Abstract
New peptide molecules with metal binding abilities proved to be active against multidrug resistant clinical isolates. One of them labeled with a dansylated lysine has been imaged inside single-multidrug resistant bacteria cells by deep ultraviolet fluorescence, showing a heterogeneous subcellular localization. The fluorescence intensity is clearly related to the accumulation of the drug inside the bacteria, being dependent both on its concentration and on the incubation time with cells.
Keywords: Multidrug resistant, amphipatic peptide, metal binding group, microspectrofluorimetry, fluorescence imaging
The dissemination of multidrug resistant (MDR) Gram-negative bacteria drastically impairs the efficacy of antibiotic families and limits their clinical uses.1,2 Actually, it is urgently needed to develop new classes of antimicrobial agents to tackle drug resistance in bacteria. Mechanisms associated with modifications of membrane permeability, decreasing passive uptake (influx) or increasing active efflux of antibiotics (efflux), are reported as key contributors of the bacterial MDR phenotype.3,4 Moreover, today, a major concern is associated with the lack of connection between the intracellular location of a molecule and its antibacterial activity.4 This defect tragically impairs our understanding of antibiotic accumulation inside the resistant bacterial cell and the possible way to combat them.
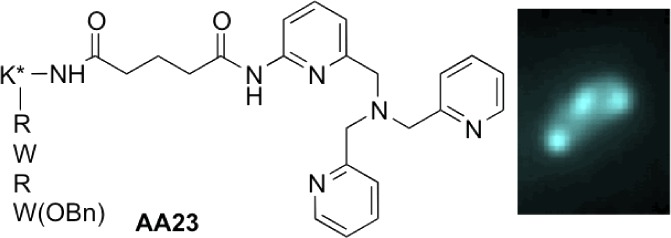
Herein, we described the activity of a new class of antibacterial agents that exhibit a noticeable activity against resistant bacterial strains. They are dual compounds based on the covalent binding between a short antimicrobial peptide analogue, to enhance the permeability, and a metal chelating group to search for a new activity. They are active against E. aerogenes, EA289, a MDR clinical isolate, which overexpresses efflux pumps5 and made this strain insensitive to classical antibiotics such as ciprofloxacin. One of these molecules (AA23) presents the original property to be strongly fluorescent after deep ultraviolet (DUV) excitation. Since resolution is directly proportional to the wavelength (Rayleigh criteria), we used this characteristic to determine its intracellular accumulation inside individual MDR EA289 bacteria.
The lead molecule AA08 is a two-component compound, which contains a tris-(pyridylmethyl)amine metal binding group (TPA), [N((6-(amino)-2-pyridyl)methyl)N,N’-bis((2-pyridyl) methyl)amine] linked via a glutaryl spacer to a small tetrapeptide analogue of antimicrobial peptide (AMP), RWRWOBn, with a benzyl ester in C-terminal position.6 AMPs are cationic, amphipathic peptides that have a broad spectrum of activity associated with membrane permeabilization.7,8 Thus, they can facilitate entry of additional molecules from the exterior. To test the efficiency of the sequence, we prepared derivative AA37 functionalized by the tetrapeptide with the alternated sequence WRWROBn, and the hexapeptides AA36 and AA38 with the sequences RWRWRWOBn and WRWRWROBn, respectively. Then, to evaluate the efficacy of the metal binding group, we also synthesized the free N-acylRWRWOBn tetrapeptide, AA19, and a derivative closely related to AA08, AA24, in which the TPA moiety has been replaced by a tris(benzyl)amine (TBA) without any chelating ability. Finally, for the fluorescence imaging studies, we prepared AA23 and AA33, fluorescent analogues of AA08 and AA24, respectively, including in the peptide sequence (K*RWRWOBn), an additional lysine residue labeled on the ε-amino group with a dansyl moiety.
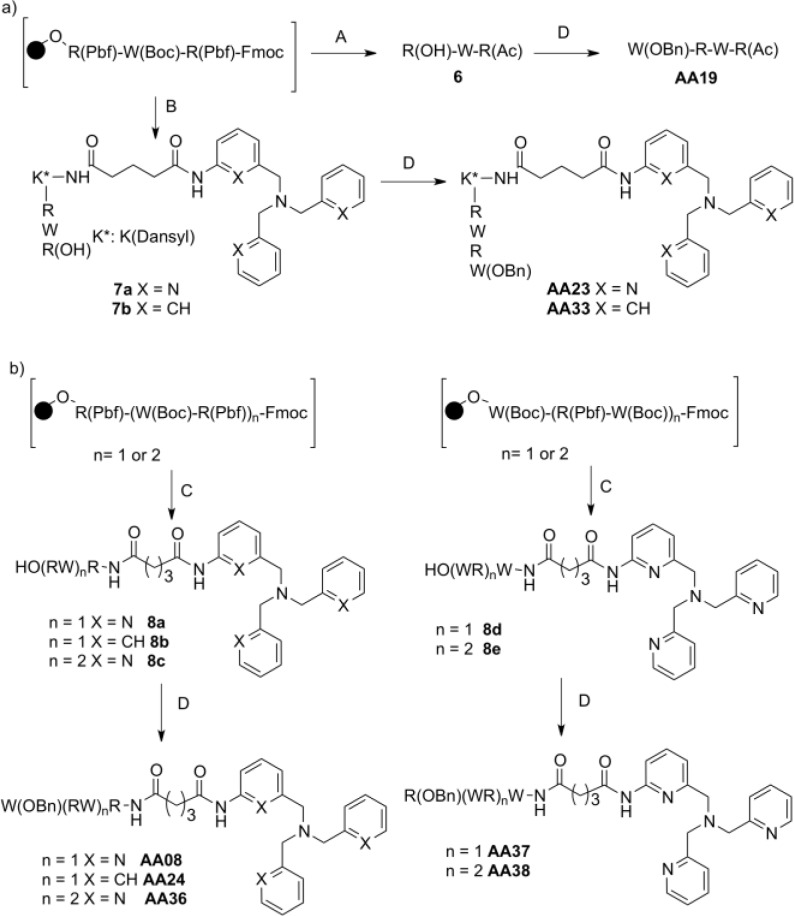
Major pathways for the synthesis of all the compounds are depicted in Scheme 1. N((6-(Amino-2-pyridyl)methyl)N,N’bis((2-pyridyl)methyl)amine (TPA-NH2) was prepared as described by Berreau et al.9 Condensation of dibenzylamine with 3-nitrobenzyl bromide, followed by reduction in EtOH with hydrazine hydrate in the presence of a catalytic amount of Pd/C afforded 3-((dibenzylamino)methyl)amine (TBA-NH2). The glutaryl spacer was introduced by reaction of TPA-NH2 or TBA-NH2 with methyl 5-chloro-5-oxopentanoate in THF, followed by saponification under mild conditions with LiOH in THF/H2O, yielding TPA-NHCO(CH2)3COOLi and TBA-NHCO(CH2)3COOLi (Schemes S1 and S2, Supporting Information). The peptide was assembled manually through solid phase peptide synthesis (SPPS) on a low load Wang resin preloaded with arginine or tryptophane (0.39 mmol·g–1) using Fmoc synthesis. Each deprotection of Fmoc groups was performed twice with 20% piperidine/DMF (v:v) and checked by UV–vis spectroscopy at 300 nm. Coupling was achieved in DMF with 3 equiv of the amino acids Fmoc-Trp(Boc)-OH, Fmoc-Arg(Pbf)-OH or Fmoc-Lys(Dansyl)-OH, and TPA-NHCO(CH2)3COOLi or TBA-NHCO(CH2)3COOLi. HOBt, and PyBOP or HBTU were used as activators in the presence of DIEA (aa/HOBt/(PyBOP or HBTU)/DIEA, 1:1:1:2). The precursor of peptide 6 was acylated with acetic anhydride in a mixture of pyridine and DMF. After cleavage of the final peptides from the resin, the side-chain protecting groups were removed by treatment with TFA/EDT/TIS/water (94/2.5/2/1.5, v:v:v:v). All the peptides in TFA solution were directly precipitated into diethyl ether. Then, after dissolution in water, they were lyophilized to afford the desired peptides 6, 7a–b, and 8a–e. The last residue Trp-COOBn or Arg-COOBn was then introduced as previously described,6 in DMF solution using HOBT and HBTU as activators in the presence of DIEA to afford AA08, AA19, AA23, AA24, AA33, AA36, AA37, and AA38. All the final compounds were thoroughly characterized by 1H NMR (Figure S1, Supporting Information), mass spectrometry, and elemental analysis, and their characteristics are described in the experimental procedures. Complete syntheses of all intermediates are detailed in the Supporting Information.
Scheme 1. Synthesis of Peptide Compounds.
(A) (i) piperidine/DMF (20/80%); (ii) DMF/pyridine/Ac2O (7/1/2, v:v:v); (iii) H2O/TIS/EDT/TFA (1.5:2:2.5:94, v:v:v:v). (B) (i) piperidine/DMF (20:80, %); (ii) Fmoc-Lys(Dansyl)-OH/PyBOP/HOBt/DIEA, DMF; (iii) piperidine/DMF (20/80, %); (iv) TPA-NHCO(CH2)3COOLi or TBA-NHCO(CH2)3COOLi/HBTU/HOBt/DIEA, DMF; (v) H2O/TIS/EDT/TFA (1.5:2:2.5:94, v:v:v:v). (C) (i) piperidine/DMF (20/80%); (ii) TPA-NHCO(CH2)3COOLi or TBA-NHCO(CH2)3COOLi/HBTU/HOBT/DIEA, DMF; (iii) H2O/TIS/EDT/TFA (1.5:2:2.5:94, v:v:v:v). (D) (i) piperidine/DMF, (20/80, %); (ii) Trp-CO2Bn or Arg-CO2Bn/HBTU/HOBt/DIEA, DMF.
The activity of the various molecules was evaluated using recommended bacteriological assays.10 While AA08 displayed some activity against wild type E. coli AG100, it showed a noticeable activity against E. aerogenes EA289, a MDR clinical isolate which has lost its sensitivity toward fleroxacin and ciprofloxacin. In contrast, the N-acyl tetrapeptide alone, AA19, as well as AA24, which possesses a tris(benzyl)amine without any chelating property, were inefficient against both AG100 and EA289. These results associated with the high binding affinity of divalent cations such as Zn(II), Cu(II), and Fe(II) for TPA11 support the hypothesis that the efficiency of AA08 is likely related to its ability either to trap cations essential to bacterial survival12 or to interact with a presently unknown target in bacteria. There is no marked difference of activity between TPA compounds containing a tetrapeptide or a hexapeptide. All derivatives presented similar activities on the tested strains (Table 1) being more active than actinonin and usual antibiotics such as ciprofloxacin on clinical MDR isolates. Moreover, at a difference of one dilution, compound AA37 with the alternate sequence WRWROBn is equipotent as AA08 toward EA289 and EA298. Introduction of a dansylated lysine in AA08 did not alter the activity, and AA23 was as active as AA08.
Table 1. Antibacterial Activity of AA Series and Antibiotics on Enterobacterial Strains.
| MIC
(μM) |
||||
|---|---|---|---|---|
|
E. colia |
E. aerogenesb |
|||
| AG100 | AG100A | EA289 | EA298 | |
| molecules | WT | AcrAB-strain | MDR isolate | TolC-strain |
| Actc | >128 | 2 | >128 | 32 |
| Cipd | 0.125 | 0.03 | 64 | 8 |
| Flee | 0.5 | 0.125 | >128 | 16 |
| AA08 | 32 | 8 | 16 | 8 |
| AA23 | 16 | 8 | 16 | 8 |
| AA19 | 64 | 16 | >128 | 128–64 |
| AA24 | 64 | 64 | 128–64 | 64 |
| AA36 | 8 | 8 | 16 | 8 |
| AA37 | 16 | 8 | 8 | 4 |
| AA38 | 8 | 8 | 16 | 8 |
Laboratory strains.
Resistant clinical isolates.
Actinonin.
Ciprofloxacin.
Fleroxacin.
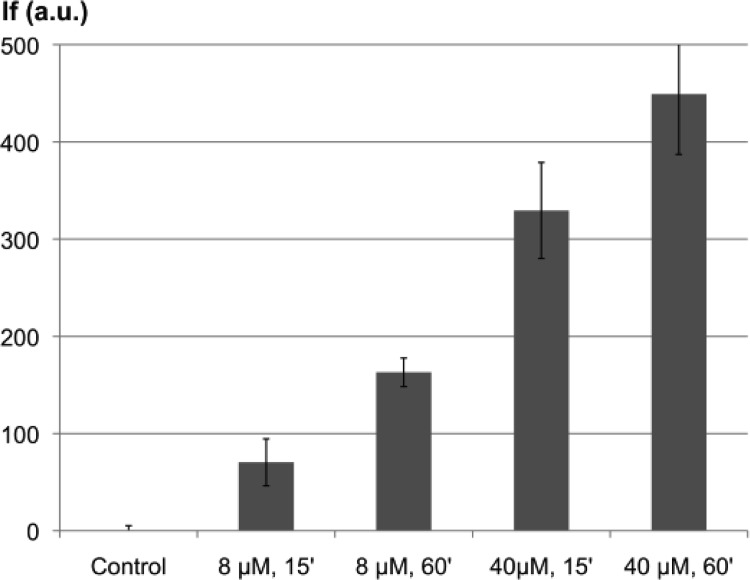
AA23 was further assayed for accumulation and location determinations inside resistant isolates. DUV microspectrofluorimetry spectra recorded with λex 280 nm from single bacteria treated or untreated with AA23 are shown in Figure S2 (Supporting Information). The emission peak of AA23 around 340 nm overlaps with the tryptophan fluorescence of bacterial proteins, while the peak centered at 510 nm exclusively corresponds to AA23 fluorescence. In these conditions, the resolution is around 180 nm,13 which enables to visualize the accumulation of the labeled compound in single bacteria cell with size in the range 0.5 to 5 μm (2 × 10–12 cm3). Thereby, for single-cell imaging, the fluorescence of AA23 was detected within a 480–550 nm wavelength range. The concentrations of AA23 ranged from 0.5 to 3 × MIC, and two incubation times of 15 and 60 min were selected. For each condition, two different determinations of localization with a minimum of 30 bacteria per plane were performed. Each bacterium (minimum size 4 pixels; maximum size 20 pixels) was individually counted after automatic recognition of bacteria patterns under ImageJ. Subcellular segregation of the compound inside individual bacteria is clearly shown in Figures 1 and S3 (Supporting Information) with biapical localization in most of the bacteria retaining the molecule and, for some, a central punctuation. Even though an absolute quantification of the molecule inside a bacterium is not yet possible, the fluorescence intensity clearly depends on the concentration of AA23 used in the incubation experiments, as well as on the incubation time, as shown in Figure 2.
Figure 1.
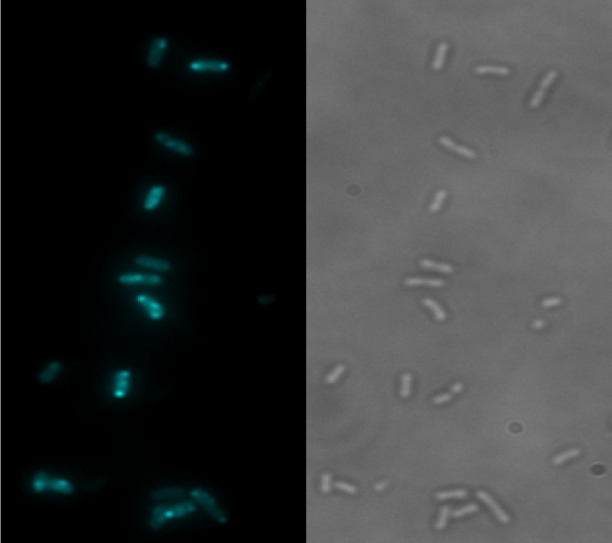
DUV fluorescence of AA23. Fluorescence (on the left) and transmission (on the right) images of EA289 bacteria after 30 min incubation with AA23 at 57.5 μM (λex 280 nm; λem 480–550 nm). The heterogeneous localization of the molecule is clearly seen between bacterial cells and inside one bacterium.
Figure 2.
Concentration and incubation time-dependent fluorescence intensity. This figure shows the fluorescence intensity of EA289 bacteria after incubation times of 15 and 60 min with AA23 at concentrations corresponding to 0.5 MIC (8 μM) and 2.5 × MIC (40 μM).
For the sake of comparison, we tried the same experiment of single-cell imaging by fluorescence with AA33, an inactive compound. Results are clearly different since we observed the formation of aggregates inside the bacteria (Figure S4, Supporting Information).
In summary, we have described synthesis and antibacterial properties of a new class of peptides with both metal complexing abilities and permeabilizing properties, which are active against multidrug resistant bacteria. For the first time, the biological activity and the location of a new antibacterial molecule are directly related using antibacterial susceptibility assays and DUV fluorescence spectroscopy. This combined approach enables to assess the relationships between the antibacterial properties of a new molecule with a fluorescent tag to its internalization. The in situ determination of drug accumulation gives new clues to dissect the membrane-associated mechanisms of resistance and to improve the diffusion profile of new antibacterial molecules.
Experimental Procedures
Chemicals
2-(1H-Benzotriazole-1-yl)-1,1,3,3-tetramethyl-uronium hexafluorophosphate (HBTU), N-hydroxybenzotriazole (HOBt), benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate (PyBOP), trifluoroacetic acid (TFA), triisopropylsilane (TIS), ethan-1,2-dithiol (EDT), anisole, N,N-diisopropylethylamine (DIEA), (N-α-Fmoc-N-γ-(2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl)-l-arginine (Fmoc-Arg(Pbf)-OH), N-α-Fmoc-N(in)-Boc-l-tryptophan, N-alpha-(9-fluorenylmethyloxycarbonyl)-N-in-tert-butyl-oxycarbonyl-l-tryptophan (Fmoc-Trp(Boc)-OH), N-α-Fmoc-N-ε-[5-(dimethylamino)naphthalene-1-sulfonyl]-l-lysine (Fmoc-Lys(Dansyl)-OH)), and tryptophan benzyl ester (Trp-OBn) were purchased from Aldrich.
All the syntheses are detailed in the Supporting Information.
Bacterial Strains, Growth and Susceptibility Determinations
E. aerogenes strains used: EA289 is a clinical multidrug resistant strain that overexpresses AcrAB-tolC efflux pumps, and EA298 is its tolC– derivative. E. coli AG100 is used as a control strain, and its derivative AG100A is deleted of AcrAB and hypersensitive to chloramphenicol, tetracycline, ampicillin, and nalidixic acid.14 Strains were routinely grown at 37 °C on Luria–Bertani (LB) agar or in LB broth, supplemented with kanamycine (50 μg·mL–1) for EA298 and AG100A. Minimal inhibitory concentrations (MICs) were determined with an inoculum of 2 × 105 cfu in 200 μL of Mueller Hinton (MH) broth containing 2-fold serial dilutions of each antibiotics or compounds.
Ciprofloxacin and fleroxacin were used as internal antibiotic reference. The results were scored after 18 h at 37 °C.
Compound Accumulation in Single Bacterial Cells
Bacteria growth and incubation have been previously described.10 Briefly, the bacterial suspension was centrifuged at 6000g for 15 min at 20 °C, and pellets were resuspended in 1/10 of the volume in a sodium phosphate buffer (50 mM) at pH 7 supplemented with MgCl2 (NaPi-MgCl2 buffer) to obtain a density of 1010 CFU·mL–1. Bacteria suspension (1.6 mL) was incubated for 15, 30, or 60 min at 37 °C (final volume 2 mL) with AA23 at different concentrations (8, 40, or 57.5 μM). Bacterial suspensions incubated without molecules were used as controls. Suspensions (800 or 400 μL) were then loaded on 1 M sucrose cushion (1100 or 550 μL, respectively) and centrifuged at 13 000 rpm for 5 min at 4 °C to eliminate extracellular-adsorbed compound and collect washed bacteria. To measure the compound uptake by EA289 and EA298 strains, we used the routine fluorimetric method previously described.10 Briefly, pellets corresponding to 800 μL of suspension were solubilized with 500 μL of 0.1 M Glycin-HCl pH 3 buffer at least 2 h at room temperature. After a centrifugation for 10 min at 13 000 rpm, 400 μL of lysates was diluted in 600 μL of 0.1 M Glycin-HCl pH 3 buffer and analyzed by spectrofluorimetry. To detect the antibiotic fluorescence from single bacteria, pellets corresponding to 400 μL of suspension were resuspended in 200 μL of NaPi-MgCl2 buffer and analyzed by DUV microspectrofluorimetry or DUV fluorescence imaging as described in Jamme et al.13
Acknowledgments
ANR-2010 BLAN METABACT, CNRS (France), Paris Descartes and Aix-Marseille Universities, IRBA, and Synchrotron Soleil are gratefully acknowledged.
Supporting Information Available
All the syntheses of peptides combined to a TPA moiety are described. 1H NMR spectra of AA08, AA19, AA23, AA24, AA33, AA36, AA37, and AA38 are shown in Figure S1. Figure S2 shows fluorescence emission spectra after excitation at λ = 280 nm recorded on a film of EA289 single bacterial cells untreated and treated with AA23. Figures S3 and S4 show DUV fluorescence imaging of single EA289 bacterial cells treated or not with AA23 and AA33, respectively (λex = 280 nm; 480 ≤ λem ≤ 550 nm). This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The three groups contributed equally to this work.
S.K., and L.M. and A.A. received postdoctoral fellowships from Soleil and ANR METABACT, respectively.
The authors declare no competing financial interest.
Supplementary Material
References
- Chopra I.; Schofield C.; Everett M.; O’Neill A.; Miller K.; Wilcox M.; Frère J.-M.; Dawson M.; Czaplewski L.; Urleb U.; Courvalin P. Treatment of health-care-associated infections caused by Gram-negative bacteria: a consensus statement. Lancet Infect. Dis. 2008, 133–139. [DOI] [PubMed] [Google Scholar]
- Gandhi T. N.; DePestel D. D.; Collins C. D.; Nagel J.; Washer L. L. Managing antimicrobial resistance in intensive care units. Crit. Care Med. 2010, 38, S315–23. [DOI] [PubMed] [Google Scholar]
- Pages J.-M.; James C. E.; Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 2008, 6, 893–903. [DOI] [PubMed] [Google Scholar]
- Nikaido H.; Pages J.-M. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 2011, 36, 340–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E.; Pages J.-M. The AcrAB-TolC efflux pump contributes to multidrug resistance in the nosocomial pathogen Enterobacter aerogenes. Antimicrob. Agents Chemother. 2002, 46, 2640–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strøm M. B.; Haug B. E.; Skar M. L.; Stensen W.; Stiberg T.; Svendsen J. S. The pharmacophore of short cationic antibacterial peptides. J. Med. Chem. 2003, 46, 1567–1570. [DOI] [PubMed] [Google Scholar]
- Findlay B.; Zhanel G. G.; Schweizer F. Cationic amphiphiles, a new generation of antimicrobials inspired by the natural antimicrobial peptide scaffold. Antimicrob. Agents Chemother. 2010, 54, 4049–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimley W. C. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 2010, 5, 905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajna E.; Makowska-Grzyska M. M.; Wasden C. C.; Arif A. M.; Berreau L. M. A deprotonated intermediate in the amide methanolysis reaction of an N 4O-ligated mononuclear zinc complex. Inorg. Chem. 2005, 44, 7595–7605. [DOI] [PubMed] [Google Scholar]
- Kaščáková S.; Maigre L.; Chevalier J.; Réfrégiers M.; Pages J.-M.; Webber M. A. Antibiotic transport in resistant bacteria: synchrotron UV fluorescence microscopy to determine antibiotic accumulation with single cell resolution. PLoS One 2012, 7, e38624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderegg G.; Hubmann E.; Podder N. G.; Wenk F. Pyridinderivate als komplexbildner. XI. Die thermodynamik der metallkomplexbildung mit bis-, tris- und tetrakis[(2-pyridyl)methyl]-aminen. Helv. Chim. Acta 1977, 60, 123–140. [Google Scholar]
- Ma Z.; Jacobsen F. E.; Giedroc D. P. Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 2009, 109, 4644–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamme F.; Villette S.; Giuliani A.; Rouam V.; Wien F.; Lagarde B.; Réfrégiers M. Synchrotron UV fluorescence microscopy uncovers new probes in cells and tissues. Microsc. Microanal. 2010, 16, 507–514. [DOI] [PubMed] [Google Scholar]
- Viveiros M.; Jesus A.; Brito M.; Leandro C.; Martins M.; Ordway D.; Molnar A. M.; Molnar J.; Amaral L. Inducement and reversal of tetracycline resistance in Escherichia coli K-12 and expression of proton gradient-dependent multidrug efflux pump genes. Antimicrob. Agents Chemother. 2005, 49, 3578–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.