Abstract
Deep hypersaline anoxic basins (DHABs) of the Mediterranean Sea are among the most extreme ecosystems on Earth and host abundant, active and diversified prokaryotic assemblages. However, factors influencing biodiversity and ecosystem functioning are still largely unknown. We investigated, for the first time, the impact of viruses on the prokaryotic assemblages and dynamics of extracellular DNA pool in the sediments of La Medee, the largest DHAB found on Earth. We also compared, in La Medee and L'Atalante sediments, the diversity of prokaryotic 16S rDNA sequences contained in the extracellular DNA released by virus-induced prokaryotic mortality. We found that DHAB sediments are hot-spots of viral infections, which largely contribute to the release of high amounts of extracellular DNA. DNase activities in DHAB sediments were much higher than other extracellular enzymatic activities, suggesting that extracellular DNA released from killed prokaryotes can be the most suitable trophic resource for benthic prokaryotes. Preserved extracellular DNA pools, which contained novel and diversified gene sequences, were very similar between the DHABs but dissimilar from the respective microbial DNA pools. We conclude that the strong viral impact in DHAB sediments influences the genetic composition of extracellular DNA, which can preserve the signatures of present and past infections.
Keywords: viruses, prokaryotes, extracellular DNA, deep hypersaline anoxic basins
1. Introduction
Deep-sea ecosystems include some of the most extreme and unique ecosystems on Earth, such as the hydrothermal vents, oxygen minimum zones (including the anoxic deep waters of the Black Sea) and the deep hypersaline anoxic basins (DHABs) of the Eastern Mediterranean Sea. The DHABs are characterized by absence of light, high hydrostatic pressure (around 35 MPa), sharp chemoclines, variable pH values, permanently anoxic conditions and nearly saturated salt concentrations [1]. A thick layer of brines, indeed, with variable ionic composition, creates a strong halocline, which prevents oxygen exchange and strongly reduces the supply of organic particles from the upper oxygenated water layers. As a consequence, the DHABs are characterized by a severe limitation of organic matter inputs [2,3]. Despite this, available information indicates that the brines of DHABs contain active and diversified microbial assemblages [1,4–6]. Previous studies also revealed the presence of new microbial metabolic/physiological functions [6–8]. Investigations conducted on the microbes inhabiting these extreme ecosystems have extended the comprehension of the limits of life [1,6,9] and the potential for microorganisms and metazoans to survive in extraterrestrial environments [10–12].
Despite evidence that viruses play a crucial role in shaping microbial assemblage composition and controlling the functioning of all aquatic ecosystems [13,14], their role in DHABs has received limited attention. Nevertheless, in anoxic and hypersaline sediments, where the abundance of eukaryotic grazers has been reported to be very low [15], viral impact on the abundance, activity and diversity of prokaryotes could be even more relevant [16].
Viral lysis of prokaryotes can have important biogeochemical implications and the cell debris released can be used for sustaining the metabolism of non-infected prokaryotes [14,17–19].
Such a cell debris also includes extracellular DNA, which has a high (phosphorus) P content and plays an important role in benthic trophodynamics [20–22]. At the same time, a fraction of extracellular DNA can escape the degradation processes and accumulate in the sediments [23,24]. Since high extracellular DNA concentrations have been previously reported in permanently anoxic and hypersaline sediments [16,25], these extreme systems can preserve extracellular DNA [20] and the genetic information contained therein [23,26–28].
In this study, we investigated the ecological role of virus-induced prokaryotic mortality in the sediments of La Medee, the largest DHAB found on Earth [29]. We provided new insights into the extent to which such a process influences the dynamics of the extracellular DNA pool. We determined DNase and other exoenzymatic activities (aminopeptidases, β-glucosidases and alkaline phosphatases) to compare the potential availability of extracellular DNA and other trophic resources for prokaryotes inhabiting DHAB sediments. We also investigated the diversity of prokaryotic gene sequences contained in the extracellular DNA to explore the potential of these extreme ecosystems to preserve the genetic imprint of microbial assemblages killed by viruses.
Our findings open new perspectives for the comprehension of the role of viral impact on the diversity of microbial assemblages and for disclosing the mechanisms, which control the dynamics and evolution of prokaryotic assemblages in one of the most extreme system existing on Earth.
2. Material and methods
(a). Study site and sampling strategy
Sediment samples were collected on June 2008 in the DHABs La Medee and L'Atalante (Eastern Mediterranean Sea; electronic supplementary material, figure S1), on board the R/V Urania.
The sampling strategy included two sampling stations within La Medee basin (Station 1 at 2952 m depth, 22°26.98′ N and 34°24.02′ E, and Station 2 at 3077 m depth, 22°33.63′ N and 34°19.62′ E), one station within L'Atalante basin (at 3352 m depth, 21°23.34′ N and 35°18.18′ E) and one oxic station between the two basins (hereafter defined as oxic station, at 2793 m depth, 22°18.12′ N and 34°26.99′ E).
The basins are filled by brines originating from the dissolution of ancient subterranean salt deposits from the Miocene period [30]. La Medee is the largest DHAB of the world oceans, located at ca 100 km southwest of Crete (see electronic supplementary material for additional details). In La Medee stations, bottom temperature was 15.4°C and salinity was more than 300 whereas in the oxic station, temperature was 14°C and salinity was 38.8. L'Atalante basin is located ca 200 km off the Western coast of Crete and is one of the smallest DBABs of the Eastern Mediterranean Sea [31] (see the electronic supplementary material for additional details).
From each station, three independent sediment cores were collected by independent deployments of box-corer (NIOZ-type), which allows the collection of hermetically closed samples. Immediately after retrieval, the sediment cores were sliced vertically (under strictly anaerobic conditions i.e. N2 atmosphere for anoxic sediments) into five sediment layers: the top 1 cm, 1–3 cm, 3–5 cm, 5–10 cm and 10–15 cm.
After sampling, sediment sub-aliquots were collected from La Medee and oxic station for the analyses of microbiological parameters and total extracellular DNA concentrations.
To provide accurate estimates of viral abundance, production and decay, sediment samples were immediately processed onboard (under anaerobic conditions for La Medee sediments), without the addition of any preservative [32,33]. This is because it is widely recognized that the use of formaldehyde or glutaraldehyde for sample storage can result in a rapid loss of viruses in sediment samples [33,34]. For prokaryotic counts, the samples from surface (0–1 cm) and subsurface layers (down to 10–15 cm) of each sediment core were fixed with buffered 2% formalin and stored at 4°C until processing (within two weeks) [35]. Samples for total extracellular DNA concentrations were stored at −80°C until laboratory analyses.
The determinations of DNase, alkaline phosphatase, β-glucosidase and aminopeptidase activities in surface and subsurface sediments were carried out immediately after sediment collection under anaerobic conditions for La Medee samples, as described below. During the time-course experiments carried out to determine viral production and decay in La Medee and oxic sediments (see below), we also collected surface sediment subsamples for the analyses of total extracellular DNA concentrations and taxonomic composition of prokaryotic assemblages associated with extracellular and microbial DNA pools by 16S rDNA pyrosequencing. The same approach was also used to investigate prokaryotic diversity associated with both DNA pools in the sediments of L'Atalante.
Sediment samples collected in L'Atalante basin were processed under the same conditions used for La Medee sediments.
(b). Viral and prokaryotic abundances
Viral and prokaryotic counts were carried out by epifluorescence microscopy using SYBR Green I dye according to Danovaro et al. [32]. Details are reported in the electronic supplementary material.
(c). Viral production and decay and extracellular DNA released by virus-induced prokaryotic mortality
Viral production in the sediments was determined by monitoring the increase of viruses over time [33], while viral decay was determined by monitoring the decrease of viruses over time [36,37].
Gross viral production was estimated as the sum of net viral production and decay rates. Further information is available in the electronic supplementary material.
Sediment subsamples were also collected during time-course experiments of viral production to test for the increase in extracellular DNA pool in the sediments, potentially released by virus-induced prokaryotic mortality. In particular, sediment subsamples were collected at the beginning of the experiment (t0) and after 12 h (t12; i.e. after the detection of the highest value of viral production) for the analyses of total extracellular DNA concentrations. At t12, sediment subsamples were also collected for pyrosequencing analyses of 16S rDNA contained in extracellular and microbial DNA pools, in order to compare their gene composition at the end of the viral infection events.
(d). Virus-induced prokaryotic mortality
Virus-induced prokaryotic mortality (VIPM) was calculated as follows:
where KP is the number of killed prokaryotes (per gram) of sediment (per hour) and TPA the total prokaryotic abundance (per gram) of sediment. KP was determined by time-course experiments on the basis of the expected number of prokaryotes produced (calculated as prokaryotic turnover multiplied by prokaryotic abundance) and the number of prokaryotes actually counted during the same time interval and in the same sediment samples used for determining viral production [17]. Prokaryotic turnover rates were calculated as the ratio between prokaryotic heterotrophic C production and biomass (see details in electronic supplementary material).
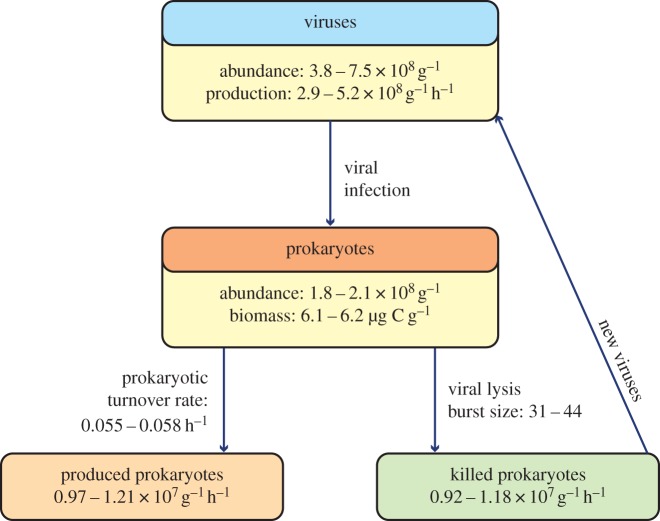
The burst size (i.e. the average number of viruses released by a single prokaryotic cell due to viral infection) values obtained as the ratio between viral production and KP were 31 and 44 at Stations 2 and 1 of the DHAB sediments, respectively, and 30 in the oxic sediments. Such values fall within the range of burst size values determined using independent approaches on deep-sea sediments collected worldwide [17]. To evaluate the quantitative importance of protists as potential top-down controllers of prokaryotes in comparison with viruses in DHAB sediments, additional microscopic analyses were carried out (see details in the electronic supplementary material).
(e). Total extracellular DNA determination
The working conditions, precautions and analytical details during the extracellular DNA analyses are reported in the electronic supplementary material. The concentrations of total extracellular DNA in the sediment were determined according to Dell'Anno et al. [22,24]. This procedure is based on the hydrolysis of the extracellular DNA (using commercial nucleases; see electronic supplementary material for details) and does not allow the recovery of the DNA for subsequent molecular studies [25,38]; therefore, separate samples were also processed to recover extracellular DNA that was suitable for molecular analyses as described below. The total extracellular DNA concentrations were expressed as nanograms of DNA per gram of dry sediment.
Estimates of the amount of DNA released by viral lysis were obtained by converting the number of killed prokaryotes per gram of sediment per hour into the amount of DNA released by viral lysis. To obtain more reliable estimates of the amount of DNA released by viral lysis we calculated both the prokaryotic DNA entirely released after viral lysis and the released prokaryotic DNA subtracted from the incorporated DNA fraction by the new virions [39,40]. These estimates were obtained considering: (i) a mean DNA content per prokaryotic cell of 3.2 fg DNA per cell, which was obtained by using the genome size and the molar GC content of Bacteria and Archaea contained in the NCBI gene bank and (ii) a mean value of 0.039 fg of DNA per virus based on the bacteriophage genome size and the molar GC content of different marine phages sequenced [41]. Such estimates were compared with the increase in extracellular DNA concentrations after 12 h of incubation experiments, to assess the quantitative relevance of the DNA released by virus-induced prokaryotic mortality to the total extracellular DNA pool.
(f). Extracellular enzymatic activities
DNase activity was determined fluorometrically using a fluorescent DNA analogue [poly(dεA): polydeoxyribo-1-N6 ethenoadenylic acid] [42], as described by Dell'Anno & Corinaldesi [22].
Aminopeptidase, β-glucosidase and alkaline phosphatase activities in the sediments were determined by the analysis of the cleavage rates of artificial fluorogenic substrates (l-Leucine-4-methylcoumarinyl-7-amide [Leu-MCA]; 4-MUF-β-d-glucopyranoside [Glu-MUF] and 4-MUF-P-phosphate [MUF-P], respectively) as described by Danovaro [35] (see details in the electronic supplementary material).
(g). Extractions of extracellular and microbial DNA pools suitable for molecular analyses
The extracellular DNA used for molecular analyses was recovered from sediment samples and purified using the procedure described by Corinaldesi et al. [38]. The robustness and reliability of this protocol for extracellular DNA extraction has been shown to exclude any possible contamination due to cell lysis or coextraction of DNA contained in the microbial cells (see the electronic supplementary material).
Before the extraction of DNA associated with microbial biomass (hereafter defined as microbial DNA) by in situ cell lysis, sediment samples were pre-treated with a chemical–physical procedure to remove as much as possible the extracellular DNA as described in Danovaro [35]. The microbial DNA was then recovered and purified by using the UltraClean soil DNA isolation kit (MoBio Laboratories Inc., CA, USA) according to the manufacturer's instructions.
(h). Pyrosequencing analyses of 16S rDNA
To analyse the genetic diversity of the prokaryotic 16S rDNA sequences associated with the extracellular and microbial DNA pools, we used the tag-encoded amplicon pyrosequencing of hypervariable regions (V5 and V6). Bacterial and archaeal 16S rDNA amplicons were generated using the universal primers 789F (5′-TAGATACCCSSGTAGTCC-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) [43] and sequenced by using a Genome Sequencer FLX Titanium (Roche). Three independent PCR analyses were carried out from each replicated extraction (n = 3) of extracellular and microbial DNA. The amplicons obtained from PCR analysis of extracellular DNA were pooled together as well as those for microbial DNA.
A total of 7705 sequences were obtained from the six libraries corresponding to microbial and extracellular DNA samples extracted from La Medee, L'Atalante and oxic sediments. Analyses of sequence data were carried out using the MOTHUR pipeline [44,45]. Reads were trimmed using the FASTA and quality files provided by the sequencer according to the following parameters: minimum length = 100 bp, average quality score = 35, maximum number of Ns = 0. Sequences passing the quality check steps were screened for chimeras using the chimera.slayer tool provided by the pipeline, using the SILVA database as a template [46]. Subsequently, clean sequences were aligned and clustered into OTUs using the farthest neighbour algorithm at 0.03 distance; representative sequences from each OTU were obtained and classified using the Silva taxonomy and Bayesian classifier [47]. The MOTHUR pipeline was used to produce rarefaction curves [44]. To compare samples with different number of sequences, random re-sampling of an equal number of sequences per sample (n = 100) was carried out by using the sub.sample tool [47,48].
(i). Statistical analyses
To test for differences in all microbiological variables, extracellular DNA concentrations and enzymatic activities between the different benthic deep-sea sites and at different depths in the sediment, univariate analyses of variance (ANOVA) were carried out.
To investigate the similarity among the bacterial OTU composition associated with extracellular and microbial DNA pools recovered from oxic and anoxic sediments, cluster analyses were carried out at the phylum and class level (see details in the electronic supplementary material).
3. Results
(a). Viral and prokaryotic abundance
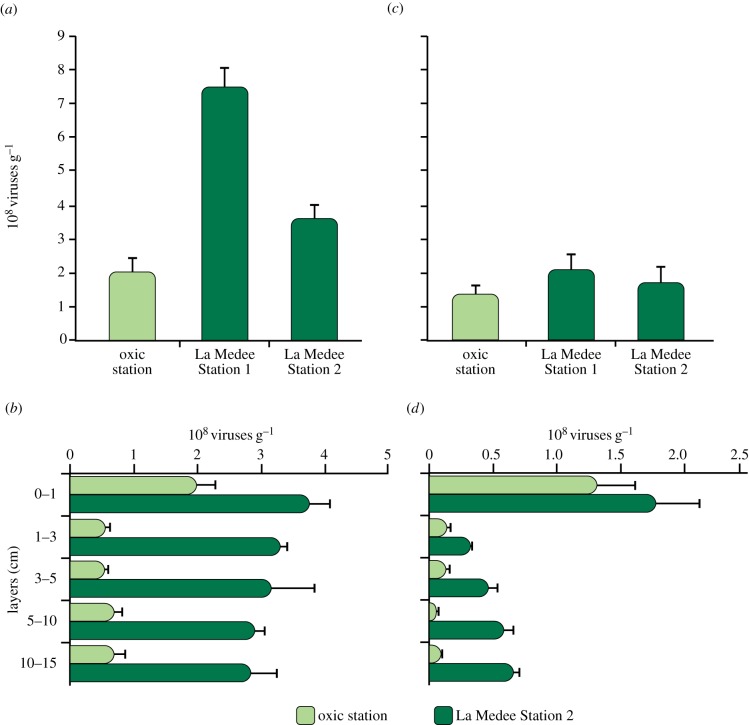
Viral abundance in surface sediments ranged from 2.0 ± 0.29 × 108 to 7.5 ± 0.58 × 108 viruses per gram (at the oxic station and Station 1 of La Medee, respectively, figure 1a) and was significantly higher in the sediments of La Medee basin than in the oxic sediments (ANOVA, p < 0.01). Within the basin (at Station 2), viral abundance did not vary significantly from the surface (3.70 ± 3.38 × 108 viruses per gram) to the subsurface sediment layers (on average, 3.06 × 108 viruses per gram, figure 1b; ANOVA, n.s.). Conversely, in the oxic sediments, it decreased significantly from the top 1 cm of the sediment to the subsurface layers (0.62 × 108 viruses per gram; ANOVA, p < 0.01; figure 1b).
Figure 1.
Viral abundance in (a) surface (0–1 cm) and (b) subsurface (down to 10–15 cm) sediments of La Medee basin and oxic station. Prokaryotic abundance in (c) surface (0–1 cm) and (d) subsurface (down to 10–15 cm) sediments of La Medee basin and oxic station. Standard deviations (n = 3) are reported.
Prokaryotic abundance in the surface sediments ranged from 1.32 ± 0.30 to 2.09 ± 0.42 × 108 cells per gram (at oxic station and Station 1 of La Medee, respectively; figure 1c). No significant differences were observed between the prokaryotic abundances determined in the oxic sediments and at Station 2 of La Medee (ANOVA, n.s.). Similarly, prokaryotic biomass in the DHAB was very close to that outside the basin (6.1 and 5.8 µgC g−1, respectively). Both within and outside the basin, prokaryotic abundances decreased significantly in the subsurface sediment layers compared with those determined in the surface sediment layers (ANOVA, p < 0.01, figure 1d).
The virus-to-prokaryote abundance ratio in surface sediments was higher within the basin than outside (on average, 2.8 versus 1.5, respectively) and, in subsurface sediments of both sampling sites, it increased up to values of more than 10.
(b). Viral production, decay and virus-induced prokaryotic mortality
Net viral production in the surface sediments ranged from 0.44 ± 0.17 to 5.17 ± 1.42 × 108 virus per gram per hour at the oxic station and at Station 1 of La Medee, respectively. Significant differences were observed among the stations with values higher within the DHAB than in the oxic sediments (ANOVA, p < 0.01, figure 2).
Figure 2.
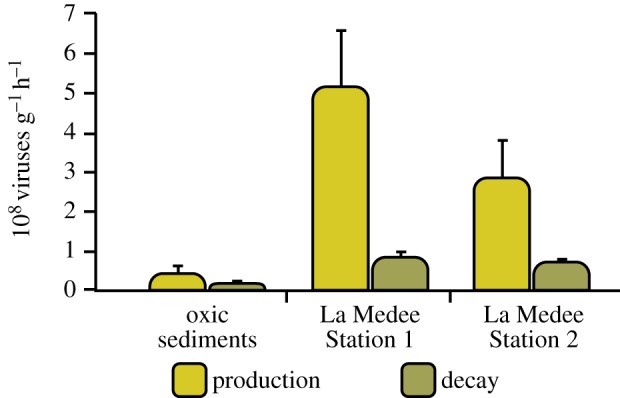
Viral production and decay in surface sediments (0–1 cm) of La Medee and oxic station. Standard deviations (n = 3) are reported.
Viral decay ranged from 1.82 ± 1.42 to 8.51 ± 1.39 × 107 viruses per gram per hour (at the oxic station and Station 1 of La Medee, respectively; figure 2). The DHAB sediments were characterized by viral decay rates significantly higher than those determined in the oxic sediments (ANOVA, p < 0.01). Viral decay contributed, on average, to the gross viral production for 29.3% and 17.2% in the oxic and DHAB sediments, respectively.
The rate of killed prokaryotes by viruses was, on average, 1.5 ± 0.6 × 106 prokaryotes per gram per hour and 10 ± 1.5 × 106 prokaryotes per gram per hour in the oxic and DHAB sediments, respectively. Virus-induced prokaryotic mortality was, on average, ca fivefold lower in the oxic than in the DHAB sediments (% of killed prokaryotes per hour: 5.4 versus 1.1). The turnover rates of prokaryotes in the DHAB were ca fourfold higher than in the oxic sediments (0.055–0.058 h−1 versus 0.015 h−1). Also the rates of produced prokaryotes were higher in the DHAB than outside the basin (0.97–1.21 × 107 prokaryotes per gram per hour versus 0.19 × 107 prokaryotes per gram per hour; figure 3).
Figure 3.
Simplified scheme showing the impact of the viral lysis on prokaryotic dynamics in DHAB sediments. The values of prokaryotes produced per gram of sediment and per hour are based on the determination of prokaryotic heterotrophic C production.
(c). Total extracellular DNA concentrations
Total extracellular DNA concentrations in the investigated surface sediments ranged from 71.1 ± 7.2 to 338.3 ± 5.4 ng DNA per gram (at the oxic station and at Station 2 of La Medee, respectively; figure 4a). These were significantly higher in the sediments within the basin than outside (ANOVA, p < 0.001). In the oxic station, total extracellular DNA concentrations increased significantly along the vertical profile of the sediment (up to 305.6 ± 10.2 ng DNA per gram at 10–15 cm sediment layer, ANOVA, p < 0.001; figure 4b). Conversely, in La Medee basin, extracellular DNA concentrations did not show a clear pattern with depth in the sediment, with the highest values at 1–3 cm sediment layer (1218.5 ± 155.9 ng DNA per gram).
Figure 4.
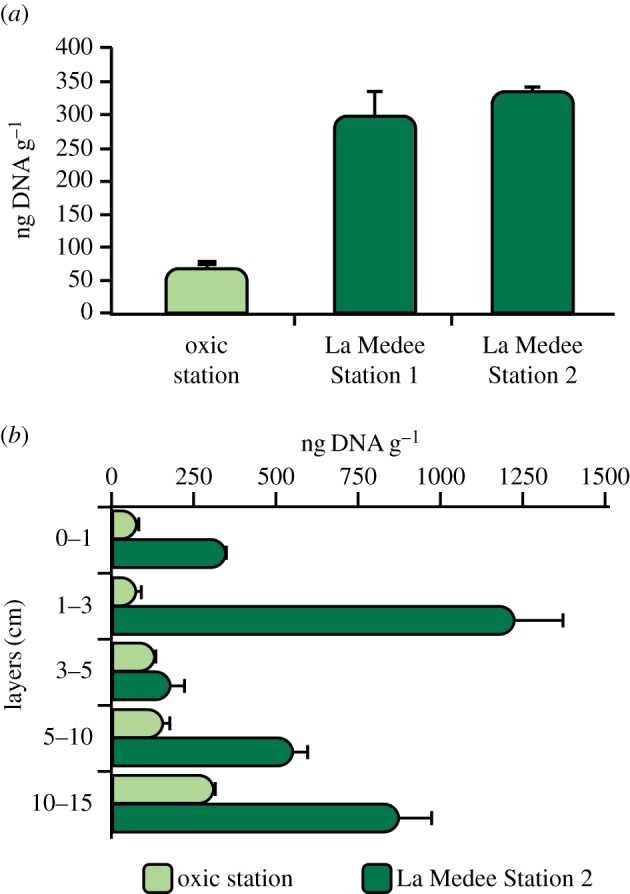
Total extracellular DNA concentrations in (a) surface (0–1 cm) and (b) subsurface (down to 10–15 cm) sediments of La Medee and oxic station. Standard deviations (n = 3) are reported.
Incubation experiments revealed a significant increase in total extracellular DNA concentrations after 12 h both in La Medee and L'Atalante and in oxic sediments (electronic supplementary material, figure S3). Such an increase was higher in the DHAB sediments (394 and 489 ng DNA per gram, in La Medee and L'Atalante sediments, respectively) than in the oxic station (130 ng DNA per gram). Moreover, in the DHAB sediments, the increase in extracellular DNA concentration measured during the sediment incubations, was similar to that estimated by transforming the number of killed prokaryotes determined experimentally into equivalents of extracellular DNA (differences between the two approaches ca 12%).
(d). Enzymatic activities
DNase activities in the surface sediments were significantly higher within the DHAB than in the oxic sediments (on average, 6.11 versus 0.20 ng DNA per gram per hour, respectively, ANOVA, p < 0.05; figure 5a).
Figure 5.
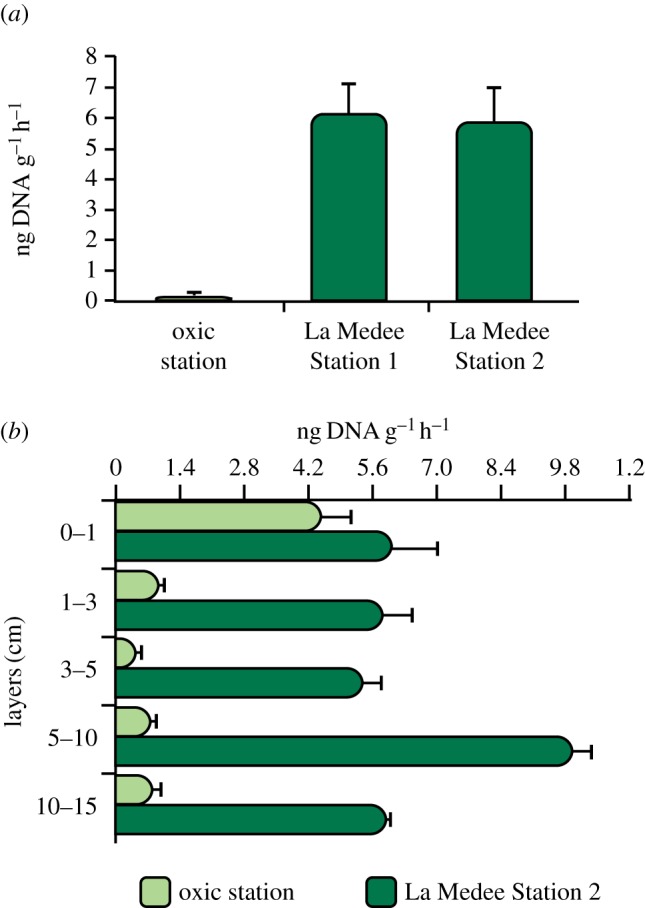
DNase activity in (a) surface (0–1 cm) and (b) subsurface (down to 10–15 cm) sediments of La Medee and oxic station. Standard deviations (n = 3) are reported.
In the subsurface sediments of La Medee, DNase activities were ca 1 order of magnitude higher than in the oxic sediments (on average, 0.7 versus 6.7 ng DNA per gram per hour, p < 0.01) and in both sites they did not show a clear pattern along the sediment vertical profile (figure 5b). Aminopeptidase, β-glucosidase and alkaline phosphatase activities were significantly higher in the oxic than in the DHAB sediments (ANOVA, p < 0.01, electronic supplementary material, figure S2).
(e). Prokaryotic assemblage composition of extracellular and microbial DNA pools
The number of prokaryotic sequences and OTUs obtained in the extracellular and microbial DNA pools are reported in electronic supplementary material, table S1.
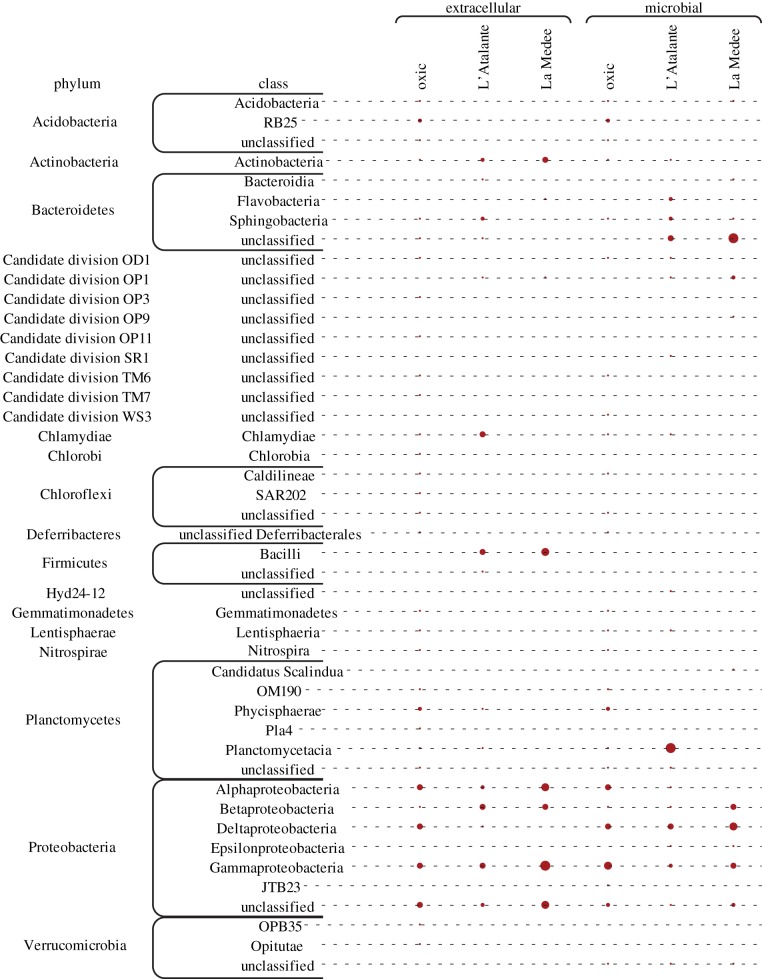
The rarefaction curves are shown in electronic supplementary material, figure S4. A large fraction of OTUs was unaffiliated with known prokaryotic taxa both in the oxic and DHAB sediments. The contribution of bacterial OTUs to the total prokaryotic OTU number was higher in the microbial (34–82%) than in the extracellular (17–31%) DNA pools both in oxic and DHAB sediments. In all benthic systems, Proteobacteria represented the dominant phylum both in the extracellular (from 34 to 56% in L'Atalante and La Medee sediments, respectively) and microbial DNA (from 45 to 68% in L'Atalante and La Medee sediments, respectively, figure 6). Additional results on other bacterial phyla are reported in the electronic supplementary material.
Figure 6.
Contribution of different bacterial classes to the total bacterial assemblages associated with extracellular DNA and microbial DNA in La Medee, L'Atalante and oxic sediments.
In all benthic systems investigated, the largest fraction of archaeal OTUs within extracellular DNA pool was affiliated with Euryarchaeota (58–96%), mainly belonging to unclassified Euryarchaeota (44–63%) and Thermoplasmata (8–36%). Marine Group I contributed for 2–21% to Crenarchaeota. Cluster analyses of bacterial OTU composition, at the class level (based on Bray–Curtis similarity), indicated a higher similarity between extracellular DNA pools of La Medee and L'Atalante sediments than between extracellular and microbial DNA pools recovered from the same environment (electronic supplementary material, figure S5a). Similar results were obtained also when randomly replicated subsamples of bacterial OTUs were considered (electronic supplementary material, figure S5b). Further analyses on the contribution of the different OTUs, at the genus level, within the dominant phylum Proteobacteria revealed clear differences between microbial and extracellular DNA pools in DHAB sediments (electronic supplementary material, table S2).
4. Discussion
Viral abundances in La Medee sediments were similar to the values previously reported for L'Atalante basin [16] and higher than values reported in the oxic sediments investigated here and in other benthic deep-sea ecosystems worldwide [17,49]. Viral abundances in La Medee sediments were ca three times higher than the prokaryotic abundances. The virus-to-prokaryote abundance ratio was double that in the oxic sediments and increased in the deeper sediment layers. Viral production rates (up to 10-times higher than in the oxic sediments) were among the highest values reported so far for marine sediments worldwide [17,49]. At the same time in hypersaline–anoxic sediments viral decay was lower and viral turnover was faster than in oxic sediments (on average, 0.7 per hour versus 0.2 per hour). These findings suggest that DHAB sediments favour a fast viral replication and a long preservation of the free viruses.
Virus-induced prokaryotic mortality in La Medee was similar to that reported for L'Atalante sediments [20]. Such values were much higher (up to more than one order of magnitude) than those found in the sediments collected at similar depths worldwide [17], including the oxic sediments investigated here. The high virus-induced prokaryotic mortality in DHAB sediments can be explained by the high metabolic activity (both autotrophic and heterotrophic) of prokaryotes inhabiting these ecosystems [9,20,50]. Indeed, the turnover rates of prokaryotes were much higher in La Medee than in oxic sediments.
The presence of eukaryotes potentially feeding on prokaryotes in anoxic marine systems has been reported [51], despite their abundance in the DHABs of the Eastern Mediterranean is extremely low (ca 20 cells per millilitre) [15]. This was also confirmed by microscopic observations, which we carried out on the sediments of La Medee (less than 10 cells per gram of sediment). Moreover, incubation experiments revealed that in the DHABs the release of DNA due to virus-induced prokaryotic mortality accounted for more than 85% of the total extracellular DNA produced in the sediments, whereas such a contribution was lower in the oxic sediments. Since almost the entire fraction of produced prokaryotes was removed by viral lysis (figure 3), all of these findings indicate that virus-induced prokaryotic mortality in DHAB sediments represents the dominant mechanism of top-down control of prokaryotic dynamics.
The sediments of La Medee were characterized by high extracellular DNA concentrations (much higher than those determined in the oxic sediments) and by high rates of extracellular DNA released by viral lysis (4.4 and 5.0 mg of extracellular DNA per square metre per day at Station 2 and Station 1 of the DHAB, respectively) as reported also for L'Atalante sediments [20]. Such rates largely exceed the fluxes of extracellular DNA from the water column to the sea floor reported for different oceanic regions, including the Mediterranean Sea, at the depths similar to those considered in our study (extracellular DNA fluxes less than 0.15 mg m−2 d−1) [52]. Since the inputs of organic particles from the upper oxygenated water layers to the DHAB sediments are strongly reduced by the thick layer of brines above the sea floor [2,3], the supply of extracellular DNA is expected to be even lower.
These findings suggest that in the DHAB sediments extracellular DNA dynamics are largely controlled by virus-induced prokaryotic mortality.
The extracellular DNA released by viral lysis can be an important trophic resource for benthic prokaryotic assemblages [21]. The main route for DNA removal in aquatic ecosystems is represented by degradation processes mediated by DNases [53]. Indeed, biologically driven processes (i.e. degradation activity mediated by DNases) have a more relevant role than chemical modification (e.g. depurination processes) in influencing extracellular DNA decay in marine sediments, independently from environmental characteristics, including anoxic conditions [25]. We found that DNase activities, even when normalized to cell abundance, were higher in the sediments of La Medee than in the oxic sediments outside the anoxic basin. Conversely, aminopeptidase, β-glucosidase and alkaline phosphatase activities (used as a proxy for potential mobilization of proteins, carbohydrates and organophosphate monoesters) [54] were up to one order of magnitude lower in La Medee than in oxic sediments. We cannot exclude the possibility that the measured rates might be influenced by sediment decompression [55,56]. However, we compared samples collected at similar depths so that the potential effect of the decompression on the measured enzymatic activities should be similar.
Overall, these findings suggest that, even though proteins and carbohydrates in DHAB sediments represent the main biochemical classes of organic compounds [16], extracellular DNA released from killed prokaryotes might be the most suitable trophic resource (for the high nitrogen and P content) for benthic prokaryotes.
Although DNase activities were higher in La Medee sediments than in oxic sediments, the fraction of extracellular DNA released by viral lysis, which escapes DNase degradation, accumulates in the DHAB to a larger extent than outside (on average 4.4 versus 0.8 mg of extracellular DNA per square metre per day). Such a higher accumulation in DHAB sediments is consistent with the higher extracellular DNA concentrations observed in subsurface sediment layers. These results suggest that, in DHAB sediments, the accumulation of extracellular DNA can be not only due to the preferential preservation favoured by the permanently anoxic conditions (as previously reported) [23,25,26,28,57] but also by the high virus-mediated release rates of extracellular DNA, which largely exceed its removal rates (on average four times).
Extracellular DNA accumulated in the sediments can contain present and past gene sequences [23,25–28]. In the DHAB sediments, the extracellular DNA pool was characterized by a higher diversity of 16S rDNA sequences than the microbial DNA. Indeed, the rarefaction curves resulting from the analyses of the microbial DNA pool were closer to saturation than those obtained from extracellular DNA, despite the lower number of sequences obtained from microbial DNA (electronic supplementary material, figure S4). Such a low bacterial diversity associated with the microbial DNA pool is consistent with the results of clone library analyses previously obtained from the sediments of La Medee [58] and brines of other DHABs, possibly as a result of the extreme environmental conditions [5,50].
Both in La Medee and L'Atalante sediments, the analyses of the bacterial 16S rDNA sequences, at the class level, revealed that the taxonomic composition of extracellular DNA pool was different from that of the microbial DNA even when the same number of sequences per sample (based on random re-sampling) was considered (electronic supplementary material, figure S5a,b). Although further investigations based on a deeper sequencing effort are needed, the results of this study suggest that the genetic diversity associated with microbial biomass only partially overlaps with that contained within the extracellular DNA pool, expanding previous findings obtained through molecular fingerprinting analyses [25,59].
The genetic imprint of extracellular DNA represents a record of processes occurring in the pelagic (through the DNA supply attached to sinking particles) and benthic (through in situ extracellular DNA release) domains on different temporal scales [22,23,52].
In this regard, the higher diversity of prokaryotic gene sequences contained in extracellular than in microbial DNA, together with the low similarity between the genetic composition of the two pools (within the same DHAB), suggests that extracellular DNA can preserve sequences belonging to prokaryotes no longer present in the sediments of these systems.
At the same time, the high similarity between bacterial OTU compositions in the extracellular DNA pools in La Medee and L'Atalante sediments suggests a common origin of the bacterial genes released by viral lysis and similar preservation mechanisms. However, although the extracellular DNA inputs from the upper brines to the DHAB sediments is expected to be very low, the preservation of genes also of allochthonous origin cannot be completely ruled out.
Overall, our results suggest that the strong viral impact in DHAB sediments can largely control the dynamics of the extracellular DNA, by supplying this pool with prokaryotic gene sequences, which represent the signatures of present and past infection events.
Supplementary Material
Supplementary Material
Acknowledgements
We would like to thank A. Vezzi for his useful suggestions on the bioinformatic analyses.
Data accessibility
All partial SSU rDNA sequences obtained in this study have been deposited in the NCBI Short Read Archive (SRA). Experiment accession nos. SRX218129, SRX218716, SRX218717, SRX218718, SRX218719, SRX218720. Run accession nos. SRR651042, SRR651099, SRR651100, SRR651101, SRR651102, SRR651103. The data used in this study are available as electronic supplementary material.
Funding statement
This work has been financially supported by the National Projects EXPLODIVE (FIRB 2008, contract no. I31J10000060001) and RITMARE coordinated by the Italian National Research Council (CNR) and funded by the Italian Ministry of Education, University and Research within the National Research Program 2011–2013. This study has been conducted during the field activities of the European project ESF Eurocore, BIOFUN (G067007N).
References
- 1.Van der Wielen PWJJ, et al. 2005. The enigma of prokaryotic life in deep hypersaline anoxic basins. Science 307, 121–123. ( 10.1126/science.1103569) [DOI] [PubMed] [Google Scholar]
- 2.De Lange GJ, et al. 1990. Composition of anoxic hypersaline brines in the Tyro and Bannock basins, Eastern Mediterranean. Mar. Chem. 31, 63–88. ( 10.1016/0304-4203(90)90031-7) [DOI] [Google Scholar]
- 3.Sass AM, Sass H, Coolen MJL, Cypionka H, Overmann J. 2001. Microbial communities in the chemocline of a hypersaline deep-sea basin (Urania Basin, Mediterranean Sea). Appl. Environ. Microbiol. 67, 5392–5402. ( 10.1128/AEM.67.12.5392-5402.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borin S, et al. 2009. Sulfur cycling and methanogenesis primarily drive microbial colonization of the highly sulfidic Urania deep hypersaline basin. Proc. Natl Acad. Sci. USA 106, 9151–9156. ( 10.1073/pnas.0811984106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daffonchio D, et al. 2006. Stratified prokaryote network in the oxic–anoxic transition of a deep-sea halocline. Nature 440, 203–207. ( 10.1038/nature04418) [DOI] [PubMed] [Google Scholar]
- 6.Eder W, Jahnke L, Schmidt M, Huber R. 2001. Microbial diversity of the brine–seawater interface of the Kebrit Deep, Red Sea, studied via 16S rRNA gene sequences and cultivation methods. Appl. Environ. Microbiol. 67, 3077–3085. ( 10.1128/AEM.67.7.3077-3085.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antunes A, Alam I, El Dorry H, Siam R, Robertson A, Bajic VB, Sting U. 2011. Genome sequence of Haloplasma contractile, an unusual contractile bacterium from a deep-sea anoxic brine. J. Bacteriol. 193, 4551–4552. ( 10.1128/JB.05461-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrer M, et al. 2005. Microbial enzymes mined from the Urania deep-sea hypersaline anoxic basin. Chem. Biol. 12, 895–904. ( 10.1016/j.chembiol.2005.05.020) [DOI] [PubMed] [Google Scholar]
- 9.Yakimov MM, La Cono V, Denaro R, Auria G, Decembrini F, Timmis KN, Golyshin PN, Giuliano L. 2007. Primary producing prokaryotic communities of brine, interface and seawater above the halocline of deep anoxic lake L'Atalante, Eastern Mediterranean Sea. ISME J. 1, 743–755. ( 10.1038/ismej.2007.83) [DOI] [PubMed] [Google Scholar]
- 10.Danovaro R, Dell'Anno A, Pusceddu A, Gambi C, Heiner I, Kristensen MR. 2010. The first metazoa living in permanently anoxic conditions. BMC Biol. 8, 1–10. ( 10.1186/1741-7007-8-30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mancinelli RL, Fahlen TF, Landheim R, Klovstad MR. 2004. Brines and evaporites: analogs for Martian life. Adv. Space Res. 33, 1244–1246. ( 10.1016/j.asr.2003.08.034) [DOI] [Google Scholar]
- 12.Rothschild LJ, Mancinelli RL. 2001. Life in extreme environments. Nature 409, 1092–1101. ( 10.1038/35059215) [DOI] [PubMed] [Google Scholar]
- 13.Danovaro R, Corinaldesi C, Dell'Anno A, Fuhrman JA, Middelburg JJ, Noble RT, Suttle CA. 2011. Marine viruses and global climate change. FEMS Microbiol. Rev. 35, 993–1034. ( 10.1111/j.1574-6976.2010.00258.x) [DOI] [PubMed] [Google Scholar]
- 14.Suttle CA. 2005. Viruses in the sea. Nature 437, 356–361. ( 10.1038/nature04160) [DOI] [PubMed] [Google Scholar]
- 15.Orsi W, Charvet S, Vd'ačný P, Bernhard JM, Edgcomb VP. 2012. Prevalence of partnerships between bacteria and ciliates in oxygen-depleted marine water columns. Front. Microbiol. 3, 314 ( 10.3389/fmicb.2012.00341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danovaro R, Corinaldesi C, Dell'Anno A, Fabiano M, Corselli C. 2005. Viruses, prokaryotes and DNA in the sediments of a deep-hypersaline anoxic basin (DHAB) of the Mediterranean Sea. Environ. Microbiol. 7, 586–592. ( 10.1111/j.1462-2920.2005.00727.x) [DOI] [PubMed] [Google Scholar]
- 17.Danovaro R, et al. 2008. Major viral impact on the functioning of benthic deep-sea ecosystems. Nature 454, 1084–1087. ( 10.1038/nature07268) [DOI] [PubMed] [Google Scholar]
- 18.Fuhrman JA. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548. ( 10.1038/21119) [DOI] [PubMed] [Google Scholar]
- 19.Middelboe M, Jørgensen NOG. 2006. Viral lysis of bacteria: an important source of dissolved aminoacids and cell wall compounds. J. Mar. Biol. Assoc. UK 86, 605–612. ( 10.1017/S0025315406013518) [DOI] [Google Scholar]
- 20.Corinaldesi C, Dell'Anno A, Danovaro R. 2007a. Viral infection plays a key role in extracellular DNA dynamics in marine anoxic systems. Limnol. Oceanogr. 52, 508–516. ( 10.4319/lo.2007.52.2.0508) [DOI] [Google Scholar]
- 21.Corinaldesi C, Dell'Anno A, Danovaro R. 2007b. Early diagenesis and trophic role of extracellular DNA in different benthic ecosystems. Limnol. Oceanogr. 52, 1710–1717. ( 10.4319/lo.2007.52.4.1710) [DOI] [Google Scholar]
- 22.Dell'Anno A, Corinaldesi C. 2004. Degradation and turnover of extracellular DNA in marine sediments: ecological and methodological considerations. Appl. Environ. Microbiol. 70, 4384–4386. ( 10.1128/AEM.70.7.4384-4386.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corinaldesi C, Barucca M, Luna GM, Dell'Anno A. 2011. Preservation, origin and genetic imprint of extracellular DNA in permanently anoxic deep-sea sediments. Mol. Ecol. 20, 642–654. ( 10.1111/j.1365-294X.2010.04958.x) [DOI] [PubMed] [Google Scholar]
- 24.Dell'Anno A, Bompadre S, Danovaro R. 2002. Quantification, base composition, and fate of extracellular DNA in marine sediments. Limnol. Oceanogr. 47, 899–905. ( 10.4319/lo.2002.47.3.0899) [DOI] [Google Scholar]
- 25.Corinaldesi C, Beolchini F, Dell'Anno A. 2008. Damage and degradation rates of extracellular DNA in marine sediments: implications for the preservation of gene sequences. Mol. Ecol. 17, 3939–3951. ( 10.1111/j.1365-294X.2008.03880.x) [DOI] [PubMed] [Google Scholar]
- 26.Coolen MJL, Overmann J. 2007. 217 000-year-old DNA sequences of green sulfur bacteria in Mediterranean sapropels and their implications for the reconstruction of the paleoenvironment. Environ. Microbiol. 9, 238–249. ( 10.1111/j.1462-2920.2006.01134.x) [DOI] [PubMed] [Google Scholar]
- 27.Coolen MJL, Volkman JK, Abbas B, Muyzer G, Schouten S, Sinninghe Damsté JS. 2007. Identification of organic matter sources in sulfidic Late Holocene Antarctic fjord sediments from fossil rDNA sequence analysis. Paleoceanography 22, PA1005, 17PP ( 10.1029/2006PA001309) [DOI] [Google Scholar]
- 28.Coolen MJL, Orsi WD, Balkema C, Quince C, Harris K, Sylva SP, Filipova-Marinova M, Giosan L. 2013. Evolution of the plankton paleome in the Black Sea from the Deglacial to Anthropocene. Proc Natl Acad. Sci. USA 110, 8609–8614. ( 10.1073/pnas.1219283110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chamot-Rooke N, Rabaute A, Kreemer C. 2005. Western Mediterranean Ridge mud belt correlates with active shear strain at the prism-backstop geological contact. Geology 33, 861–864. ( 10.1130/G21469.1) [DOI] [Google Scholar]
- 30.Wallmann K, Suess E, Westbrook GH, Winckler G, Cita MB. 1997. Salty brines on the Mediterranean sea floor. Nature 387, 31–32. ( 10.1038/387031a0) [DOI] [Google Scholar]
- 31.Van der Wielen PWJJ, Heijs SK. 2007. Sulfate-reducing prokaryotic communities in two deep hypersaline anoxic basins in the Eastern Mediterranean deep sea. Environ. Microbiol. 9, 1335–1340. ( 10.1111/j.1462-2920.2006.01210.x) [DOI] [PubMed] [Google Scholar]
- 32.Danovaro R, Dell'Anno A, Trucco A, Vannucci S. 2001. Determination of virus abundance in marine sediments. Appl. Environ. Microbiol. 67, 1384–1387. ( 10.1128/AEM.67.3.1384-1387.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dell'Anno A, Corinaldesi C, Magagnini M, Danovaro R. 2009. Determination of viral production in aquatic sediments using the dilution-based approach. Nat. Protoc. 4, 1013–1022. ( 10.1038/nprot.2009.82) [DOI] [PubMed] [Google Scholar]
- 34.Helton RR, Liu L, Wommack KE. 2006. Assessment of factors influencing direct enumeration of viruses within estuarine sediments. Appl. Environ. Microbiol. 72, 4767–4774. ( 10.1128/AEM.00297-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Danovaro R. 2010. Methods for the study of deep-sea sediments, their functioning and biodiversity. Boca Raton, FL: CRC Press. [Google Scholar]
- 36.Corinaldesi C, Dell'Anno A, Magagnini M, Danovaro R. 2010. Viral decay and viral production rates in continental-shelf and deep-sea sediments of the Mediterranean Sea. FEMS Microbiol. Ecol. 72, 208–218. ( 10.1111/j.1574-6941.2010.00840.x) [DOI] [PubMed] [Google Scholar]
- 37.Noble RT, Fuhrman JA. 1997. Virus decay and its causes in coastal waters. Appl. Environ. Microbiol. 63, 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corinaldesi C, Danovaro R, Dell'Anno A. 2005. Simultaneous recovery of extracellular and intracellular DNA suitable for molecular studies from marine sediments. Appl. Environ. Microbiol. 71, 46–50. ( 10.1128/AEM.71.1.46-50.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steward GF, Montiel JL, Azam F. 2000. Genome size distributions indicate variability and similarities among marine viral assemblages from diverse environments. Limnol. Oceanogr. 45, 1697–1706. ( 10.4319/lo.2000.45.8.1697) [DOI] [Google Scholar]
- 40.Wikner J, Vallino JJ, Steward GF, Smith DC, Azam F. 1993. Nucleic acids from the host bacterium as a major source of nucleotides for three marine bacteriophages. FEMS Microbiol. Ecol. 12, 237–248. ( 10.1111/j.1574-6941.1993.tb00036.x) [DOI] [Google Scholar]
- 41.Paul JH, Sullivan MB. 2005. Marine phage genomics: what have we learned? Curr. Opin. Biotech. 16, 299–307. ( 10.1016/j.copbio.2005.03.007) [DOI] [PubMed] [Google Scholar]
- 42.Cazenave C, Toulme` JJ, Helene C. 1983. Binding of recA protein to single-stranded nucleic acids: spectroscopic studies using fluorescent polynucleotides. EMBO J. 2, 2247–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Qian PY. 2009. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE 4, e7401 ( 10.1371/journal.pone.0007401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao H, Auguet JC, Gu JD. 2013. Global ecological pattern of ammonia-oxidizing Archaea. PLoS ONE 8, e52853 ( 10.1371/journal.pone.0052853) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schloss PD, et al. 2009. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. ( 10.1128/AEM.01541-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quast C, Pruesse E, Yilmaz P, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. ( 10.1093/nar/gks1219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Campbell BJ, Yu L, Heidelberg JF, Kirchman DL. 2011. Activity of abundant and rare bacteria in a coastal ocean. Proc. Natl Acad. Sci. USA 108, 12 776–12 781. ( 10.1073/pnas.1101405108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zinger L, et al. 2011. Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS ONE 6, e24570 ( 10.1371/journal.pone.0024570) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danovaro R, Corinaldesi C, Filippini M, Fischer UR, Gessner MO, Jacquet S, Magagnini M, Velimirov B. 2008. Viriobenthos in freshwater and marine sediments: a review. Freshwater Biol. 53, 1186–1213. ( 10.1111/j.1365-2427.2008.01961.x) [DOI] [Google Scholar]
- 50.La Cono V, et al. 2011. Unveiling microbial life in new deep-sea hypersaline Lake Thetis. Part I: prokaryotes and environmental settings. Environ. Microbiol. 13, 2250–2268. ( 10.1111/j.1462-2920.2011.02478.x) [DOI] [PubMed] [Google Scholar]
- 51.Edgcomb V, Orsi W, Leslin C, Epstein SS, Bunge J, Jeon S, Yakimov MM, Behnke A, Stoeck T. 2009. Protistan community patterns within the brine and halocline of deep hypersaline anoxic basins in the eastern Mediterranean Sea. Extremophiles 1, 151–167. ( 10.1007/s00792-008-0206-2) [DOI] [PubMed] [Google Scholar]
- 52.Dell'Anno A, Danovaro R. 2005. Extracellular DNA play a key role in deep-sea ecosystem functioning. Science 309, 2179 ( 10.1126/science.1117475) [DOI] [PubMed] [Google Scholar]
- 53.Lorenz MG, Wackernagel W. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58, 563–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luna GM, Bianchelli S, Decembrini F, De Domenico E, Danovaro R, Dell'Anno A. 2012. The dark portion of the Mediterranean Sea is a bioreactor of organic matter cycling. Global Biogeochem. Cycles 26, GB2017 ( 10.1029/2011GB004168) [DOI] [Google Scholar]
- 55.Poremba K. 1995. Hydrolytic enzymatic activity in deep-sea sediments. FEMS Microbiol. Ecol. 16, 213–222. ( 10.1016/0168-6496(94)00085-B) [DOI] [Google Scholar]
- 56.Tamburini C, Boutrif M, Garel M, Colwell RR, Deming JW. 2013. Prokaryotic responses to hydrostatic pressure in the ocean—a review. Environ. Microbiol. 15, 1262–1274. ( 10.1111/1462-2920.12084) [DOI] [PubMed] [Google Scholar]
- 57.Coolen MJL, Cypionka A, Sass M, Sass H, Overmann J. 2002. Ongoing modification of Mediterranean Pleistocene sapropels mediated by prokaryotes. Science 296, 2401–2410. ( 10.1126/science.1071893) [DOI] [PubMed] [Google Scholar]
- 58.Akoumianaki I, Nomaki H, Pachiadaki M, Kormas KA, Kitazato H, Tokuyama H. 2012. Low bacterial diversity and high labile organic matter concentrations in the sediments of the Medee deep-sea hypersaline anoxic basin. Microbes Environ. 25, 504–508. ( 10.1264/jsme2.ME12045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agnelli A, Ascher J, Corti G, Ceccherini MT, Nannipieri P, Pietramellara G. 2004. Distribution of microbial communities in a forest soil profile investigated by microbial biomass, soil respiration and DGGE of total and extracellular DNA. Soil Biol. Biochem. 36, 859–868. ( 10.1016/j.soilbio.2004.02.004) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All partial SSU rDNA sequences obtained in this study have been deposited in the NCBI Short Read Archive (SRA). Experiment accession nos. SRX218129, SRX218716, SRX218717, SRX218718, SRX218719, SRX218720. Run accession nos. SRR651042, SRR651099, SRR651100, SRR651101, SRR651102, SRR651103. The data used in this study are available as electronic supplementary material.