Abstract
A series of analogues of the natural product sinefungin lacking the amino acid moiety was synthesized and probed for their ability to inhibit EHMT1 and EHMT2. This study led to inhibitors 3b and 4d of methyltransferase activity of EHMT1 and EHMT2 and it demonstrates that such analogues constitute an interesting scaffold to develop selective methyltransferase inhibitors. Surprisingly, the inhibition was not competitive toward AdoMet.
Keywords: Sinefungin, methyltransferase inhibitors, EHMT1, EHMT2
Histone modifications have a central role in regulating gene expression1 and it has been suggested that they constitute an epigenetic code.2 The enzymes capable of transferring acetyl, methyl, and other groups are referred to as writers, whereas enzymes responsible for removing the modifications are referred to as erasers. Furthermore, a range of domains capable of recognizing the modified substrates are termed readers. Protein lysine methyltransferases (PKMTs) are a group of epigenetic writers, responsible for the transfer of one to three methyl groups from S-adenosyl-l-methionine(s) (AdoMet, 1, Figure 1) to the ε-amino group of the target lysine residues in histone tails3 and some nonhistone targets.4 To date, more than 50 PKMTs have been identified and EHMT1 (GLP, G9a like proteins) and EHMT2 (G9a) are among the most well-studied PKMTs. They are capable of mono- and dimethylating lysine 9 residue of histone 3 (H3K9) in euchromatin5 but have also been found to methylate nonhistone substrates such as tumor suppressor p53,6 Wiz, and reptin.7
Figure 1.
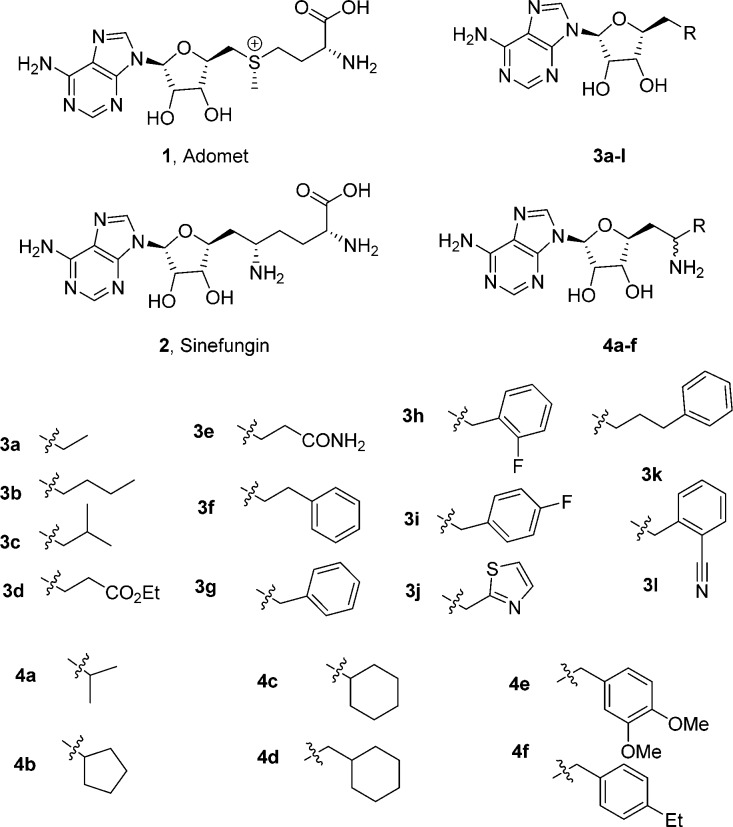
Chemical structures of AdoMet (1), sinefungin (2), and the analogues 3a–l and 4a–f.
EHMT1/2-mediated methylation has been implicated to be pivotal for processes such as embryo development,8 germ cell development and meiosis,9 and lymphocyte functioning,10,11 as well as cognition and adaptive behaviors.12 Genetic variations of EHMT1/2 have been associated with human diseases such as cancer, inflammatory diseases, and neurogenerative disorders.12−17 Removal of EHMT1 causes subtelomeric deletion syndrome with severe mental retardation,13,14 along with reduced locomotor activity and exploration.12 Increased expression of EHMT2 has been associated with lung, prostate, and hepatocellular carcinoma.15,16 Interestingly, studies also suggest that EHMT2 deficiency impairs immune response6 while knock down of EHMT1/2 causes severe growth defects inducing embryonic lethality.8 There has been a growing interest to identify potent inhibitors of these enzymes. Since the discovery of the first PKMT inhibitor, chaetocin,17 in 2005, several selective and nonselective inhibitors of PKMTs have been reported. BIX 01294,18 E72,19 and UNC032120 are some of the potent substrate (N-terminal peptides of H3) competitive inhibitors of EHMT1/2. Later, in vivo active analogues of UNC0321 were also reported.21,22 Some inhibitors of PKMTs are natural products such as SAH (the reaction product of methylation) and sinefungin, which compete with AdoMet instead. In this study, we employed sinefungin as a lead structure for the synthesis of a series of methyltransferase inhibitors and tested them for inhibition of EHMT1/2.
Sinefungin is an amino acid and a natural product from cultures of Streptomyces incamatus and S. griseolus, which is structurally related to AdoMet but has an amino methylene group instead of the methylated sulfonium group. In this study, two different series of compounds 3a–l and 4a–f were designed that exchanged the terminal α-amino acid moiety of sinefungin. One series possessed the additional α-amino group of sinefungin, and the other series had no amino group (Figure 1).
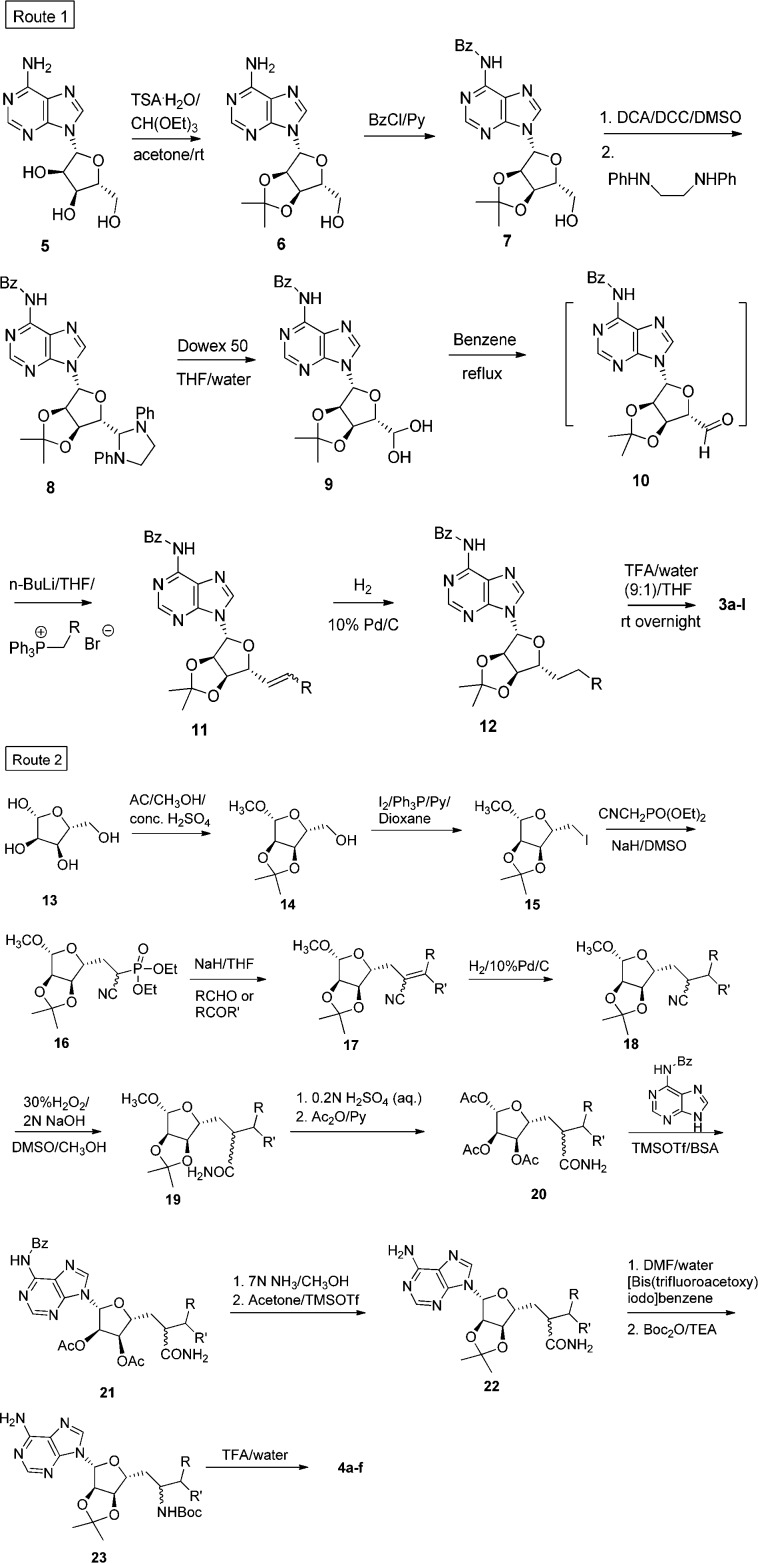
The two series were synthesized via two different routes (Figure 2). Route 1 was employed for analogues 3a–l, and route 2 for analogues 4a–f. Route 1 started with adenosine 5 that was protected to form N6-benzoyl-2′,3′-O-isopropylideneadenosine 7 by two steps. Oxidation of 7 was achieved by treatment with dimethyl sulfoxide and dicyclohexylcarbodiimide in the presence of dichloroacetic acid. The addition of N,N′-diphenylethylenediamine formed 1,3-diphenylimidazolidene derivative of adenosine-5′-aldehyde 8,23 which was treated with Dowex 50 (H+) resin in aqueous tetrahydrofuran at room temperature to regenerate adenosine-5′-aldehyde as its stable monohydrate form 9.
Figure 2.
Synthetic routes 1 and 2 to analogues 3a–l and 4a–f, respectively.
Compound 9 was then dehydrated in benzene and directly reacted with respective Wittig reagents to yield alkenes 11. Hydrogenation yielded compound 12 that was deprotected in one step giving 3a–l. Route 2 started with β-d-ribofuranoside 13, which was subsequently protected and iodinated giving compound 15. Reaction between diethyl cyanomethylphosphonate and 15 in dimethyl sulfoxide gave ethyl phosphonate 16,24 which was then treated with various aldehydes and ketones under basic conditions to yield conjugated nitriles 17. Hydrogenation of the alkene gave 18 that was followed by transformation of the nitrile to amide 19 using alkaline hydrogen peroxide. Protecting groups were exchanged in two steps giving acetyl protected 20. Coupling of N6-benzoyl-N6,N9-bis(trimethylsilyl)adenine with 1′,2′,3′-tri-O-acetyl-β-d-ribofuranosides at high temperature (90 °C) in a microwave produced the more thermodynamically stable 9-isomer 21. Removal of the benzyl group and reestablishing the acetonide in 22 was followed by a Hofmann rearrangement yielding Boc-protected amine 23(25) that could be completely deprotected in one step to give the final products 4a–f as 1:1 mixture of diastereomers.
Initially, the compounds were screened in a FRET-based LANCE ultra G9a histone H3-lysine N-methyltransferase assay26 that measures the dimethylation of a biotinylated histone H3 (1–21) peptide at lysine 9. Previous studies have suggested that EHMT1 and EHMT2 both can mono-, di-, and trimethylate histone H3 at lysine 9. This assay is based on the capture of a modified peptide by the Europium labeled antibody (Eu–H3K9me2, donor) and ULight-Streptavidin (SA, acceptor dye), ultimately bringing them in close proximity. When irradiated at 320 or 340 nm, the energy transfer from the donor to the acceptor emits light at 665 nm, which is directly proportional to the amount of modified substrate. (Constructs for SET domain of EHMT1 and EHMT2 and expression and purification are described in Supporting Information.)
Initial characterization of the compounds was performed at 100 μM at EHMT1 and EHMT2 (see Supporting Information for assay procedure), and the results are shown in Figure 3. All the assay characterization and inhibition studies were done in triplicates, using the same batch of purified proteins. Most of the compounds tested showed weak inhibitory activity at this concentration; however, the inhibition varied and 4 analogues 3b, 3f, 4d, and 4e showed more than 40% inhibition toward EHMT1 when screened at 100 μM, and the results were followed up with further inhibition studies. Thus, dose–response curves were established for the inhibition of these compounds and sinefungin toward methylation activity of EHMT1 and EHMT2, and these results are shown in Figure 4. Furthermore, IC50 values are reported in Table 1.
Figure 3.
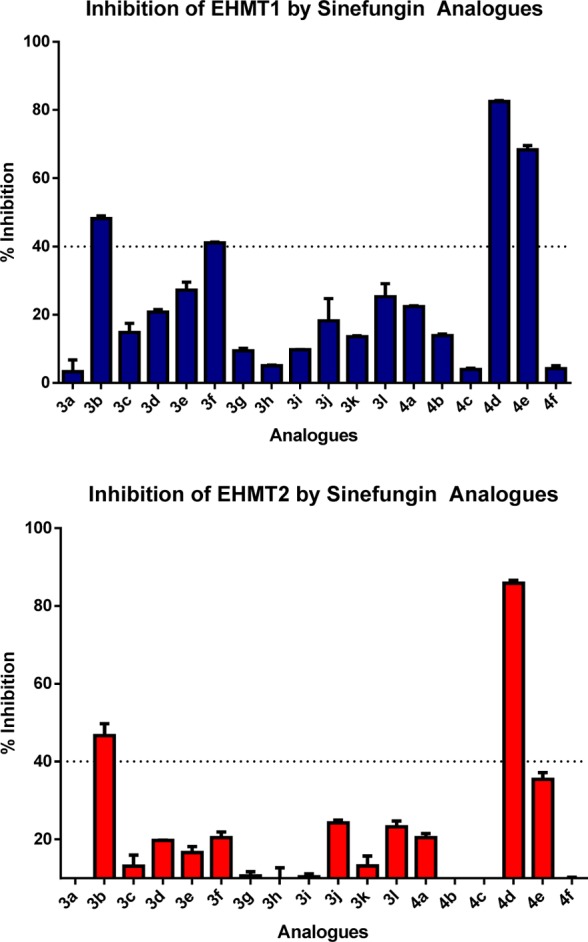
Inhibition of methyltransferase activity of EHMT1 and EHMT2 by compounds 3a–l and 4a–4f at 100 μM determined in a LANCE Ultra assay.
Figure 4.
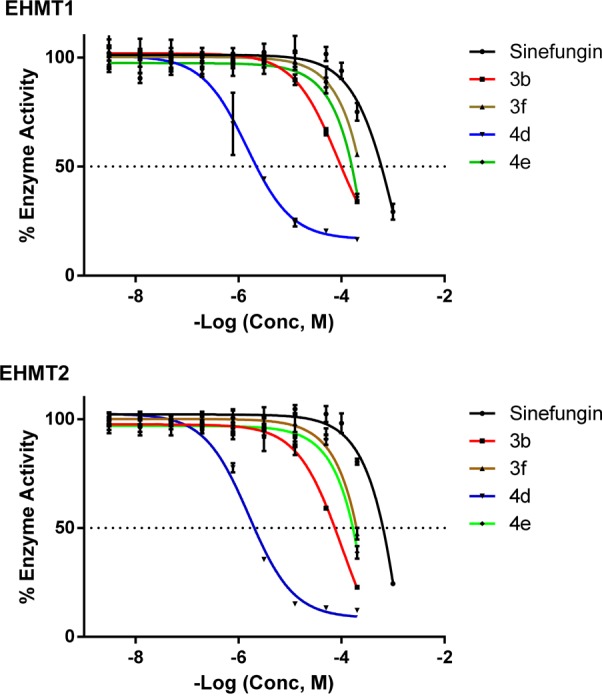
Dose–response curves of analogues 3b, 3f, 4d, 4e, and sinefungin for EHMT1 and EHMT2 determined by LANCE Ultra assay. All the presented compounds have lower IC50 values compared to the natural scaffold sinefungin.
Table 1. IC50 Values of Analogues 3b, 3f, 4d, 4e, and Sinefungin on EHMT1 and EHMT2a.
| compds | EHMT1 IC50 (μM) | EHMT2 IC50 (μM) |
|---|---|---|
| 3b | 87 (57–130)b | 103 (68–156)b |
| 3f | 235 (196 −281)b | 190 (174–206)b |
| 4d | 1.5 (1.1–1.8) | 1.6 (1.0–2.0) |
| 4e | 147 (119–181)b | 157 (120–203)b |
| sinefungin | 513 (397–662)b | 509 (434–595)b |
IC50 values are average of experiment done in triplicates. Numbers in brackets indicate 95% confidence interval data.
Since a full dose–response curve was not achieved, these are estimated values. Thus, a fairly potent analogue 4d was obtained when changing this part. In addition, the second β-amino group can be removed and still show weak inhibitory activity as demonstrated with analogue 3b. The compounds did not discriminate EHMT1 and EHMT2, but the selectivity changes compared to sinefungin since 4d and 3b did not show inhibition of DNMT1, PRMT1, and SET7/9 at 200 μM, except for 4d that is a very weak inhibitor of DNMT1. We cannot exclude possible activity at other methyltransferases, but the study demonstrates that selectivity can be achieved among methyltransferases with this compound series. An inhibition kinetic study of both compounds showed noncompetitive inhibition kinetics, which is surprising since they could be expected to compete with SAM.
As seen in Table 1, the four analogues are more potent than sinefungin with compound 4d as the best inhibitor on EHMT1 and EHMT2 (IC50 values of 1.5 and 1.6 μM, respectively) in this study.
We then performed a kinetic study of the two most potent compounds 3b and 4d. Both of the compounds demonstrated AdoMet noncompetitive inhibition in this study (see Figure S3 and Table S2, Supporting Information), and thus the measured IC50 values represent Ki values.
Neither of the compounds seems to discriminate significantly between EHMT1 and EHMT2. However, when probed against three other methyltransferases (DNMT1, PRMT1, and SET7/9), 4d and 3b showed no significant inhibition at 200 μM and below (except for 4d that has an estimated IC50 of 179 μM at DNMT1) (see Table S3, Supporting Information).
We have presented the synthesis and characterization of a series of analogues of sinefungin. Recently, a series of N-alkylated sinefungin analogues were reported27 to suppress methyltransferase activities leading to a potent N-propyl substituted SETD2 inhibitor. In this study, we exchanged the α-amino acid moiety and demonstrated that it is not essential for inhibitory activity at EHMT1 and EHMT2.
Thus, these scaffolds appear to be promising for further modification to increase affinity of these selective noncompetitive inhibitors of EHMTs. While there is a risk for achieving unselective inhibitors, the benefit of this compound class is that it may be possible to find selective inhibitors toward different methyltransferases in this compound class. The recent discovery of a sinefungin analogue as a SETD2 inhibitor emphasizes this notion. Furthermore, compounds with a therapeutically favorable profile against several methyltransferases could be envisioned.
Future studies will focus on improving the synthetic route of the compounds to extend the series and enable a broader assessment of the activity of the compound class at various methyltransferases. Furthermore, it will be important to verify that the compounds bind as presumed in the SAM binding pocket despite the observed noncompetitive inhibition and to perform in vivo studies.
Acknowledgments
We are grateful to the Novo Nordisk Foundation Center for Protein Research and Department of Health Sciences, University of Copenhagen for the Ph.D. fellowship, Dr. Thomas Frimurer for valuable inputs in designing the analogues, Ms. Huili Lu and Ms. Jie Zhang for technical assistance, the Danish Cancer Society, the University of Copenhagen Programme of Excellence and the Ministry of Science and Technology of China (2009ZX09302-001, 2012ZX09304011, and 2013ZX09507002) for grant support, and the EU COST-action TD0905, for providing a vital Epigenetic focused network.
Glossary
ABBREVIATIONS
- TSA
p-toluenesulfonic acid
- BzCl
benzoylchloride
- DCA
dichloroacetic acid
- DCC
dicyclohexylcarbodiimide
- BSA
N,O-bis(trimethylsilyl)acetamide
- TFA
trifluoroactic acid
- TMS
trimethylsilyl
- AC
acetone
- TEA
triethylamine
Supporting Information Available
Detailed information about compound synthesis, characterization, protein purification, assay procedure, and data analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
∥ (J.B.) Department of International Health, Immunology and Microbiology, University of Copenhagen, Blegdamsvej 3, DK-2200 Copenhagen, Denmark.
Funding provided by University of Copenhagen for the Ph.D. fellowship and the Danish Cancer Society, the University of Copenhagen Programme of Excellence, and the Ministry of Science and Technology of China (2009ZX09302-001, 2012ZX09304011, and 2013ZX09507002) for grant support.
The authors declare no competing financial interest.
This paper was published ASAP on February 4, 2014 with errors in the Supporting Information. A corrected Supporting Information file was reposted with the manuscript on February 18, 2014.
Supplementary Material
References
- Kouzarides T. Chromatin modifications and their function. Cell 2007, 1284693–705. [DOI] [PubMed] [Google Scholar]
- Jenuwein T.; Allis C. D. Translating the histone code. Science 2001, 29355321074–80. [DOI] [PubMed] [Google Scholar]
- Rea S.; Eisenhaber F.; O’Carroll D.; Strahl B. D.; Sun Z. W.; Schmid M.; Opravil S.; Mechtler K.; Ponting C. P.; Allis C. D.; Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000, 4066796593–9. [DOI] [PubMed] [Google Scholar]
- Huang J.; Berger S. L. The emerging field of dynamic lysine methylation of non-histone proteins. Curr. Opin. Genet. Dev. 2008, 182152–8. [DOI] [PubMed] [Google Scholar]
- Tachibana M.; Ueda J.; Fukuda M.; Takeda N.; Ohta T.; Iwanari H.; Sakihama T.; Kodama T.; Hamakubo T.; Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Gene Dev. 2005, 197815–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.; Dorsey J.; Chuikov S.; Perez-Burgos L.; Zhang X.; Jenuwein T.; Reinberg D.; Berger S. L. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J. Biol. Chem. 2010, 285139636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S.; Kim Y.; Kim I. S.; Kim B.; Choi H. J.; Lee J. M.; Shin H. J.; Kim J. H.; Kim J. Y.; Seo S. B.; Lee H.; Binda O.; Gozani O.; Semenza G. L.; Kim M.; Kim K. I.; Hwang D.; Baek S. H. Negative regulation of hypoxic responses via induced Reptin methylation. Mol. Cell 2010, 39171–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M.; Sugimoto K.; Nozaki M.; Ueda J.; Ohta T.; Ohki M.; Fukuda M.; Takeda N.; Niida H.; Kato H.; Shinkai Y. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Gene Dev. 2002, 16141779–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M.; Nozaki M.; Takeda N.; Shinkai Y. Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J. 2007, 26143346–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L. R.; Miyashita H.; Cobb R. M.; Pierce S.; Tachibana M.; Hobeika E.; Reth M.; Shinkai Y.; Oltz E. M. Functional analysis of histone methyltransferase g9a in B and T lymphocytes. J. Immunol. 2008, 1811485–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B.; Northrop J. P.; Antignano F.; Burrows K.; Hadidi S.; Mullaly S. C.; Rossi F. M.; Zaph C. Activating and inhibitory functions for the histone lysine methyltransferase G9a in T helper cell differentiation and function. J. Exp. Med. 2010, 2075915–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A.; Sampath S. C.; Intrator A.; Min A.; Gertler T. S.; Surmeier D. J.; Tarakhovsky A.; Greengard P. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron 2009, 645678–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T.; Smidt M.; Banning M. J.; Oudakker A. R.; Van Esch H.; de Brouwer A. P.; Nillesen W.; Sistermans E. A.; Hamel B. C.; de Bruijn D.; Fryns J. P.; Yntema H. G.; Brunner H. G.; de Vries B. B.; van Bokhoven H. Disruption of the gene Euchromatin Histone Methyl Transferase1 (Eu-HMTase1) is associated with the 9q34 subtelomeric deletion syndrome. J. Med. Genet. 2005, 424299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T.; Brunner H. G.; Amiel J.; Oudakker A. R.; Nillesen W. M.; Magee A.; Genevieve D.; Cormier-Daire V.; van Esch H.; Fryns J. P.; Hamel B. C.; Sistermans E. A.; de Vries B. B.; van Bokhoven H. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am. J. Hum. Genet. 2006, 792370–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H.; Soejima K.; Yasuda H.; Kawada I.; Nakachi I.; Yoda S.; Naoki K.; Ishizaka A. Deregulation of histone lysine methyltransferases contributes to oncogenic transformation of human bronchoepithelial cells. Cancer Cell Int. 2008, 8, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. W.; Hua K. T.; Kao H. J.; Chi C. C.; Wei L. H.; Johansson G.; Shiah S. G.; Chen P. S.; Jeng Y. M.; Cheng T. Y.; Lai T. C.; Chang J. S.; Jan Y. H.; Chien M. H.; Yang C. J.; Huang M. S.; Hsiao M.; Kuo M. L. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 2010, 70207830–40. [DOI] [PubMed] [Google Scholar]
- Greiner D.; Bonaldi T.; Eskeland R.; Roemer E.; Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat. Chem. Biol. 2005, 13143–5. [DOI] [PubMed] [Google Scholar]
- Kubicek S.; O’Sullivan R. J.; August E. M.; Hickey E. R.; Zhang Q.; Teodoro M. L.; Rea S.; Mechtler K.; Kowalski J. A.; Homon C. A.; Kelly T. A.; Jenuwein T. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell 2007, 253473–81. [DOI] [PubMed] [Google Scholar]
- Chang Y.; Ganesh T.; Horton J. R.; Spannhoff A.; Liu J.; Sun A.; Zhang X.; Bedford M. T.; Shinkai Y.; Snyder J. P.; Cheng X. Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases. J. Mol. Biol. 2010, 40011–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F.; Chen X.; Allali-Hassani A.; Quinn A. M.; Wigle T. J.; Wasney G. A.; Dong A.; Senisterra G.; Chau I.; Siarheyeva A.; Norris J. L.; Kireev D. B.; Jadhav A.; Herold J. M.; Janzen W. P.; Arrowsmith C. H.; Frye S. V.; Brown P. J.; Simeonov A.; Vedadi M.; Jin J. Protein lysine methyltransferase G9a inhibitors: design, synthesis, and structure activity relationships of 2,4-diamino-7-aminoalkoxy-quinazolines. J. Med. Chem. 2010, 53155844–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedadi M.; Barsyte-Lovejoy D.; Liu F.; Rival-Gervier S.; Allali-Hassani A.; Labrie V.; Wigle T. J.; Dimaggio P. A.; Wasney G. A.; Siarheyeva A.; Dong A.; Tempel W.; Wang S. C.; Chen X.; Chau I.; Mangano T. J.; Huang X. P.; Simpson C. D.; Pattenden S. G.; Norris J. L.; Kireev D. B.; Tripathy A.; Edwards A.; Roth B. L.; Janzen W. P.; Garcia B. A.; Petronis A.; Ellis J.; Brown P. J.; Frye S. V.; Arrowsmith C. H.; Jin J. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat. Chem. Biol. 2011, 78566–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F.; Barsyte-Lovejoy D.; Allali-Hassani A.; He Y.; Herold J. M.; Chen X.; Yates C. M.; Frye S. V.; Brown P. J.; Huang J.; Vedadi M.; Arrowsmith C. H.; Jin J. Optimization of cellular activity of G9a inhibitors 7-aminoalkoxy-quinazolines. J. Med. Chem. 2011, 54176139–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton D. H. R.; Gero S. D.; Quiclet-Sire B.; Samadi M. Expedient synthesis of natural (S)-sinefungin and of its C-6[prime or minute] epimer. J. Chem. Soc., Perkin Trans. I 1991, 05981–985. [Google Scholar]
- Hampton A.; Sasaki T.; Paul B. Synthesis of 6′-cyano-6′-deoxyhomoadenosine-6′-phosphonic acid and its phosphoryl and pyrophosphoryl anhydrides and studies of their interactions with adenine nucleotide utilizing enzymes. J. Am. Chem. Soc. 1973, 95134404–14. [DOI] [PubMed] [Google Scholar]
- Geze M.; Blanchard P.; Fourrey J. L.; Robert-Gero M. Synthesis of sinefungin and its C-6′ epimer. J. Am. Chem. Soc. 1983, 105267638–7640. [Google Scholar]
- Gauthier N.; Caron M.; Pedro L.; Arcand M.; Blouin J.; Labonte A.; Normand C.; Paquet V.; Rodenbrock A.; Roy M.; Rouleau N.; Beaudet L.; Padros J.; Rodriguez-Suarez R. Development of homogeneous nonradioactive methyltransferase and demethylase assays targeting histone H3 lysine 4. J. Biomol. Screening 2012, 17149–58. [DOI] [PubMed] [Google Scholar]
- Zheng W.; Ibanez G.; Wu H.; Blum G.; Zeng H.; Dong A.; Li F.; Hajian T.; Allali-Hassani A.; Amaya M. F.; Siarheyeva A.; Yu W.; Brown P. J.; Schapira M.; Vedadi M.; Min J.; Luo M. Sinefungin derivatives as inhibitors and structure probes of protein lysine methyltransferase SETD2. J. Am. Chem. Soc. 2012, 1344318004–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.