Abstract
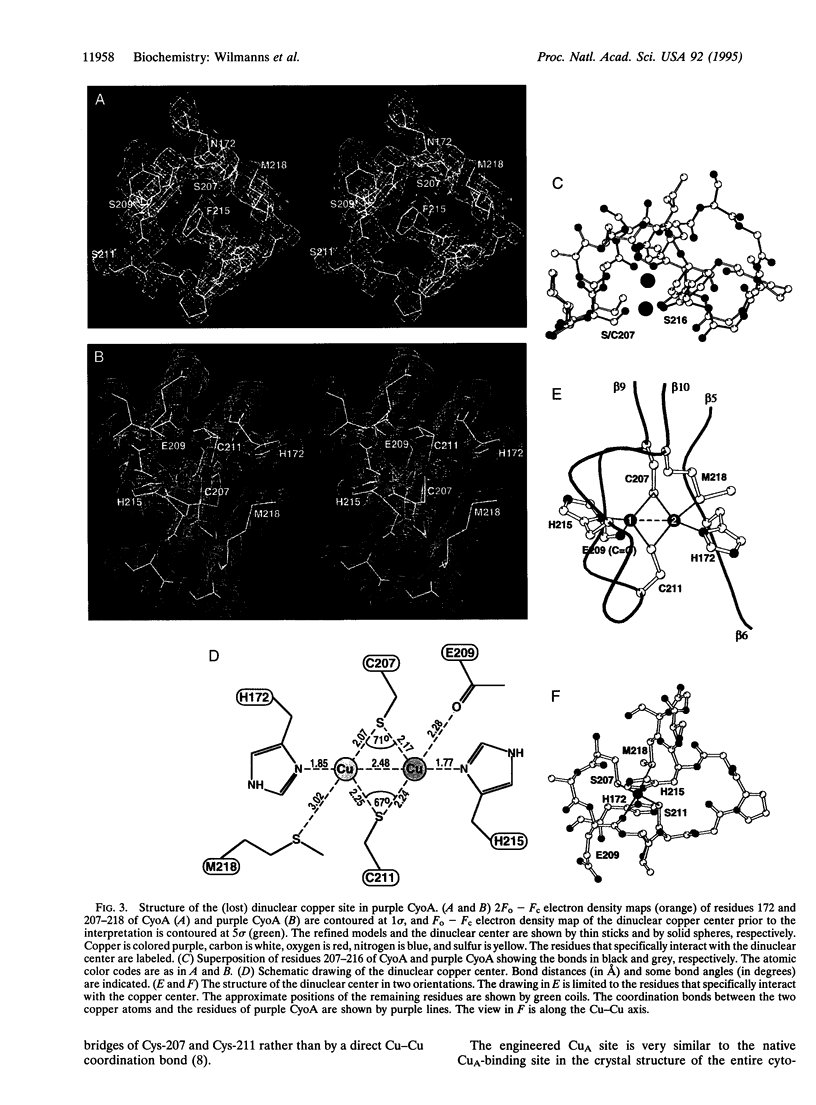
Cytochrome oxidase is a membrane protein complex that catalyzes reduction of molecular oxygen to water and utilizes the free energy of this reaction to generate a transmembrane proton gradient during respiration. The electron entry site in subunit II is a mixed-valence dinuclear copper center in enzymes that oxidize cytochrome c. This center has been lost during the evolution of the quinoloxidizing branch of cytochrome oxidases but can be restored by engineering. Herein we describe the crystal structures of the periplasmic fragment from the wild-type subunit II (CyoA) of Escherichia coli quinol oxidase at 2.5-A resolution and of the mutant with the engineered dinuclear copper center (purple CyoA) at 2.3-A resolution. CyoA is folded as an 11-stranded mostly antiparallel beta-sandwich followed by three alpha-helices. The dinuclear copper center is located at the loops between strands beta 5-beta 6 and beta 9-beta 10. The two coppers are at a 2.5-A distance and symmetrically coordinated to the main ligands that are two bridging cysteines and two terminal histidines. The residues that are distinct in cytochrome c and quinol oxidases are around the dinuclear copper center. Structural comparison suggests a common ancestry for subunit II of cytochrome oxidase and blue copper-binding proteins.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adman E. T. Copper protein structures. Adv Protein Chem. 1991;42:145–197. doi: 10.1016/s0065-3233(08)60536-7. [DOI] [PubMed] [Google Scholar]
- Antholine W. E., Kastrau D. H., Steffens G. C., Buse G., Zumft W. G., Kroneck P. M. A comparative EPR investigation of the multicopper proteins nitrous-oxide reductase and cytochrome c oxidase. Eur J Biochem. 1992 Nov 1;209(3):875–881. doi: 10.1111/j.1432-1033.1992.tb17360.x. [DOI] [PubMed] [Google Scholar]
- Babcock G. T., Wikström M. Oxygen activation and the conservation of energy in cell respiration. Nature. 1992 Mar 26;356(6367):301–309. doi: 10.1038/356301a0. [DOI] [PubMed] [Google Scholar]
- Blackburn N. J., Barr M. E., Woodruff W. H., van der Oost J., de Vries S. Metal-metal bonding in biology: EXAFS evidence for a 2.5 A copper-copper bond in the CuA center of cytochrome oxidase. Biochemistry. 1994 Aug 30;33(34):10401–10407. doi: 10.1021/bi00200a022. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Castresana J., Lübben M., Saraste M., Higgins D. G. Evolution of cytochrome oxidase, an enzyme older than atmospheric oxygen. EMBO J. 1994 Jun 1;13(11):2516–2525. doi: 10.1002/j.1460-2075.1994.tb06541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar J. A., Lappalainen P., Zumft W. G., Saraste M., Thomson A. J. Spectroscopic and mutagenesis studies on the CuA centre from the cytochrome-c oxidase complex of Paracoccus denitrificans. Eur J Biochem. 1995 Aug 15;232(1):294–303. doi: 10.1111/j.1432-1033.1995.tb20811.x. [DOI] [PubMed] [Google Scholar]
- García-Horsman J. A., Barquera B., Rumbley J., Ma J., Gennis R. B. The superfamily of heme-copper respiratory oxidases. J Bacteriol. 1994 Sep;176(18):5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill B. C. Modeling the sequence of electron transfer reactions in the single turnover of reduced, mammalian cytochrome c oxidase with oxygen. J Biol Chem. 1994 Jan 28;269(4):2419–2425. [PubMed] [Google Scholar]
- Iwata S., Ostermeier C., Ludwig B., Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995 Aug 24;376(6542):660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kelly M., Lappalainen P., Talbo G., Haltia T., van der Oost J., Saraste M. Two cysteines, two histidines, and one methionine are ligands of a binuclear purple copper center. J Biol Chem. 1993 Aug 5;268(22):16781–16787. [PubMed] [Google Scholar]
- Kroneck P. M., Antholine W. A., Riester J., Zumft W. G. The cupric site in nitrous oxide reductase contains a mixed-valence [Cu(II),Cu(I)] binuclear center: a multifrequency electron paramagnetic resonance investigation. FEBS Lett. 1988 Dec 19;242(1):70–74. doi: 10.1016/0014-5793(88)80987-6. [DOI] [PubMed] [Google Scholar]
- Lamzin V. S., Wilson K. S. Automated refinement of protein models. Acta Crystallogr D Biol Crystallogr. 1993 Jan 1;49(Pt 1):129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- Lappalainen P., Watmough N. J., Greenwood C., Saraste M. Electron transfer between cytochrome c and the isolated CuA domain: identification of substrate-binding residues in cytochrome c oxidase. Biochemistry. 1995 May 2;34(17):5824–5830. doi: 10.1021/bi00017a014. [DOI] [PubMed] [Google Scholar]
- Malmström B. G., Aasa R. The nature of the CuA center in cytochrome c oxidase. FEBS Lett. 1993 Jun 28;325(1-2):49–52. doi: 10.1016/0014-5793(93)81411-r. [DOI] [PubMed] [Google Scholar]
- Rydén L. G., Hunt L. T. Evolution of protein complexity: the blue copper-containing oxidases and related proteins. J Mol Evol. 1993 Jan;36(1):41–66. doi: 10.1007/BF02407305. [DOI] [PubMed] [Google Scholar]
- Saraste M. Structural features of cytochrome oxidase. Q Rev Biophys. 1990 Nov;23(4):331–366. doi: 10.1017/s0033583500005588. [DOI] [PubMed] [Google Scholar]
- Steinrücke P., Steffens G. C., Panskus G., Buse G., Ludwig B. Subunit II of cytochrome c oxidase from Paracoccus denitrificans. DNA sequence, gene expression and the protein. Eur J Biochem. 1987 Sep 15;167(3):431–439. doi: 10.1111/j.1432-1033.1987.tb13356.x. [DOI] [PubMed] [Google Scholar]
- van der Oost J., Lappalainen P., Musacchio A., Warne A., Lemieux L., Rumbley J., Gennis R. B., Aasa R., Pascher T., Malmström B. G. Restoration of a lost metal-binding site: construction of two different copper sites into a subunit of the E. coli cytochrome o quinol oxidase complex. EMBO J. 1992 Sep;11(9):3209–3217. doi: 10.1002/j.1460-2075.1992.tb05398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J., Musacchio A., Pauptit R. A., Ceska T. A., Wierenga R. K., Saraste M. Crystallization and preliminary X-ray analysis of the periplasmic fragment of CyoA-a subunit of the Escherichia coli cytochrome o complex. J Mol Biol. 1993 Feb 5;229(3):794–796. doi: 10.1006/jmbi.1993.1083. [DOI] [PubMed] [Google Scholar]