Abstract
Traditional knowledge is influenced by ancestry, inter-cultural diffusion and interaction with the natural environment. It is problematic to assess the contributions of these influences independently because closely related ethnic groups may also be geographically close, exposed to similar environments and able to exchange knowledge readily. Medicinal plant use is one of the most important components of traditional knowledge, since plants provide healthcare for up to 80% of the world's population. Here, we assess the significance of ancestry, geographical proximity of cultures and the environment in determining medicinal plant use for 12 ethnic groups in Nepal. Incorporating phylogenetic information to account for plant evolutionary relatedness, we calculate pairwise distances that describe differences in the ethnic groups' medicinal floras and floristic environments. We also determine linguistic relatedness and geographical separation for all pairs of ethnic groups. We show that medicinal uses are most similar when cultures are found in similar floristic environments. The correlation between medicinal flora and floristic environment was positive and strongly significant, in contrast to the effects of shared ancestry and geographical proximity. These findings demonstrate the importance of adaptation to local environments, even at small spatial scale, in shaping traditional knowledge during human cultural evolution.
Keywords: ethnobotany, cultural evolution, phylogeny
1. Introduction
The ability to learn from others and to transmit knowledge and skills has shaped human history [1–3]. Transmission of traditional knowledge underpins both long-term conservation and rapid change in cultural traits, enabling humans to refine survival strategies and occupy diverse habitats [4,5]. Two modes of transmission of traditional knowledge have been described. Traditional knowledge is passed from generation to generation, and so from ancestral to descendant cultures, in what is termed ‘vertical transmission’. Selective diffusion or borrowing is referred to as ‘horizontal transmission’, and serves to modify traditional knowledge, although innovation in the absence of horizontal transmission may also modify an ethnic group's knowledge. The combination of modification and vertical and horizontal transmission establishes bodies of traditional knowledge unique to each culture, but still reflecting the traditional knowledge of their ancestors [4,6].
The study of the evolution of human culture is a matter of considerable current interest, particularly as phylogenetic methods borrowed from biology are finding wide application. Phylogenetic methods have shed light on the transmission of human culture, treating cultural traits as analogous to biological traits [6–8], and modes of inheritance of cultural traits are beginning to be ascertained for aspects of human culture. Those passed on vertically include some aspects of material culture [9,10] and family and kinship organization [4,11]; traits transmitted horizontally include cases of technical innovations [12], music [13] and other aspects of material culture [14].
Some behavioural and cultural traits show correlation with environmental factors, revealing the adaptation of traditional knowledge to the environment [15,16]. Two patterns of cultural trait distribution are suggestive of trait evolution in response to environment: traits differing between closely related cultures found in different environments, and traits similar between unrelated cultures sharing an environment may be traits which are adapted to the environment. Ecological correlates of behavioural and cultural traits reveal adaptation of this kind [15,16]. The environment may necessitate cultural innovation, but it also influences cultural traits by imposing functional constraints [17].
Medicine is an important element of traditional knowledge which includes indigenous healthcare traditions, beliefs and various practices. Well over half of the world's population depend on traditional medicine for healthcare, up to 80% in countries of the developing world [18]. Between 10 000 and 53 000 plant species are used in traditional medicine, and use of plants in medicine is a ubiquitous and important cultural trait [19,20]. Because not all plants are found everywhere, the floristic environment constrains plant use. The adaptation of traditional medicine when migrations expose cultures to new floristic environments may occur through horizontal transmission of plant use and homogenization of practices [21–23]. In this study, we focus on 12 moderately to closely related ethnic groups from Nepal. With approximately 75 ethnic groups and approximately 7000 plant species, Nepal has remarkable cultural [24,25] and floristic diversity (http://www.floraofnepal.org) [26,27]. One in seven plants is or has been used in some sort of medicinal preparation [24]. By performing comparisons of traditional plant use among closely related cultures, the aim of this study is to shed light on the evolutionary processes that have shaped this body of traditional knowledge.
Our study investigates how similarities among the medicinal floras of 12 Nepalese ethnic groups reflect affinities of the floristic environments to which these ethnic groups are exposed, their cultural affinities and their geographical proximities (figure 1). Studies of closely related groups are ‘potentially the most informative for testing cross-cultural hypotheses’ [7], since both vertical and horizontal transmission can occur when closely related cultures are found within a region. For the 12 Nepalese ethnic groups, we use linguistic affinities as a proxy for ancestry, and calculate geographical distances based on their distribution in the country to investigate the effect of geographical separation on the composition of medicinal floras. By performing comparisons of traditional plant use among closely related cultures, this study aims to shed light on the evolutionary processes that have shaped this body of traditional knowledge while explicitly evaluating the role of the environment in shaping the evolution of traditional medicine. In the context of burgeoning research aimed at elucidating the modes of inheritance of cultural traits, we put forward and test an approach we develop to disentangle the spatial, environmental and historical determinants of traditional knowledge.
Figure 1.
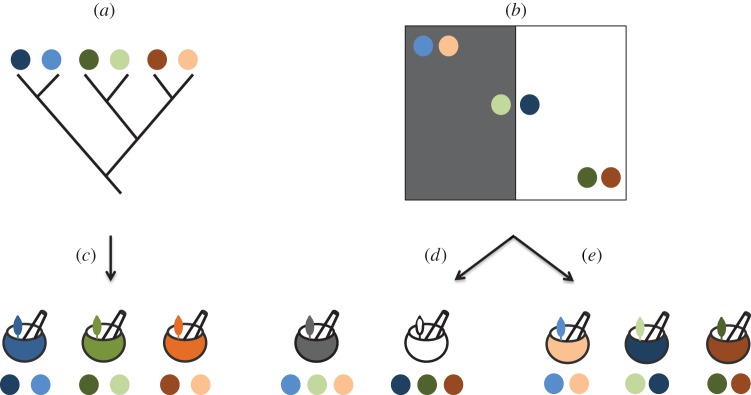
Cross-cultural similarities in traditional medicine can be determined by shared ancestry, geography or the environment. The relationships of six hypothetical cultures, shown in circles of different colours, are represented using a cultural phylogenetic tree (a), and (b) shows their distribution in a hypothetical region, where two types of environment (black and white) are present. Mortars and pestles represent traditional medicinal systems. If cultural ancestry determines similarities in traditional medicine, closely related cultures will end up with similar traditional medicinal systems (c). However, if those similarities are determined by the environment, those medicinal systems will reflect environmental similarities (d). Finally, if horizontal transmission shapes traditional medicinal systems, then cultures in close geographical proximity will share traditional medicinal systems (e).
2. Material and methods
(a). Cultural distances
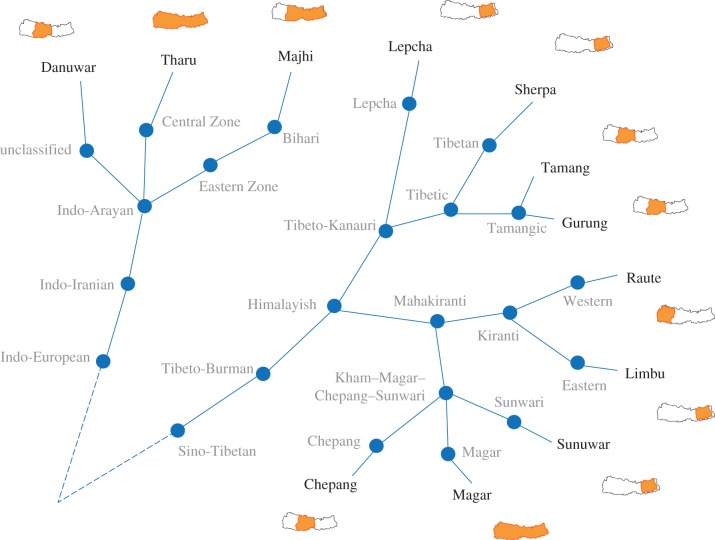
The 12 ethnic groups from Nepal under study were Chepang, Danuwar, Gurung, Lepcha, Limbu, Magar, Majhi, Raute, Sherpa, Sunuwar, Tamang and Tharu. These groups represent more than a quarter of the total population in the country and the two main language families in Nepal, namely Indo-European (Danuwar, Majhi and Tharu) and Sino-Tibetan (Chepang, Gurung, Lepcha, Limbu, Magar, Raute, Sherpa, Sunuwar and Tamang). Languages from these two groups are spoken by approximately 98% of the Nepalese population. To calculate cultural distances among these ethnic groups, we used linguistic data, demonstrated to be a good proxy for relationships of human groups [28,29]. We selected ethnic groups with language retention (strong one-to-one relationship between ethnic communities and language) [30] and therefore correspondence between language and cultural ancestry is maximized. A matrix of pairwise distances of the languages was created based on language grouping information extracted from the Ethnologue [31], a worldwide dataset and classification of languages. Pairwise cultural distance was calculated using the number of hierarchical levels of language groupings separating the ethnic groups' respective languages, summarized in figure 2. Cultural distances are shown in the electronic supplementary material, table S1.
Figure 2.
Classification of the 12 languages of ethnic groups under study and their distribution in the floristic areas of Nepal. Classification based on language data acquired from the Ethnologue [31]. Pairwise cultural distances among ethnic groups were calculated by counting the language subgroupings (shown in circles) that any given language pair does not share. For example, the distance between Tamang and Gurung is zero, as they are placed in same language group (Tamangic) and share all language subgroupings. The distance between Tamang and Lepcha is three, as there are three language subgroupings they do not share (Tamangic, Tibetic, Lepcha).
(b). Medicinal floras and floristic environments
Ethnomedicinal information was collated from nationwide compilations of ethnobotanical plants [24] (http://www.eson.org.np/database/index.php). All plant species used by each of the 12 ethnic groups were recorded and presence or absence of usage was scored at the genus level for each ethnic group. To assign floristic environments to ethnic groups, each ethnic group was located in one or more of the three major biogeographical regions of Nepal (western, central and eastern) depending on its distribution in the country [25], as shown in figure 2. The floristic environment of each group was all the species found across its range over these three biogeographical regions in Nepal. Data on the distribution of plant species in the biogeographical regions were collated from a checklist of the flora of Nepal [26].
(c). Phylogenetic analyses
To estimate phylogenetic distances between all pairs of medicinal floras and all pairs of floristic environments, we used a genus-level phylogeny of the flora of Nepal from a previous study, which included 1335 genera, representing more than 85% of the Nepalese flora [32]. The phylogeny includes one exemplar species per genus. Where possible, that species was from the local flora, but in cases where a DNA sequence or plant material was not available, species of the same genus from other localities were used. The tree is based on sequence data from the plastid DNA marker rbcL, which were analysed under the maximum likelihood (ML) criterion [33]. For more details on taxon sampling, molecular techniques and phylogenetic reconstruction, see [32].
Pairwise distances for floristic environments and medicinal floras were calculated using the ‘comdistnn’ command in Phylocom v. 4.1 [34] on the phylogenetic tree of the flora of Nepal. This command calculates the phylogenetic distance between two samples based on the nearest-neighbour phylogenetic distance. For each taxon in one sample, the algorithm finds the closest relative in the other sample and records the phylogenetic distance between them. The final value for the two samples is the average of these distances for all taxa in both samples. Our samples were either the floristic environment of a region or the medicinal flora of an ethnic group. Large values acquired from ‘comdistnn’ denote that the two samples do not include many taxa that are particularly closely related, whereas small values indicate close relationships between two samples. Genera included in medicinal floras, but not sampled in the phylogeny were excluded from these analyses. The distance matrices for floristic environments and medicinal floras are shown in the electronic supplementary material, tables S2 and S3, respectively.
(d). Geography
We calculated geographical distance between all pairs of cultures using the haversine formula [35] on the midpoint locations of each culture. The haversine formula calculates the great-circle distance between two points on the globe, i.e. the shortest distance between them on the surface of the globe. In our study, it is the great-circle distance between midpoint locations of cultures. The geographical distributions of cultures were obtained from the Ethnologue [31]. When there were multiple cultural variants (e.g. Gurung, Magar, Tamang and Tharu), we calculated the distance from each variant to the relevant culture and averaged these distances. The distance matrix is shown in the electronic supplementary material, table S4.
(e). Statistics and simulations
The four distance matrices (1, culture; 2, floristic environment; 3, medicinal flora and 4, geography) were used to perform five correlations: 1, floristic environment versus medicinal flora; 2, culture versus geography; 3, geography versus floristic environment; 4, culture versus medicinal flora; 5, geography versus floristic environment. Pearson product-moment correlation coefficient (r) and significance of correlations between these matrices were estimated with Mantel tests [36] in the ‘vegan’ package in R [37]. Table 1 summarizes the results of these correlations.
Table 1.
Pearson product-moment correlation coefficient (r) and significance (p) of correlations among distance matrices. Floristic environment refers to the distance matrix between the total floras found in each floristic region. Medicinal flora refers to the distance matrix between the medicinal floras used by each ethnic group. Culture refers to the distance matrix describing cultural relatedness, and based on linguistic affinities. Geography refers to the distance matrix describing geographical proximity of different cultures. n.s., non-significant.
| distance matrix 1 | distance matrix 2 | correlation coefficient (r) | significance (p value) |
|---|---|---|---|
| floristic environment | medicinal flora | 0.73 | < 0.001 |
| culture | geography | 0.13 | n.s. |
| geography | medicinal flora | 0.48 | n.s. |
| culture | medicinal flora | 0.21 | n.s. |
| geography | floristic environment | 0.58 | < 0.01 |
| culture | medicinal flora (simulated) | 0.95 | < 0.001 |
To test whether the absence of signal of vertical transmission of medicinal floras in our study can be attributed to methodological artefact, we generated an artificial dataset that represents an extremely conservative mode of vertical transmission. We generated a random ‘medicinal flora’ composed of a set of 66 genera (similar size to the medicinal floras in our dataset). This was used as the ancestral medicinal flora of all cultural groups. Starting from an ‘ancestral’ language node, we ‘evolved’ the medicinal flora in the following way: every time we came across a language bifurcation, we substituted one of the genera in the medicinal flora with its closest relative from the phylogenetic tree of the flora of Nepal. The resulting medicinal floras associated with the extant languages were between 3 and 17% different from one another (the range of distances in observed medicinal floras was 18.5–87%). Pairwise phylogenetic distances were calculated with the ‘comdistnn’ option in Phylocom v. 4.1, as described above, and the correlation of these distances with linguistic distances was assessed with a Mantel test.
3. Results and discussion
Five tests for correlations between four matrices of pairwise distances show unequivocally that the floristic environment to which a culture is exposed exerts the strongest influence on the medicinal flora adopted (table 1). The statistically significant correlation between floristic environment and medicinal flora (r = 0.73, p < 0.001; table 1) provides strong evidence for this link. The distance matrices used in this test, one to describe relatedness of floristic environments and the other relatedness of medicinal floras, were both calculated using a genus-level phylogenetic tree of the flora of Nepal encompassing 85% of the total flora. For the floristic environment, relatedness was calculated using the phylogeny and published Nepalese plant distribution data. Medicinal floras of the 12 ethnic groups were superimposed onto the phylogenetic tree to calculate the relatedness of the medicinal floras. Correlation between these distances was assessed using a Mantel test [36], an approach used in similar studies [6,38].
The correlation between floristic environment and medicinal flora could be attributed to spatial autocorrelation, Galton's problem of closely related cultures being spatially proximate, a problem which has long bedevilled comparative cultural studies [39]. To evaluate whether Galton's problem is confounding our results, our second Mantel correlation test is crucial: using language as a proxy for cultural relatedness [(8, 16), we show there is no significant effect of geographical structure of ethnic groups in Nepal (r = 0.13, p > 0.05; table 1). To perform this test, cultural relatedness among the 12 ethnic groups was calculated based on language affinities (figure 2), and the geographical proximities of pairs of cultures were calculated between the midpoints of their ranges. The lack of geographical clustering of related cultural groups shows that in Nepal we do not conflate cultural relatedness and geographical proximity, overcoming spatial autocorrelation when we seek to assess the determinants of a culture's selection of medicinal plants. The dispersal of related cultures, alongside Nepal's cultural diversity and rich medicinal flora, makes the Nepalese case an exceptionally powerful one for teasing apart the factors influencing the adoption of a medicinal flora.
Further Mantel tests also contribute to the robust interpretation of the correlation between floristic environment and medicinal flora. Background signal of shared ancestry could contribute to our finding of environmental convergence of medicinal floras. However, we found culture and medicinal flora not to be strongly correlated (r = 0.21, p > 0.05; table 1), revealing that vertical transmission over the time scale at which ethnic groups diverge is not the main determinant of traditional medicinal knowledge. Even when taking into account the possibility of using close relatives to substitute for unavailable plants in different environments, presumed ancestral use—as would be revealed by related cultures selecting the most similar sets of plants—is not the main determinant of plant use by a culture.
Environmental convergence of medicinal floras could also be because of horizontal exchange between cultures. Nevertheless, we found that pairwise distances describing the geographical proximities of cultures and the relatedness of medicinal floras are not significantly correlated (r = 0.48, p > 0.05; table 1), indicating that horizontal transmission, or borrowing of traditional knowledge, which would be most probable between cultures with overlapping distributions, is in fact not important here.
Overall, our findings indicate that through their history, the 12 Nepalese ethnic groups under investigation have adopted a flexible approach and incorporated new, unrelated plants into their medicinal floras. Although signal of both vertical and horizontal transmission was found in the medicinal floras of the 12 ethnic groups under investigation, this was very weak and not significant. Instead, we found strong human responses to similar floristic environments. Our finding that related cultures are not geographically structured also indicates that closely related cultures can occupy different floristic environments, therefore driving innovation in medicinal plant use.
The spatial level used to delineate floristic regions was the three-zone longitudinal division of Nepal (western, central, eastern biogeographical regions), as finer botanical distribution data across the whole of Nepal are not yet available. This delimitation of floristic regions could weaken our study. Nevertheless, there was a strong correlation (r = 0.58, p < 0.01; table 1) between geographical proximity of cultures and their floristic environments. This shows that our delimitation of floristic regions is meaningful: our data recover cultures in close geographical proximity being exposed to similar plants, even though cultures have distributions that do not map to our crude floristic regions. Of course, there is considerable latitudinal variation for both the plants and the ethnic groups within those regions, but this issue cannot be tackled until finer distribution data are available.
Our study depends on the recognition of ethnic groups, and the accurate estimation of the relationships between them. Here, we use language affinities to infer relationships between ethnic groups. Language affinities have often been used as a proxy for intra-cultural relationships, but increasingly quantitative linguistic models are used [40–42]. Detailed linguistic studies do not exist for the Nepalese groups in our study, so our estimates of relatedness might be unreliable. Studies comparing traditional language classifications, as used here, and Bayesian phylogenetic estimates, reveal striking congruence between the two classifications [42,43]. Whether this would be true in our case is not known, and comparative linguistic studies are needed to consolidate our understanding of historical intra-cultural relationships in Nepal. In terms of the recognition of ethnic groups, anthropologists are revealing ethnic group identities in Nepal to be very fluid, challenging the idea of fixed ethnic groups [44]. While a particular village may have historical depth in a place, the knowledge carried by an individual informant may well derive from a complex ancestry and social heritage [44,45]. If the ethnic groups we recognize have in fact experienced cultural exchange, then we would expect a signal from horizontal transmission. Our study does not reveal a strong relationship between the geographical proximity of cultures and their medicinal flora, so if this horizontal exchange is occurring it does not overwhelm the impact of the floristic environment. Cultural exchange may be occurring between cultures that are not proximate, due to long-distance migration of individuals to new cultures. Cultural exchange of this kind, where individuals migrate to either geographically distant cultures or cultures that are not linguistically related, would not be revealed by our approach. Further detailed studies of individuals' ethnobotanical knowledge and heritage would be needed. This shortcoming also highlights limitations with the sourcing of ethnobotanical knowledge for this study. Our ethnobotanical data were sourced from publically available resources, which do not take into account the number of informants or the personal heritage of informants. Thus, the ethnobotanical data used may introduce biases into our study. Meta-analyses of the kind presented here highlight the importance of further, detailed ethnobotanical work on the ground.
Finally, a limitation lies in the interpretation of results from Mantel tests, which have been shown to be prone to type-I and type-II errors [46,47]. Moreover, if one variable is poorly measured, its correlation with other variables might be reduced. We have already highlighted that our use of language affinities as a proxy for intra-cultural relationships may be a weakness. If our estimates of ancestry are misleading, this could explain the absence of significant correlation between ancestry and other traits. Despite the pitfalls of Mantel tests, they are still used very commonly in comparative cultural studies, as other similar methods, such as independent contrasts, are oversensitive to horizontal transmission [47].
In spite of the caveats discussed above, we reveal an experimental, flexible approach in the macro-evolution of traditional knowledge, and the strong influence of the natural environment. Similarly strong environmental influences have been observed for other cultural characters [15], reflecting geographical barriers to cultural transmission [38] and adaptation of human populations to different environments [11]. Our study highlights independent adoption of similar medicinal plants when human are exposed to similar floristic environments. Other studies have attributed independent adoption to independent discovery of plant bioactivity [32], but between much more distantly related cultures, and therefore over a much longer time scale than is inferred here. The experimental and flexible approach revealed here is especially intriguing given the importance of traditional medicine for healthcare, and previous research showing that functional cultural characters are conserved [17,48]. However, the role of traditional medicinal knowledge is to contribute to the fitness of human populations in different environments through healthcare [49]. As availability of resources varies among environments, medicinal floras need to be adapted to local environments in order to respond to healthcare needs [21,23,50].
Our study is unique among studies of bio-cultural evolution in incorporating both a cultural and a biological phylogenetic tree to investigate the transmission of a key cultural trait. We use the phylogeny of Nepalese flowering plants to calculate similarities between the floristic environments that ethnic groups are exposed to—a proxy for environmental similarity. We then calculate similarities between medicinal floras using the same phylogeny. Phylogenetic measures are particularly appropriate for our study, since closely related ethnic groups exposed to novel floristic environments often use close relatives of their ancestral medicinal plant species [21,51]. Selecting relatives in this way is a form of vertical transmission. The strength of this study lies in the use of biological phylogenies to estimate the relatedness of medicinal floras. Conventional taxonomic approaches based on the number of species in common will not capture underlying similarities between medicinal floras. Some studies have shown that during human migrations, closely related plants are selected to substitute for plants that are not available in the new floristic environment [50–52]. If, as has been suggested, phylogeny underlies peoples' selection of medicinal plants [32], taxonomic approaches could overemphasize the differences between medicinal floras, while phylogenetic measures would consider substitution by a closely related plant to be attributed to vertical transmission of knowledge (see electronic supplementary material, figure S1). To test whether the use of a plant phylogenetic tree is capable of identifying this type of signal, we created a simulated dataset where differences in medicinal floras are the result of vertical transmission, but which accommodates the selection of closest relatives by sister cultures exposed to different plants (see Material and methods). Our simulated data revealed a correlation between phylogenetic distance of medicinal floras and cultural distance which was highly significant (r = 0.82, p < 0.001; table 1), showing that the approach used here is capable of recovering signal of vertical transmission of medicinal floras, and that absence of this signal in our real dataset is not a methodological artefact.
4. Conclusion
Intriguing patterns are emerging from the increasing numbers of studies using phylogenetic approaches to characterize cultural evolution [4,9–14]. While many studies have identified cultural traits which are predominantly vertically or horizontally inherited, we develop a method able to elucidate the effects of the environment while acknowledging both of these evolutionary processes. Using this method, we demonstrate that ethnic groups resemble each other in medicinal plant use simply because they exist in similar floristic environments, regardless of whether they are geographically proximate or share common ancestors. Future work could focus on evolutionary change in other aspects of traditional knowledge which might be influenced by environment [53], providing insights into the interplay between the environment and traditional knowledge. Further work could also investigate the rates of this evolutionary change [54–56] and the coevolution between traditional knowledge and different aspects of culture [7,57] in different environmental settings, shedding more light into the tempo and mode of the of the interaction between culture and the environment.
Supplementary Material
Acknowledgement
We thank the staff at the herbarium of the Royal Botanic Garden, Edinburgh (E), the University of Reading Statistical Advisory Service, Lynsey Bunnefeld, Yael Kisel and Camile Moray.
Data accessibility
All DNA sequences associated with the phylogenetic tree used in this study are given in http://dx.doi.org/10.1073/pnas.1202242109.
Funding statement
This study was financially supported by a John Spedan Lewis Fellowship to support the doctoral studies of C.H.S.-L. V.S. thanks the European Research Council and the Royal Society for financial support.
References
- 1.Tomasello M. 1999. The human adaptation for culture. Annu. Rev. Anthropol. 28, 509–529. ( 10.1146/annurev.anthro.28.1.509) [DOI] [Google Scholar]
- 2.Pagel M. 2012. Evolution: adapted to culture. Nature 482, 297–299. ( 10.1038/482297a) [DOI] [PubMed] [Google Scholar]
- 3.Pagel M. 2012. Wired for culture, pp. 406–415. New York, NY: Norton. [Google Scholar]
- 4.Guglielmino CR, Viganotti C, Hewlett B, Cavalli-Sforza LL. 1995. Cultural variation in Africa: role of mechanisms of transmission and adaptation. Proc. Natl Acad. Sci. USA 92, 7585–7589. ( 10.1073/pnas.92.16.7585) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd R, Richerson PJ, Henrich J. 2011. The cultural niche: why social learning is essential for human adaptation. Proc. Natl Acad. Sci. USA 108(Suppl. 2), 10 918–10 925. ( 10.1073/pnas.1100290108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagel M, Mace R. 2004. The cultural wealth of nations. Nature 428, 275–278. ( 10.1038/428275a) [DOI] [PubMed] [Google Scholar]
- 7.Mace R, Pagel M. 1994. The comparative method in anthropology. Curr. Anthropol. 35, 549–564. ( 10.1086/204317) [DOI] [Google Scholar]
- 8.Mace R, Holden CJ. 2005. A phylogenetic approach to cultural evolution. Trends Ecol. Evol. 20, 116–121. ( 10.1016/j.tree.2004.12.002) [DOI] [PubMed] [Google Scholar]
- 9.Tehrani J, Collard M. 2002. Investigating cultural evolution through biological phylogenetic analyses of Turkmen textiles. J. Anthropol. Archaeol. 21, 443–463. ( 10.1016/S0278-4165(02)00002-8) [DOI] [Google Scholar]
- 10.Jordan P, O'Neill S. 2010. Untangling cultural inheritance: language diversity and long-house architecture on the Pacific northwest coast. Phil. Trans. R. Soc. B 365, 3875–3888. ( 10.1098/rstb.2010.0092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulder-Borgerhoff M, George-Cramer M, Eshleman J, Ortolani A. 2001. A study of East African kinship and marriage using a phylogenetically based comparative method. Am. Anthropol. 103, 1059–1082. ( 10.1525/aa.2001.103.4.1059) [DOI] [Google Scholar]
- 12.Rogers EM. 1995. Diffusion of innovations. New York, NY: Free Press. [Google Scholar]
- 13.Rzeszutek T, Savage PE, Brown S. 2012. The structure of cross-cultural musical diversity. Proc. R. Soc. B 279, 1606–1612. ( 10.1098/rspb.2011.1750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cochrane EE, Lipo CP. 2010. Phylogenetic analyses of Lapita decoration do not support branching evolution or regional population structure during colonization of Remote Oceania. Phil. Trans. R. Soc. B 365, 3889–3902. ( 10.1098/rstb.2010.0091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nettle D. 2009. Ecological influences on human behavioural diversity: a review of recent findings. Trends Ecol. Evol. 24, 618–624. ( 10.1016/j.tree.2009.05.013) [DOI] [PubMed] [Google Scholar]
- 16.Mace R, Jordan FM. 2011. Macro-evolutionary studies of cultural diversity: a review of empirical studies of cultural transmission and cultural adaptation. Phil. Trans. R. Soc. B 366, 402–411. ( 10.1098/rstb.2010.0238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rogers DS, Ehrlich PR. 2008. Natural selection and cultural rates of change. Proc. Natl Acad. Sci. USA 105, 3416–3420. ( 10.1073/pnas.0711802105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farnsworth NR, Akerele O, Bingel AS, Soejarto DD, Guo Z. 1985. Medicinal plants in therapy. Bull. World Health Organ. 63, 965–981. [PMC free article] [PubMed] [Google Scholar]
- 19.McChesney JD, Venkataraman SK, Henri JT. 2007. Plant natural products: back to the future or into extinction? Phytochemistry 68, 2015–2022. ( 10.1016/j.phytochem.2007.04.032) [DOI] [PubMed] [Google Scholar]
- 20.Schippmann U, Leaman DJ, Cunningham AB. 2002. Impact of cultivation and gathering of medicinal plants on biodiversity: global trends and issues. In Biodiversity and the ecosystem approach in agriculture, forestry and fisheries satellite event on the occasion of the Ninth Regular Session of the Commission on Genetic Resources for Food and Agriculture, Rome, 12–13 October, Inter-Departmental Working Group on Biological Diversity for Food and Agriculture. Rome: Food and Agriculture Organization. [Google Scholar]
- 21.Inta A, Shengji P, Balslev H, Wangpakapattanawong P, Trisonthi C. 2008. A comparative study on medicinal plants used in Akha's traditional medicine in China and Thailand, cultural coherence or ecological divergence? J. Ethnopharmacol. 116, 508–517. ( 10.1016/j.jep.2007.12.015) [DOI] [PubMed] [Google Scholar]
- 22.Mustafa B, Hajdari A, Krasniqi F, Hoxha E, Ademi H, Quave C, Pieroni A. 2012. Medical ethnobotany of the Albanian Alps in Kosovo. J. Ethnobiol. Ethnomed. 8, 6 ( 10.1186/1746-4269-8-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medeiros PMD, Soldati GT, Alencar NL, Vandebroek I, Pieroni A, Hanazaki N, de Albuquerque UP. 2012. The use of medicinal plants by migrant people: adaptation, maintenance, and replacement. Evid. Based Complement. Altern. Med. 2012, 11 ( 10.1155/2012/807452) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manandhar NP. 2002. Plants and people of Nepal. Portland, OR: Timber Press. [Google Scholar]
- 25.Shrestha DB, Singh CB. 1992. Ethnic groups of Nepal and their ways of living. Kathmandu, Nepal: Mandala Book Point. [Google Scholar]
- 26.Press JR, Shrestha KK, Sutton DA. 2000. Annotated checklist of the flowering plants of Nepal. London, UK: The Natural History Museum. [Google Scholar]
- 27.Watson MF, Akiyama S, Ikeda H, Pendry CA, Rajbhandari KR, Shrestha KK. 2011. Flora of Nepal, vol. 3 Edinburgh, UK: Royal Botanic Garden Edinburgh. [Google Scholar]
- 28.Cavalli-Sforza LL, Piazza A, Menozzi P, Mountain J. 1988. Reconstruction of human evolution: bringing together genetic, archaeological, and linguistic data. Proc. Natl Acad. Sci. USA 85, 6002–6006. ( 10.1073/pnas.85.16.6002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poloni ES, Semino O, Passarino G, Santachiara-Benerecetti AS, Dupanloup I, Langaney A, Excoffier L. 1997. Human genetic affinities for Y-chromosome P49a,f/TaqI haplotypes show strong correspondence with linguistics. Am. J. Hum. Genet. 61, 1015–1035. ( 10.1086/301602) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yadava YP. 2007. Linguistic diversity in Nepal; perspectives on language policy. In Proceedings from the Conference of Constitutionalism and Diversity in Nepal Kathmandu, Nepal: Centre for Nepal and Asian Studies, Tribhuvan University. [Google Scholar]
- 31.Lewis MP. 2009. Ethnologue: languages of the world, 16th edn Dallas, TX: SIL International. [Google Scholar]
- 32.Saslis-Lagoudakis CH, Savolainen V, Williamson EM, Forest F, Wagstaff SJ, Baral SR, Watson MF, Pendry CA, Hawkins JA. 2012. Phylogenies reveal predictive power of traditional medicine in bioprospecting. Proc. Natl Acad. Sci. USA 109, 15 835–15 840. ( 10.1073/pnas.1202242109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. ( 10.1080/10635150802429642) [DOI] [PubMed] [Google Scholar]
- 34.Webb CO, Ackerly DD, Kembel SW. 2008. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24, 2098–2100. ( 10.1093/bioinformatics/btn358) [DOI] [PubMed] [Google Scholar]
- 35.Sinnott RW. 1984. Virtues of the haversine. Sky Telesc. 68, 159. [Google Scholar]
- 36.Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220. [PubMed] [Google Scholar]
- 37.Oksanen J, Kindt R, Legendre P, O'Hara B, Simpson GL, Solymos P, Stevens HH, Wagner H. 2009. Vegan: community ecology package. R package version 1.15–4. See http://cran.r-project.org, http://vegan.r-forge.r-project.org/.
- 38.Ross RM, Greenhill SJ, Atkinson QD. 2013. Population structure and cultural geography of a folktale in Europe. Proc. R. Soc. B 280, 20123065 ( 10.1098/rspb.2012.3065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dow MM, Burton ML, White DR, Reitz KP. 1984. Galton's Problem as network autocorrelation. Am. Ethnol. 11, 754–770. ( 10.1525/ae.1984.11.4.02a00080) [DOI] [Google Scholar]
- 40.Currie TE, Greenhill SJ, Gray RD, Hasegawa T, Mace R. 2010. Rise and fall of political complexity in island South-East Asia and the Pacific. Nature 467, 801–804. ( 10.1038/nature09461) [DOI] [PubMed] [Google Scholar]
- 41.Gray RD, Bryant D, Greenhill SJ. 2010. On the shape and fabric of human history. Phil. Trans. R. Soc. B 365, 3923–3933. ( 10.1098/rstb.2010.0162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gray RD, Drummond AJ, Greenhill SJ. 2009. Language phylogenies reveal expansion pulses and pauses in Pacific settlement. Science 323, 479–483. ( 10.1126/science.1166858) [DOI] [PubMed] [Google Scholar]
- 43.Greenhill SJ, Drummond AJ, Gray RD. 2010. How accurate and robust are the phylogenetic estimates of Austronesian language relationships? PLoS ONE 5, e9573 ( 10.1371/journal.pone.0009573) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaenszle M, Turin M, Tuladhar-Douglas W. In press. Peoples of Nepal. In Nepal. An introduction to the natural history, ecology and the human environment in the Himalayas (eds Miehe G, Pendry CA, Chaudhary RP.). Edinburgh, UK: Royal Botanic Garden Edinburgh. [Google Scholar]
- 45.Caplan L. 1973. Inter-caste marriages in a Nepalese town. In Contributions to the anthropology of Nepal (ed. von Führer-Haimendorf C.), pp. 40–61. Surrey: Aris and Philips. [Google Scholar]
- 46.Harmon LJ, Glor RE. 2010. Poor statistical perfomance of the Mantel test in phylogenetic comparative analyses. Evolution 64, 2173–2178. ( 10.1111/j.1558-5646.2010.00973.x) [DOI] [PubMed] [Google Scholar]
- 47.Nunn CL, Mulder MB, Langley S. 2006. Comparative methods for studying cultural trait evolution: a simulation study. Cross-Cultur. Res. 40, 177–209. ( 10.1177/1069397105283401) [DOI] [Google Scholar]
- 48.Rogers DS, Feldman MW, Ehrlich PR. 2009. Inferring population histories using cultural data. Proc. R. Soc. B 276, 3835–3843. ( 10.1098/rspb.2009.1088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDade TW, Reyes-Garcia V, Blackinton P, Tanner S, Huanca T, Leonard WR. 2007. Ethnobotanical knowledge is associated with indices of child health in the Bolivian Amazon. Proc. Natl Acad. Sci. USA 104, 6134–6139. ( 10.1073/pnas.0609123104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voeks R. 1990. Sacred leaves of Brazilian Candomblé. Geogr. Rev. 80, 118–131. ( 10.2307/215476) [DOI] [Google Scholar]
- 51.Leporatti ML, Ivancheva S. 2003. Preliminary comparative analysis of medicinal plants used in the traditional medicine of Bulgaria and Italy. J. Ethnopharmacol. 87, 123–142. ( 10.1016/S0378-8741(03)00047-3) [DOI] [PubMed] [Google Scholar]
- 52.Akerreta S, Cavero R, Lopez V, Calvo M. 2007. Analyzing factors that influence the folk use and phytonomy of 18 medicinal plants in Navarra. J. Ethnobiol. Ethnomed. 3, 16 ( 10.1186/1746-4269-3-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beheim BA, Bell AV. 2011. Inheritance, ecology and the evolution of the canoes of east Oceania. Proc. R. Soc. B 278, 3089–3095. ( 10.1098/rspb.2011.0060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Atkinson QD, Meade A, Venditti C, Greenhill SJ, Pagel M. 2008. Languages evolve in punctuational bursts. Science 319, 588 ( 10.1126/science.1149683) [DOI] [PubMed] [Google Scholar]
- 55.Greenhill SJ, Atkinson QD, Meade A, Gray RD. 2010. The shape and tempo of language evolution. Proc. R. Soc. B 277, 2443–2450. ( 10.1098/rspb.2010.0051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reyes-García V, Guèze M, Luz AC, Paneque-Gálvez J, Macía MJ, Orta-Martínez M, Pino J, Rubio-Campillo X. 2013. Evidence of traditional knowledge loss among a contemporary indigenous society. Evol. Hum. Behav. 34, 249–257. ( 10.1016/j.evolhumbehav.2013.03.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holden CJ, Mace R. 2003. Spread of cattle led to the loss of matrilineal descent in Africa: a coevolutionary analysis. Proc. R. Soc. Lond. B 270, 2425–2433. ( 10.1098/rspb.2003.2535) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All DNA sequences associated with the phylogenetic tree used in this study are given in http://dx.doi.org/10.1073/pnas.1202242109.