Abstract
Many active compounds may be excluded from biological assays due to their low aqueous solubility. In this study, a simple method for the determination of the solubility of compounds containing aromatic rings is proposed. In addition to DMSO, five organic solvents for screening experiments of TNF-α inhibitors were explored. DMSO and PEG3350 were the most suitable for both protein stability and ligand-binding experiments. In addition, glycerol is a promising solvent for the screening of other compounds for which it might provide acceptable solubilization, due to its strong tendency to preserve the protein. Moreover, a fluorescence binding assay was developed using the TNF-α/SPD304 system, and a Kd of 5.36 ± 0.21 μM was determined. The results of this study could be used for the future screening of potential TNF-α inhibitors, while the protocols developed in this work could be applied to other proteins.
Keywords: solvent selection, insoluble ligands, aqueous solubility, protein stability, fluorescence binding assay
In recent years, compound solubility in aqueous solutions has emerged as an important issue in the field of drug discovery.1 Poorly soluble compounds not only create problems for in vitro and in vivo assays in drug discovery but also carry a higher risk of failure during development.2,3 Although several solvents are available to dissolve lipophilic (hydrophobic) drugs, poor solubility poses a challenge for biological assays, where hydrophobic compounds are usually dissolved at concentrations up to 30 mM in DMSO.4 Improvements in the solubility of small molecules can often be achieved through structural modification designed to reduce molecular planarity and symmetry so as to disrupt crystal packing (lattice energy of the molecule) and/or by introducing solubilizing groups at positions on the core structure of a lead series without being detrimental to other properties (see ref (3) and references within).
These modifications are not possible with commercial collections of compounds where approximately 10–20% are not soluble in DMSO at high (30 mM) concentration. In addition, lowering the storage temperature further reduces the solubility.5 Thus, these structural modifications cannot be performed in the early stages of discovery when lead and hit compounds are sought from an existing collection. Nevertheless, the use of alternative solvents might overcome these solubility issues in early discovery. These solvents should not affect protein stability or protein/ligand-binding affinity. Thus, there is a need to find further protein friendly solvents for lipophilic molecule solubilization.
Proteins with shallow ligand-binding sites that exhibit hydrophobic characteristics are the most challenging. The existence of hydrophobic binding sites in a protein implies similar characteristics for potential ligands. Tumor necrosis factor α (TNF-α), a pleiotropic cytokine involved in inflammation, immunity, and cellular organization,6 and its newly discovered ligand SDP304 represent a typical case.7
Dysregulation of TNF-α has been implicated in cases of tumorigenesis, diabetes, and especially in autoinflammatory diseases such as psoriatic arthritis and Crohn's disease.8 In addition, TNF-α plays an insidious role in the pathogenesis of rheumatoid arthritis.9 Therefore, the development of relatively low toxicity, small molecule inhibitors of TNF-α for therapeutic applications remains a highly desirable goal.10 Even though a plethora of compounds from chemical libraries is available, poor aqueous solubility of most potential inhibitors for TNF-α still remains a problem.
The initial objective of our research was the discovery of new potential inhibitors for TNF-α to provide successful candidate leads for clinical evaluation. However, from the beginning, we have faced the dual problems of low aqueous solubility of potential ligands (many compounds were insoluble in 100% DMSO) and the negative effect of organic solvents on both protein stability and ligand binding. Thus, the objective of our work orientated towards the selection of solvents that would overcome both of these problems.
Many solubility methodologies have been developed over the years to measure solubility, including kinetic, semiequilibrium (close to equilibrium), and equilibrium methods (see ref (3) and references within). However, a rapid and inexpensive solubility assay is needed. Therefore, in this work, we propose a simple, straightforward protocol to estimate the solubility of compounds containing aromatic rings that can be carried out in almost any laboratory equipped with a spectrophotometer and a centrifuge. We are aware that the calculated solubility values probably lack accuracy. However, for accurate measurements, significantly more expensive equipment is required (e.g., HPLC). Our method has been developed using SPD304, a commercially available inhibitor of TNF-α, and as such should be applicable to other aromatic compounds.
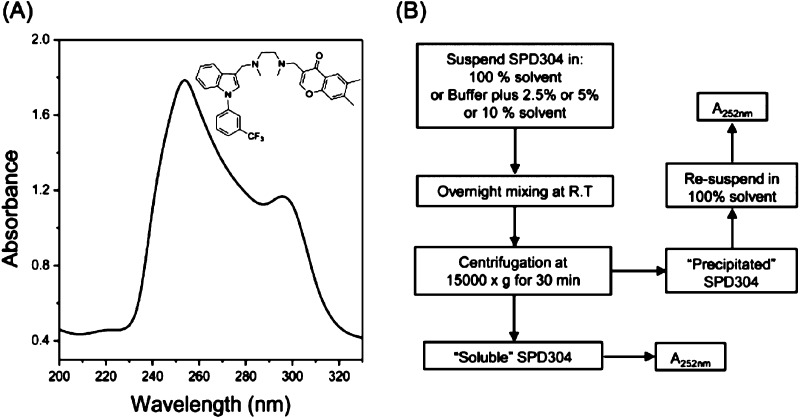
SPD304 contains three aromatic rings: one indole, one benzene, and one chromone (Figure 1A, inset) and, therefore, has two absorption bands, a strong one at approximately 252 nm and a weaker one at approximately 300 nm (Figure 1A). On the basis of this observation, we developed a simple method for the determination of SPD304 solubility (Figure 1B). One of the advantages of our method is the measurement of both the soluble and the insoluble amounts of the compound from the same sample (as described in the Supporting Information). Measurement of the absorbance (at 252 nm in the case of SPD304) of the resuspended insoluble material is very important as a check of whether our determinations are correct. Moreover, in the case of extremely insoluble compounds where the UV absorbance is close to the detection limit of the spectrophotometer, the exact solubility of the compound can be easily determined by measuring the UV absorbance of the precipitate, after it was resuspended in pure solvent (as described in the Supporting Information). The main disadvantage of our method is that it cannot be applied to small molecules that do not contain suitable chromophores.
Figure 1.
(A) SPD304 absorption spectra analysis. (B) Schematic representation of the solubility screen of SPD304 in aqueous solutions containing various concentrations of different organic solvents.
In addition to DMSO, the solubility of SPD304 in various solvents like cryoprotectants (e.g., glycerol and PEG), methanol, and DMF was investigated (Table 1). Cryoprotectants commonly used in protein crystallography could be excellent candidates for solubilization of lipophilic compounds since they are widely used for protein storage.11 Moreover, it is well known that many proteins are stable in some organic solvents.12 Thus, the solubility of SPD304 in two commonly used solvents, methanol and DMF, was also tested.
Table 1. Solubility of SPD304 in 10 mM Citrate-Phosphate Buffer (pH 6.5) Containing Various Concentrations of Different Organic Solventsa.
| solubility of SPD304 (μM) | |||
|---|---|---|---|
| solvent/concn | 2.5% (v/v) | 5% (v/v) | 10% (v/v) |
| DMSO | 33 | 52 | 79 |
| DMF | 35 | 58 | 82 |
| glycerol | 15 | 19 | 28 |
| MeOH | 37 | 58 | 83 |
| PEG3350 | 71 | 84 | 96 |
| PEG5000 | 90 | 97 | >100 |
The solubility of SPD 304 in 0% solvent is 10 μM. The mean values of four independent measurements are presented.
Initially, a stock solution of SPD304·HCl in dd-H2O was prepared. The pH of this solution (10 mM) was approximately 1, and the ligand was completely soluble. However, when SPD304 was diluted in citrate-phosphate buffer (pH 6.5, which is protein “friendly”), its solubility significantly decreased, and its solubility under these conditions was only 10 μM. In contrast, the solubility of this ligand increased up to 10 times when an organic solvent was added in the buffer (Table 1).
As illustrated in Table 1, the solubility of SPD304 was increased with increasing nonaqueous solvent concentration. Nevertheless, SPD304 was still only partially soluble in solutions containing DMSO, methanol, or DMF even at concentrations of 10% v/v, since the maximum solubility values obtained were approximately 80 μM (Table 1). SPD304 exhibited higher (84–97 μM) solubility when it was dissolved in citrate-phosphate buffer (pH 6.5) containing 5 or 10% PEG3350 or 5% PEG5000 while it was soluble at least to 100 μM in 10% PEG5000 solution. In contrast, the solubility of SPD304 was very low (<30 μM) in glycerol solution (Table 1).
According to Ungnade13 solvents can affect the intensities and wavelengths of the absorbance maxima of aromatic compounds. Although SPD304 contains three aromatic rings, the presence of all solvents in all concentrations tested did not significantly affect the absorption maxima of the compound (data not shown). It is worthwhile mentioning that this simple method has the sensitivity and reproducibility to determine solubilities as low as 5 μM (data not shown). The correlation between the solubility determined using the UV method and the HPLC method (in the range of 1–1000 μM) is currently in progress in our laboratory. It should be noted that a multiwavelength UV plate reader has been used previously to measure the solubility of small molecules.14
Following the solubility screening of SPD304, the stability of TNF-α in the presence of the above solvents was investigated. The tyrosine fluorescence properties of human TNF-α were exploited initially to investigate the stability of protein in the presence of these solvents. As shown in Figure 2, the extracellular domain TNF-α (77-233) contains seven Tyr and two Trp residues. The X-ray structure (PDB ID: 2az5) of TNF-α with the SPD304 ligand clearly shows hydrophobic interactions with Tyr 59, Tyr119, and Tyr 151 (Figure 2A). In addition, Tyr119 is located close to the three-fold symmetry axis (Figure 2B) of the TNF-α trimer and is making contact with its counterparts from the other monomeric subunits.7 From Figure 2B, it is obvious that dissociation of the TNF-α trimer to a dimer would significantly change the environment of the above tyrosines. Those close contacts of SPD304 with the above Tyr residues (Figure 2A) should affect the intrinsic Tyr fluorescence intensity.
Figure 2.
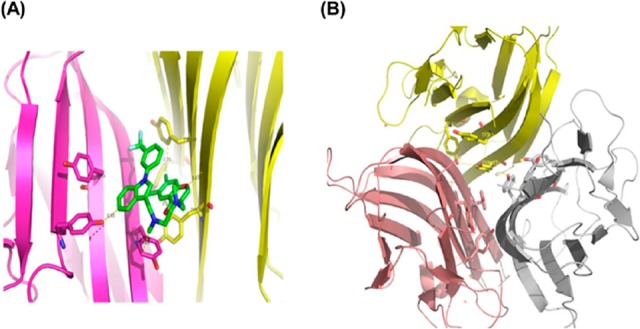
(A) Ribbon diagram of TNF-α dimer interaction with SPD304 (green) (PDB ID: 2AZ5). (B) Ribbon diagram of the TNF-α trimer (PDB ID: 3ALQ) showing the interface contacts for each monomer. Side chains of Tyr59, Tyr119, and Tyr151 (shown in stick form) of each monomer are located at the center of the interface. Removing one of the monomers would significantly affect the environment of the Tyr residues in the remaining monomers.
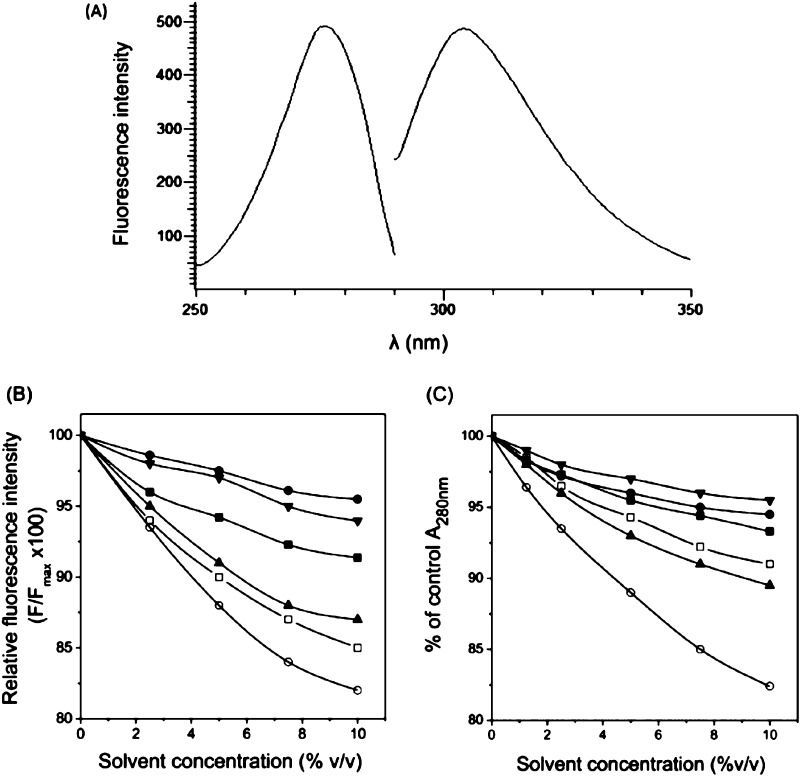
Because these particular Tyr residues represent a significant fraction (three out of seven per monomer) of the total Tyr content of the molecule, it suggested that they might form a suitable “chromophore” to study the binding of small molecules to TNF-α. The above hypothesis was tested further by fluorescence titration experiments with SPD304 (as described in the next paragraph). Excitation and emission scans of TNF-α indicated that respective maxima were 274± 2 and 304 ± 1 nm (Figure 3A), which are typical for buried tyrosyl side chains.
Figure 3.
TNF-α tyrosine emission spectra analysis. The excitation (250–300 nm) and emission (290–350 nm) spectra of TNF-α were recorded using λem = 303 nm and λext = 270 nm, respectively (excitation/emission slit width 5 and 20 nm, respectively). The protein concentration was 0.25 μM in 10 mM citrate-phosphate buffer, pH 6.5 (A). Changes of relative fluorescence intensities (B) and absorbance at 280 nm (C) of TNF-α (1.0 μM) solutions in 10 mM citrate-phosphate buffer (pH 6.5) containing various concentrations of one of the following organic solvents: DMSO (■), DMF (□), glycerol (●), MeOH (▲), PEG3350 (▼), and PEG5000 (○).
The influence of the various solvents on TNF-α stability was determined by monitoring the changes of fluorescence emission intensities (at 304 nm) of protein solutions containing 0–10% solvent concentration (Figure 3B). TNF-α exhibited stability levels above 90% of the aqueous protein in solutions containing PEG3350, DMSO, or glycerol, while in DMF, methanol, or PEG5000 solutions, the stability decreased below 90 % (Figure 3B). PEG5000 also gave a high background fluorescence emission signal and led to TNF-α precipitation. Therefore, PEG5000 can be used only in concentrations lower than 2.5% in fluorescence binding experiments. The use of PEG5000 in the concentration range 0.1–2 % is currently in progress in our laboratory.
In addition, the stability of TNF-α in the presence of the various organic solvents was studied by monitoring the changes of absorbance at 280 nm (A280nm) (Figure 3C). The results showed that TNF-α is remarkably stable in the presence of high (10%) concentrations of DMSO, glycerol, and PEG3350 since the decrease of absorbance at 280 nm was approximately 5%. In the presence of 5 and 10% methanol or DMF, the absorbance decreased by approximately 7 and 10%, respectively, indicating that the protein can tolerate both solvents at concentrations higher than 10%. These results also confirmed that a significant percentage (>20%) of TNF-α precipitated in the presence of PEG5000, as the A280nm of the protein solution decreased almost linearly with increasing PEG5000 concentration (Figure 3C).
For the determination of the dissociation constant (Kd) of SPD304/TNF-α, an approach was used based on the analysis performed by Eftink15 and Bujalowski and Lohman16 with some modifications (the derivation of our model is presented in the Supporting Information). It should be noted that in this study we analyzed the case of binding of a small molecule to a fluorescent protein and that our approach cannot be applied to a nonfluorescent one.
It has previously been reported that the presence of DMSO is favorable for TNF-α/SPD304 binding.7 Thus, to test the accuracy of our approach to Kd determination, we initially performed the titration experiments in 10 mM citrate-phosphate buffer (pH 6.5) containing 5% DMSO.
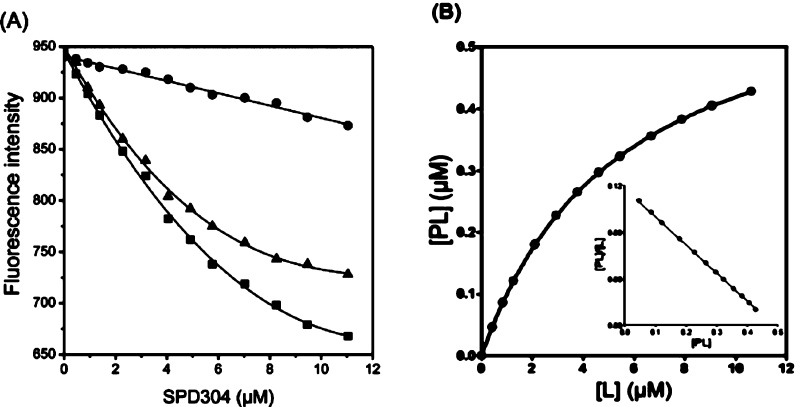
As SPD304 has micromolar affinity for TNF-α7, a significant decrease of the fluorescence signal (about 35% quenching) was observed as the ligand concentration increased from 0.1 to 12 μM as compared to the blank experiment (Figure 4A), resulting in a Kd value of 5.36 ± 0.21 μM. Using the raw data of Figure 4A, the saturation curve was drawn (Figure 4B), and subsequently, the Scatchard analysis was performed (Figure 4B, inset graph). The linearity of the Scatchard plot resulted in an “n” value of 1, suggesting that there is only one SPD304 binding site per protein molecule. It has been shown elsewhere that SPD304 inhibits TNF-α activity in biochemical and cell-based assays with median inhibitory concentrations of 22 and 4.6 μM, respectively.7
Figure 4.
(A) Dissociation constant (Kd) determination of TNF-α with SPD304. The experiment was performed in 10 mM citrate-phosphate (pH 6.5) containing 5% DMSO. (B) Direct plot of fluorescence intensity against ligand total concentration. Sequential additions of ligand were made into a cuvette containing TNF-α (■) or Tyr (●). The dissociation constant was calculated by fitting the fluorescence intensity corrected values (▲) to a quadratic equation. (B) Saturation plot after calculation of free (L) and bound (PL) ligand concentrations. Inset: Scatchard plots. The mean values of three independent measurements are presented.
The effect of solvent concentration on the SPD304/TNF-α binding interaction was studied by varying the concentration of DMSO, methanol, or PEG3350 in citrate-phosphate buffer (10 mM, pH 6.5) (Figure 5). Hydrophobic interactions play the dominant role in dissociation of the TNF-α trimer.7 Thus, organic solvents that can replace water in the hydration shell of the protein molecule without significant distortion of hydrophobic interactions in the protein globule are needed. In other words, the organic solvents used should possess the ability to maintain solvophobic interactions to a sufficient degree.
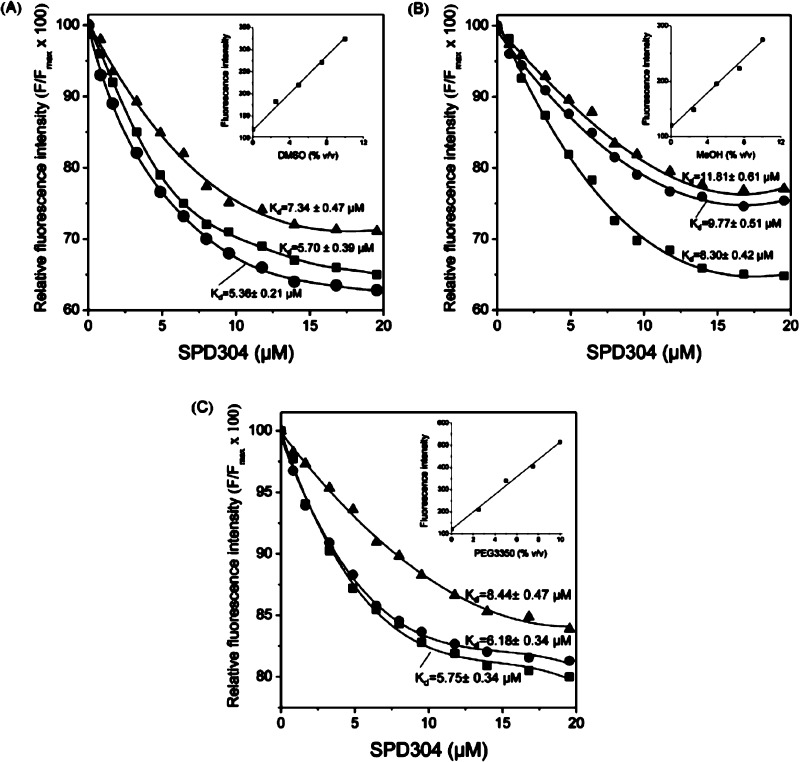
Figure 5.
Effect of DMSO (A), MeOH (B), and PEG3350 (C) concentration on TNF-α/SPD304 interaction. Changes of TNF-α fluorescence (λex = 274 nm/λem = 304 nm) were measured after incubation of TNF-α with SPD304 at 25 °C. Tested were the following concentrations of each solvent: 2.5% (■), 5% (●), and 10% (▲). Insets: changes in fluorescence intensities of 10 mM citrate-phosphate buffer with solvents concentration. The mean values of three independent measurements are presented.
As shown in Figure 5A, in the range of 2.5–5% DMSO, similar binding curves were obtained when TNF-α was titrated with SPD304, resulting in Kd values of ∼5.5 μM. In contrast, the quenching of Tyr fluorescence intensities was decreased in the presence of 10% DMSO, resulting in a Kd value of 7.34 ± 0.47 μM (Figure 5A). When titration of a TNF-α solution was performed in the presence of 2.5% DMF, a Kd value of 7.51 ± 0.52 μM was obtained by curve-fitting analysis (data not shown).
In protein solutions containing methanol, quenching of fluorescence intensities was decreased at methanol concentrations above 5% (Figure 5B). Curve fitting produced Kd values that were significantly different as illustrated in Figure 5B. It is notable that Kd values obtained from protein solutions containing methanol were almost 1.5-fold higher than the values obtained from protein solutions containing DSMO. Titration of TNF-α with SPD304 in the presence of 2.5 or 5% PEG3350 resulted in similar binding curves with Kd values of approximately 5 μM (Figure 5C). When 10% PEG3350 was used, a Kd value of 8.44 ± 0.47 μM was obtained (Figure 5C). However, PEG3350 has a strong fluorescence emission background in concentrations higher than 5% so that higher amounts of both protein and ligand are needed.
On the basis of the above results, it can be assumed that DMSO and PEG3350 are the most suitable solvents for fluorescence ligand-binding experiments on TNF-α since they enhance ligand solubility without disturbing protein structure significantly. In addition, glycerol is a promising solvent for other potential inhibitors of TNF-α since the protein is extremely stable in the presence of high (10%) glycerol concentrations. According to Ray's17 classification, all solvents can be grouped into three classes based on their capacity for solvophobic interactions. Examples of these groups are (i) water, glycerol, ethylene glycol, aminoethanol and formamide; (ii) methylformamide and dimethylformamide; and (iii) methanol, ethanol and toluene. Solvophobic interactions are best realized in solvents of class i, much less in class ii, and are practically absent in class iii. According to this classification, this occurs because the molecules of the solvents of class i contain at least two centers capable of forming hydrogen bonds. As a result of intermolecular interactions, hydrogen bonds form a rigid (thermodynamically unfavorable) framework around the dissolved solvophobic molecules and thus induce solvophobic interactions. Among the solvents of class i, solvophobic interactions are most effective in water and glycerol. Thus, it can be assumed that the presence of DMSO and PEG3350 promotes TNF-α/SPD304 binding due to the most effective solvophobic interactions of these solvents as compared to that of DMF or methanol. In addition, DMSO and water are completely miscible and interact via hydrogen bonding between the basic oxygen atom of DMSO and the acidic protons of water that can result in extreme deviations in physicochemical properties.18
To conclude, in this work, a simple method for the determination of solubility of small compounds containing aromatic rings was implemented. The unique Tyr fluorescence properties of human TNF-α were successfully used to study its stability in the presence of organic solvents. In addition, the interaction of TNF-α with SPD304 was studied by developing a Tyr fluorescence binding assay and the appropriate mathematical model for Kd determination. These methods will be used for the future screening of potential inhibitors of TNF-α.
Acknowledgments
We are grateful to Emeritus Professor Lindsay Sawyer from Edinburgh University, United Kingdom, for the fruitful discussion on the manuscript and his helpful remarks.
Supporting Information Available
TNF-α expression host and media, expression and purification of recombinant human TNF-α, solubility screening of SPD304, stability of TNF-α in the presence of various solvents, fluorescence binding assay, and determination of dissociation constant (Kd) from fluorescence measurements. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was funded by project TheRAlead (09ΣYN-21-784), which is cofinanced by the European Union and Greece, Operational Program “Competitiveness & Entrepreneurship”, NSFR 2007-2013 in the context of GSRT-National action “Cooperation”.
The authors declare no competing financial interest.
Supplementary Material
References
- Kennedy T. Managing the drug discovery/development interface. Drug Discovery Today 1997, 2, 436–444. [Google Scholar]
- Alsenz J.; Kansy M. High throughput solubility measurement in drug discovery and development. Adv. Drug Delivery Rev. 2007, 59, 546–567. [DOI] [PubMed] [Google Scholar]
- Di L.; Fish P. V.; Mano T. Bridging solubility between drug discovery and development. Drug Discovery Today 2012, 17, 486–495. [DOI] [PubMed] [Google Scholar]
- Di L.; Kerns E. H. Biological assay challenges from compound solubility: Strategies for bioassay optimization. Drug Discovery Today 2006, 11, 446–451. [DOI] [PubMed] [Google Scholar]
- Balakin V. DSMO solubility and bioscreening. Curr. Opin. Struct. Biol. 2003, 4, 499–512. [Google Scholar]
- Locksley R. M.; Killeen N.; Lenardo M. J. The TNF and TNF receptor superfamilies: Integrating mammalian biology. Cell 2001, 104, 487–501. [DOI] [PubMed] [Google Scholar]
- He M. M.; Smith A. S.; Oslob J. D.; Flanagan W. M.; Braisted A. C.; Whitty A.; Cancilla M. T.; Wang J.; Lugovskoy A. A.; Yoburn J. C.; Fung A. D.; Farrington G.; Eldredge J. K.; Day E. S.; Cruz L. A.; Cachero T. G.; Miller S. K.; Friedman J. E.; Choong I. C.; Cunningham B. C. Small-molecule inhibition of TNF-alpha. Science 2005, 310, 1022–1025. [DOI] [PubMed] [Google Scholar]
- Aggarwal B. B. Signalling pathways of the TNF superfamily: a double-edged sword. Nat. Rev. Immunol. 2003, 3, 745–756. [DOI] [PubMed] [Google Scholar]
- Feldmann M. Development of anti-TNF therapy for rheumatoid arthritis. Nat. Rev. Immunol. 2002, 2, 364–371. [DOI] [PubMed] [Google Scholar]
- Yang J. S.; Chun K.; Park J. E.; Cho M.; Seo J.; Song D.; Yoon H.; Park C. H.; Joe B. Y.; Choi J. H.; Kim M. H.; Han G. Structure based optimization of chromen-based TNF-alpha converting enzyme (TACE) inhibitors on S1′ pocket and their quantitative structure-activity relationship (QSAR) study. Bioorg. Med. Chem. 2010, 18, 8618–8629. [DOI] [PubMed] [Google Scholar]
- Shah B. N.; Chinte U.; Tomanicek S. J.; Hanson B. L.; Schall C. A. Flash Cooling Protein Crystals: Estimate of Cryoprotectant Concentration Using Thermal Properties. Cryst. Growth Des. 2011, 11, 1493–1501. [Google Scholar]
- Mattos C.; Ringe D. Proteins in organic solvents. Curr. Opin. Struct. Biol. 2001, 11, 761–764. [DOI] [PubMed] [Google Scholar]
- Ungnade H. E. The Effect of Solvents on the Absorption Spectra of Aromatic Compounds. J. Am. Chem. Soc. 1953, 75, 432–434. [Google Scholar]
- Bard B.; Martel S.; Carrupt P. A. High throughput UV method for the estimation of thermodynamic solubility and the determination of the solubility in biorelevant media. Eur. J. Pharm. Sci. 2008, 33, 230–240. [DOI] [PubMed] [Google Scholar]
- Eftink M. R. Fluorescence methods for studying equilibrium macromolecule-ligand interactions. Methods Enzymol. 1997, 278, 221–257. [DOI] [PubMed] [Google Scholar]
- Bujalowski W.; Lohman T. M. A general method of analysis of ligand-macromolecule equilibria using a spectroscopic signal from the ligand to monitor binding. Application to Escherichia coli single-strand binding protein-nucleic acid interactions. Biochemistry 1987, 26, 3099–3106. [DOI] [PubMed] [Google Scholar]
- Ray A. Solvophobic interactions and micelle formation in structure forming nonaqueous solvents. Nature 1971, 231, 313–315. [DOI] [PubMed] [Google Scholar]
- Mancers R. L.; Chalaris M.; Refson K.; Samios J. Molecular dynamics simulation of dilute aqueous DMSO solutions. A temperature-dependence study of the hydrophobic and hydrophilic behaviour around DMSO. Phys. Chem. Chem. Phy. 2004, 6, 94–102. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.