Abstract
Sibling care is a hallmark of social insects, but its evolution remains challenging to explain at the molecular level. The hypothesis that sibling care evolved from ancestral maternal care in primitively eusocial insects has been elaborated to involve heterochronic changes in gene expression. This elaboration leads to the prediction that workers in these species will show patterns of gene expression more similar to foundress queens, who express maternal care behaviour, than to established queens engaged solely in reproductive behaviour. We tested this idea in bumblebees (Bombus terrestris) using a microarray platform with approximately 4500 genes. Unlike the wasp Polistes metricus, in which support for the above prediction has been obtained, we found that patterns of brain gene expression in foundress and queen bumblebees were more similar to each other than to workers. Comparisons of differentially expressed genes derived from this study and gene lists from microarray studies in Polistes and the honeybee Apis mellifera yielded a shared set of genes involved in the regulation of related social behaviours across independent eusocial lineages. Together, these results suggest that multiple independent evolutions of eusociality in the insects might have involved different evolutionary routes, but nevertheless involved some similarities at the molecular level.
Keywords: bumblebees, cooperative brood care, eusociality, gene expression, microarrays
1. Introduction
One of the hallmarks of the insect societies is sibling care, where the adult female offspring of the queen, called workers, remain in their natal nest and care for their siblings. Sibling care in the social insects is multifaceted and involves tasks such as provisioning the nest, direct feeding of larvae and cleaning of brood cells [1]. Ultimate explanations for the evolution of sibling care have been thoroughly explored in social insect research [2,3]. However, the exploration of proximate mechanisms involved in the evolution of sibling care is a relatively young area of research.
A prominent hypothesis that focuses on proximate mechanisms states that sibling care is evolutionarily derived from ancestral maternal care [4–6]. According to this idea, reproductive (e.g. egg laying) and maternal care (e.g. brood provisioning) behaviour were uncoupled temporally across an evolutionary timescale, eventually occurring in the separate female castes, the queens and workers. A molecular perspective on this hypothesis posits that sibling care originated via heterochronic changes in gene expression, such that the molecular architecture underlying maternal care behaviour was co-opted to be used in a new developmental context, the worker caste [7]. The hypothesis that sibling care evolved from maternal care (hereafter, the ‘molecular heterochrony hypothesis’) predicts that sibling care and maternal care behaviours are regulated by similar patterns of gene expression. This hypothesis is particularly compelling in social insect lineages where both maternal care and sibling care are present and involve strikingly similar behaviours in queens and workers. More broadly, the molecular heterochrony hypothesis is supported by studies on the evolution of development in a variety of taxa, which together suggest that evolution is often conservative, and evolutionary novelty can be achieved through the co-option of pre-existing genetic architecture [8].
There are alternatives to the molecular heterochrony hypothesis, including the idea that sibling care evolved de novo [9], and therefore is not evolutionarily rooted in maternal care behaviour. Support for the ‘de novo hypothesis’ may be found in the complete absence of maternal care in some lineages of eusocial insects. However, recent studies on the honeybee Apis mellifera, a highly eusocial species that lacks maternal care, suggest that there may nonetheless be an evolutionary link between sibling care and maternal care in this species [10]. Perhaps, the loss of maternal care has repeatedly been a component of the evolution of highly eusocial lineages, in which case the de novo hypothesis is difficult to explore in these lineages. Additional support for the de novo hypothesis has recently been discovered using transcriptome sequencing in Polistes canadensis, which revealed an abundance of putatively novel or rapidly evolving sequences among those differentially expressed between queens and workers [11]. Given that complex sociality evolved 11 or more times in the insects [12–16], it is possible, and perhaps even likely [17], that sibling care evolved via multiple evolutionary routes in different social insect lineages.
A related question is whether similar genes and molecular pathways have been involved in the evolution of sibling care across independent social insect lineages, regardless of the routes or evolutionary mechanisms through which sibling care evolved [8,17]. There is a growing body of evidence that suggests that there are many commonalities in genes and molecular pathways associated with various behavioural states across the independently evolved social insect lineages, referred to as ‘genetic toolkits’ [8]. The insulin signalling pathway appears to be one such genetic toolkit, as it has been implicated in the regulation of various aspects of worker division of labour in all of the major groups of social insects (the ants, bees, wasps and termites) [18]. An important goal in the study of genetic toolkits in the social insects is to determine how widespread this phenomenon is, as well as to further refine our understanding of how the same genes and pathways operate in the regulation of social behaviours in disparate lineages [8].
The so-called primitively eusocial insects (which include some bees and wasps) possess an annual colony cycle and tend to have simpler forms of social organization than the ‘highly’ eusocial insects, such as honeybees and stingless bees [1]. Primitively eusocial species are excellent subjects to explore the evolution of sibling care, because queens and workers often perform similar brood-care-related tasks, albeit during different stages in the colony cycle. For example, prior to the emergence of the first workers in a colony, nest-founding queens (‘foundresses’) must provision, feed and otherwise tend to their offspring, tasks that are assumed by workers following their emergence in the nest.
Toth et al. [19] used the similarity between maternal and sibling care behaviour in the primitively eusocial wasp Polistes metricus to perform the first test of the molecular heterochrony hypothesis. Using patterns of brain gene expression for a set of 32 genes associated with worker division of labour in the honeybee A. mellifera, Toth et al. [19] showed that foundresses and foraging workers, the two groups that forage and provision the nest, share the most similar patterns of brain gene expression relative to other females in the nest, the queens and ‘gynes’ (non-foraging and non-reproductive individuals destined to become queens the following season). This clustering of foundresses and foraging workers based on brain gene expression data occurred despite the fact that there are major differences in reproductive status between these two groups; Polistes foundresses are mated, have developed ovaries and lay eggs, and foraging workers are entirely non-reproductive. Using a more extensive set of genes (approx. 3200) in a microarray study, Toth et al. [20] showed that the above pattern was specific to the subset of genes that was tested, because for most genes in the Polistes genome, foundresses and queens show more similar patterns of brain gene expression to each other than to workers.
Building on the studies of Toth et al. [19,20], an important question to address in social insect research is whether support for the molecular heterochrony hypothesis can be found in other social insect lineages in addition to Polistes. This is important because sociality evolved independently in the Hymenoptera (ants, bees and wasps) about a dozen times [12–16], and molecular evolutionary analyses suggest that a mixture of common and distinct genes have been involved in these independent evolutions [17]. If the molecular heterochrony hypothesis holds in other lineages, then this would suggest that there might be unifying principles of insect social evolution at the level of gene regulatory changes. Alternatively, independent evolutions of eusociality in insects may have involved primarily distinct heterochronic or other types of gene regulatory changes. Distinguishing these two possibilities is currently one of the most important issues in the study of the evolution of eusociality.
We tested the molecular heterochrony hypothesis for the evolution of sibling care in the bumblebee lineage using an Agilent microarray based on expressed sequence tag (EST) sequence for the bee Bombus terrestris [21]. Despite evolving eusociality independently, both Bombus and Polistes share an annual colony cycle that is characteristic of the primitively eusocial lifestyle. Each spring, foundresses initiate new nests that will persist until the end of the summer. Following the emergence of the first offspring in the nest, foundresses are considered true queens. The first female offspring to emerge will develop into workers who perform brood care and other work-related tasks for the nest, but later in the season, some female offspring develop into gynes; these gynes will mate, overwinter and become the foundresses (and later on, queens) of the following season.
As in Toth et al. [19,20], we explored patterns of brain gene expression in the four female groups in a primitively eusocial society (foundresses, queens, gynes and workers) to identify similarities in brain gene expression between these groups. Unlike Toth et al. [19,20], who focused on nest-provisioning behaviour, which involves flight- and foraging-related activities, our experiments in Bombus instead focused on the direct feeding of brood (hereafter, ‘brood-feeding behaviour’) as a component of sibling and maternal care. In bumblebee colonies, developing brood are progressively fed during the larval stage, a task that involves the direct placement of food (pollen and honey) in larval cells [22]. As with nest provisioning, brood-feeding behaviour is performed by foundresses during the nest-founding phase of the colony cycle and also later on by workers upon their emergence in the colony. Although Polistes nests openly and all individuals are exposed to light, Bombus are cavity nesters, and queens and workers highly specialized on brood-feeding behaviour typically spend much of their time inside the nest. By focusing on brood-feeding behaviour in B. terrestris, we could collect individuals that differed in their brood care behaviour, but were kept under constant, similar light conditions and unable to freely forage, conditions that control for effects of light exposure and flight activity on brain gene expression [23].
We performed an additional, comparative analysis to identify genes that may have repeatedly played a role in the evolution of sociality in insects, across three putatively independent eusocial insect lineages [12]. Here, we explored the overlap between lists of genes potentially involved in reproduction and/or brood care in bumblebees (generated from this study) and lists of genes associated with brood care and reproduction in Polistes [19,20] and with worker division of labour in the honeybee A. mellifera [23,24]. Genes found in the overlap of two or more of these gene lists may represent components of a genetic toolkit for the evolution of complex social behaviours in the insects [8].
2. Material and methods
(a). Bees
The bee experiments were performed in Israel during a two month period in summer 2009. Bumblebees (B. terrestris) were purchased from Polyam Industries (Kibbutz Yad-Mordechai, Israel) and reared under the conditions described in references [25,26]. The four Bombus groups (figure 1) were identified and collected as follows: (i) foundresses (n = 14) were individuals in the queen caste who had recently initiated nests, and had larvae in their nests and were necessarily performing all brood care, because adult workers had not yet emerged; (ii) queens (n = 16) were individuals in the queen caste with mature nests (approx. 50 workers) who had ceased to perform direct brood feeding, but were still actively laying eggs and were observed to be dominant over workers in the colony; (iii) gynes (n = 11) were newly emerged (3 days old) queens collected from three separate colonies; (iv) workers (n = 14) were 7–9 days old, non-reproductive workers who were specialized on feeding brood in the nest. To identify workers specialized on feeding brood, all workers in three source colonies were individually marked with coloured number tags on the day of their emergence and the colonies were observed for a two week period. Each day of observation, each colony was observed for 1 h and both the total number of brood-feeding events observed and the identity of the brood-feeding bee were recorded. Brood feeding is an easily observable behavioural sequence that lasts approximately 5–15 s; larvae are clumped together spatially, and brood-feeding bees go to these clumps of larvae, open and inspect larval cells, and then regurgitate food into the cells [21,22]. Bees collected as workers were observed feeding larvae on three of five observation days, including the day of or day before collection. For each colony, a pollen feeder was kept in a lighted foraging arena attached to the colony with an approximately 0.3 m tube. Before and after observations, the lighted foraging chambers were scanned, and the identity of any bees in the foraging arenas was recorded; no workers included in the analysis were ever observed in the lighted foraging arenas.
Figure 1.
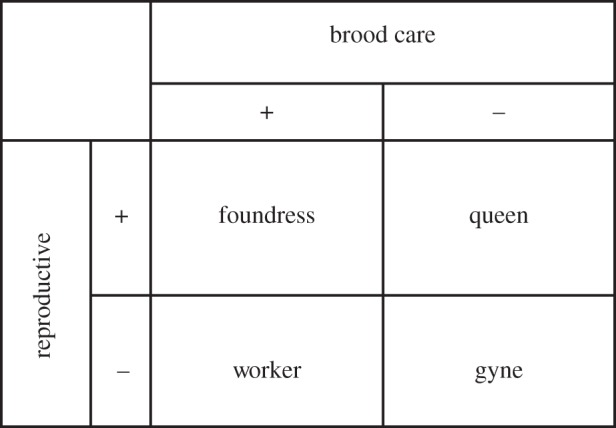
Factorial design of B. terrestris microarray experiment. ‘Reproductive’ refers to whether an individual had developed ovaries (+) or did not (−); ‘brood care’ refers to whether an individual belonged to a group that feeds brood (+) or a group that does not actively feed brood (−).
(b). Ovary dissections
The ovaries of all collected bees were dissected to confirm reproductive status. The abdomens were collected and stored at −20°C until dissections were performed using methods described in reference [25]. For workers, the average lengths of the two largest terminal oocytes (one per ovariole) were measured using an optical scale, and these values were averaged as a measure of ovary development, as in reference [27]. All workers in the study had values less than 1.2 mm. For all other groups (foundresses, queens and gynes), ovaries were classified as developed or undeveloped based on gross morphology. As has previously been reported [26,28], there was little variation in ovary development within these three groups; all foundresses and queens had fully developed ovaries, and all gynes had completely undeveloped ovaries.
(c). RNA preparation
All bees were collected directly into dry ice, and their heads were immediately removed, placed in liquid nitrogen and stored at −80°C to ensure RNA preservation. For brain dissections, whole bee heads were partially lyophilized, and the dissections were performed over dry ice [29]. RNA was isolated from dissected brains using an RNeasy mini kit (Qiagen, Valencia, CA, USA), following the kit protocol except that the initial homogenization was performed in a 500 μl microfuge tube using 100 μl extraction buffer.
(d). Microarrays
Agilent 4 × 44K microarrays designed from B. terrestris EST sequence [21] were used to examine brain gene expression. Each microarray slide contained four identical arrays, each containing approximately 44 K EST-based probes corresponding to a total of 4503 putative A. mellifera genes; additional details of array design in reference [21]. Total RNA per bee sample (250–1000 ng) was reverse transcribed and linear amplified according to the manufacturer's instructions (Agilent Technologies, Santa Clara, CA, USA). Samples (one per array) were hybridized on the microarray slide and washed according to the Agilent protocol. Slides were scanned using an Axon 4000B scanner, and images were analysed with genepix 6.1 software (Molecular Devices, Sunnyvale, CA, USA). Microarray experimental methods are described in greater detail in references [24,30]. The microarray results from this study can be accessed at the ArrayExpress website (http://www.ebi.ac.uk/arrayexpress/).
(e). Statistical analysis of microarray data
Microarray data pre-processing and statistical analyses were performed with the limma package [31] in R [32]. Treatment of manually flagged spots and background correction was performed as previously described in reference [21]. Because only one sample was hybridized to each microarray slide, the single-channel expression values were normalized with the quantile method [33] then log2-transformed. Coefficient of variation (CV) values were calculated using the 55 samples, and removal of control spots and probes with CV < 0.015 was performed as in reference [26], leaving 36 876 spots out of 45 220.
A 2 × 2 factorial model [34] (main effects: reproduction, feeding; figure 1) was fitted in limma, taking into account variation owing to possible differences in each microarray slide [35]. The overall ANOVA F-test, the main effects of reproduction and feeding, the interaction term and mean expression level for the four groups were estimated. Multiple test adjustment was performed separately for each contrast using the false discovery rate (FDR) method [36]. Applying probe annotation methods based on BLAST to the Official Honey Bee Gene Set version 2 for A. mellifera [37] (described in [21]), 10 037 probes were annotated, putatively representing 4503 unique A. mellifera genes. Lists of genes potentially associated with brood-feeding behaviour and with reproduction (the ‘feeding gene list’ and reproduction gene list’, respectively) were created from the lists of annotated probes that were significant at an FDR-corrected p < 0.05 for each of these main effects in the ANOVA. When multiple probes corresponding to the same A. mellifera gene were significant for a main effect but regulated in opposite directions, the corresponding genes were removed from the feeding (n = 71 genes removed) and reproduction (n = 178 genes removed) gene lists. As is customary in the field of transcriptomics, a small subset of genes that were significantly differentially expressed in microarray analysis were also measured with real-time, quantitative reverse transcription polymerase chain reaction to provide a general sense of the strength of the microarray results (the electronic supplementary material, figures S1–S6, Methods and results).
(f). Gene functional analysis
To identify gene functional terms enriched in the feeding and reproduction gene lists, these lists were transformed into lists of Drosophila melanogaster orthologues using a previously published A. mellifera–D. melanogaster orthologue list [37], and analysed using the gene ontology (GO) functional annotation tool (GOFat level) on the DAVID website [38]. Genes present in both the feeding and reproduction gene lists, as well as genes significant for the interaction term in the ANOVA, were removed from the lists prior to identifying D. melanogaster orthologues; these filtered genes were combined to create a third list (the ‘both gene list’). For each of the three gene lists, enriched (p < 0.05; more than five genes per term) biological process and molecular function terms were identified using a background list consisting of 3686 D. melanogaster genes corresponding to the 4503 putative A. mellifera genes on the microarray.
(g). Patterns of gene expression across the four Bombus groups
To explore patterns of brain gene expression across the four Bombus groups, hierarchical clustering analysis (HCA) was performed on multiple sets of probes using the stats and gplots [39] R packages. To limit our clustering analysis to genes that were differentially expressed between the four Bombus groups, all HCA was limited to genes with significant p-values from the ANOVA. First, to identify overall patterns, HCA was performed on the four mean group estimates for all 8044 probes significant from the ANOVA. This analysis was performed to characterize general patterns of similarity in brain gene expression across the four groups, and served as a ‘null’ pattern to be contrasted with our functional subsets of genes. As a specific comparison with the heterochronic changes previously detected in Polistes [19,20], we next approximated the analyses used by Toth et al. [19,20] by performing HCA on two subsets of probes: (i) the 45 B. terrestris probes corresponding to the 32 feeding- and reproduction-related genes originally used to measure patterns of brain gene expression in Polistes [19] (‘test 1’); and (ii) a broader subset (‘test 2’), which included an amalgam of probes on the microarray corresponding to genes that met any of the following criteria:
(1) Feeding- and/or nutrition-related function in D. melanogaster (D. melanogaster orthologues have associated biological process or molecular function GO terms that include the words ‘food’, ‘feed’, ‘nutrient’ or ‘foraging’; n = 17).
(2) Reproduction-related functions in D. melanogaster (D. melanogaster orthologues have associated biological process or molecular function GO terms that include the words ‘reproductive’, ‘reproduction’, ‘oocyte’, ‘oogenesis’ or ‘gamete’; n = 222).
(3) Used in Toth et al. [19] (n = 19).
(4) Differential expression associated with brood care (the 27 genes differentially expressed between A. mellifera workers specialized on in-hive tasks versus foraging tasks in [23] and/or [24], and also differentially expressed between provisioning and non-provisioning Polistes wasps in [20] and between brood-feeding and non-brood-feeding individuals in this study).
(5) Differential expression associated with reproduction (35 genes differentially expressed between reproductive and non-reproductive Polistes wasps in [20] and also between reproductive and non-reproductive bumblebees in this study).
Some probes used for test 2 met more than one of the criteria. Finally, to discern whether the patterns from tests 1 and 2 were specific to these functional subsets, HCA was performed using 200 random subsets of 45 and 1022 probes, and the topologies resulting from this analysis were compared with the topologies resulting from tests 1 and 2.
(h). Cross-species comparisons
Both pairwise Fisher's exact tests (one-tailed) and a simulation-based method (also used and described in 20) were used to determine whether a statistically significant (p < 0.05) degree of overlap exists between the Bombus reproduction and feeding gene lists, and lists of genes associated with reproduction and feeding generated from a brain microarray experiment in Polistes [20] and lists of genes differentially expressed in A. mellifera nurse and forager brains, also generated from microarray experiments [23,24]. Both nurse and forager honeybees undertake feeding-related brood care tasks, with nurses directly interacting with and feeding larvae, and foragers leaving the nest to collect food provisions. Nurse and forager honeybees are not typically reproductive, although nurses may possess some ‘reproductive-like’ phenotypic traits [10]. All cross-species comparisons were performed using A. mellifera orthologues, and background lists for each comparison were limited to genes represented on both microarrays. Genes were counted as overlapping regardless of the direction of change (up- or downregulated) in the two lists.
3. Results
(a). ANOVA of microarray data
Of the 10 037 probes included in the statistical analysis, 80% (n = 8044) had overall ANOVA F-tests with FDR-corrected p < 0.05, putatively representing 4040 unique genes differentially expressed across the four Bombus groups. Many genes (2563) had levels of brain expression associated with differences in brood-feeding behaviour identified using ANOVA (‘feeding gene list’). This list included the gene insulin receptor substrate 1, which was upregulated in brood-feeding individuals, and was also upregulated in nest-provisioning Polistes in Toth et al.'s study [19], and the gene kruppel homolog 1. In the filtered feeding gene list (n = 525 genes), in which genes also significant for other main effect and/or interaction were removed, the term ‘response to hormone stimulus’ was enriched (electronic supplementary material, table S1). Many genes (3112) had expression levels associated with differences in reproductive status identified using ANOVA (‘reproduction gene list’). This list included an additional insulin signalling-related gene, target of rapamycin (tor), which was upregulated in reproductive individuals. In the filtered reproduction gene list (n = 1697 genes), one term related to reproduction (‘gamete generation’) was enriched, as well as terms related to ageing (e.g. ‘ageing’) and brain function (e.g. ‘axon guidance’; the electronic supplementary material, table S1). Within the ‘both gene list’ (n = 2875 genes), several biological process terms related to oocyte development (e.g. ‘oocyte fate determination’) were enriched (the electronic supplementary material, table S1).
(b). Patterns of expression across the four Bombus groups
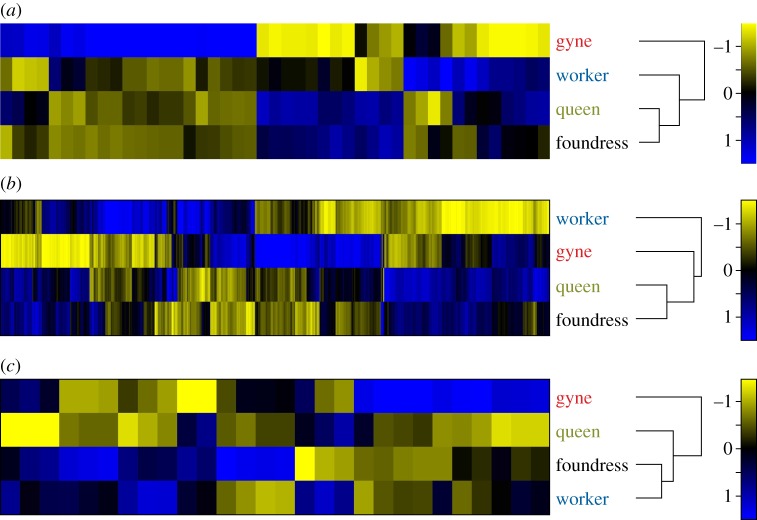
HCA performed using expression data from all probes significant in the overall ANOVA (n = 8044) yielded the following relationships between the four Bombus groups: (worker (gyne (foundress, queen))) (figure 2a). Neither main effect in the ANOVA appears to have been driving this clustering, as the distributions of absolute values of fold changes for the sets of probes significant for the main effects were both similarly negatively skewed (feeding: mean = 0.21, σ = 0.10; reproduction: mean = 0.28, σ = 0.20; the electronic supplementary material, figures S7 and S8). For comparison, figure 2b shows relationships between the analogous four groups in Polistes, which Toth et al. [20] derived from HCA of brain expression data using 447 probes significant in their Polistes microarray experiment using ANOVA.
Figure 2.
HCA of Bombus and Polistes brain gene expression data using all probes significant for the ANOVA F-test. (a) Heat map derived from HCA of Bombus microarray data (n = 8044 probes). (b) Heat map derived from HCA of Polistes microarray data (n = 447 probes) [20]. Blue colour represents upregulation, yellow represents downregulation and black represents no difference in expression. The number scale represents the strength of the difference in expression.
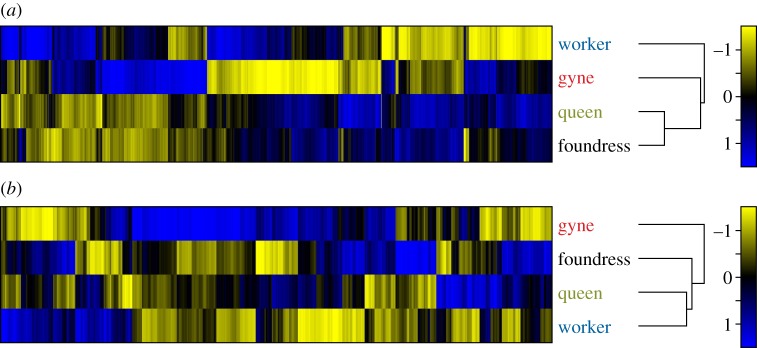
For test 1, HCA yielded the following relationships between the four Bombus groups: (gyne (worker (foundress, queen))) (figure 3a). Table S2 in the electronic supplementary material provides a summary of expression data for the 32 genes in Toth et al.'s [19] original Polistes study, including results from both Polistes studies [19,20] and this study. For test 2, HCA yielded the same relationships obtained from HCA of all probes significant for the overall ANOVA: (worker (gyne (foundress, queen))) (figure 3b). For comparison, figure 3c also includes a heat map recreated from the Polistes data in reference [19]. In 100% of the 200 random subsets of 45 and 1022 probes, foundresses and queens clustered together, with workers and gynes clustering in various ways (data not shown).
Figure 3.
HCA of Bombus and Polistes brain gene expression data using functional subsets of genes related to feeding and reproduction. (a) Heat map derived from HCA in test 1 (45 B. terrestris probes corresponding to the 32 feeding- and reproduction-related genes originally used to measure patterns of brain gene expression in Polistes [19]). (b) Heat map derived from HCA in test 2 (1022 B. terrestris probes corresponding to genes implicated in reproduction and/or feeding in insects in other studies). (c) Heat map derived from HCA of Polistes qPCR data [19]. Blue colour represents upregulation, yellow represents downregulation, and black represents no difference in expression. The number scale represents the strength of the difference in expression.
(c). Cross-species comparisons
For the tests of overlap between the Bombus (this study), Polistes [20] and Apis [23,24] gene lists, only the comparison between the Bombus feeding gene list and the list of genes associated with nest provisioning in Polistes [20] showed a significant (p < 0.05 for Fisher's exact test; p < 0.01 for simulation-based test) degree of overlap (the electronic supplementary material, figure S7). Tables S3 and S4 in the electronic supplementary material list genes present in the overlap of two or more studies. Among the seven genes present in the brood-care-related lists from all three species was the gene inositol 1,4,5,-tris-phosphate receptor (itpr1), which was downregulated in brood-feeding Bombus (this study), nest-provisioning Polistes [19] and Apis foragers [24].
4. Discussion
Discovering the proximate mechanisms involved in the evolutionary origins of sibling care is a major topic of research on the social insects. We found differences and similarities in patterns of brain gene expression associated with brood care and reproduction in bumblebees (B. terrestris) compared with paper wasps (P. metricus). These results suggest that the multiple independent evolutions of eusociality in the insects might have involved different evolutionary routes, but nevertheless involved some similarities at the molecular level.
We found that foundresses and queens share the most similar patterns of brain gene expression in B. terrestris for most genes on the microarray, as well as for functional subsets of genes related to feeding and reproduction. By contrast, studies on the wasp P. metricus have shown that foundresses and workers share the most similar patterns of brain gene expression for a functional subset of genes related to provisioning, reproduction and division of labour [19], whereas queens and workers show the most similar patterns for approximately 3200 genes analysed by microarray [20]. In both Bombus and Polistes, foundresses and queens are reproductive, but differ in their brood care behaviour, suggesting that in Bombus, but not in Polistes, reproductive status has a stronger influence on overall patterns of brain gene expression than brood-feeding behaviour.
The strong, shared patterns of expression that we detected in foundress and queen bumblebees may also suggest that at the level of brain gene expression, fewer changes occur during the transition from foundress to queen in the Bombus life cycle relative to Polistes, although in both lineages a complex suite of physiological and behavioural changes are involved in this transition [28,40]. A recent study in bumblebees demonstrated that queens have considerable control over their reproduction, and therefore they appear to have retained some plasticity resembling that which is seen in solitary insects, and is also seen in foundresses [41]. Caste differentiation is also more profound in bumblebees compared with Polistes. For example, bumblebee workers are much smaller and variable in size than queens and have lost their spermathecae. In natural contexts, the transition from foundress to queen in Bombus may be viewed as more involved than in Polistes, as in Bombus this transition involves cessation of flight and exposure to light, whereas in Polistes, which nest openly, foundresses and queens are both exposed to light and queens may occasionally fly from the nest. Additionally, foundresses in many Polistes species (including P. metricus) may found nests in small groups, in which case the dominant foundress behaves ‘queen-like’ [42], suggesting that there may be greater flexibility in the transition from foundress to queen in Polistes relative to Bombus.
The results of our HCA suggest that perhaps in the bumblebees, sibling care is not evolutionarily rooted in ancestral maternal care behaviour, and therefore may have evolved de novo. Alternatively, sibling care may have evolved from maternal care in this lineage, but predictions consistent with the molecular heterochrony hypothesis were not upheld, perhaps because evolutionary diversification in brain gene expression patterns in queens and workers precludes detecting similarities in brain transcription associated with sibling and maternal care.
An additional, methodological explanation for the different patterns observed in Bombus and in Polistes [19,20] is the focus here on the direct brood-feeding component of brood care, rather than on nest provisioning, as was focused on in Polistes. Foraging and other flight-related behaviours have strong, documented effects on brain gene expression in bees [23], whereas the relationship between brain gene expression and brood-feeding behaviour in the social insects is relatively unknown [21], and may be weaker than for flight-related behaviours. In bumblebees, workers can transition between specialization on foraging and brood-feeding over the course of a day [27], suggesting great plasticity in these behaviours, which therefore may not involve large-scale changes in brain gene expression [43].
The results of this study also suggest that several candidate genes and molecular pathways previously implicated as important in eusocial insect evolution have also been important in the evolution of sociality in the bumblebees. These include: the insulin signalling pathway, which has repeatedly been implicated as playing an important role in social feeding and foraging behaviours [18,44]; the transcription factor kruppel homolog 1, a gene that appears to be regulated by queen mandibular pheromone in honeybees [45] and also by both the queen and juvenile hormone in B. terrestris [46]; and the inositol signalling pathway, which is involved in both larval feeding [47] and flight behaviour [48] in Drosophila. The gene inositol 1,4,5,-tris-phosphate receptor is a particularly promising candidate for future studies on the shared molecular basis of social evolution in insects, as this gene was differentially expressed in studies of feeding-related behaviours in Bombus, Polistes [19] and Apis [23], which represent three putatively independent evolutions of eusociality [12]. Our finding of genes of shared evolutionary involvement across multiple social insect lineages does not preclude the importance of novel and rapidly evolving genes in social evolution, as these and other molecular changes were likely also involved in the evolution of sociality in these lineages [11,17].
In the cross-species comparisons of gene lists derived from Bombus, Polistes [20] and Apis [23,24] microarray studies, only the feeding-related lists from Bombus and Polistes showed a significant degree of overlap. This finding suggests that in regard to brain gene expression, there may be more similarity in the regulation of feeding-related social behaviours relative to reproductive behaviours in Bombus and Polistes. The greater similarity between the Bombus and Polistes gene lists relative to Apis may also be related to the fact that the Bombus and Polistes lists were both generated from studies that included different castes (i.e. queens and workers), whereas the Apis lists were generated from studies on workers performing different feeding-related tasks (i.e. nurses and foragers). Toth et al. [20] similarly found a significant degree of overlap between Apis nurse/forager [23,24] and Polistes feeding-related lists, but not between the Apis nurse/forager and Polistes reproduction-related lists. In relation to this study, this suggests that there may be greater similarity in the molecular regulation of feeding-related behaviours in Bombus and Polistes, which are both primitively eusocial, than exists between Bombus and Apis, which differ more in their degree of social complexity but are more closely related to one another than to Polistes. Additionally, a recent phylogenetic analysis suggests the possibility that Bombus and Apis shared a common eusocial ancestor [49], which makes the similar pattern between Bombus and Polistes more difficult to explain.
Some caveats exist in relation to our use of microarrays to explore social evolution in bumblebees. Although our requirement that sequences be annotated in order to be included in our analyses enabled us to make inferences about the functional importance of genes, this requirement also likely biased our analyses towards the inclusion of genes that are more highly conserved between insect lineages. There is growing evidence that other types of molecular evolutionary changes, including rapidly evolving and novel genes, have also played important roles in social evolution in many insect lineages [11,50], including bumblebees [17]. Additionally, analyses that include information about alternative splicing, which was not addressed in this study, may provide a more refined picture of how individual genes can be differentially regulated in social contexts [51,52]. In the future, additional genomic methods, for example the direct sequencing of RNA to explore gene expression, will be important for exploring the role of these and other types of molecular evolutionary changes in social insect evolution.
When integrated with other studies on the molecular basis for social evolution in insects, our study provides additional support that the multiple independent evolutions of eusociality that occurred in the insects were multifaceted, involving a variety of molecular changes, both shared and unique across lineages [50]. For example, here, we found evidence that bumblebees might not have evolved sibling care via the same heterochronic changes in gene regulation as it appears to have in Polistes [19,20], and yet our results also strengthen the idea that genetic toolkits have played a widespread role in social insect evolution, with feeding- or nutrition-related genetic pathways playing a prominent role [8,18]. A remaining challenge is to unify our understanding of the diverse proximate mechanisms involved in insect social evolution with the selective forces and evolutionary pressures that ultimately led to complex social life in the insects.
Supplementary Material
Acknowledgements
Thanks to H. Shpigler, Y. Gruber, M. Tamarkin, M. Cohen, and other members of the Bloch laboratory for comments on experimental design and assistance with collections; A. Toth for assistance with experimental design; B. Fischman, other members of the Robinson laboratory, and two anonymous reviewers for comments on the manuscript; M. Majewski and J. Drnevich for assistance with microarrays; Yad-Mordechai for the bees; K. Varala, M.E. Hudson, and A. Venkat for bioinformatic assistance; and A. Bourke for comments on the manuscript.
Data acquisition
The microarray results from this study can be accessed at the ArrayExpress website (http://www.ebi.ac.uk/arrayexpress/).
Funding statement
This study was supported by 454 Life Sciences (Roche Diagnostics Corporation) via the Roche 1GB contest; the National Science Foundation (grant number DEB07-43154 to G.E.R.); the National Institutes of Health–University of Illinois Sensory Neuroscience Training Grant (grant number PHS2T32DC006612 to A. Feng); and the US–Israel Binational Agricultural Research and Development (BARD) fund (IS-4418-11 to G.B., G.E.R. and M.R.B.).
References
- 1.Wilson EO. 1971. The insect societies. Cambridge, MA: Harvard University Press. [Google Scholar]
- 2.Darwin CR. 1866. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life, 4th edn London, UK: J. Murray. [PMC free article] [PubMed] [Google Scholar]
- 3.Queller DC, Strassmann JE. 1988. Kin selection and social insects. Bioscience 48, 165–175. ( 10.2307/1313262) [DOI] [Google Scholar]
- 4.Evans HE, West-Eberhard MJ. 1970. The wasps. Ann Arbor, MI: University of Michigan Press. [Google Scholar]
- 5.West-Eberhard MJ. 1987. Flexible strategy and social evolution. In Animal societies: theories and facts (eds Ito Y, Brown JL, Kikkawa J.), pp. 35–51. Tokyo, Japan: Scientific Society Press. [Google Scholar]
- 6.West-Eberhard MJ. 1996. Wasp societies as microcosms for the study of development and evolution. In Natural history and evolution of paper wasps (eds Turillazzi S, West-Eberhard MJ.), pp. 290–317. New York, NY: Oxford University Press. [Google Scholar]
- 7.Linksvayer T, Wade M. 2005. The evolutionary origin and elaboration of sociality in the aculeate Hymenoptera: maternal effects, sib social effects, and heterochrony. Q. Rev. Biol. 80, 317–336. ( 10.1086/432266) [DOI] [PubMed] [Google Scholar]
- 8.Toth AL, Robinson GE. 2007. Evo-devo and the evolution of social behaviour. Trends Genet. 23, 334–341. ( 10.1016/j.tig.2007.05.001) [DOI] [PubMed] [Google Scholar]
- 9.Tautz D, Domazet-Loso T. 2011. The evolutionary origin of orphan genes. Nat. Rev. Genet. 12, 692–702. ( 10.1038/nrg3053) [DOI] [PubMed] [Google Scholar]
- 10.Amdam GV, Csondes A, Fondrk MK, Page RE. 2006. Complex social behaviour derived from maternal reproductive traits. Nature 439, 76–78. ( 10.1038/nature04340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira PG, Patalano S, Chauhan R, Richard Ffrench-Constant R, Gabaldón T, Guigó R, Sumner S. 2013. Transcriptome analyses of primitively eusocial wasps reveal novel insights into the evolution of sociality and the origin of alternative phenotypes. Genome Biol. 14, R20 ( 10.1186/gb-2013-14-2-r20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cameron SA, Mardulyn P. 2001. Multiple molecular data sets suggest independent origins of highly eusocial behavior in bees (Hymenoptera: Apinae). Syst. Biol. 50, 194–214. ( 10.1080/10635150151125851) [DOI] [PubMed] [Google Scholar]
- 13.Hines HM, Hunt JH, O'Connor TK, Gillespie JJ, Cameron SA. 2007. Multigene phylogeny reveals eusociality evolved twice in vespid wasps. Proc. Natl Acad. Sci. USA 104, 3295–3299. ( 10.1073/pnas.0610140104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brady SG, Sipes S, Pearson A, Danforth BN. 2006. Recent and simultaneous origins of eusociality in halictid bees. Proc. R. Soc. B 273, 1643–1649. ( 10.1098/rspb.2006.3496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes WOH, Oldroyd BP, Beekman M, Ratnieks FLW. 2008. Ancestral monogamy shows kin selection is key to the evolution of eusociality. Science 320, 1213–1216. ( 10.1126/science.1156108) [DOI] [PubMed] [Google Scholar]
- 16.Wilson EO, Hölldobler B. 2005. Eusociality: origin and consequences. Proc. Natl Acad. Sci. USA 102, 13 367–13 371. ( 10.1073/pnas.0505858102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodard SH, Fischman BJ, Venkat A, Hudson MA, Varala K, Cameron SA, Clark AG, Robinson GE. 2010. Genes involved in convergent evolution of eusociality in bees. Proc. Natl Acad. Sci. USA 108, 7472–7477. ( 10.1073/pnas.1103457108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith CR, Toth AL, Suarez AV, Robinson GE. 2008. Genetic and genomic analyses of the division of labour in insect societies. Nat. Rev. Genet. 9, 735–748. ( 10.1038/nrg2429) [DOI] [PubMed] [Google Scholar]
- 19.Toth AL, et al. 2007. Wasp gene expression supports an evolutionary link between maternal behaviour and eusociality. Science 318, 441–444. ( 10.1126/science.1146647) [DOI] [PubMed] [Google Scholar]
- 20.Toth AL, Varala K, Henshaw MT, Rodriguez-Zas SL, Hudson ME, Robinson GE. 2010. Brain transcriptomic analysis in paper wasps identifies genes associated with behaviour across social insect lineages. Proc. R. Soc. B 277, 2139–2148. ( 10.1098/rspb.2010.0090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodard SH, Bloch G, Band MR, Robinson GE. 2013. Social regulation of maternal traits in nest-founding bumble bee (Bombus terrestris) queens. J. Exp. Biol. 216, 3474–3482. ( 10.1242/jeb.087403) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Free JB, Butler CG. 1959. Bumblebees. London, UK: Collins. [Google Scholar]
- 23.Whitfield CW, Cziko AM, Robinson GE. 2003. Gene expression profiles in the brain predict behavior in individual honey bees. Science 302, 296–299. ( 10.1126/science.1086807) [DOI] [PubMed] [Google Scholar]
- 24.Alaux C, et al. 2009. Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc. Natl Acad. Sci. USA 106, 15 400–15 405. ( 10.1073/pnas.0907043106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geva S, Hartfelder K, Bloch G. 2005. Reproductive division of labor, dominance, and ecdysteroid levels in hemolymph and ovary of the bumble bee Bombus terrestris. J. Insect Physiol. 51, 811–823. ( 10.1016/j.jinsphys.2005.03.009) [DOI] [PubMed] [Google Scholar]
- 26.Yerushalmi S, Bodenhaimer S, Bloch G. 2006. Developmentally determined attenuation in circadian rhythms links chronobiology to social organization in bees. J. Exp. Biol. 209, 1044–1051. ( 10.1242/jeb.02125) [DOI] [PubMed] [Google Scholar]
- 27.Duchateau MJ, Velthuis HHW. 1989. Ovarian development and egg-laying in workers of Bombus terrestris. Entomol. Exp. Appl. 51, 199–213. ( 10.1111/j.1570-7458.1989.tb01231.x) [DOI] [Google Scholar]
- 28.Röseler PF, van Honk CGJ. 1990. Castes and reproduction in bumblebees. In Social insects: an evolutionary approach to castes and reproduction (ed. Engels W.), pp. 147–166. Berlin, Germany: Springer. [Google Scholar]
- 29.Schulz DJ, Robinson GE. 1999. Biogenic amines and division of labor in honey bee colonies: behaviorally related changes in the antennal lobes and age-related changes in the mushroom bodies. J. Comp. Physiol. A 184, 481–488. ( 10.1007/s003590050348) [DOI] [PubMed] [Google Scholar]
- 30.Sen Sarma M, Rodriguez-Zas S, Hong F, Zhong S, Robinson GE. 2009. Transcriptomic profiling of central nervous system regions in three species of honey bee during dance communication behavior. PLoS ONE 4, e6408 ( 10.1371/journal.pone.0006408) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyth GK. 2005. Limma: linear models for microarray data. In Bioinformatics and computational biology solutions using R and bioconductor (eds Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W.), pp. 397–420. New York, NY: Springer. [Google Scholar]
- 32.R Development Core Team. 2011. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 33.Smyth GK, Speed TP. 2003. Normalization of cDNA microarray data. Methods 31, 265–273. ( 10.1016/S1046-2023(03)00155-5) [DOI] [PubMed] [Google Scholar]
- 34.Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. App. Genet. Mol. Biol. 3 ( 10.2202/1544-6115.1027) [DOI] [PubMed] [Google Scholar]
- 35.Smyth GK, Michaud J, Scott H. 2005. The use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21, 2067–2075. ( 10.1093/bioinformatics/bti270) [DOI] [PubMed] [Google Scholar]
- 36.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing . J. R. Stat. Soc. B 57, 289–300. [Google Scholar]
- 37.HBGSC. 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443, 931–949. ( 10.1038/nature05260) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang DW, Sherman BT, Lempicki RA. 2008. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. ( 10.1038/nprot.2008.211) [DOI] [PubMed] [Google Scholar]
- 39.Warnes GR. 2010. gplots: various R programming tools for plotting data. R package version 2.8.0.
- 40.Toth AL, Bilof KBJ, Henshaw MT, Hunt JH, Robinson GE. 2009. Lipid stores, ovary development, and brain gene expression in Polistes metricus females. Insectes Soc. 56, 77–84. ( 10.1007/s00040-008-1041-2) [DOI] [Google Scholar]
- 41.Holland JG, Guidat FS, Bourke AFG. 2013. Queen control of a key life-history event in a eusocial insect. Biol. Lett. 9, 20130056 ( 10.1098/rsbl.2013.0056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reeve HK. 1991. Polistes. In The social biology of wasps (eds Ross KG, Matthews RG.), pp. 99–148. Ithaca, NY: Cornell University Press. [Google Scholar]
- 43.Burmeister SS, Jarvis ED, Fernald RD. 2005. Rapid behavioral and genomic responses to social opportunity. PLoS Biol. 3, e363 ( 10.1371/journal.pbio.0030363) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ament SA, Corona M, Pollock HS, Robinson GE. 2008. Insulin signaling is involved in the regulation of worker division of labor in honey bee colonies. Proc. Natl Acad. Sci. USA 105, 4226–4231. ( 10.1073/pnas.0800630105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grozinger CM, Sharabash NM, Whitfield CW, Robinson GE. 2003. Pheromone-mediated gene expression in the honey bee brain. Proc. Natl Acad. Sci. USA 100, 14 519–14 525. ( 10.1073/pnas.2335884100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shpigler H, Patch HM, Cohen M, Fan Y, Grozinger CM, Bloch G. 2010. The transcription factor Krüppel homolog 1 is linked to hormone mediated social organization in bees. BMC Evol. Biol. 10, 120 ( 10.1186/1471-2148-10-120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agrawal N, Padmanabhan N, Hasan G. 2009. Inositol 1,4,5-trisphosphate receptor function in Drosophila insulin producing cells. PLoS ONE 4, e6652 ( 10.1371/journal.pone.0006652) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agrawal N, Venkiteswaran G, Sadaf S, Padmanabhan N, Banerjee S, Hasan G. 2010. Inositol 1,4,5-trisphosphate receptor and dSTIM function in Drosophila insulin-producing neurons regulates systemic intracellular calcium homeostasis and flight. J. Neurosci. 30, 1301–1313. ( 10.1523/JNEUROSCI.3668-09.2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cardinal S, Danforth BN. 2011. The antiquity and evolutionary history of social behaviour in bees. PLoS ONE 6, e21086 ( 10.1371/journal.pone.0021086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fischman BJ, Woodard SH, Robinson GE. 2011. Molecular evolutionary analyses of insect societies. Proc. Natl Acad. Sci. USA 108, 10 847–10 854. ( 10.1073/pnas.1100301108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Foret S, Kucharski R, Pellegrini M, Feng S, Jacobsen SE, Robinson GE, Maleszka R. 2012. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proc. Natl Acad. Sci. USA 109, 4968–4973. ( 10.1073/pnas.1202392109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li-Byarlay H, et al. 2013. RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee. Proc. Natl Acad. Sci. USA 110, 12 750–12 755. ( 10.1073/pnas.1310735110) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.