Abstract
Purpose.
Autosomal dominant Stargardt-like macular dystrophy (STGD3) is a juvenile-onset disease that is caused by mutations in Elovl4 (elongation of very long fatty acids-4). The Elovl4 catalyzes the first step in the conversion of C24 and longer fatty acids (FAs) to very long-chain FAs (VLC-FAs, ≥C26). Photoreceptors are particularly rich in VLC polyunsaturated FAs (VLC-PUFA). To explore the role of VLC-PUFAs in photoreceptors, we conditionally deleted Elovl4 in the mouse retina.
Methods.
Proteins were analyzed by Western blotting and lipids by gas chromatography (GC)–mass spectrometry, GC-flame ionization detection, and tandem mass spectrometry. Retina function was assessed by electroretinography (ERG), and structure was evaluated by bright field, immunofluorescence, and transmission electron microscopy.
Results.
Conditional deletion (KO) of retinal Elovl4 reduced RNA and protein levels by 91% and 96%, respectively. Total retina VLC-PUFAs were reduced by 88% compared to the wild type (WT) levels. Retinal VLC-PUFAs incorporated in phosphatidylcholine were less abundant at 12 months compared to 8-week-old levels. Amplitudes of the ERG a-wave were reduced by 22%, consistent with photoreceptor degeneration (11% loss of photoreceptors). Analysis of the rod a-wave responses gave no evidence of a role for VLC-PUFA in visual transduction. However, there were significant reductions in rod b-wave amplitudes (>30%) that could not be explained by loss of rod photoreceptors. There was no effect of VLC-PUFA reduction on cone ERG responses, and cone density was not different between the WT and KO mice at 12 months of age.
Conclusions.
The VLC-PUFAs are important for rod, but not cone, function and for rod photoreceptor longevity.
Keywords: VLC-PUFA, Elovl4, STDG3
Comprehensive ERG and histologic analysis revealed that VLC-PUFAs are vital for rod function and longevity.
Introduction
Patients with autosomal dominant Stargardt-like macular dystrophy (STGD3), a juvenile form of macular degeneration, develop a loss of central vision at an early age. Mutations in the elongation of very long-chain fatty acids-4 (Elovl4) gene have been associated with the disease in patients with STGD3.1–5 These mutations cause a frame shift in the Elovl4 transcript, introducing a premature stop codon, resulting in the synthesis of a truncated protein that has lost an ER retention/retrieval signal. The truncated protein is not targeted to the endoplasmic reticulum, the site of synthesis of very long-chain polyunsaturated fatty acids (VLC-PUFAs; 26–40 carbons6–8). Because the mutant protein has no enzymatic activity,9 the loss of VLC-PUFAs may be involved in the STGD3 disease pathogenesis. Expression of the Elovl4 gene is limited mainly to the brain, testis, skin, and retinal photoreceptor cells.10 While the skin contains very long chain saturated fatty acids (VLC-FAs),11,12 sperm cells and the retina are enriched in VLC-PUFAs.13,14
Retinal VLC-PUFAs are incorporated into phosphatidylcholine in photoreceptor outer segment membranes15 and have been suggested to have a role in disk curvature and plasma membrane fluidity, thus aiding in phototransduction.6 The role of VLC-PUFAs in the neural retina still is under debate.7,8 The purpose of this study was to elucidate the role of VLC-PUFA in the retina by conditionally deleting Elovl4 expression in rod and cone photoreceptor cells, thus, removing VLC-PUFAs from both cell types. Here we showed that these FAs are important for rod, but not cone, survival in 12-month-old mice. We found no evidence from our electroretinographic (ERG) analysis to support a role for VLC-PUFA in visual transduction in rod or cone photoreceptors. However, we did find significant reduction in rod b-wave amplitudes in conditional deletion (KO) mice that could not be explained by rod cell death. Also, there were significant reductions in the oscillatory potentials (OPs) and scotopic threshold responses in KO mice. These findings are presented and discussed in the companion paper.
Materials and Methods
Materials
Primary antibodies used were anti-Elovl46 (1:1000) and anti-β-actin (1:1000; ABCAM, Cambridge, MA, USA). The Elovl4 antibody used here has been shown by Agbaga et al.6 to label protein only in the inner segments and outer nuclear layer of whole rat retina. Specificity of the antibody was shown by preadsorbtion of the Elovl4 antigen by immunohistochemistry and by Western blotting resulting in the absence of Elovl4 staining or the absence of the 32 kDa immunospecific band, respectively.6 Horseradish-conjugated secondary antibodies (rabbit polyclonal and mouse monoclonal) were from Pierce Scientific (Rockford, IL, USA). Fluorescein-conjugated antibodies were anti-rabbit antibody 488 (Invitrogen, Grand Island, NY, USA), and peanut agglutinin-594 (PNA) and 4′6-dimidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA, USA). All solvents for lipid analysis were HPLC grade. Lipid internal standards used for tandem MS analysis were 14:0/14:0 phosphatidylcholine (PC), 14:0/14:0 phosphatidylethanolamine (PE), and 14:0/14:0 phosphatidylserine (PS; Avanti Polar Lipids, Alabaster, AL, USA).
Animals
Mice with the Elovl4 gene containing LoxP sites flanking exons 2 and 3 were mated with a transgenic mouse line expressing Cre recombinase driven by the ChX10 promoter (Jackson Laboratories, Bar Harbor, ME, USA) to delete Elovl4 before photoreceptor differentiation (Elovl4 floxed mice were a gift from Kang Zhang, University of Southern California, San Diego, CA, USA).7 Three genotypes of mice were generated after several backcrosses: Cre−/−/Elovl4f/f (wild type [WT]), Cre±/Elovl4f/f (KO), and Cre±/Elof/WT (Het). The Hets were used as a control for Cre expression. The mice were housed in a facility with a 12-hour light on/off cycle, with on light equal to 5 lux.
Mice were killed with CO2 inhalation or perfusion. Perfusion involved placing a single mouse in a bell jar containing isoflurane until the mouse was deeply anesthetized and unresponsive to intense pinching of the footpad. The heart was exposed, a cannula was placed in the left ventricle, and an incision was made in the right atrium to allow fluid to escape. Paraformaldehyde (2%) and glutaraldehyde (2.5%) in phosphate buffer (PB, 0.1 M, pH 7.4) were perfused at 120 mm Hg for 5 minutes. Eyeball orientation was marked by cauterization, harvested intact, and submerged in the same perfusion fixative cocktail for later analysis.
All procedures were performed according to the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals in Ophthalmic and Vision Research, and the University of Oklahoma Health Sciences Center Guidelines for Animals in Research. All protocols were reviewed and approved by the Institutional Animal Care and Use Committees of the University of Oklahoma Health Sciences Center and the Dean A. McGee Eye Institute.
Western Blot Analysis
Whole retinas were homogenized in T-Per (Pierce Scientific) buffer and 30 μg of protein were loaded from three different mice per genotype for verification of Elovl4 deletion. SDS-PAGE and Western blotting (WB) were performed using standard methods. Primary antibodies were used at 1:500 or 1:1000 dilutions, and secondary antibodies were applied at 1:5000 dilution. Blots were visualized with a Kodak Imager (Kodak Image Station 4000R; Eastman Kodak Company, Rochester, NY, USA). Carestream imaging software (Carestream Health, Inc., Rochester, NY, USA) was used to densitometrically quantify protein expression relative to β-actin expression.
Immunofluorescence
Immunofluorescence labeling was performed on frozen sections using standard procedures described previously16 to visualize the Elovl4 (1:100) expression. Anti-rabbit-594 (Invitrogen) was used at 1:1000 and DAPI was used at 1:5000. Slides were viewed with an FV-500 Olympus Confocal Microscope (Olympus, Tokyo, Japan) and the images were acquired with Flow View software (Olympus America, Center Valley, PA, USA). Contrast and brightness were adjusted with Photoshop (Adobe Systems, Mountain View, CA, USA). Immunofluorescence for cone quantification was performed on paraffin-embedded sections stained with PNA (1:100), visualized with a Nikon epifluorescence microscope (Nikon Eclipse E800 microscope; Nikon, Tokyo, Japan), and processed with ImageJ (National Institutes of Health [NIH], available in the public domain at http://imagej.nih.gov/ij/). Cones were quantified by the threshold of PNA fluorescence relative to the total area of the photoreceptor nuclei and presented as number of cones per unit area.
Quantitative RT-PCR (qRT-PCR)
RNA was extracted from whole retinas using the Trizol reagent (Sigma-Aldrich, St. Louis, MO, USA). The cDNA was reverse transcribed from 1 μg of RNA using the Reverse Transcription kit (Promega, Madison, WI, USA). The qRT-PCR was performed in triplicate according to standard procedures. The Elovl4 message expression was normalized to Hprt expression. Primers used to detect Elovl4 were 5′-TCCAGAAATATCTTTGGTGG-3′ and 5′-GTTAAGGCCCAGTTCAATT-3′. The primers used to detect Hprt were 5′-CTTTGCTGACCTGCTGGATTAC-3′ and 5′-TTGGGGCTGTACTGCTTAACC-3′.
Fatty Acid Analysis
Retinas were pooled from three mice of the same genotype, and extracted by the method of Bligh and Dyer.17 Fatty acids were transesterified to methyl esters18 and analyzed using gas chromatography-mass spectrometry (GC-MS) and gas chromatography-flame ionization detection (GC-FID).19 The chromatographic peaks were integrated and processed with ChemStation software (Agilent Technologies, Santa Clara, CA, USA). Each genotype had 4 distinct samples containing 6 retinas each (n = 4). Lipids were measured in 16-week-old mice.
Tandem Mass Spectrometry Analysis of Retina Lipids (MS/MS)
Retinal lipids were extracted as described previously.20 The PC lipids were identified by using precursor ion (PI) scanning of m/z 184. The PE lipids were identified by scanning for a neutral loss (NL) of 141 m/z, and PS lipids were identified using an NL of 185 m/z. The isolation windows of quadrupoles 1 and 3 were maintained at 0.5 Da, while the collision gas pressure of quadrupole 2 was maintained at 0.5 mtorr of argon. Collision energies were optimized for the lipid class of interest. Automated peak finding, correction for 13C isotope effects, and quantitation of lipid molecular species against internal standards were performed by using the Lipid Mass Spectrum Analysis (LIMSA) software version 1.021 peak model fit algorithm.
ERG Studies
Mice were dark-adapted overnight and ERGs were performed as described previously.22 Rod a-wave amplitude was determined at 8 ms after the flash to exclude intrusion from inner retinal neuron responses. The b-waves were calculated by subtracting the minimum voltage (trough) from maximum voltage (peak) that was generated after the flash. To obtain the cone b-waves, mice were light-adapted for 5 minutes to bleach the rod response and given 15 consecutive flashes at 2000 cd.s/m2 from white, green, and blue light. Cone flicker ERG was performed on light-adapted mice at 5, 10, 20, and 30 Hz. The implicit times for the b-waves were determined as the time (t) after the flash minus an intrinsic delay of 1 ms (t − 1) in which the maximum amplitude was recorded.
The ERG responses were obtained over 6 log units of retinal illuminances and fit to a mathematical model developed by Hood and Birch,23 based on the biochemistry of phototransduction, to determine the saturated response of an individual rod (Rmp3) and the sensitivity (s), which represents amplification of the phototransduction cascade.24 The td is the time delay after flash onset and was predetermined by pairwise optimization of the variables Rmp3, S, and td.25 S was set to 25 s−2(td – s)−1 and td was determined from 9 different 12-month-old WT mice. The average of the optimized td was 2.84 seconds and was used thereafter as a fixed parameter for all 12-month-old mice to determine S and Rmp3. Parameters of this analysis exclude time at which the b-wave intrusion occurs so that the derived a-max is exclusively reflective of the photoreceptor response. Data were analyzed by using The MathWorks software (MatLab, Inc., Natick, MA, USA).
The maximum rod b-wave (Vmax) amplitude was determined as the least squares fit by nonlinear log intensity versus amplitude response analysis. To avoid underestimation of Vmax, responses from light intensities greater than −0.5 cd.s/m2 were fitted according to the Naka-Rushton equation.26 The sensitivity of the b-wave, k, is the retinal illuminance at ½Vmax. The implicit time (IT) was calculated by linear regression analysis of b-wave response time as a function of light intensity, where IT was determined at ½Vmax. Data were analyzed with GraphPad Prism (GraphPad Software, Inc., La Jolla, CA, USA).
Histology
Intact eyeballs were prepared as described previously27 for quantifying the photoreceptor nuclei in the outer nuclear layer (ONL). One eye was used per mouse.
Electron Microscopy
Plastic-embedded eyes were prepared after perfusion using reagents purchased from Electron Microscopy Sciences (EMS; Hatfield, PA, USA). After cauterization to mark orientation, the cornea and lens were removed, and the eyeball was placed in fresh fixative for 2 hours. Eyes were incubated in 1% osmium tetroxide in 0.1 M phosphate buffer (pH 7.4) for 1 hour followed by dehydration in a graded series of ethanol up to 100%. Eyes were embedded in epoxy resin Epon-Araldite (5:3) plus accelerators BDMA and DP30. Ultra-thin (100 nm) sections were lead-stained and viewed with and Hitachi H-7600 transmission electron microscope (Hitachi High Technologies America, Inc., Pleasanton, CA, USA). Images were taken at ×10,000.
Statistical Analyses
Statistical analyses were performed by using GraphPad Prism 5.0 software (GraphPad Software, Inc.). Student's 2-tailed t-test was performed for GC-FID, Rmp3, Vmax, ONL area, cone percentage, and implicit time analysis. Two-way ANOVAs were performed for rod and cone ERG analysis. One-way ANOVA was performed for analysis of lipid species by MS/MS Bonferroni's multiple comparison post hoc test was performed with a 95% confidence interval to determine statistical significance at P < 0.05.
Results
Elovl4 Was Deleted in Photoreceptors
We conditionally deleted Elovl4 in mouse rod and cone photoreceptors using floxed Elovl4 mice bred to mice expressing Cre-recombinase driven by the Chx10 promoter. Chx10 is expressed briefly, but uniformly, in neuroprogenitor cells28 (E9.5–E16.5) and is retained in only a subset of bipolar cells in the adult mouse retina. Thus, we used Chx10-driven Cre to efficiently delete Elovl4 from photoreceptors without constitutively expressing Cre in these cells. Since Chx10 is expressed in neuroprogenitor cells, which precedes differentiation to the other retinal cell types (horizontal, ganglion, amacrine, Müller glial, and bipolar cells), the Chx10-cre would delete Elovl4 if it were expressed in those cells as well. Immunofluorescence microscopy (Fig. 1A) shows that Elovl4 protein expression (red) was located primarily in the inner segments and around the photoreceptor nuclei of Cre−/Elovl4f/f (WT) mice, whereas only background staining was detected in the photoreceptor ONL in the conditional KO (Cre+/Elovl4f/f) retinas.
Figure 1.
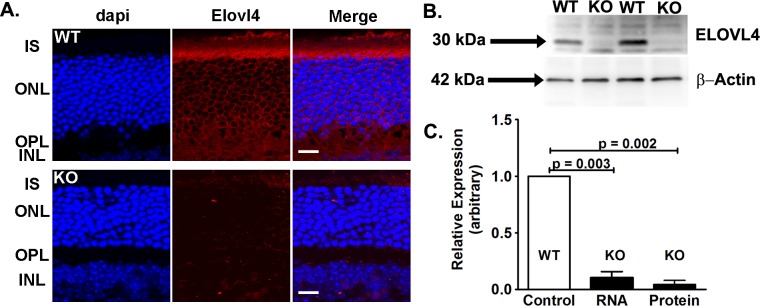
Conditional deletion of Elovl4. (A) Immunofluorescence of WT and KO retinas showed Elovl4 (red) was located in the perinuclear area and the inner segments (IS) in WT, with only background fluorescence in the KO retinas. Scale bars: 20 μm. (B) Immunoblots showed that Elovl4 was deleted in the KO, but not in the WT retina. (C) Elovl4 mRNA in KO retinas relative to Hprt was 9% of the mRNA levels in WT retinas. Elovl4 protein expression relative to β-actin in the KO retina was 4% of the WT levels (n = 7 WT mRNA and 7 WT protein; n = 4 KO mRNA and 5 KO protein). Data are expressed as mean ± SEM.
We measured Elovl4 protein and RNA expression in the WT and KO retinas to assess the efficiency of Cre-recombinase excision of the Elovl4 gene. Representative immunoblots of protein extracts from whole retinas showed that Elovl4 protein levels in the KO retina were barely detectable compared to the protein in the WT retina (Fig. 1B). Quantification of Elovl4 deletion indicated that protein expression was reduced by 96% in the KO mouse retina compared to the WT (Fig. 1C). The Elovl4 mRNA levels were reduced by 91% in the KO compared to the WT retina (Fig. 1C).
To determine if this effective deletion of Elovl4 resulted in changes in retinal lipid biochemistry, we analyzed the content of VLC-PUFAs in total lipids from KO and control retinas by GC-MS, GC-FID, and MS/MS. GC-MS was run first to identify the VLC-PUFA, which then were quantified by GC-FID. The most abundant retinal fatty acid, docosahexaenoic acid (DHA; 22:6n3), was not significantly affected by Elovl4-deletion (Fig. 2A). However, VLC-PUFAs and 24:6n3 were significantly lower in the KO retina compared to control retinas (Fig. 2B). The Elovl4 deletion resulted in 88% less VLC-PUFAs in the total lipid content of the KO retinas compared to WT. Mice heterozygous for the Elovl4 floxed allele (Cre+/Elovl4f/w, Het) had VLC-PUFA levels indistinguishable from WT mice, demonstrating that this reduction in VLC-PUFAs was specific for Elovl4 deletion and not a consequence of Cre transgene expression (Fig. 2B). However, VLC-PUFA precursor 24:6n3 was increased in the heterozygous (Het) mice compared to WT and KO mouse retina.
Figure 2.
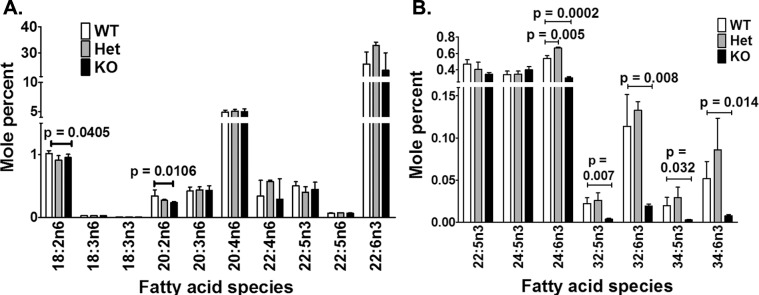
Elovl4 KO mice had reduced VLC-PUFAs. (A) GC-FID showed that C16 to C22 FAs in whole retinas were not affected by Elovl4 ablation (n = 4). (B) The FAs containing more than 26 carbons were barely detectable in KO retinas. Precursor 24:6n3 in KO retina was lower compared to WT (n = 4). Data expressed as the mean ± SD.
Analysis of the molecular species of PC, PE, and PS by tandem mass spectrometry confirmed and expanded the GC-FID results. Comparing 8-week-old WT and KO mice, there were significantly less retinal VLC-PUFA in PC species in Elovl4 KO retinas (Fig. 3), while lipid species containing fatty acids with less than 24 carbons were not grossly altered (Supplementary Fig. S1A). As expected,11 there were no VLC-PUFAs in PE or PS (Supplementary Figs. S1B, S1C, respectively). After 1 year, all VLC-PUFA PC species in the Elovl4 KO retinas trended toward further reductions relative to their levels at 8 weeks of age, whereas VLC-PUFA PC species ≥ 54 total fatty acyl carbons trended to increase in the WT retinas at 12 months of age. The net effect of the observed age-related changes in VLC-PUFA abundances was an exacerbation of the disparity in VLC-PUFA PC levels between WT and Elovl4 KO with age. Histograms of detected PC, PE, and PS are included in Supplementary Figure S1.
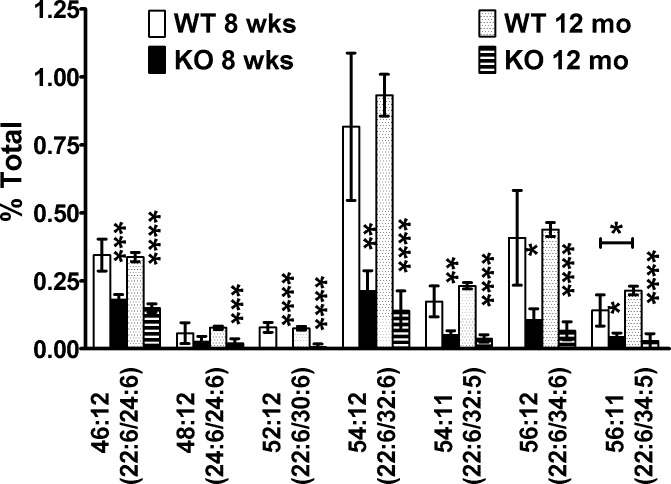
Figure 3.
The VLC-PUFA PC is not replenished in 12-month-old Elovl4 KO retina. The PC molecular species containing VLC-PUFAs (52:12 to 56:11) were not replenished in the retinas of KO mice at 12 months of age compared to 8-week-old KO mice. The VLC-PUFA values were lower in the KO mice than in the age-matched WT mice.
Rod Function Was Impaired in Retinal VLC-PUFA–Deficient Mice
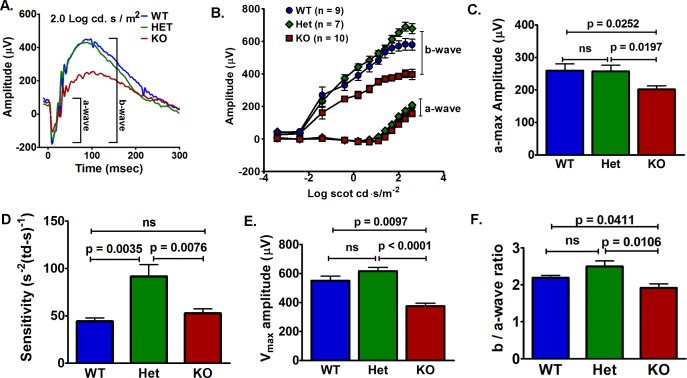
To determine the effects of VLC-PUFAs on rod function, we measured rod responses by ERG. The rod a-wave is a measure of phototransduction cascade triggered by photon capture in the rod outer segments and is indicative of rod function. The rod b-wave reflects the summed bipolar cells response to rod-mediated glutamate concentration in the synaptic cleft. Although there were no differences in rod responses of 5-week-old WT, Het, and KO mice (Supplementary Fig. S2A), we found reductions in rod-mediated function (a- and b-waves) in 12-month-old KO mice compared to congenic controls (Fig. 4A). The KO responses to increasing light intensities failed to reach the same amplitudes as the WT and Het mice (Fig. 4B). It is important to note that the a-wave was measured at a time (8 ms) before intrusion of the b-wave, so that voltage contribution from secondary neurons would not mask the rod response. The ERG responses were fit to a model of phototransduction that also excludes b-wave intrusion to determine the maximum rod photocurrent and the sensitivity of the phototransduction cascade.23 The VLC-PUFA–deficient mice had a 22% decrease in maximum rod response amplitude (Rmp3) compared to WT and Het mice (Fig. 4C). The sensitivity (S) of Het rod response was higher compared to WT and KO mice, but was not different between the WT and KO mice (Fig. 4D).
Figure 4.
Rod-mediated function deteriorated in 12-month-old VLC-PUFAs–deficient mice. (A) Representative ERG response to Log 2.0 cd.s/m2 from WT, Het, and KO mice. (B) The KO rod (a-wave) and bipolar (b-wave) responses to increasing light intensities had lower amplitudes compared to WT and Het mice. (C) Maximum rod response (Rmp3) from the KO (201.3 ± 11.21) mice was lower than the responses from the WT (259.5 ± 20.73) and Het (257.0 ± 19.58) mice. (D) Rod Rmp3 sensitivity was increased in Het (91.6 ± 12.61) mice, but was not different between the WT (44.2 ± 3.57) and KO (52.8 ± 4.8) mice. (E) Maximum bipolar response (Vmax) from the KO mice was lower than the responses from WT and Het mice. (F) The b-/a-wave ratio was lower in KO (1.92 ± 0.11) compared to WT (2.19 ± 0.06) and Het (2.45 ± 0.13) mice (n = 8 WT, 6 Het, and 6 KO). Data are represented as the mean ± SEM. ns, not significant.
The b-wave was analyzed to determine the maximum bipolar cell amplitude (Vmax) and the implicit time (IT), which is time after the flash when the maximum b-wave amplitude occurs. At 5 weeks, there were no differences in Vmax, sensitivity, or IT (Supplementary Fig. S2B). At 12 months, the KO mice had a 32% lower Vmax than WT and 39% lower Vmax amplitude than Het mice (Fig. 4E). At 12 months of age, the VLC-PUFA–deficient mice had a 16.9 and 24.4 msec delay in the IT compared to WT and Het mice, respectively (Supplementary Fig. S3). At 12 months, the KO mice had significantly reduced b/a-wave ratio (1.92 ± 0.11) compared to WT (2.19 ± 0.06) and Het (2.45 ± 0.13) mice (Fig. 4F), which indicated that the KO b-wave had decreased disproportionately to the decreased a-wave. To avoid b-wave intrusion, the maximum responses (Rmp3 and Vmax) were used to calculate the b/a-wave ratio. Additionally, the b-wave response appears 2 orders of magnitude before the a-wave response so ratios to specific light intensities would be limited by the occurrence of the a-wave response. The reduction of b-wave in the KO mice was observed at low light intensities before the a-wave amplitude was evident and exceeded the reduction of the a-wave indicated by the b-/a-wave ratio (Fig. 4B). These results indicated that the reduced rod-mediated bipolar cell responses could not be attributed to the loss of rod cells, because the a- and b-waves would have decreased proportionally if this were the case.
Mice With Reduced VLC-PUFA Had a Loss of Photoreceptors
To determine if the loss of rod cells could explain the decreased rod responses in the VLC-PUFA–deficient mice, we examined the retinal morphology and found no apparent defects in the younger mice (Supplementary Fig. S4). There was, however, a small but significant loss of rod photoreceptor nuclei in the 12-month-old KO mice (Fig. 5A). The total area occupied by photoreceptor nuclei was 11% smaller in the Elovl4 KO retina than in the control retinas at 12 months (Fig. 5B), which could account for the smaller a-wave responses in the KO mice.
Figure 5.
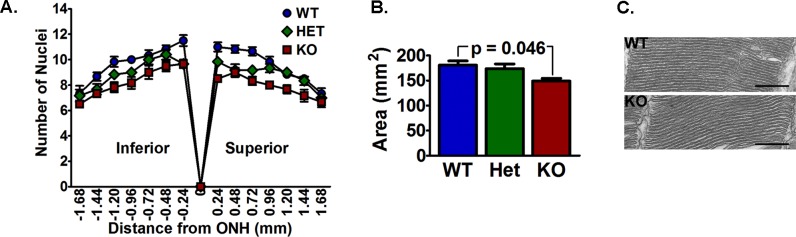
The VLC-PUFA–deficient mice had a loss of photoreceptors. (A) Histogram of the number of photoreceptor nuclei in a column at indicated distances from the optic nerve head to the peripheral retina. (B) Area occupied by photoreceptor nuclei measured from hematoxylin and eosin–stained sections was smaller in the KO mice than in the WT mice, but was not different compared to the Het retina (n = 5 WT, 7 Het, and 6 KO). (C) Representative micrographs of rod outer segments were similar between the WT and KO mice. Scale bars: 500 nm.
The VLC-PUFAs have been suggested to provide disk structure and membrane fluidity to photoreceptor outer segments (OS) and, thus, influence phototransduction efficiency.6,14 Therefore, we examined rod outer segment disk ultrastructure in 12-month-old WT and KO mice. Micrographs showed that the disks appeared similar between the two groups (Fig. 5C). This supported that the reduction of the a-wave was due to a loss of photoreceptor cells.
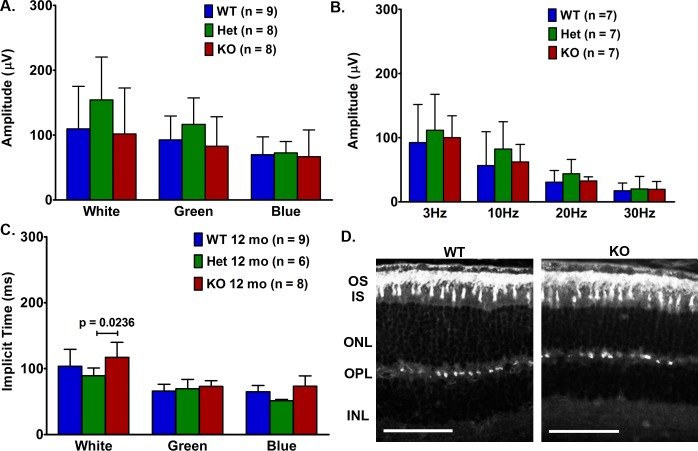
Reduced VLC-PUFA Did Not Affect Cone Photoreceptors
To investigate the role of VLC-PUFAs in cone photoreceptors, we used ERG to test the function of light-adapted Elovl4 WT, HET, and KO mice at 12 months of age. We found similar cone responses for all groups of mice to white, blue, and green light stimuli (Figs. 6A, 6B). The KO cone response time to white light was increased slightly compared to the Het mice, but all other cone response times were comparable between the mice (Fig. 6B). Additionally, we measured cone responses to different frequencies of light stimuli (flicker) in 12–month-old mice at 3, 10, 20, and 30 Hz pulse frequencies (Fig. 6C). We found no effects of retinal VLC-PUFA depletion on cone function. Furthermore, the presence and distribution of cones were similar between the groups of mice as indicated by PNA immunofluorescence (white) labeling of cone OS sheaths and the cone pedicles located in the outer plexiform layer (OPL; Fig. 6D). The percent of cones relative to total area occupied by the photoreceptor nuclei (density of fluorescence per mm2) in the 12-month-old mice was comparable between the Elovl4 WT and KO mice (8.284 ± 0.3637, n = 4 and 8.859 ± 0.6373, n = 5, respectively).
Figure 6.
Analysis of cones in Elovl4 WT and KO mice. (A) Responses to white light, medium wavelength green light, and short wavelength blue light were similar in age-matched WT and KO mice at 5 weeks and 12 months of age. (B) Cone response times were not different between age-matched WT and KO mice. (C) Cone flicker responses (b-wave) to indicated frequencies showed no differences between WT and KO mice at 12 months of age. (D) The PNA immunolabeling of cone outer segment sheaths and cone pedicles indicated similar cone distribution between 12-month-old WT and KO mice. Scale bars: 50 μm; 5-week (n = 10 WT, 5 Het, and 10 KO). 12-month (n = 8 WT, 7 Het, and 8 KO). The ERG response data are expressed as mean ± SEM.
Discussion
Photoreceptor-specific deletion of Elovl4 significantly reduced the levels of VLC-PUFAs in the retina, but did not alter the overall composition of the major retinal fatty acids (Figs. 2, 3; Supplementary Figs. S2, S3). These results supported previously published work showing that Elovl4 elongates long chain PUFAs to VLC-PUFA in Elovl4-transduced cells and in vivo where Elovl4 was conditionally deleted independently in either rod or cone cells.6,7,29 In contrast with these reports, however, we have demonstrated that reduction of retinal VLC-PUFA PC in Elovl4 KO mice is maintained over time, and by 12 months the differences in the abundance of retinal PC VLC-PUFAs between the WT and Elovl4 KO mice were amplified (Fig. 3). Our results implied that these fatty acids are synthesized locally and remain stable over time, and that the effects of retinal Elovl4 deletion are likely to be compounded with age. Furthermore, the very low levels of VLC-PUFAs at 12 months indicated that other Elovl4-expressing tissues are not able to replenish photoreceptors deficient in these fatty acids. It currently is unclear as to why the Het mice did not have half the total amount of VLC-PUFAs compared to WT or twice as much as the KO because they express one functional copy of Elovl4. One explanation could be that elongation catalysis is regulated by specific fatty acid concentration.19,30 Reducing elongase expression could alter fatty acid concentrations resulting in compensentory upregulation of Elovl4 catalytic activity.
Extensive cone function analyses at multiple time points (not shown) revealed no differences in cone b-wave amplitudes to different wavelengths or frequencies of light stimulation (Figs. 6A, 6B). There were only minor changes in cone b-wave implicit time and no signs of cone loss even after 12 months of VLC-PUFA deprivation (Fig. 6C). These results agreed with those of Barabas et al.,29 but are contrary to those of Harkewicz et al.,7 who found that cone-specific deletion of Elovl4 resulted in the dysfunction of cone flicker responses at 7 months of age compared to generic WT mice. We did not find cone flicker dysfunction in our 12-month-old KO mice in which rods and cones were targeted for Elovl4 deletion, compared to congenic control WT and Het mice. It currently is unclear why cone-only deletion of Elovl4 would result in cone dysfunction in one study, while our mice with Elovl4 deletion in rods and cones did not have cone structural or functional deficits. One possibility for the difference could be different mouse backgrounds and/or the absence of Cre-expressing controls in the other studies. A recent study reported that transgenic mice expressing a mutated Elovl4 had comparable lipid profiles with WT mice, but had rod loss beginning at 2 months followed by a late onset of cone degeneration at 24 months.31 Thus, we cannot rule out cone degeneration in the VLC-PUFA–deficient mice as late onset cone degeneration or as a secondary late event resulting from the rod degeneration.
We found that mice with reduced VLC-PUFAs had diminished rod function (a-wave, Rmp3) at 12 months of age compared to controls, which correlated with a loss of photoreceptor nuclei (Figs. 4, 5), in agreement with Harkewicz et al.,7 who also reported that rod-specific Elovl4 conditional KO (R-cKO) adult mice had retinal degeneration at 10 and 15 months of age. Barabas et al.29 did not find photoreceptor degeneration in R-cKO mice, but their mice were younger, which may explain the difference between the studies, since time exacerbates the detrimental effects of VLC-PUFA reduction as we have shown here (Figs. 4, 5).
Loss of rod-mediated bipolar cell responses (b-wave; Vmax) first became evident at 6 months of age (not shown), and was greater in Elovl4 KO mice by 12 months of age compared to WT and Het mice (Fig. 4). These results agreed with those of Harkewicz et al.,7 who reported that R-cKO mice had decreased b-wave responses, but disagreed with those of Barabas et al.,8 who did not find rod-mediated deficits in their 6-month-old R-cKO mice. There are several possible explanations for the contradictory results. First, the genotype of the mice used in the previous studies was Rod Opsin-Cre+/Elovl4f/f, which resulted in Elovl4 ablation only from rod cells (with a 77% Cre-efficiency32), whereas the Chx10-Cre+/Elovl4f/f mice used here had Elovl4 deleted in rods and cones (with more than 95% Cre-efficiency33). Additionally, in the previous studies, Cre expression remained in the photoreceptors of adult mice, unlike the Chx10-Cre mice that did not express Cre in adult photoreceptors. The Elovl4f/f mice used in all three studies were from the same founders and had intronic LoxP sites.7,8 In the absence of Cre-recombinase, WT Elovl4 protein would be expressed and, therefore, is considered a good generic control.8 However, this strategy does not control for potential consequences of Cre transgene expression itself. In the current study, we included mice that expressed Cre and were heterozygous for the floxed allele. Given that VLC-PUFAs were not reduced in these mice and other measured parameters were not different, our results unequivocally showed the consequences of tissue-specific Elovl4 ablation, and assured that the resulting structural and functional abnormalities are not due to off-target effects of Cre expression. Whether there is a masking effect and Elovl4 is expressed elsewhere in the retina still could be the case and the Chx10 deletion would result in loss of Elovl4 in these cells. However, our antibody can be peptide blocked as shown by Agbaga et al.6 and does not recognize a protein in the Elovl4 KO retina. Since we do not detect Elovl4 in other retinal cells it is unlikely that the phenotypes presented here result from deletion of the gene in other retinal cells.
The Elovl4 protein has been well established as the elongase involved in the initial rate limiting step in the production of VLC-PUFAs.6–8 There has been controversy, however, surrounding the exact role of VLC-PUFAs in the retina. The results presented here clearly showed that VLC-PUFAs are vital for rod function and rod longevity. In the companion paper, we showed a reduction in rod ERG oscillatory potentials and scotopic threshold responses in KO mice, and presented biochemical and morphologic evidence that the ERG changes are correlated with reduced VLC-PUFAs and synaptic architecture.
Acknowledgments
The authors thank members of the Dean Bok laboratory (University of California, Los Angeles [UCLA], Los Angeles, CA, USA) for their help in the perfusion experiments, and Mark Dittmar for his invaluable assistance with the animals.
Supported by NIH Grants EY00871, EY04149, P30EY021725, and P20RR017703 (REA); and EY019494 (MHE); and by the Foundation Fighting Blindness (REA); Research to Prevent Blindness (Departmental); and Grants GM103508 (GER, JVB) and EY016077 (JVB, GER).
Disclosure: L.D. Bennett, None; R.S. Brush, None; M. Chan, None; T.A. Lydic, None; K. Reese, None; G.E. Reid, None; J.V. Busik, None; M.H. Elliott, None; R.E. Anderson, None
References
- 1. Edwards AO, Donoso LA, Ritter R III. A novel gene for autosomal dominant Stargardt-like macular dystrophy with homology to the SUR4 protein family. Invest Ophthalmol Vis Sci. 2001; 42: 2652–2663 [PubMed] [Google Scholar]
- 2. Zhang K, Kniazeva M, Han M, et al. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet. 2001; 27: 89–93 [DOI] [PubMed] [Google Scholar]
- 3. Donoso LA, Frost AT, Stone EM, et al. Autosomal dominant Stargardt-like macular dystrophy: founder effect and reassessment of genetic heterogeneity. Arch Ophthalmol. 2001; 119: 564–570 [DOI] [PubMed] [Google Scholar]
- 4. Griesinger IB, Sieving PA, Ayyagari R. Autosomal dominant macular atrophy at 6q14 excludes CORD7 and MCDR1/PBCRA loci. Invest Ophthalmol Vis Sci. 2000; 41: 248–255 [PubMed] [Google Scholar]
- 5. Kniazeva M, Chiang MF, Morgan B, et al. A new locus for autosomal dominant stargardt-like disease maps to chromosome 4. Am J Hum Genet. 1999; 64: 1394–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Agbaga MP, Brush RS, Mandal MNA, Henry K, Elliott MH, Anderson RE. Role of Stargardt-3 macular dystrophy protein (ELOVL4) in the biosynthesis of very long chain fatty acids. Proc Nat Acad Sci U S A. 2008; 105: 12843–12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harkewicz R, Du H, Tong Z, et al. Essential role of ELOVL4 protein in very long chain fatty acid synthesis and retinal function. J Biol Chem. 2012; 287: 11469–11480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barabas P, Liu A, Xing W, et al. Role of ELOVL4 and very long-chain polyunsaturated fatty acids in mouse models of Stargardt type 3 retinal degeneration. Proc Natl Acad Sci U S A. 2013; 110: 5181–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Logan S, Agbaga MP, Chan MD, et al. Deciphering mutant ELOVL4 activity in autosomal-dominant Stargardt macular dystrophy. Proc Natl Acad Sci U S A. 2013; 110: 5446–5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mandal MNAR, Wong PW, Gage PJ, Sieving PA, Ayyagari R. Characterization of mouse orthologue of ELOVL4: genomic organization and spatial and temporal expression. Genomics. 2004; 83: 626–635 [DOI] [PubMed] [Google Scholar]
- 11. Vasireddy V, Uchida Y, Salem N Jr, et al. Loss of functional ELOVL4 depletes very long-chain fatty acids (> or =C28) and the unique omega-O-acylceramides in skin leading to neonatal death. Hum Mol Genet. 2007; 16: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li W, Sandhoff R, Kono M, et al. Depletion of ceramides with very long chain fatty acids causes defective skin permeability barrier function, and neonatal lethality in ELOVL4 deficient mice. Int J Biol Sci. 2007; 3: 120–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Furland NE, Oresti GM, Antollini SS, Venturino A, Maldonado EN, Aveldano MI. Very long-chain polyunsaturated fatty acids are the major acyl groups of sphingomyelins and ceramides in the head of mammalian spermatozoa. J Biol Chem. 2007; 282: 18151–18161 [DOI] [PubMed] [Google Scholar]
- 14. Aveldaño MI, Sprecher H. Very long chain (C24 to C36) polyenoic fatty acids of the n-3 and n-6 series in dipolyunsaturated phosphatidylcholines from bovine retina. J Biol Chem. 1987; 262: 1180–1186 [PubMed] [Google Scholar]
- 15. Aveldaño MI. A novel group of very long chain polyenoic fatty acids in dipolyunsaturated phosphatidylcholines from vertebrate retina. J Biol Chem. 1987; 262: 1172–1179 [PubMed] [Google Scholar]
- 16. Ueki Y, Wang J, Chollangi S, Ash JD. STAT3 activation in photoreceptors by leukemia inhibitory factor is associated with protection from light damage. J Neurochem. 2008; 105: 784–796 [DOI] [PubMed] [Google Scholar]
- 17. Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Canad J Biochem Physiol. 1959; 37: 911–917 [DOI] [PubMed] [Google Scholar]
- 18. Morrison WR, Smith LM. Preparation of fatty acid methyl esters and dimethylacetals from lipids with boron fluoride--methanol. J Lipid Res. 1964; 5: 600–608 [PubMed] [Google Scholar]
- 19. Yu M, Benham A, Logan S, et al. ELOVL4 protein preferentially elongates 20:5n3 to very long chain PUFAs over 20:4n6 and 22:6n3. J Lipid Res. 2012; 53: 494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Julia V, Busik GER, Lydic TA. Global analysis of retina lipids by complementary precursor ion and neutral loss mode tandem mass spectrometry. Methods Mol Biol. 2009; 579: 33–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haimi P, Uphoff A, Hermansson M, Somerharju P. Software tools for analysis of mass spectrometric lipidome data. Anal Chem. 2006; 78: 8324–8331 [DOI] [PubMed] [Google Scholar]
- 22. Li F, Marchette LD, Brush RS, et al. DHA does not protect ELOVL4 transgenic mice from retinal degeneration. Mol Vis. 2009; 15: 1185–1193 [PMC free article] [PubMed] [Google Scholar]
- 23. Hood DC, Birch DG. Rod phototransduction in retinitis pigmentosa: estimation and interpretation of parameters derived from the rod a-wave. Invest Ophthalmol Vis Sci. 1994; 35: 2948–2961 [PubMed] [Google Scholar]
- 24. Lamb TD, Pugh EN Jr. A quantitative account of the activation steps involved in phototransduction in amphibian photoreceptors. J Physiol. 1992; 449: 719–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weymouth AE, Vingrys AJ. Rodent electroretinography: methods for extraction and interpretation of rod and cone responses. Prog Retin Eye Res. 2008; 27: 1–44 [DOI] [PubMed] [Google Scholar]
- 26. Naka KI, Rushton WA. S-potentials from luminosity units in the retina of fish (Cyprinidae). J Physiol. 1966; 185: 587–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marchette LD, Wang H, Li F, Babizhayev MA, Kasus-Jacobi A. Carcinine has 4-hydroxynonenal scavenging property and neuroprotective effect in mouse retina. Invest Ophthalmol Vis Sci. 2012; 53: 3572–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu IS, Chen JD, Ploder L, et al. Developmental expression of a novel murine homeobox gene (Chx10): evidence for roles in determination of the neuroretina and inner nuclear layer. Neuron. 1994; 13: 377–393 [DOI] [PubMed] [Google Scholar]
- 29. Barabas P, Liu A, Xing W, et al. Role of ELOVL4 and very long-chain polyunsaturated fatty acids in mouse models of Stargardt type 3 retinal degeneration. Proc Natl Acad Sci U S A. 2013; 110: 5181–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hubbard AF, Askew EW, Singh N, Leppert M, Bernstein PS. Association of adipose and red blood cell lipids with severity of dominant Stargardt macular dystrophy (STGD3) secondary to an ELOVL4 mutation. Arch Ophthalmol. 2006; 124: 257–263 [DOI] [PubMed] [Google Scholar]
- 31. Kuny S, Filion M-A, Suh M, Gaillard F, Sauvé Y. Long-term retinal cone survival and delayed alteration of the cone mosaic in a transgenic mouse model of Stargardt-like dystrophy (STGD3). Invest Ophthalmol Vis Sci. 2013; 55: 424–439 [DOI] [PubMed] [Google Scholar]
- 32. Le YZ, Zheng L, Zheng W, et al. Mouse opsin promoter-directed Cre recombinase expression in transgenic mice. Mol Vis. 2006; 12: 389–398 [PubMed] [Google Scholar]
- 33. Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol. 2004; 271: 388–402 [DOI] [PubMed] [Google Scholar]