Abstract
Resistance to antibiotics used in the treatment of bacterial infectious diseases is a global health problem. More than a decade ago, two-component systems such as WalKR were proposed as ideal targets for the development of new antibiotics. Biochemical screens for WalKR inhibitors using compound libraries have identified many hits, some of which were shown to have non-specific effects. The recently published structures of the S. mutans and B. subtilis WalK provide the opportunity to study inhibitors of WalK autophosphorylation at the atomic level and means to design compounds with improved specificity and affinity using a structure-based approach.
Recently scientists have been watching in awe as bacteria develop resistance to the latest antibiotics. Multidrug resistant tuberculosis (MDR-TB), penicillin-resistant Streptococcus pneumoniae, and hospital-acquired infections with vancomycin-resistant enterococci (VRE) or methicillin-resistant Staphylococcus aureus (MRSA) are regularly making headline news. The problem is not only one of resistance to front line drugs and the fear that one day, infections will be incurable, but also the increased cost of patient care and the prospect of future inability to perform trivial medical procedures without significant risk.
The scarcity of new antibiotics has been put down to dwindling returns on screening natural sources but also poor incentives for companies to invest in this sector as opposed to treatment for cancer and chronic diseases. However, the challenge of developing anti-infective drugs has been taken up by small enterprises and academia with the support of governments and other funding organizations.
The genomics revolution has accelerated finding new targets for anti-infective drugs, but designing drug-like inhibitors against specific targets remains highly challenging, costly, and not without significant risk of failure.
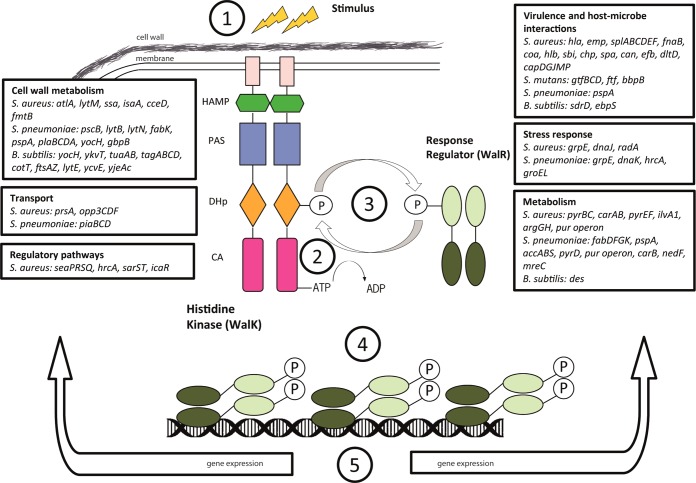
Against this somewhat gloomy perspective, there are some promising developments unfolding, one of which concerns bacterial two-component systems (TCSs) as drug targets. Bacteria respond and adapt to a large variety of environmental and intracellular signals via TCS signal transduction. The minimum components of a TCS are a sensor histidine kinase (HK) and an effector response regulator (RR)8 (Figure 1). Typically, the sensor histidine kinase (HK) is membrane-bound, and signal recognition alters the phosphorylation state of a cognate response regulator (RR). HK sensing of specific signals occurs via a variable domain that is commonly exposed to the extracellular milieu, whereas the remaining protein domains are generally conserved, cytoplasmic, and required for signal transduction8 (Figure 1). RRs are usually transcription factors of which the DNA-binding capacity and consequently, gene transcription is determined by their phosphorylation state. TCSs may directly or indirectly control numerous genes, including those involved in the regulation of metabolism, cell physiology, virulence, persistence, and resistance to antibiotics or antimicrobial peptides. Targeting virulence is a promising antibacterial-drug discovery strategy as it exerts less selective pressure on the pathogen and resistance development will be slower.
Figure 1.
TCS signaling. TCS signaling is triggered by the recognition of the signal (1) by the HK sensor variable domain, which regulates the autophosphorylation of a histidine residue in the conserved portion of HK (2).8 Signal is then transduced to the cognate RR by the transfer of the phosphoryl group from the histidine to a conserved aspartic residue in the RR (3). Typically, RRs are transcription factors, and their phosphorylation state regulates their DNA-binding capacity (4) and, consequently, gene transcription (5)1−4
TCSs were proposed as attractive targets more than 20 years ago because they are absent in mammals and essential or conditionally essential for viability in several important bacterial pathogens. WalKR system (a.k.a., YycGF, VicKR, MicAB) is an obligate essential regulatory system in Firmicutes including MRSA, VRE, and some other notorious pathogens. Other TCSs might not be essential for growth in vitro, but in vivo they can be necessary for survival or persistence (e.g., the DosRS system in Mycobacterium). Besides, some bacteria possess as many as 200 TCSs depending on their lifestyle and requirements for adaptation and metabolism in different environments. Presumably the combined effect of inhibiting all or multiple TCSs and thus the ability of a bacterium to adapt to changing physiological conditions would also greatly weaken their ability to cause infections. Given that the first TCS inhibitors were described more than a decade ago, one might think that finding TCS inhibitors is an intractable problem, and it might be better to focus on other essential pathways in bacteria. Nevertheless, there is no doubt that TCSs are good antibacterial drug targets because they have a high degree of conservation in the active sites of their catalytic domains. This fact implies that inhibitors of one TCS may in fact block multiple TCS regulatory networks, effectively incapacitating the ability of bacteria to adapt to environmental and physiological changes.
Perhaps the lack of drug candidates against WalKR or other TCSs is due to the limitations of the previously adopted approach.
The pioneering work by the Utsumís group identified imidazole and zerumbone derivatives as the first inhibitors of WalK. Afterward, the same group developed biochemical and genetic high-throughput (HTP) screening methods resulting in the discovery of a number of WalKR inhibitors.5 However, some of these inhibitors were not exclusively selective to WalKR suggesting that HTP screening approach may favor the identification of compounds that inhibit through mechanisms that are not specific for TCSs and may be toxic. A more promising avenue of research seems to be structure-based virtual screenings (SBVS) with tailored libraries or rational structure-based drug design. Targeting WalKR following structure-based approach is expected to help identify specific inhibitors of WalKR and general inhibitors of TCSs. Both these will expedite the urgently needed development of novel antibacterial drugs.
The more druggable component of TCS is the HK due to the presence of the catalytic ATP-binding domain (CA domain). The CA domain contains a well-defined and partially conserved pocket, which accommodates the ATP required for autokinase activation, i.e., initiation of signal transduction (Figure 2A,B). This pocket is hydrophobic in nature but presents two conserved polar residues, aspartic acid (Asp) and asparagine (Asn), which are responsible for ATP selectivity and Mg2+ chelation, respectively. In addition, three conserved structural water molecules increase the pocket polarity and are involved in hydrogen bonds between the ATP and the conserved Asp and Asn.
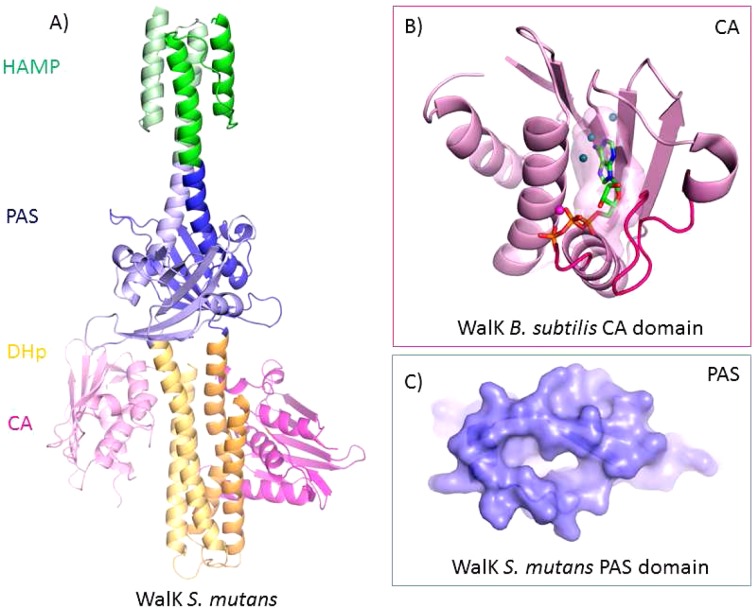
Figure 2.
Structure of WalK. (A) S. mutans WalK (PDB: 4I5S) is a long-rod dimer anchoring a HAMP signal-transducer domain (green) and a PAS sensor domain (blue) directly connected to the catalytic DHp (yellow) and CA (pink) domains. (B) B. subtilis WalK CA domain (PDB: 3SL2). The ATP-binding site (semitransparent surface, ATP shown as sticks) is generally conserved except for the variable ATP-lid (magenta). More potent and specific WalK inhibitors can be redesigned by optimizing the interactions of previously identified hits with the ATP-binding site, particularly with the conserved structural water molecules (blue spheres) and the ATP-lid. (C) Close up view of S. mutans WalK PAS domain shows the putative ligand-binding pockets with a large cavity and a unique tunnel; these structural characteristics can be exploited to identify WalK PAS domain ligands.
The major bottleneck for structure-based discovery of WalK inhibitors has been the lack of high-resolution structures of WalK. Instead, structural homology models of the CA domain of S. pneumoniae and S. epidermidis WalK based on the structures of Thermotoga maritima HK853 (PDB: 2C2A) and E. coli EnvZ (PDB: 1BXD) were used for SBVS. These screenings yielded WalK autophosphorylation inhibitors belonging to different classes of chemical structures, such as imidazole analogues and derivatives of furan, thiophene, thiazolidinone, benzamide, and pyrimidinone.6,7 This suggests that the ATP-binding pocket can accommodate a variety of ligands, a feature that can be exploited to design potent and selective HK inhibitors with a desired ADMET (absorption, distribution, metabolism, elimination, and toxicology) profile. Notably, unfavorable ADMET properties are one of the major reasons for high attrition rates of candidate molecules in drug discovery.
WalK autophosphorylation inhibitors identified by SBVS inhibited the growth of S. pneumoniae and S. epidermidis and showed bactericidal effects toward both planktonic and biofilm cells. Furthermore, some of them decreased the mortality of mice infected with S. pneumoniae in an in vivo sepsis model, supporting the idea that SBVS is a viable tool for the identification of WalK inhibitors with therapeutic effect. Nevertheless, the design of potent ATP-competitive inhibitors specific for WalK is challenging, even more so when structural homology models are used. This issue is crucial when attempting to generate a WalK inhibitor with higher specificity and affinity in the hit-to-lead optimization phase. Although the overall fold of the CA domain as well as the catalytic residues is generally conserved, there are large variations in size and sequence in the ATP-lid, a HK distinctive flexible loop that covers the ATP-binding pocket and is crucially involved in the autophosphorylation reactions8 (Figure 2A,B).
Recent elucidation of WalK structures of the entire intracellular portion from Streptococcus mutans (PDB: 4I5S) (Figure 2A,C) and the CA domain from Bacillus subtilis (PDB: 3SL2) (Figure 2B) might provide the key to design improved WalK inhibitors. These structures have not only revealed specific characteristics of the WalK CA domain but also insights into the molecular mechanism of WalK autokinase activation. These WalK structures might be used to generate potent and selective WalK inhibitors by the rational redesign of the previously identified hits by optimizing interactions with the ATP-binding site and particularly with the ATP-lid. Moreover, the structure of S. mutans WalK has also revealed specific folds of the HAMP transducer, the PAS sensor, and the DHp catalytic domains of WalK (Figure 2), opening up new possibilities to explore these domains for structure-based drug design.
PAS (Per-Arnt-Sim) domains are small sensor modules that bind a chemically diverse range of small molecules and present a conserved three-dimensional architecture but a divergent primary sequence. The B. subtilis PAS domain is required for WalK activity and localization to the divisome during cell division. The structure of S. mutans WalK shows that the putative ligand binding site forms unique pockets for a variety of ligands to bind (Figure 2C) suggesting that WalK PAS domain might bind ligands differently from other HK PAS domains. Therefore, the WalK PAS domain appears to be a promising target for novel antimicrobials, and its structure might also facilitate the rational design of specific and potent WalK inhibitors. Furthermore, WalK PAS ligands remain unknown, and their identification might generate chemical reagents to facilitate mechanistic studies.
In summary, we argue that discovery of new WalKR, and in general TCS, inhibitors, which will be further developed into antibacterial drugs, is still a realistic scenario. However, the focus should be on structure-based approaches. Certainly, ongoing efforts to solve the structures of HK from other important pathogens will help to design inhibitors with optimized activity. The high homology between TCSs and their wide distribution among bacteria imply that such inhibitors can be developed into broad-spectrum antibiotics. The road ahead is still challenging and arduous, but given the importance of the problem of drug resistance, we are convinced it is a road we should take.
Acknowledgments
We thank all colleagues from the STARS ITN (EU FP7 Marie Curie ITN grant no. 238490) for their valuable comments during meetings.
Author Contributions
The manuscript was written through equal contributions of all authors. All authors have given approval to the final version of the manuscript.
This work was supported by the European Union Framework Programme 7-funded Marie Curie Initial Training Nework STARS (Contract No. PITN-GA-2009-238490, to N.V and A.B) and by grants BIO2010-15424 from the Ministerio de Ciencia e Innovación and ACOMP2013/031 from Generalitat Valenciana (to A.M).
Views expressed in this editorial are those of the author and not necessarily the views of the ACS.
The authors declare no competing financial interest.
Notes
We apologize to authors whose work has not been cited due to space constraints.
Due to a production error, this paper was published ASAP on September 18, 2013 without its required corrections. The revised version was reposted on September 19, 2013.
References
- Dubrac S.; Boneca I. G.; Poupel O.; Msadek T. New insights into the WalK/WalR (YycG/YycF) essential signal-transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 2007, 189228257–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delauné A.; Dubrac S.; Blanchet C.; Poupel O.; Mäder U.; Hiron A.; Leduc A.; Fitting C.; Nicolas P.; Cavaillon J.-M.; Adib-Conquy M.; Msadek T. The WalKR system controls major staphylococcal virulence genes and is involved in triggering host inflammatory response. Infect. Immun. 2012, 80103438–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohedano M. L.; Overweg K.; De La Fuente A.; Reuter M.; Altabe S.; Mulholland F.; De Mendoza D.; López P.; Wells J. M. Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane-composition. J. Bacteriol. 2005, 18772357–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden B. P.; McEvoy C. R. E.; Allen D. L.; Bell J.; Coombs G.; Bennett-Wood V.; Porter J. L.; Robins-Browne R.; Davies J. K.; Seemann T.; Stinear T. P. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog. 2011, 711e1002359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada A.; Gotoh Y.; Watanabe T.; Furuta E.; Yamamoto K.; Usumi R. Targeting two-component signal transduction: a novel drug discovery system. Methods Enzymol. 2007, 422, 386–395. [DOI] [PubMed] [Google Scholar]
- Qin Z.; Zhang J.; Xu B.; Chen L.; Wu Y.; Yang X.; Shen X.; Molin S.; Danchin A.; Jiang H.; Qu D. Structure-based discovery of inhibitors of the YycG histidine kinase: new chemical leads to combat Staphyloccocus epidermidis infections. BMC Microbiol. 2006, 6, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N.; Wang F.; Niu S.; Cao J.; Wu K.; Li Y.; Yin N.; Zhang X.; Zhu W.; Yin Y. Discovery of novel inhibitors of Streptococcus pneumoniae based on the virtual screening with the homology-modelled structure of histidine kinase (VicK). BMC Microbiol. 2009, 9, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casino P.; Rubio V.; Marina A. The mechanism of signal transduction by two-component systems. Curr. Opin. Struct. Biol. 2010, 206763–771. [DOI] [PubMed] [Google Scholar]