Abstract
Currently, large-scale transmissions of infectious diseases are becoming more closely associated with accelerated globalization and climate change, but quantitative analyses are still rare. By using an extensive dataset consisting of date and location of cases for the third plague pandemic from 1772 to 1964 in China and a novel method (nearest neighbour approach) which deals with both short- and long-distance transmissions, we found the presence of major roads, rivers and coastline accelerated the spread of plague and shaped the transmission patterns. We found that plague spread velocity was positively associated with wet conditions (measured by an index of drought and flood events) in China, probably due to flood-driven transmission by people or rodents. Our study provides new insights on transmission patterns and possible mechanisms behind variability in transmission speed, with implications for prevention and control measures. The methodology may also be applicable to studies of disease dynamics or species movement in other systems.
Keywords: third plague pandemic, spread route, spread velocity, climate, transportation, movement ecology
1. Introduction
Frequent population movement and goods exchange, caused by accelerated globalization and climate change during the past decades, have significantly facilitated spatial transmission of infectious diseases worldwide [1–7]. However, the effects of environmental and social factors on spatial transmission patterns and velocity of diseases have rarely been tested. In particular, the role of human- or wildlife-driven, long-distance transmission of diseases is poorly understood due to limitations inherent in available research approaches, which constrain our capacity to prevent and control infectious diseases globally.
Transmission of pandemic diseases is often classified into contact and relocated diffusions which are caused, respectively, by short- and long-distance transmissions [8–12]. Contact diffusion refers to the short-distance spread of a disease outward from a central focus continuously in space. The disease often spreads in circles or along lines away from the central focus. Relocated diffusion refers to a long-distance discontinuous jump of an infection from one region to another, which then produces contact diffusion in a new site. Trend-surface analysis (TSA) has been used to study spatial contact diffusion of many diseases [11,13–15]. However, TSA does not work well for diseases with many long-distance transmissions, which have become increasingly common with the rapid advance of human transportation. This creates an urgent need to develop novel approaches to better understand both contact and relocated diffusions of global pandemics.
Plague has caused three pandemics, which killed about 200 million people [16], and have greatly shaped human societies [17–21]. Currently, plague outbreaks occur frequently in many countries in the Americas, Asia and Africa, and plague remains an important threat to human health as a re-emerging infectious disease [17,20,22–24]. Though plague dynamics have been well studied in the time domain [25–32], the spatial transmission of plague and its underlying influencing factors are rarely studied (but see [14,15,33]). There is a need for studies on the large-scale transmission patterns of plague, and the roles of human transportation, climate and landscape factors in shaping these patterns.
The third plague pandemic started in 1772 in Yunnan province in China (figure 1a; electronic supplementary material, figure S1) and was spread worldwide from Hong Kong in 1894 [34,35]. It spread to 541 counties in 21 provinces in China, infecting 2.5 million people and causing 2.2 million deaths [17,36]. The occurrences of human plague in both time and space were well recorded in China, which provides a good opportunity for studying the transmission pattern and the influences of both social and environmental factors. The aim of this study was to investigate the climate and transportation routes on spread velocity and patterns of human plague from 1772 to 1964 in China by using a novel nearest neighbour approach (NNA).
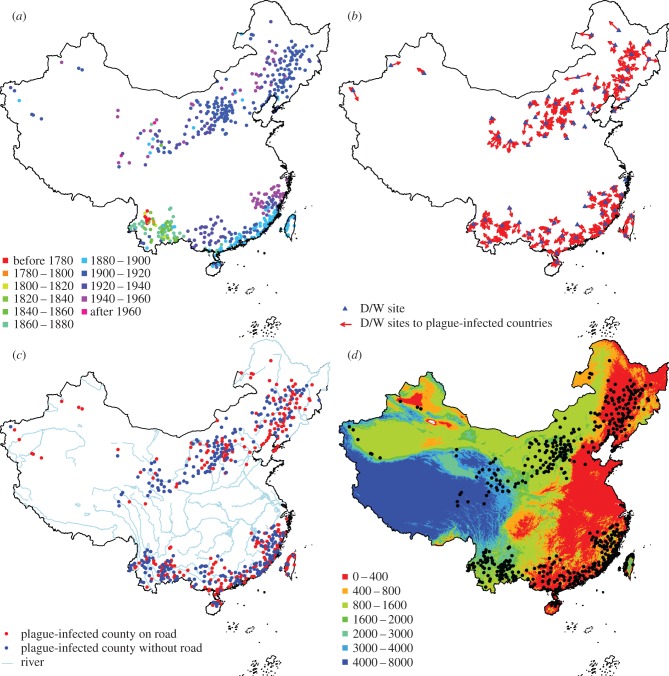
Figure 1.
(a) Plague invasion process in China during the third plague pandemic from 1772 to 1964. Points show counties where human plague was observed, with the colour showing the year when the first case was reported within the county boundary. (b) Location of hydrological stations with D/W data (blue triangle). Arrows indicate the hydrological station used for analysis of how climate affects plague spread for counties where plague manifested. (c) Red points indicate where the plague-infected county had a road going through, and blue points shows where the infected county had no major road going through. Blue lines indicate the largest rivers in China. (d) Elevation map (unit: metre) and political central location of counties with plague outbreaks. Only counties with confirmed plague cases are shown.
2. Results
(a). Analysis of plague spread paths and patterns
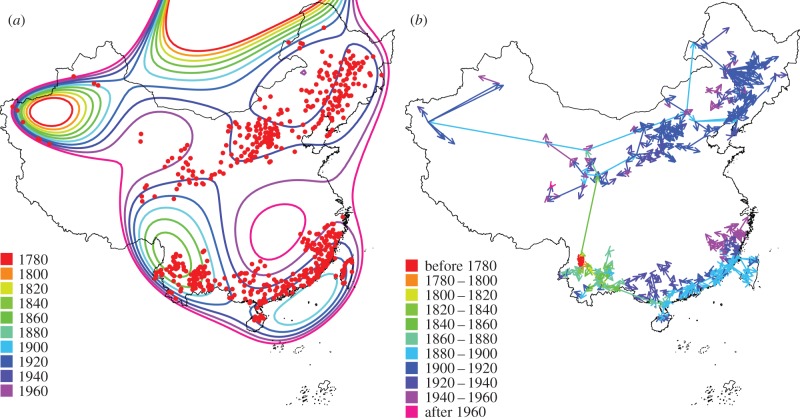
NNA revealed clearly distinguishable spread paths and showed that there were three transmission stages (figure 2b, also see figure 1a and the electronic supplementary material, figure S1). Plague was confined to a small area (red arrows) in Yunnan prior to 1800. The first transmission stage occurred between 1800 and 1880 (yellow and green arrows), during which plague spread to larger parts of Yunnan of South China, in a manner resembling short-distance transmission (contact diffusion). During this transmission stage, there were also two long-distance transmission events (perhaps early indicators of the start of the second transmission stage), which resulted in the arrival of plague in Qinghai of North China in 1854 and the arrival of plague in Guangdong of southeast China in 1867, both examples of relocated diffusion. The second transmission stage occurred between 1880 and 1900 (light-blue arrows), during which period plague spread to several sites through predominantly long-distance transmission within North China (relocated diffusion). At the same time, plague spread in southeast China along the coastline, in a mixture of both long-distance (along the coastline) and short-distance (toward inlands) transmissions. In the third stage from 1900 to 1960 (deep blue and purple arrows), plague expanded to the neighbouring regions through short-distance contact diffusion from the bases of plague sites of the second stage in both North and South China. TSA revealed one typical travelling wave (spreading outward from a central focus continuously in space) representing contact diffusions in Yunnan and another wave along the coastline of South China, but no clear travelling waves were detected in North China, perhaps due to the many long-distance transmissions (figure 2b).
Figure 2.
Two methods were used to estimate plague spread speed during the third pandemic in China. (a) TSA. Contours were drawn based on the trend surface, which presented the predicted plague invasive year. The slope of the trend surface represents the reciprocal plague spread speed. Red points indicate the location of counties with cases of plague infection. (b) NNA. Arrows were drawn to the plague-invaded counties from the nearest neighbour counties where plague appeared earlier. Plague spread speed was the length of arrows divided by the difference in the first plague-invaded year between the counties. Only counties with confirmed plague cases are shown.
(b). Analysis of plague spread velocity
We estimated plague spread velocity by using both TSA and NNA (figure 2; electronic supplementary material, table S2 and figures S2–S11). According to TSA, the median rate of plague spread was 40.1 km yr−1 in the whole of China, 112.5 km yr−1 in North China and 19.3 km yr−1 in South China (electronic supplementary material, table S2). According to NNA, the median velocity was 11.8 km yr−1 in the whole of China, 15.5 km yr−1 in North China and 9.4 km yr−1 in South China (electronic supplementary material, table S2). Notable is that plague spreads faster in North China than in South China according to both TSA and NNA, and that in general the velocity estimated by NNA is lower than that by TSA (electronic supplementary material, figure S8).
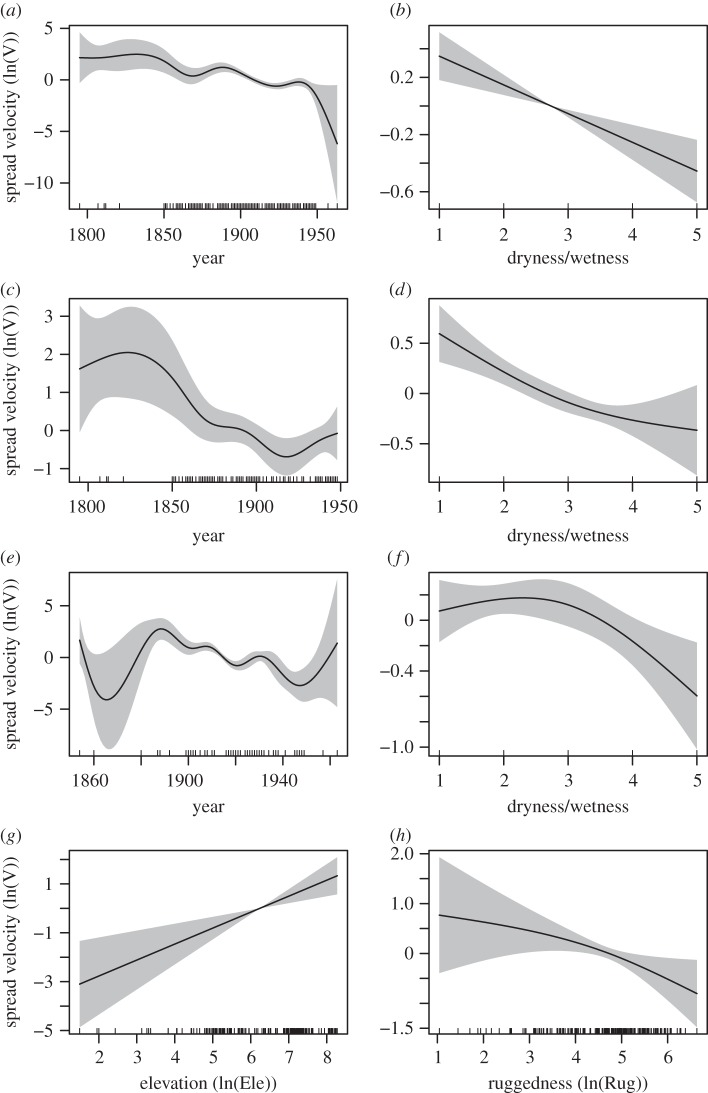
During the main epidemic periods (i.e. the periods with most case reports), plague spread velocity decreased through time for the whole China model (figure 3a, from 1850 to 1950), South China model (figure 3c, from 1850 to 1950) and North China model (figure 3e, from 1900 to 1940).
Figure 3.
Partial effects on plague spread velocities calculated by NNA methods. Partial effects are estimated from the GAM by correcting for effects of other variables. (a) Temporal trend in the whole of China. (b) Dryness/wetness effects in the whole of China. (c) Temporal trend in South China. (d) Dryness/wetness effects in South China. (e) Temporal trend in North China. (f) Dryness/wetness effects in North China. (g) Elevation effect in North China. (h) Ruggedness effect in North China. In addition, effects of location, plague prevalence, roads, rivers and coastline were accounted for (see text and electronic supplementary material).
Of the effects of transportation routes, generalized additive model (GAM) analysis suggested that roads, coastline and rivers promoted plague spread in China. Road presence showed a positive association with spread velocity in the model for the whole of China: (estimated increase in ln(velocity (km yr−1) ± s.e. = 0.38 ± 0.08, t82.56 = 4.7, p < 0.01)) in the North China model (0.22 ± 0.10, t67.36 = 2.3, p < 0.05) and in the South China model (0.63 ± 0.14, t31.95 = 4.49, p < 0.01). River presence showed positive association with spread velocity in the North China model (0.29 ± 0.12, t67.36 = 2.46, p < 0.05) and in the whole China model (0.20 ± 0.09, t82.56 = 2.13, p < 0.05). Coastline presence showed a positive association with spread velocity in the whole China model (0.44 ± 0.19, t82.56 = 2.34, p < 0.05) and marginally significantly in the South China model (0.38 ± 0.20, t31.95 = 1.94, p = 0.054).
Of the effects of geographical factors, GAM analysis revealed a positive association of elevation with spread velocity in the North China model (F1,67.36 = 12.34, p < 0.01; figure 3g) but not in the South- or whole China models. In addition, ruggedness exhibited a significant negative association with spread velocity in the North China model (F1.54, 67.36 = 3.96, p < 0.05; figure 3h) but not the South- or whole China models. Because of correlation between elevation and ruggedness (r = 0.52), their relative contributions should be interpreted with some caution; we found, however, that a model with both terms included was favoured in terms of the generalized cross validation (GCV) criterion (a measure of predictive power) compared with models with one or both terms omitted.
Of the effects of climate, GAM analysis revealed that dry conditions (high values of the dryness/wetness index, measuring the spatial and temporal variation in droughts and floods) were associated with low spread velocity in all three models: in the North China model, F1.83,67.36 = 3.68, p < 0.05 (figure 3f), in the South China model F1.6, 31.95 = 6.64, p < 0.01 (figure 3d) and in the whole China model F1, 82.56 = 17.41, p < 0.01 (figure 3b).
3. Discussion
(a). Plague spread paths and patterns
Owing to frequent long-distance human transportation activities, the spread process of diseases often involves both short- and long-distance transmission [9,10]. The traditional approaches, including TSA, are inadequate for dealing simultaneously with these two spread processes of diseases. Three stages of spatial transmission during the third plague pandemic were identified using our NNA method. After an initial short-distance transmission (contact diffusion) phase within the Yunnan province in the first stage, plague spread in North China as a clear long-distance transmission (relocated diffusion) and in South China as both short-distance and long-distance transmissions during the second stage. Spread of plague in the third stage was mostly identified as short-distance transmission.
Adjemian et al. [15] reported that the velocity of plague spread increased during the invasion of plague into the western US (1900–1969). This stands in contrast to this study, which indicates that the velocity of plague spread consistently decreased during the main pandemic period for both North and South China (figure 3a,c,e). While difference in estimation methods could be partly responsible for this difference in temporal trend between China and the United States, we note that there are also important differences between the systems, for example, plague being native to China but not the United States [17,37]. Phylogenetic analysis suggests that Yersinia pestis evolved in or near China and spread through multiple radiations to Europe, South America, Africa and Southeast Asia [19].
(b). Effects of climate
Previous studies indicated inconsistent associations between spread velocity of plague and precipitation. In a study from the western US, the spread velocity of both animal and human plague was found to be negatively associated with annual precipitation [15], but the underlying mechanism was not discussed. However, in another study, the spread velocity of plague among prairie dogs (Cynomys ludovicianus) was found to be positively associated with precipitation [33]; this observation was explained by the conventional cascade hypothesis that increased precipitation increases primary productivity, which is followed by increased rodent/flea density, and then plague transmission [25,26].
Contrary to the results on human plague by Adjemian et al. [15], we found that the association between plague spread velocity and wetness was positive in both regional models and the whole China model. Our results suggest that wet conditions, as measured by flood and drought events, facilitated spatial spread of human plague in China. Wet weather may have increased human contact with plague vectors, for example rodents [38], and caused the migration of people who had lost their homes in floods. All of these factors may have promoted the spatial transmission of plague. In a previous study, we showed that wet weather increased plague occurrences in North China with a 1-year time-lag, probably through increased rodent abundance in response to improved food or vegetation [32]. By contrast, wet weather in South China was associated with reduced plague occurrence, possibly a result of flooding events killing rodents [32]. The effects of wet conditions on plague prevalence and spread velocity thus appear to be of opposite sign in South China. We propose that in the wet South China, although floods may decrease plague occurrences by killing rodents within a location, floods may promote the speed of the transmission between locations due to increased migration of plague-infected people and rodents. It should be noted that the dryness/wetness index considered in this study is not equal to precipitation as measured using modern meteorological instruments. It represents the extreme conditions as measured by the drought and flood events in China (electronic supplementary material, section 5.1). Thus, caution is required when making comparisons of our results with previous ones using precipitation data.
(c). Effects of transportation routes
Although human transportation systems have frequently been hypothesized as causes of disease transmission [8], their effects are rarely tested. We provided quantitative evidence that roads, rivers and coastlines are positively associated with spread velocity of human plague of the third plague pandemic. Furthermore, using NNA, we identified many long-distance transmissions closely related to human transportation routes. The long-distance dispersal of plague from Yunnan to the southeast coastline was likely mediated by river ships, not by land routes because plague was not reported in Guangxi before this relocation spread diffusion. Plague in North China was caused by a long-distance transmission, possibly from Yunnan to Qinghai. The jump in the spread of plague from Yunnan to Qinghai province was likely facilitated by the Tea Road (Chama Gudao) from Yunnan, through Sichuan, to Qinghai, Shanxi and Inner Mongolia, Xinjiang, Tibet and other locations (electronic supplementary material, figure S14). Dali and Lijiang counties of Yunnan, which had severe plague occurrences, were important tea ports along this ancient route. Plague-infected humans, rodents or fleas carried by rodents or people might have occasionally transmitted the disease along this route. The long-distance transmission of plague during the second stage in China was likely promoted by the trading activities. The plague that appeared in Xinjiang in western China appears to have been transmitted from Gansu along the ancient trade network known as the Silk Road (electronic supplementary material, figures S13 and S14). The discovery of plague along the Silk Road may complement the conventional view that the international spread of the third pandemic was via ship from Hong Kong [35].
Roads might have changed during the study period, especially during the 1900s. Using another available road map published in 1946, we found similar results to those using the 1910 road map, strengthening the confidence in the findings (electronic supplementary material, figure S13). However, there is still limitation of using road maps of 1910 and 1946 to represent the road structure during the study period covering nearly 200 years. The pre-1900 road structure should be especially important in affecting spread of plague in the south, where many infections occurred before 1910. Thus, it is necessary to assess the impact of road structure changes on plague transmission in future studies.
TSA results on plague spread velocities during the third plague pandemic in China (40.1 km yr−1 for the whole of China) were comparable to corresponding estimates of plague spread in western US (45–87 km yr−1 from 1900 to 1966) [15]. However, this contrasts with the spread velocity (341.9–643.7 km yr−1 from 1347 to 1350) in Europe during the time of the Black Death [14]. The advanced sea transportation in Europe might have contributed to the much higher spread velocity of plague during the Black Death. Besides, the much higher spread velocity in Europe estimated by Noble [14] was caused by the pneumonic plague which was transmitted among people [15].
(d). Effects of geographical factors
In North America, both the southern Rockies and the northwestern forested mountains are areas where the velocity of plague spread is high [15]. The observed positive association between plague spread velocity and elevation is likely due to the fact that natural plague foci are found mainly in mountainous regions where rodent hosts and flea vectors are abundant [17,39]. Such a relationship between plague foci and elevation has been found in China for the main plague host Marmota himalayana in the Qinghai–Tibetan plateau, which inhabits various types of meadow-steppes at elevation levels between 2700 and 5450 m [17,39]. Similarly, the main rodent plague hosts in Inner Mongolia are Marmota sibirica, Spermophilus dauricus and Meriones unguiculatu [17,39], which inhabit grasslands at elevation levels of over 1000 m. The observed negative association between spread velocity and ruggedness suggests that ruggedness may reduce plague transmission owing to transportation barriers for humans and/or rodents. This may also explain why spread velocity in South China with more mountainous areas is lower than that in North China with flat plateau regions.
Results of both the TSA and NNA methods indicated that the spread velocity in North China was greater on average than in South China (all p < 0.05, ANOVA; electronic supplementary material, table S2 and figures S6–S9). This may have been caused by the differences in the regional transportation systems. In North China, transportation is relatively easy in the plains and high plateaus, whereas in South China, transportation is often made difficult by rugged mountainous regions. During the third pandemic, transportation systems (especially roads and railways) were much more advanced in North China. Indeed, NNA revealed more long-distance transmissions of human plague in North China, whereas in South China the long-distance transmissions were very rare (and those that occurred seem likely to have been caused by sea transportation).
(e). Differences in estimation of spread velocity
The spread velocities in both North and South China estimated using TSA were all higher than NNA estimates (electronic supplementary material, figures S7–S9 and table S2). This is caused by the difference in estimation methods between TSA and NNA. NNA estimates the spread velocity based on only the nearest site, whereas TSA uses all available sites nearby. Besides, we note that the differences in velocity estimates of TSA and NNA might partly reflect ability of dealing with long-distance transmissions of the two methods (electronic supplementary material, figures S5 and S8, and table S2). Though TSA works well for short-distance transmissions uniformly out from a central point [13], it is easily affected by long-distance transmission. In North China, there were many long-distance transmissions. Using TSA, the locations in North China are then estimated to be infected at approximately the same time, resulting in a flat TSA surface and high estimates of spread velocity for the entire region. The finer scale dynamics are not captured. Using NNA, the long-distance transmissions only influence the velocity estimates for the sites that are directly affected by long-distance transmission, while the velocity estimates for the majority of sites are mainly influenced by the local-scale dynamics.
NNA uses only simple data consisting of date and location, and the method could potentially have broader applications, for example, for studying the transmission ecology of other kinds of diseases or biological invasion of alien species, especially under the frequent disturbances of human or wildlife interventions. Besides, at least for low-dimensional transmission patterns as in our system (each infected site was the source of transmission to few new sites; electronic supplementary material, figure S10), the NNA appears not to be very sensitive to missing data. The estimated transmission patterns did not show much change when 10–50% of the original plague-infected counties were randomly removed as ‘missing data’ (electronic supplementary material, figure S5). Further, the calculation of transmission speed among the counties was not strongly affected by missing data (electronic supplementary material, figure S6).
(f). Implications for plague prevention and management
We identified three epidemic stages of plague transmission and showed that both contact and relocated diffusion processes can be detected by using NNA. Using NNA, we revealed the finer scale transmission pattern of plague in both time and space. This knowledge is expected to contribute to plague control and prevention as well as to the assessment of control efficiency.
Our results indicated special attention must be paid to newly opened transportation routes, for example the Qinghai–Tibet railway in China, that connect the natural foci of plague. It is essential to detect long-distance transmissions as early as possible, at the beginning of disease outbreaks, and then control local contact diffusions.
Accelerated climate change and globalization is likely increasing risk of infectious diseases [40]. Our results revealed that high levels of precipitation, especially flooding events, increased the spread velocity of plague. With accelerated impact of climate warming, some regions (e.g. northwest China) are expected to have more rainfall [41], and thus may have more chance of plague spread. Therefore, plague surveillance ought to be enhanced in regions experiencing sustained humid weather or flooding.
4. Material and methods
(a). Plague data
Extensive data on human plague cases from 1772 to 1964 were compiled by a team led by the Institute of Epidemiology and Microbiology, Chinese Academy of Medical Sciences [36]. To analyse the spread of plague, we identified the year of the first recorded human plague case for each county of China (here, we define it as the ‘first plague-invaded year’) and the locations of the political centres of the counties (figure 1a; electronic supplementary material, figure S1).
(b). Climate proxy data
Spatio-temporal dryness/wetness data were derived from the book ‘Yearly Charts of Dryness/wetness in China for the Last 500-Year Period’ [42]. The dryness/wetness index (D/W), classified into five categories from 1 to 5, represents the climate conditions ranging from extreme wet to extreme dry. We used the dryness/wetness index of the nearest region for each county as a measure of both the spatial and the temporal variation in dryness and wetness (figure 1b). Counties with no region having dryness/wetness data within a 200 km range were not included in the statistical analysis (figure 1b). For detailed information about the climate proxy data, see Central Meteorological Bureau [42].
(c). Road, river, coastline and elevation data
Road data were extracted from Chinese historical maps produced during the late Qing Dynasty (around 1910) [43], which was the best available data matching the study period. River, coastline and grid elevation data (1 × 1 km) were obtained from the National Fundamental Geographical Information System of China (http://ngcc.sbsm.gov.cn/). River, coastline and road data were coded as binary variables indicating presence (1) or absence (0) within the county (figure 1c). The river data we used in this study included rivers above ‘third-order stream’ (stream order is a measure of the relative size of streams), most of them are navigable. We note that these variables were only rough indices of the transportation system, and that in particular the road network changed during the studied period. Because of the noise in these variables, the effects of transportation factors on transmission speed were likely underestimated in the statistical analysis. Average elevation of each county was calculated as the arithmetic mean of all elevation values within each county using ArcGIS v. 9.2 (ESRI). We used the standard deviation of the elevations within each county as a ruggedness index.
(d). Trend-surface analysis
TSA has been widely used to estimate spread velocity of travelling waves of diseases including human plague over large spatial scales [14,15] (figure 2a). TSA is a smoothing method based on a random-walk model in which the spread of a disease is approximated by considering invasive year (dependent variable) as polynomial functions of the geographical coordinates (independent variables) of the infected locations [44,45]. The speed of the plague wave was then calculated from the estimated slope of the interpolated surface through the partial differentials of the polynomial model (electronic supplementary material, table S1 and figure S2).
TSA works well for short-distance (contact diffusive) transmission of diseases, but a limitation of TSA is that it may be biased when frequent long-distance transmissions of the disease obscures travelling waves of disease spread [13]. Furthermore, we found the spread velocities estimated using TSA are highly autocorrelated in space, which can limit subsequent statistical analysis (electronic supplementary material, figure S7A). Besides, TSA cannot reveal detailed transmission routes which are important for targeting disease control efforts.
(e). Nearest neighbour approach
To overcome the limitations of TSA, we propose a new method, NNA, to estimate the spread velocity and reveal transmission paths (figure 2b). Our NNA works by following the rule of ‘earlier in time, then closest in space’ (electronic supplementary material, figure S4). We assumed that plague in a given county (sink county) was transmitted from the spatially nearest county (source county) with plague-invaded time (year) earlier than that of the sink county. First, we listed the earliest plague-invaded years of plague of all counties and the location using a table. Second, for a given sink county (plague-infected county), we identified the source county as the county that was closest to the sink county in space among the counties having earlier first plague-invaded time (year) than the sink county. The spread velocity was calculated as the distance (km) between the source and sink counties, divided by the time interval (years) between the first plague-invaded years of the two counties (electronic supplementary material, figures S4 and S5). Although the nearest neighbour principle has been used in identifying disease sources for estimating basic reproductive number [46], it has, to our knowledge, not been used for reconstructing the transmission paths of diseases (or other organisms) and estimating spread velocity based on information of date and location.
NNA has three advantages over TSA. First, we found the spread velocity values estimated by NNA to be less autocorrelated in space (electronic supplementary material, figure S7). Second, NNA can reveal detailed spread routes of plague at both fine scale and large scale under the parsimonious assumption that it spreads from the nearest location where plague previously occurred (figure 2). Third, NNA functions well with both short- and long-distance transmissions compared with TSA (figure 2). Long-distance transmission in North China seems to have obscured travelling waves shown by TSA (figure 2b). However, NNA also has some disadvantages. For example, if the disease does not transmit from the nearest neighbour, the spread velocity may be underestimated. Thus, NNA and TSA may be complementary methods for unravelling the spatial transmissions of diseases.
It should be noted that for the quantification of local-scale transmission speed, the number of sink sites infected per source site may be equally relevant as the distance per time between source and sink sites (the metric considered in the current analysis). The NNA allows the quantification of both metrics, showing that in our system, the majority of the sites were only identified as source for one sink site (electronic supplementary material, figure S10). Because of the limited variability in the number of sink sites per source site, we focused the statistical analysis on the spread distance per time.
(f). Generalized additive models
To estimate the effects of potential factors on spread velocity of human plague, we analysed the velocity data using generalized additive models with a quasi-Poisson error distribution (logarithmic link function) [47]. The ‘mgcv’ package (v. 1.7–6) of R was used for these analyses [48]. We used the GCV value as a model selection criterion. The GCV of a model is a proxy for the model's out-of-sample predictive mean squared error and can be used to compare alternative model formulations [47,49]. A model with lower GCV has more predictive power and was hence preferred to ones with higher GCV. Because there are two distinct regions of plague occurrence in North and South China, where both climate and plague dynamics differ (figure 1 and electronic supplementary material, figure S1), we modelled the spread patterns of North and South China separately, in addition to one analysis covering all China (electronic supplementary material, tables S4 and S5, and figures S15–S27).
The initial candidate models of spread velocity included variables of year, road, river, coastline, elevation, ruggedness, location (longitude and latitude), plague prevalence and dryness/wetness index (D/W). Because there was spatial autocorrelation in the model residuals, we introduced an interaction effect between plague prevalence and location into the model, which not only was preferred by GCV model selection criteria but also reduced residual spatial autocorrelation (electronic supplementary material, tables S4 and S5, and figures S15–S27). The full model formula was
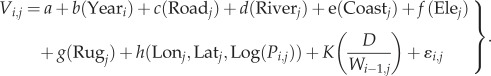 |
4.1 |
Here, Vi,j is the natural logarithm of plague spread velocity in year i and county j. Parameter a is the overall intercept. b(Yeari) is a smooth function of invasive year of plague into county j (cubic regression spline function). c(Roadj), d(Riverj) and e(Coastj) are effects of binary variables indicating whether roads, rivers and coast are present or absent in county j. f(Elej) is a smooth function of log-transformed (plus 1 to avoid logarithm of zero values) elevation in county j (cubic regression spline function with maximally 2 d.f., i.e. three knots). g(Rugj) is a smooth function of log-transformed (plus 1 to avoid zero logarithm) ruggedness in county j (cubic regression spline function with maximally 2 d.f.). h(Lonj, Latj, Log(Pi,j)) is a tensor-product anisotropic smooth function of h′(Log(Pi,j)) with h″(Lonj, Latj). h′(Log(Pi,j)) is a smooth function of the total human plague cases (log-transformed, plus 1 to avoid zero logarithm) in the assumed source county of plague transmitted to county j (i.e. the nearest neighbour with recorded plague) for all years prior to year i (cubic regression spline function with maximally 2 d.f.). h″(Lonj, Latj) is a two-dimensional smooth function of longitude and latitude (thin-plate regression spline with maximally 20 d.f. when modelling North and South China separately, and maximally 30 d.f. when modelling whole China). K(D/Wi−1,j) is a smooth function of the dryness/wetness index in the year preceding the invasive year of plague into county j (cubic regression spline function with 2 d.f.). We only included dryness/wetness of sink counties into the model because prior analyses indicate that dryness/wetness of the source counties shows no significant effect (electronic supplementary material, table S3). We used backward stepwise model selection separately for North China, South China and all of China (electronic supplementary material, table S4). The velocity estimated by NNA was used as a response variable in this analysis because these velocity estimates had minor spatial autocorrelation, while TSA velocity estimates were highly spatially autocorrelated (electronic supplementary material, figure S7).
Supplementary Material
Acknowledgements
We are grateful to Prof. Marcel Holyoak, University of California, and anonymous reviewers for their valuable comments on the manuscript. We thank Terry Boyd-Zhang for improving the English of this manuscript. Z.Z. and N.C.S. designed the study. L.X., L.C.S. and K.L.K. contributed to modelling analysis and the writing of the paper. Q.L., X.F. and S.W. contributed to data collection and editing. All co-authors contributed to discussion and writing of the manuscript.
Funding statement
This work was supported by a grant (XDA05080701) from Chinese Academy of Sciences, a grant (31370440) from the National Natural Science Foundation, the National Basic Research Program (2007CB109101; 2012CB955504) of Ministry of Science & Technology (MOST), and support from the Centre for Ecological and Evolutionary Synthesis (CEES) of the University of Oslo. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Gubler DJ, Reiter P, Ebi KL, Yap W, Nasci R, Patz JA. 2001. Climate variability and change in the United States: potential impacts on vector- and rodent-borne diseases. Environ. Health Perspect. 109, 223–233. ( 10.2307/3435012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duplantier J, Duchemin J, Chanteau S, Carniel E. 2005. From the recent lessons of the Malagasy foci towards a global understanding of the factors involved in plague reemergence. Vet. Res. 36, 437–453. ( 10.1051/vetres:2005007) [DOI] [PubMed] [Google Scholar]
- 3.McMichael A, Woodruff R, Hale S. 2006. Climate change and human health: present and future risks. Lancet 367, 859–869. ( 10.1016/S0140-6736(06)68079-3) [DOI] [PubMed] [Google Scholar]
- 4.Gage KL, Burkot TR, Eisen RJ, Hayes EB. 2008. Climate and vectorborne diseases. Am. J. Prev. Med. 35, 436–450. ( 10.1016/j.amepre.2008.08.030) [DOI] [PubMed] [Google Scholar]
- 5.Semenza JC, Menne B. 2009. Climate change and infectious diseases in Europe. Lancet Infect. Dis. 9, 365–375. ( 10.1016/S1473-3099(09)70104-5) [DOI] [PubMed] [Google Scholar]
- 6.Ostfeld RS. 2009. Climate change and the distribution and intensity of infectious diseases. Ecology 90, 903–905. ( 10.1890/08-0659.1) [DOI] [PubMed] [Google Scholar]
- 7.Rohr JR, Dobson AP, Johnson PTJ, Kilpatrick AM, Paull SH, Raffel TR, Ruiz-Moreno D, Thomas MB. 2011. Frontiers in climate change-disease research. Trends Ecol. Evol. 26, 270–277. ( 10.1016/j.tree.2011.03.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benedict C. 1996. Bubonic plague in nineteenth-century China, pp. 73–100. Stanford, CA: Stanford University Press. [DOI] [PubMed] [Google Scholar]
- 9.Brown JK, Hovmoller MS. 2002. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297, 537–541. ( 10.1126/science.1072678) [DOI] [PubMed] [Google Scholar]
- 10.Mundt CC, Sackett KE, Wallace LD, Cowger C, Dudley JP. 2009. Long-distance dispersal and accelerating waves of disease: empirical relationships. Am. Nat. 173, 456–466. ( 10.1086/597220) [DOI] [PubMed] [Google Scholar]
- 11.Pfeiffer D, Robinson T, Stevenson M, Stevens KB, Rogers DJ, Clements ACA. 2008. Spatial analysis in epidemiology, pp. 32–44. Great Britain, UK: Oxford University Press. [Google Scholar]
- 12.Riley S. 2007. Large-scale spatial-transmission models of infectious disease. Science 316, 1298–1301. ( 10.1126/science.1134695) [DOI] [PubMed] [Google Scholar]
- 13.Pioz M, Guis H, Calavas D, Durand B, Abrial D, Ducrot C. 2011. Estimating front-wave velocity of infectious diseases: a simple, efficient method applied to bluetongue. Vet. Res. 42, 60–73. ( 10.1186/1297-9716-42-60) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noble JV. 1974. Geographic and temporal development of plagues. Nature 250, 726–728. ( 10.1038/250726a0) [DOI] [PubMed] [Google Scholar]
- 15.Adjemian JZ, Foley P, Gage KL, Foley JE. 2007. Initiation and spread of travelling waves of plague, Yersinia pestis, in the western united states. Am. J. Trop. Med. Hyg. 76, 365–375. [PubMed] [Google Scholar]
- 16.Perry RD, Fetherston JD. 1997. Yersinia pestis-etiologic agent of plague. Clin. Microbiol. Rev. 10, 35–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Tan J. 2000. The atlas of plague and its environment in the People's Republic of China, pp. 48–79. Beijing, China: Science press. [Google Scholar]
- 18.Little LK. 2007. Plague and the end of antiquity: the pandemic of 541–750, pp. 3–33. Cambridge, NY: Cambridge University Press. [Google Scholar]
- 19.Morelli G, et al. 2010. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat. Genet. 42, 1140–1146. ( 10.1038/ng.705) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stenseth NC, Atshabar BB, Begon M, Belmain SR, Bertherat E, Carniel E, Gage KL, Leirs H, Rahalison L. 2008. Plague: past, present, and future. PLoS Med. 5, 9–13. ( 10.1371/journal.pmed.0050003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohn GC. 2008. Encyclopedia of plague and pestilence: from ancient times to the present. New York, NY: Facts on File. [Google Scholar]
- 22.World Health Organization. 1999. Plague manual: epidemiology, distribution, surveillance and control See http://www.who.int/csr/resources/publications/plague/WHO_CDS_CSR_EDC_99_2_EN/en/index.html.
- 23.Gage KL, Kosoy MY. 2005. Natural history of plague: perspectives from more than a century of research. Annu. Rev. Entomol. 50, 505–528. ( 10.1146/annurev.ento.50.071803.130337) [DOI] [PubMed] [Google Scholar]
- 24.Bevins SN, Baroch JA, Nolte DL, Zhang M, He H. 2012. Yersinia pestis: examining wildlife plague surveillance in China and the USA. Integr. Zool. 7, 99–109. ( 10.1111/j.1749-4877.2011.00277.x) [DOI] [PubMed] [Google Scholar]
- 25.Parmenter RR, Yadav EP, Parmenter CA, Ettestad P, Gage KL. 1999. Incidence of plague associated with increased winter–spring precipitation in New Mexico. Am. J. Trop. Med. Hyg. 61, 814–821. [DOI] [PubMed] [Google Scholar]
- 26.Enscore RE, et al. 2002. Modeling relationships between climate and the frequency of human plague cases in the southwestern United States, 1960–1997. Am. J. Trop. Med. Hyg. 66, 186–196. [DOI] [PubMed] [Google Scholar]
- 27.Collinge SK, et al. 2005. Testing the generality of a trophic-cascade model for plague. EcoHealth 2, 102–112. ( 10.1007/s10393-005-3877-5) [DOI] [Google Scholar]
- 28.Stenseth NC, et al. 2006. Plague dynamics are driven by climate variation. Proc. Natl Acad. Sci. USA 103, 13 110–13 115. ( 10.1073/pnas.0602447103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kausrud KL, Viljugrein H, Frigessi A, Begon M, Davis S, Leirs H, Dubyanskiy V, Stenseth NC. 2007. Climatically driven synchrony of gerbil populations allows large-scale plague outbreaks. Proc. R. Soc. B 274, 1963–1969. ( 10.1098/rspb.2007.0568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Li Z, Tao Y, Chen M, Wen X, Xu L, Tian H, Stenseth NC. 2007. Relationship between increase rate of human plague in China and global climate index as revealed by cross-spectral and cross-wavelet analyses. Integr. Zool. 2, 144–153. ( 10.1111/j.1749-4877.2007.00061.x) [DOI] [PubMed] [Google Scholar]
- 31.Pham HV, Dang DT, Minh NNT, Nguyen D, Nguyen TV. 2009. Correlates of environmental factors and human plague: an ecological study in Vietnam. Int. J. Epidemiol. 38, 1634–1641. ( 10.1093/ije/dyp244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu L, Liu QY, Stige LC, Ben Ari T, Fang XY, Chan KS, Wang SC, Stenseth NC, Zhang Z. 2011. Nonlinear effect of climate on plague during the third pandemic in China. Proc. Natl Acad. Sci. USA 108, 10 214–10 219. ( 10.1073/pnas.1019486108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snäll T, O'Hara RB, Ray C, Collinge SK. 2008. Climate-driven spatial dynamics of plague among prairie dog colonies. Am. Nat. 171, 238–248. ( 10.1086/525051) [DOI] [PubMed] [Google Scholar]
- 34.Anonymous. 1904. The spread of plague. Nature 70, 616–617. ( 10.1038/070616b0) [DOI] [Google Scholar]
- 35.Wu L, Chen YH, Pollitzer R, Wu CY. 1936. Plague: a manual for medical and public health workers, pp. 1–421. Shanghai, China: Weishengshu National Quarantine Service Shanghai Station. [Google Scholar]
- 36.Institute of Epidemiology and Microbiology, Chinese Academy of Medical Sciences. 1980. The history of spread of plague in China, pp. 33–1794. [In Chinese.] [Google Scholar]
- 37.Ben-Ari T, et al. 2012. Identification of Chinese plague foci from long-term epidemiological data. Proc. Natl Acad. Sci. USA 109, 8196–8201. ( 10.1073/pnas.1110585109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madsen T, Shine R. 1999. Rainfall and rats: climatically-driven dynamics of a tropical rodent population. Aust. J. Ecol. 24, 80–89. ( 10.1046/j.1442-9993.1999.00948.x) [DOI] [Google Scholar]
- 39.Fang XY. 1990. Natural focuses of plague in China, pp. 1–276. Beijing, China: People's Medical Publishing House; [In Chinese.] [Google Scholar]
- 40.Zhang Z. 2013. Biological consequences of global change: past and future. Integr. Zool. 8, 123 ( 10.1111/1749-4877.12043) [DOI] [PubMed] [Google Scholar]
- 41.Shi Y, et al. 2003. Discussion on the present climate change from warm-dry to warm-wet in northwest china. Q. Sci. 23, 152–164. [Google Scholar]
- 42.Central Meteorological Bureau. 1981. Yearly charts of dryness/wetness in China for the last 500-year period, pp. 321–332. Beijing, China: Cartographic Publishing House. [Google Scholar]
- 43.Tan Q. 1996. The historical atlas of China, the Qing dynasty period, pp. 3–65. Beijing, China: China Cartographic Publishing House; [In Chinese.] [Google Scholar]
- 44.Mollison D. 1977. Spatial contact models for ecological and epidemic spread. J. R. Stat. Soc. B. Met. 39, 283–326. [Google Scholar]
- 45.Abramson G, Kenkre VM, Yates TL, Parmenter RR. 2003. Traveling waves of infection in the hantavirus epidemics. Bull. Math. Biol. 65, 519–534. ( 10.1016/S0092-8240(03)00013-2) [DOI] [PubMed] [Google Scholar]
- 46.Ward MP, Maftei D, Apostu C, Suru A. 2009. Estimation of the basic reproductive number (R0) for epidemic, highly pathogenic avian influenza subtype H5N1 spread. Epidemiol. Infect. 137, 219–226. ( 10.1017/S0950268808000885) [DOI] [PubMed] [Google Scholar]
- 47.Wood SN. 2006. Generalized additive models: an introduction with R, pp. 121–270. Boca Raton, FL: Chapman and Hall/CRC. [Google Scholar]
- 48.R Development Core Team. 2006. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 49.Stige LC, Ottersen G, Brander K, Chan KS, Stenseth NC. 2006. Cod and climate: effect of the North Atlantic oscillation on recruitment in the North Atlantic. Mar. Ecol. Prog. Ser. 325, 227–241. ( 10.3354/meps325227) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.