target RNAi using artificial miRNAs. However, the rational design of artificial miRNAs based primarily on seed matches is an inefficient process and is not currently accurate enough for practical use. An alternative approach that may be more effective would involve combining rational design with functional screening. Small libraries of artificial miRNAs could be designed to target several genes of interest and then functionally screened to identify highly active artificial miRNA sequences that provide combined gene repression. In the future, as additional non-seed factors responsible for mRNA targeting efficacy are defined, an improved understanding of design principles may eventually enable highly effective rational design of artificial miRNAs to target sets of related genes or to inhibit several pathways in concert, thereby significantly expanding the experimental toolkit.
ACCESSION NUMBER
GenBank GSE50249.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including Supplementary Tables 1–7 and Supplementary Figures 1–2.
Acknowledgments
We thank the Fred Hutchinson Genomics Shared Resource for microarray processing. We also thank E. Knouf, P. Paddison and D. Hockenbery for advice and helpful discussion.
Footnotes
Present address: Department of Internal Medicine, University of Michigan Medical School, 4029 BSRB, SPC 2200, 109 Zina Pitcher Place, Ann Arbor, MI 48109-2200
FUNDING
American Cancer Society [Postdoctoral Fellowship to J.D.A.]; National Institutes of Health [Transformative R01: R01DK085714 to M.T.]; and Damon Runyon-Rachleff Innovation Award [to M.T.]. Funding for open access charge: National Institutes of Health [Transformative R01: R01DK085714 to M.T.].
REFERENCES
- 1.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doench J.G., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai E.C., Tam B., Rubin G.M. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 7.Schirle N.T., MacRae I.J. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Didiano D., Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 9.Hausser J., Landthaler M., Jaskiewicz L., Gaidatzis D., Zavolan M. Relative contribution of sequence and structure features to the mRNA binding of Argonaute/EIF2C-miRNA complexes and the degradation of miRNA targets. Genome Res. 2009;19:2009–2020. doi: 10.1101/gr.091181.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 12.Long D., Lee R., Williams P., Chan C.Y., Ambros V., Ding Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 13.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chi S.W., Hannon G.J., Darnell R.B. An alternative mode of microRNA target recognition. Nat. Struct. Mol. Biol. 2012;19:321–327. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 17.Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 18.Satoh J., Tabunoki H. Comprehensive analysis of human microRNA target networks. BioData Mining. 2011;4:17. doi: 10.1186/1756-0381-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivey K.N., Muth A., Arnold J., King F.W., Yeh R.F., Fish J.E., Hsiao E.C., Schwartz R.J., Conklin B.R., Bernstein H.S., et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anokye-Danso F., Trivedi C.M., Juhr D., Gupta M., Cui Z., Tian Y., Zhang Y., Yang W., Gruber P.J., Epstein J.A., et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 22.Passioura T., Gozar M.M., Goodchild A., King A., Arndt G.M., Poidinger M., Birkett D.J., Rivory L.P. Interfering ribonucleic acids that suppress expression of multiple unrelated genes. BMC Biotechnol. 2009;9:57. doi: 10.1186/1472-6750-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent W.J. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer L.R., Zweig A.S., Hinrichs A.S., Karolchik D., Kuhn R.M., Wong M., Sloan C.A., Rosenbloom K.R., Roe G., Rhead B., et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R Core Team. R Foundation for Statistical Computing. Vienna, Austria: 2012. [Google Scholar]
- 27.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer; 2009. [Google Scholar]
- 29.Wickham H. The split-apply-combine strategy for data analysis. J. Stat. Softw. 2011;40:1–29. [Google Scholar]
- 30.Ding Y., Lawrence C.E. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003;31:7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Y., Chan C.Y., Lawrence C.E. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 2005;11:1157–1166. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kruger J., Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia D.M., Baek D., Shin C., Bell G.W., Grimson A., Bartel D.P. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., Muller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fellmann C., Zuber J., McJunkin K., Chang K., Malone C.D., Dickins R.A., Xu Q., Hengartner M.O., Elledge S.J., Hannon G.J., et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol. Cell. 2011;41:733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 38.Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 39.Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 40.Vella M.C., Choi E.Y., Lin S.Y., Reinert K., Slack F.J. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen J., Parker B.J., Jacobsen A., Krogh A. MicroRNA transfection and AGO-bound CLIP-seq data sets reveal distinct determinants of miRNA action. RNA. 2011;17:820–834. doi: 10.1261/rna.2387911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 43.Tominaga K., Srikantan S., Lee E.K., Subaran S.S., Martindale J.L., Abdelmohsen K., Gorospe M. Competitive regulation of nucleolin expression by HuR and miR-494. Mol. Cell. Biol. 2011;31:4219–4231. doi: 10.1128/MCB.05955-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young L.E., Moore A.E., Sokol L., Meisner-Kober N., Dixon D.A. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol. Cancer Res. 2012;10:167–180. doi: 10.1158/1541-7786.MCR-11-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Epis M.R., Barker A., Giles K.M., Beveridge D.J., Leedman P.J. The RNA-binding protein HuR opposes the repression of ERBB-2 gene expression by microRNA miR-331–3p in prostate cancer cells. J. Biol. Chem. 2011;286:41442–41454. doi: 10.1074/jbc.M111.301481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim H.H., Kuwano Y., Srikantan S., Lee E.K., Martindale J.L., Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glorian V., Maillot G., Poles S., Iacovoni J.S., Favre G., Vagner S. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death Differ. 2011;18:1692–1701. doi: 10.1038/cdd.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lebedeva S., Jens M., Theil K., Schwanhausser B., Selbach M., Landthaler M., Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 49.Mukherjee N., Corcoran D.L., Nusbaum J.D., Reid D.W., Georgiev S., Hafner M., Ascano M., Jr, Tuschl T., Ohler U., Keene J.D. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kedde M., Strasser M.J., Boldajipour B., Oude Vrielink J.A., Slanchev K., le Sage C., Nagel R., Voorhoeve P.M., van Duijse J., Orom U.A., et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.