Abstract
RNA-sequencing was performed on three tenosynovial giant cell tumors (TSGCT) in an attempt to elicit more information on the mechanisms of CSF1 expression in this tumor type. A novel CSF1-S100A10 fusion gene was found in a TSGCT that carried the translocation t(1;1)(q21;p11) as the sole karyotypic abnormality. In this fusion gene, the part of CSF1 coding for the CSF1 protein (exons 1–8 in sequences with accession nos. NM_000757 and NM_172212) is fused to the 3′-part of S100A10. Since the stop codon TAG of CSF1 is present in it, the CSF1-S100A10 fusion gene’s predominant consequence seems to be the replacement of the 3′-untranslated region (UTR) of CSF1 (exon 9; nt 2092–4234 in sequence with accession no. NM_000757 or nt 2092–2772 in NM_172212) by the 3′-end of S100A10 (exon 3; nt 641–1055 in sequence with accession no. NM_002966). In the other two TSGCT, a novel CSF1 transcript was detected, the same in both tumors. Similar to the occurrence in the CSF1-S100A10 fusion gene, the novel CSF1 transcript 3′-UTR is replaced by a new exon located ∼48 kb downstream of CSF1 and 11 kb upstream of AHCYL1. Although only 3 TSGCT were available for study, the finding in all of them of a novel CSF1-S100A10 fusion gene or CSF1 transcript indicates the existence of a common pathogenetic theme in this tumor type: the replacement of the 3′-UTR of CSF1 with other sequences.
Keywords: tenosynovial giant cell tumor, RNA sequencing, CSF1, CSF1-S100A10 fusion gene, 3′-untranslated region
Introduction
Tenosynovial giant cell tumors (TSGCT) usually occur in tendon sheaths and in the synovia of joints and bursae (1). The tumor is also known under a variety of other names including pigmented villonodular synovitis (PVNS) (1,2). TSGCT is more common in women than in men, may occur at any age, and may be more or less diffuse in growth pattern (3). The localized tumors are the most common, they are found predominantly in fingers, develop slowly over years, and manifest themselves as small, circumscribed tumors that recur in 10–20% of the cases. Most diffuse TSGCT are larger than their localized counterparts, occur in large joints (in particular the knee), have an expansive growth pattern, and up to 50% recur locally. TSGCT are often moderately cellular with mononuclear cells, scattered giant cells, xanthoma cells and some degree of collagenization.
The pathogenetic mechanisms underlying TSGCT have been debated; some authors see them as inflammatory in nature (1,4), whereas the predominating view at present holds that they are neoplastic (2,3,5), something that also concurs with the opinion held by the original describers of the entity (6). The description of TSGCT-like lesions after injection of pro-inflammatory agents (7) and the finding of polyclonality in lyonization studies (8,9) argue for an inflammatory disease mechanism, whereas the finding of monoclonality in another X-inactivation study (10) as well as the detection of recurrent clonal chromosomal aberrations (11–15) suggest a neoplastic pathogenesis. Partly because of the uncertainty regarding the nature of this disease, the existence of malignant TSGCT has been questioned, but it seems that such tumors do exist, rare though they are (16–18). The preferred treatment for TSGCT is surgery with free margins to avoid recurrence, and the prognosis is good (3). At present, no adjuvant therapy is recommended for most lesions.
It seems that over 50% of TSGCT have an abnormal karyotype, but this can of course be a misrepresentation because there is a general tendency towards reporting abnormal findings more often than normal ones (19). Most TSGCT with chromosomal aberrations are near-diploid or pseudodiploid and ∼50% carry balanced chromosomal rearrangements. So far, 28 cases of the diffuse type and 18 localized TSGCT have been reported with chromosomal aberrations (http://cgap.nci.nih.gov/Chromosomes/Mitelman). Gains of chromosome 7 and/or chromosome 5 have been the most common numerical aberrations by karyotyping but only in the diffuse type of TSGCT. Structural rearrangements preferably involve chromosomal areas 1p11–13, 2q35–37, and 16q22–24 (14) and two distinct subgroups of TSGCT can be recognized based on the above-mentioned structural aberrations: those with 1p11–13 rearrangements and those with 16q22–24 rearrangements. Several translocation partners have participated in the changes affecting 1p11–13 (http://cgap.nci.nih.gov/Chromosomes/Mitelman) of which 2q35–37 is the most common (14). These results were confirmed in a study by West et al (20) who additionally identified the colony-stimulating factor-1 (CSF1 or M-CSF1) locus at 1p13 as a molecular target of chromosomal rearrangements in 20 of 23 TSGCT. Chromosome 2 was the translocation partner in a subset (3 of 10) of tumors and collagen type VI α-3 (COL6A3) was then identified as the partner gene involved at 2q37. Moreover, combined interphase FISH and CSF1 immunohistochemistry demonstrated that only a minority (2–16%) of the cells in the tumor samples carried the t(1;2)(p13;q37) and that only those cells expressed CSF1, while tissue microarray analyses showed that the CSF1 receptor gene CSF1R is highly overexpressed in TSGCT (20). To explain these findings, it was suggested that the translocation involving CSF1 and COL6A3 results in a high level of CSF1 expression in the neoplastic parenchyma cells, which in turn recruit also non-neoplastic CSF1R-expressing cells. In the words of West et al (20): ‘The CSF1-COL6A3 translocation in TSGCT and PVNS is reminiscent of the translocation that defines DFSP (dermatofibrosarcoma protuberans). In this malignancy, t(17;22) brings PDGF-B under control of the strong COL1A1 promoter. The posttranslationally processed form of the fusion protein is a fully functional PDGF-B protein that may stimulate oncogenesis through its receptor, PDGFRB. The PDGFRB receptor is also upregulated in DFSP, suggesting an autocrine loop’. Based on the suggestion that a fusion of these genes through the translocation would result in overexpression of CSF1 due to a strong COL6A3 promoter, Möller et al (21) performed RT-PCR on six TSGCT cases with t(1;2) to search for a putative COL6A3-CSF1 fusion gene. Such fusion transcripts were detected in three cases and in one of them it was in-frame. In all cases, however, the breakpoints in CSF1 appeared downstream of exon 5, indicating that the amino-terminal part of CSF1, which interacts with its receptor CSF1R, was not encoded by the identified chimeric transcripts (21,22). The authors concluded that ‘the COL6A3-CSF1 fusion transcripts identified in the present study are unlikely to produce a functional protein containing the receptor-binding part of CSF1, which is required for the mechanism described earlier’ (21). In another study, Cupp et al (23) examined 57 TSGCT/PVNS and in all of them found expression of CSF1 mRNA and/or CSF1 protein, including 22 tumors (39%) that lacked a CSF1 translocation, suggesting that alternative mechanisms may lead to upregulation of CSF1.
In the present study we performed RNA-sequencing of three TSGCT in an attempt to elicit more information on the mechanisms of altered CSF1 expression in this tumor type.
Materials and methods
Ethics statement
The study was approved by the regional ethics committee (Regional komité for medisinsk forskningsetikk Sør-Øst, Norge, http://helseforskning.etikkom.no), and written informed consent was obtained from the patients.
Patients
Case 1 was a localized TSGCT. Cases 2 and 3 were both diffuse type tumors and were previously reported (11). Culturing and cytogenetic analysis were done as previously described (11).
High-throughput paired-end RNA-sequencing
Total RNA was extracted from the three tumors using TRIzol reagent according to the manufacturer’s instructions (Invitrogen, Life Technologies, Oslo, Norway) and its quality was checked by Experion (Bio-Rad Laboratories, Oslo, Norway). Total RNA (3 μg) was sent for high-throughput paired-end RNA-sequencing to the Norwegian Sequencing Centre at Ullevål Hospital (http://www.sequencing.uio.no/). The Illumina software pipeline was used to process image data into raw sequencing data and only sequence reads marked as ‘passed filtering’ were used in the downstream data analysis. A total of 60 million reads were obtained. The FASTQC software was used for the quality control of the raw sequence data (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The software FusionMap and the associated pre-built Human B37 and RefGene from the FusionMap website were used for the discovery of fusion transcripts (24), release date 2012-04-16 (http://www.omicsoft.com/fusionmap/).
PCR and 3′-RACE
The primers used for PCR amplification and sequencing are listed in Table I. The Human Universal Reference Total RNA was used as control (Clontech Laboratories, Takara Bio Group, France). According to the company’s information it is a ‘mixture of total RNAs from a collection of adult human tissues, chosen to represent a broad range of expressed genes. Both male and female donors are represented’. Expression analysis was performed using the human cell line MTC panel (Clontech Laboratories, Takara Bio Group), which contains cDNA from the cell lines Ad5, SKOV-3, Saos2, A431, Du145, H1299, HeLa, and MCF7.
Table I.
Primers used for PCR amplifications and sequencing.
| Oligo name | Sequence (5′→3′) |
|---|---|
| CSF1-1886F | GCA GCT CCA GGA GTC TGT CTT CCA C |
| CSF1-2006F | GGA TTC TCC CTT GGA GCA ACC AGA |
| CSF1-2057F | ACA GGT GGA ACT GCC AGT GTA GA |
| CSF1-2176R | AGC TCT GGT GGA GGG CAG ACC A |
| CSF1-3end-R1out | ATT CAC AGA CCA ACA AAT GCT GCC A |
| CSF1-trans4-2130R | GCT GGG CGT CAC ATT TTC AGA GG |
| CSF1-3end-R2out | TGG GAT GTT TGC AGA ATC CTT GTC A |
| CSF1-trans1-2123R | GTG GAC GCC CCA TAA TGT CTC |
| CSF1-trans4-2120R | CAC ATT TTC AGA GGG ACA TTG ACA |
| CSF1-transNew-Rq | ACA CCA TGC TCA TGG GGT TTA G |
| S100A10-555F | TTC ACA AAT TCG CTG GGG ATA AAG G |
| S100A10-664R | TCC AGG TCC TTC ATT ATT TTG TCC |
| S100A10-809R | TAT CAG GGA GGA GCG AAC TGC TCA T |
| S100A10-840R | GAT TCC TTA AGC GAC CCT TTG GGA C |
| A3RNV-RACE | ATC GTT GAG ACT CGT ACC AGC AGA GTC ACG AGA GAG ACT ACA CGG TAC TGG TTT TTT TTT TTT TTT |
| A3R-1New | TCG TTG AGA CTC GTA CCA GCA GAG TCA C |
| A3R-2New | GAG TCA CGA GAG AGA CTA CAC GGT ACT GGT T |
Total RNA (2 μg) was reverse-transcribed in a 20 μl reaction volume using iScript Advanced cDNA Synthesis kit for RT-qPCR according to the manufacturer’s instructions (Bio-Rad Laboratories, Oslo, Norway). The cDNA was diluted to 20 ng equivalent of RNA/μl and 2 μl were used as templates in subsequent PCR assays. The 25-μl PCR volume contained 12.5 μl of Premix Taq (Takara-Bio), 2 μl of diluted cDNA (or 2 μl of cDNA from the cDNA panels which corresponds to 4 ng of cDNA), and 0.4 μM of each of the forward and reverse primers. The PCRs were run on a C-1000 Thermal cycler (Bio-Rad Laboratories). The PCR conditions were: an initial denaturation at 94˚C for 30 sec followed by 35 cycles of 7 sec at 98˚C, 120 sec at 68˚C and a final extension for 5 min at 68˚C.
For detection of the CSF1-S100A10 fusion transcript the primer set CSF1-1886F/S100A10-840R was used. For the amplification of the new CSF1 transcript (transcript 5, see below) the primer set CSF1-1886F/CSF1-3end-R1out was used. For the amplification of CSF1 transcript 1 (NM_000757) the primer set CSF1-1886F/CSF1-2176R was used. This primer combination and the primer set CSF1-2057F and CSF1-trans1-2123R (which is used for real-time PCR) also detect the transcript variant 2 (NM_172210) which lacks an alternate in-frame segment in exon 6 compared to transcript 1, resulting in a shorter isoform b compared to isoform a. No investigation was done for the expression of transcript 2, and for simplicity we refer in this report to primer set CSF1-1886F/CSF1-2176R as amplifying transcript 1 (in reality 1 and 2). For the amplification of CSF1 transcript 4 (NM_172212), the primer set CSF1-1886F/CSF1-trans4-2130R was used. For amplification of S100A10 wild-type transcript, the primer set S100A10-555F/S100A10-840R was used.
For 3′-RACE, 1 μg of total RNA was reverse-transcribed in a 20 μl reaction volume with the A3RNV-RACE primer (Table I) using iScript Select cDNA Synthesis kit according to the manufacturer’s instructions (Bio-Rad Laboratories). A 1 μl template was amplified using the outer primer combination CSF1-1886F/A3R-1New. The amplified products were diluted 1:1,000 and 1 μl of the diluted amplified PCR product was used as template in nested PCR with the primers CSF1-2006F/A3R-2New. For both PCRs the 25 μl reaction volume contained 12.5 μl of Premix Taq (Takara-Bio), template, and 0.4 μM of each of the forward and reverse primers. PCR cycling consisted of an initial step of denaturation at 94˚C for 30 sec followed by 35 cycles of 7 sec at 98˚C, 30 sec at 60˚C, 3 min at 72˚C (2 min for nested PCR), and a final extension for 5 min at 72˚C.
PCR products (4 μl) was stained with GelRed (Biotium), analyzed by electrophoresis through 1.0% agarose gel and photographed. The amplified fragments were purified using the NucleoSpin Gel and PCR clean-up kit (Macherey-Nagel), and direct sequencing was performed using the light run sequencing service of GATC Biotech (http://www.gatcbiotech.com/en/sanger-services/lightrun-sequencing.html). The BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat) programs were used for computer analysis of sequence data.
Real-time PCR
To quantify the expression of CSF1 transcript 1 (NM_000757), CSF1 transcript 4 (NM_172212), the new CSF1 transcript described here, and the CSF1-S100A10 fusion transcript, SYBR Green-based gene expression real-time PCR was used. The 20 μl reaction volume contained 1X SsoAdvanced SYBR Green supermix (Bio-Rad), 0.5 μM of each of the forward and reverse primers, and 2 μl cDNA (40 ng equivalent of RNA). Four replicates of each sample were used to ensure statistical representativity. For CSF1 transcript 1 the primer set was CSF1-2057F and CSF1-trans1-2123R, for CSF1 transcript 4 the primer set was CSF1-2057F and CSF1-trans4-2120R, for the newly identified CSF1 transcript 5 the primer set was CSF1-2057F and CSF1-transNew-Rq, and for the CSF1-S100A10 fusion transcript the primer set was CSF1-2057F and S100A10-664R. To evaluate the efficiency of the real-time PCR reactions, standard curves were generated using serial dilutions of purified PCR products which were amplified with Premix Taq and the primer set described above in ‘PCR and 3′-RACE’. To generate standard curves seven 10-fold dilutions were prepared for each PCR product starting with 0.1 ng. The efficiency (E), correlation coefficient (R^2), slope and y-intercept are given in Table II.
Table II.
Standard curve analyses of the real-time PCR for the quantification of the expression of CSF1 transcripts 1, 4 and 5, and CSF1-S100A10.
| Template (purified PCR) product | Gene-transcript | Accession no. | Primer set for real-time PCR | Efficiency (E %) | Correlation coefficient (R^2) | Slope | y-intercept |
|---|---|---|---|---|---|---|---|
| CSF1-1886F/CSF1-2176R | CSF1-transcript 1 | NM_000757 version 5 | CSF1-2057F/CSF1-trans1-2123R | 107.1 | 0.998 | 3.164 | 29.532 |
| CSF1-1886F/CSF1-trans4-2130R | CSF1-transcript 4 | NM_172212 version 2 | CSF1-2057F/CSF1-trans4-2120R | 98.3 | 0.997 | 3.364 | 29.690 |
| CSF1-1886F/CSF1-3end-R1out | CSF1-transcript 5 | - | CSF1-2057F/CSF1-transNew-Rq | 97.6 | 0.997 | 3.380 | 30.662 |
| CSF1-1886F/S100A10-840R | CSF1-S100A10 | - | CSF1-2057F/S100A10-664R | 110.1 | 0.991 | 3.102 | 29.282 |
Real-time PCR was run on CFX96 Touch™ Real-Time PCR Detection system (Bio-Rad). The thermal cycling included an initial step at 95˚C for 30 sec, followed by 40 cycles of 10 sec at 95˚C and 30 sec at 60˚C. Melting curve analysis at the end of the PCR was performed in order to confirm whether or not a single product was amplified and that no primer dimers interfered with the reaction. There was an initial denaturation step at 95˚C followed by temperature rising from 65 to 95˚C with 0.5˚C increment for 0.05 sec. The data were analyzed using the Bio-Rad CFX Manager Software (Bio-Rad).
For quantification of the expression of CSF1 and CSF1R transcript two assays were performed which were supplied by Applied Biosystems. Assay Hs00174164_m1 was used for the expression of CSF1. This assay was specific for exon 4/5 boundary and detects all the four reported transcripts of CSF1 (accession nos. NM_000757, NM_172210, NM_172211 and NM_172212). Assay Hs00911250_m1 was used for CSF1R end of the transcript and was specific for exon 20/21 boundary (NM_005211). The assay Hs99999901_s1 18S (Applied Biosystems) was used as endogenous control for relative gene expression quantification. Four replicates of each sample and endogenous control were again used to ensure statistical representativity. The 20 μl reaction volume contained 1X TaqMan Universal Mix, 1X 20X TaqMan Gene Expression Mix and 2 μl cDNA (40 ng equivalent of RNA). Real-time PCR was run on CFX96 Touch™ Real-Time PCR Detection system (Bio-Rad). The thermal cycling included an initial step at 50˚C for 2 min, followed by 10 min at 95˚C and 40 cycles of 15 sec at 95˚C and 1 min at 60˚C. The data were analyzed using the Bio-Rad CFX Manager Software (Bio-Rad).
Results
The cytogenetic analysis of case 1 showed that there were two unrelated abnormal clones: 46,XX,dic(2;13)(q11;p11),der(11) t(2;11)(q11;p15)[3]/46,XX,r(11)[2]/46,XX[33]. The karyotypes of cases 2 and 3 were reported previously (11). Case 2 had the karyotype 46,XY,t(1;22)(p13;q12)[20], while the karyotype of case 3 was 46,XX,t(1;1)(q21;p11)[9]/47,XX,+7[2]/46,XX[14].
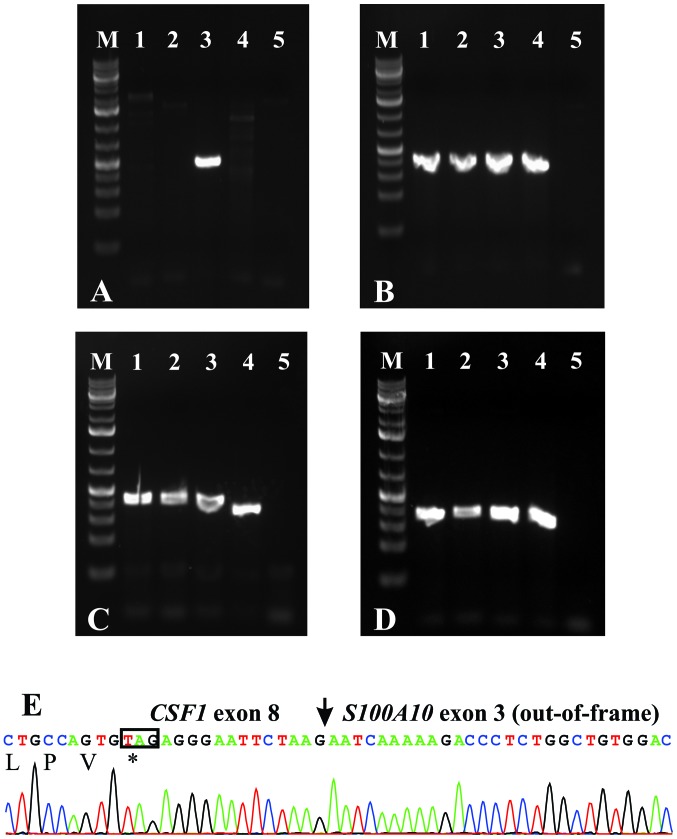
Using the FusionMap on the raw sequencing data obtained from the Norwegian Sequencing Centre, the CSF1-S100A10 fusion transcript, ranked 1st with 599 seed counts, was found in case 3, which carried the translocation t(1;1)(q21;p11), whereas no CSF1 fusion transcript was found in the other two tumors. Because CSF1 and S100A10 map to chromosome bands 1p13.3 and 1q21.3, respectively, we decided to study the CSF1-S100A10 fusion transcript. RT-PCR with the CSF1-1886F/S100A10-840R primer combination amplified a single cDNA fragment in case 3, but not in cases 1 and 2 (Fig. 1A). The wild-type S100A10 cDNA, the CSF1 transcript 1 (NM_000757) and the CSF1 transcript 4 (NM_172212) were amplified in all cases (Fig. 1B–D). Sequencing of the fragment amplified with the CSF1-1886F/S100A10-840R primer combination showed that exon 8 of CSF1 (nt 2091 in sequence with accession no. NM_000757 version 5) was fused to exon 3 of S100A10 (nt 641 in sequence with accession no. NM_002966 version 2) (Fig. 1E).
Figure 1.
RT-PCR analysis of TSGCT. (A) cDNA fragment amplification using the primer set CSF1-1886F/S100A10-840R. (B) cDNA amplification of the wild-type S100A10 using the primer set S100A10-555F/S100A10-840R. (C) cDNA amplification of the CSF1 transcript 1 using the primer set CSF1-1886F/CSF1-2176R. (D) cDNA amplification of the CSF1 transcript 4 using the primer set CSF1-1886F/CSF1-trans42130R. Lane M, 1 kb Plus DNA ladder (GeneRuler, Fermentas); lane 1, cDNA from case 1; lane 2, cDNA from case 2; lane 3, cDNA from case 3; lane 4, cDNA synthesized from Human Universal Reference Total RNA (Takara); lane 5, blank, no RNA in cDNA synthesis. (E) Partial sequence chromatogram of the cDNA fragment amplified with the primer set CSF1-1886F/S100A10-840R showing the fusion of exon 8 of CSF1 with exon 3 of S100A10. The stop codon TAG is in the box.
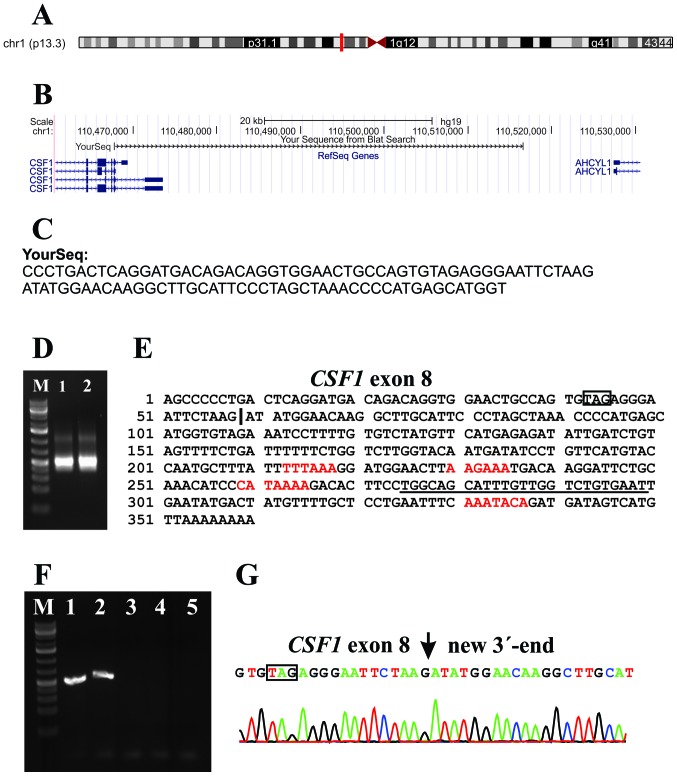
Because Cupp et al (23) found expression of CSF1 in all studied TSGCT, including those that lacked the CSF1 translocation, we decided to investigate further the expression of CSF1 in cases 1 and 2 which did not have CSF1-fusion transcripts. No other fusion transcripts or genes were examined. We retrieved reads from the raw sequencing data which contained the last 20 nt of exon 8 of CSF1 (agtgtagag ggaattctaag; nt 2072–2091 in sequences with accession nos. NM_000757 and NM_172212) from both cases 1 and 2 and performed a database search by applying the BLAST and BLAT algorithms (Fig. 2A and B). The analyses showed that exon 8 of CSF1 was fused to a sequence with features, according to BLAST, ‘48731 bp at 5′ side: macrophage colony-stimulating factor 1 isoform a precursor and 11081 bp at 3′ side: putative adenosylhomocysteinase 2 isoform a’ (Fig. 2C). 3′-RACE amplified a single fragment in cases 1 and 2 (Fig. 2C). Sanger sequence analysis of the amplified fragment verified the data obtained by RNA-Seq, i.e., the fusion of CSF1 exon 8 with the new sequence, and showed that the latter had a poly-adenylation signal, AAATACA, close to the polyA tail (Fig. 2E). Three other poly-adenylation signals were found in this sequence (Fig. 2E). PCR with the CSF1-1886F/CSF1-3end-R1out primer combination (Table I) amplified a single cDNA fragment in cases 1 and 2 (Fig. 2F). Sequence analysis of these fragments verified the results obtained by RNA-Seq and 3′-RACE (Fig. 2G). Expression analysis of 8 cell lines showed that none of them expressed the new CSF1 transcript whereas both transcripts 1 and 4 were expressed (Fig. 3). Because the new non-genic sequence fused to exon 8 of CSF1 is 48 kb downstream of the currently known CSF1 locus, we consider it as a new alternative exon and we call this novel sequence CSF1 transcript 5.
Figure 2.
Analysis of the new CSF1 transcript 5 in TSGCT. (A) Localization of the BLAT results on the chromosome 1 as it is obtained from genome browser. (B) Results of the BLAT search with one of the reads (YourSeq) which contained the last 20 nt of exon 8 of CSF1 (agtgtagagggaattctaag; nt 2081–2091 in sequence with accession no. NM_000757) and extracted from the raw sequencing data. (C) The sequence of the YourSeq which was used for BLAT search. (D) 3′-RACE on the cases 1 and 2 amplified a single cDNA fragment. (E) Sequence of the 3′-RACE-amplified cDNA fragment. The stop codon TAG is in box. The vertical line is the junction between exon 8 of CSF1 and the new sequence on 1p13 which is 41 kb downstream from the currently known CSF1 locus. The red letters are the polyadenylation signals. The primer CSF1-3end-R1out is underlined. (F) RT-PCR amplification using CSF1-1886F/CSF1-3 end-R1out primer combination. In case 1 (lane 1) and case 2 (lane 2), a single cDNA fragment is amplified. In case 3 (lane 3), the control cDNA (lane 4) and blank (no RNA in cDNA) (lane 5), no fragments are amplified. M is 1 kb Plus DNA ladder (GeneRuler, Fermentas). (G) Partial sequence chromatogram of the cDNA fragment amplified with primers CSF1-1886F/CSF1-3end-R1out showing the fusion of exon 8 of CSF1 with the new 3′-end.
Figure 3.
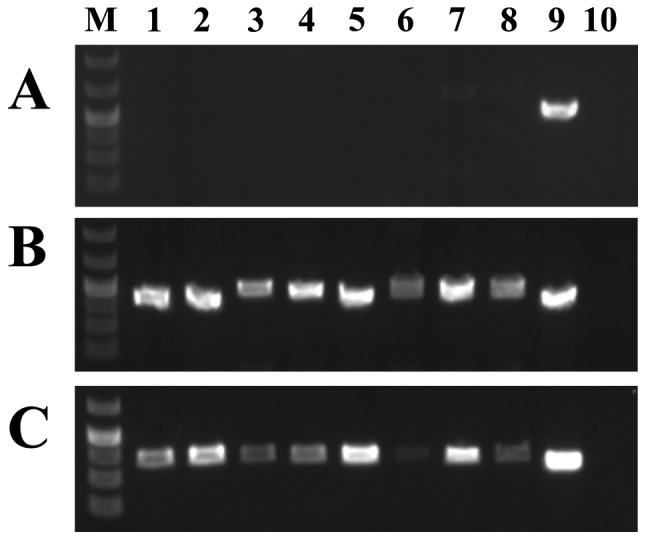
Expression analysis of the various CSF1 transcripts in a cDNA panel of eight cell lines. (A) Expression of CSF1 transcript 5. (B) Expression of CSF1 transcript 1. (C) Expression of CSF1 transcript 4. Expression analysis was performed using the human cell line MTC cDNA panel (Clontech). Lane 1, Ad5 cell line; lane 2, SKOV-3; lane 3, Saos2; lane 4, A431; lane 5, Du145; lane 6, H1299; lane 7, HeLa; lane 8, MCF7; lane 9, case 1 which was used as positive control; lane 10, blank (no RNA in cDNA). Lane M is 1 kb Plus DNA ladder (GeneRuler, Fermentas).
Real-time PCR to quantify the expression of the CSF1 transcripts and CSF1-S100A10 showed that in cases 1 and 2, the new transcript 5 was the most highly expressed followed by transcripts 1 and 4 (Fig. 4A). In case 1, the mean quantification cycle (Cq mean) was 25.74, 29.65 and 31.31 for transcript 5, transcript 1 and transcript 4, respectively. In case 2, the respective values for Cq mean were 28.2, 29.92, and 32.94. In case 3, the highest expression was observed for the fusion CSF1-S100A10 transcript (Cq mean = 24.77) followed by CSF1 transcripts 1 (Cq mean = 30.47) and 4 (Cq mean = 31.15).
Figure 4.
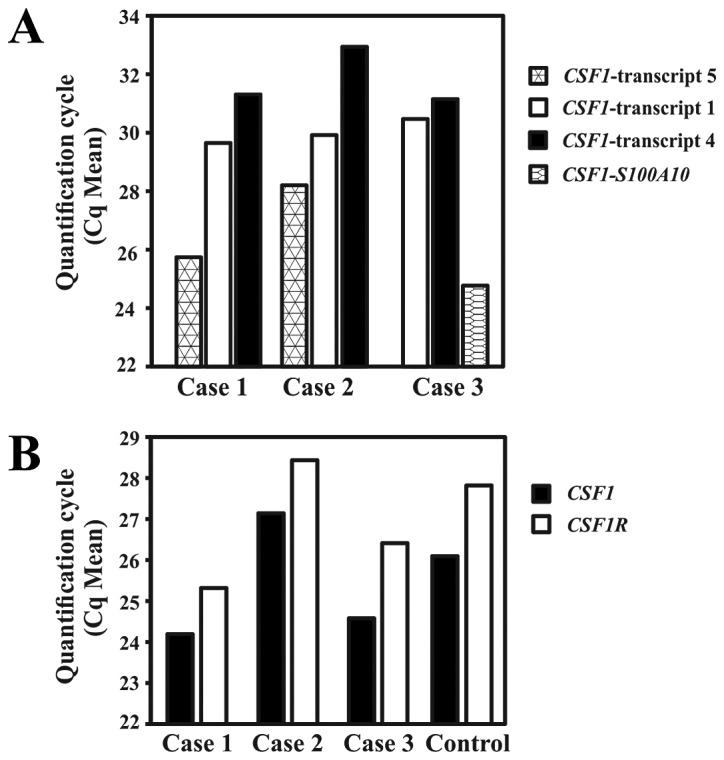
Real-time PCR to quantify the transcripts 1, 4 and 5 of CSF1, CSF1-S100A10 and CSF1R in TSGCT. (A) Quantification of the CSF1 transcripts 5, 1 and 4 in cases 1 and 2, and CSF1-S100A10, and CSF1 transcripts 1 and 4 in case 3. (B) Quantification of the expression of the CSF1 gene (all transcripts together) and its receptor CSF1R. The expression is presented as quantification cycle (Cq mean).
Real-time PCR to quantify the expression of CSF1 (all transcripts) and CSF1R showed that CSF1 was slightly higher expressed than CSF1R in all cases, including the control Human Universal Reference Total RNA. The Cq means for CSF1/CSF1R were 24.19/25.32, 27.1/28.44, 24.58/26.41 and 26.09/27.82 for cases 1, 2, 3 and the control, respectively (Fig. 3B).
Discussion
We have identified a novel CSF1-S100A10 fusion gene in a TSGCT carrying the translocation t(1;1)(q21;p11). In this fusion gene, the part of CSF1 coding for the CSF1 protein (exons 1–8 in sequences with accession nos. NM_000757 and NM_172212) is fused to the 3′-part of S100A10. Since the stop codon TAG of CSF1 is present in the fusion gene (Fig. 1C), the consequence of the CSF1-S100A10 seems to be replacement of the 3′-untranslated region (UTR) of CSF1 (exon 9; nt 2092–4234 in sequence with accession no. NM_000757 or nt 2092–2772 in NM_172212) by the 3′-end of S100A10 (exon 3; nt 641–1055 in sequence with accession no. NM_002966).
The CSF1-S100A10 fusion gene is reminiscent of the HMGA2-fusions in benign connective tissue tumors. Chromosomal rearrangements involving 12q13–15 and targeting HMGA2 result in mostly out-of-frame fusion genes in which a stop codon is encountered quickly so that only a few amino acids are added to the AT-hook of HMGA2 (25). The fusion also has another result, namely the removal of the 3′-UTR of HMGA2 which contains multiple let-7 binding sites. Let-7 miRNA might act as a repressor of HMGA2 and miRNA-directed repression of an oncogene could provide a mechanism of tumorigenesis (25).
S100A10 has been reported as the 3′-partner gene in the HDGF/S100A10 fusion gene which was found in the UACC-812 breast cancer cell line (26). S100A10 codes for a member of the S100 family of proteins containing 2 EF-hand calcium-binding motifs. S100 proteins are located in the cytoplasm and/or nucleus of a wide range of cells and are involved in the regulation of a number of cellular processes such as cell cycle progression and differentiation. S100 genes include at least 13 members which are located as a cluster in chromosome band 1q21 (http://www.ncbi.nlm.nih.gov/gene/6281). S100A10 plays a role in oncogenesis by regulating the plasmin proteolytic activity of cancer cells and by regulating the migration of macrophages to the tumor site (27).
In the other two TSGCT we were able to examine, no fusions of CSF1 with known genes were found. Instead, a novel fusion of the coding region of CSF1 with a roughly 300 bp sequence located 48 kb downstream of the CSF1 locus was detected, identical in both cases. Based on the location of the new sequence which is fused to exon 8 of CSF1 (48 kb downstream of the currently known CSF1 locus), we call this novel sequence CSF1 transcript 5. However, the possibility that this transcript is a product of the chromosome aberrations of case 1 and the chromosome translocation t(1;22) found in case 2 cannot be ruled out. Similar to the CSF1-S100A10 fusion gene, the novel CSF1 transcript 5 has the 3′-UTR of CSF1 (exon 9; nt 2092-4234 in sequence with accession no. NM_000757 or nt 2092–2772 in NM_172212) replaced by a new exon located 48 kb downstream of CSF1 and 11 kb upstream of AHCYL1 (adenosylhomocysteinase-like 1) (Fig. 2C). The novel CSF1 transcript 5 was not found in the Human Universal Reference Total RNA which is a mixture of total RNAs from adult human tissues and a panel of 8 well known cell lines (Figs. 2F and 3). At the moment, both its frequency and its specificity for TSGCT are unknown.
Cupp et al (23) described two groups of TSGCT/PVNS defined by CSF1 biology. The first group had CSF1 rearrangements and high levels of CSF1 RNA expression. In the second group of TSGCT, there were no CSF1 rearrangements as determined by FISH, but the same characteristic CSF1 RNA and CSF1 protein expression pattern was nevertheless present. Thus, an alternative mechanism must exist leading to CSF1 upregulation in this tumor subset. An altered CSF1-CSF1R signaling pathway still appears to be a critical tumorigenic event in these TSGCT/PVNS, but the putative translocation or mutation seems to involve a gene whose product leads to upregulation of CSF1. An alternative mechanism for at least some cases could be the novel CSF1 transcript presented here.
Although we studied only 3 TSGCT, a common pathogenetic theme is discernible shared by the CSF1-S100A10 fusion gene and the novel CSF1 transcript: the replacement of the 3′-UTR of CSF1 with new sequences. This replacement results in expression of the protein-coding part of CSF1. Thus, the reported t(1;2) found in TSGCT might not bring CSF1 under control of the promoter of COL6A3 as has been proposed (20,21) but might instead result in the replacement of the 3′-UTR of CSF1 with sequences from COL6A3 in a similar way to what we describe here.
While there is no information on the 3′-UTR of CSF1 transcript 4 (sequence with accession no. NM_172212), there is ample information on the 3′-UTR region of CSF1 transcript 1 (sequence with accession no. NM_000757). The exon 9 of CSF1 mRNA (accession no. NM-000575) contains microRNA targets (miRNA), a non-canonical G-quadruplex, and AU-rich elements (AREs) which control the expression of CSF1 (28–31). It has at least 14 miRNA sites (30) and it was shown that both miRNA-128 and miRNA-152 downregulate the expression of CSF1 and its protein (30). Two other miRNAs, miR-130a and miR-301a, in the presence of a nucleolin also downregulate the expression of CSF1 (29). AREs are known to dictate mRNA (32). For the AREs of CSF1, glyceraldehyde-3-phosphate dehydrogenase was shown to bind to the AU-rich elements and regulates the stability and decay of CSF1 mRNA (28,31). Recently, Woo et al (29) showed that 3′-UTR of CSF1 contains a non-canonical G-quadruplex which is involved in the post-transcriptional regulation of CSF1 and that nucleolin is the interacting protein. In the same study, using a luciferase reporter system fused to CSF1 mRNA 3′-UTR, they showed that the disruption of the miRNA target region, G-quadruplex, and AREs together dramatically increased reporter RNA levels, suggesting important roles for these cis-acting regulatory elements in the downregulation of CSF1 mRNA.
The CSF1-CSF1R signaling pathway seems to be critically involved in TSGCT/PVNS tumorigenesis something that has led to the trial of alternative therapies based on tyrosine kinase inhibition with imatinib mesylate. In addition to its inhibitory activity on BCR-ABL, KIT, and PDGFRA, imatinib mesylate has been reported to block CSF1R activation at therapeutic concentrations (33). After the first report of complete response obtained with imatinib in a single patient with TSGCT (34), imatinib mesylate has been tried in several patients who were affected with locally advanced and/or metastatic TSGCT, and the response has been promising (34–36). Also for this reason a better understanding of the mechanisms of CSF1 activation in TSGCT and the role played by the CSF1-CSF1R signaling pathway are important. More tumors must be studied in order to determine the frequency and specificity of the new transcript as well as the possible ubiquity of the replacement of the 3′-UTR of CSF1 in aberrations targeting the CSF1 gene.
Acknowledgments
The authors thank Jim Thorsen for help with bioinformatics. This study was supported by grants from the Norwegian Cancer Society and The South-Eastern Norway Regional Health Authority.
References
- 1.Jaffe HL, Lichtestein L, Sutro CJ. Pigmented villonodular synovitis, bursitis and tenosynovitis. A discussion of the synovial and bursal equivalents of the tenosynovial lesion commonly denoted as xanthoma, xanthogranuloma, giant cell tumor or myeloplaxoma of the tendon sheath, with some consideration of this tendon sheath lesion itself. Arch Pathol. 1941;31:731–764. [Google Scholar]
- 2.Somerhausen NS, Fletcher CD. Diffuse-type giant cell tumor: clinicopathologic and immunohistochemical analysis of 50 cases with extraarticular disease. Am J Surg Pathol. 2000;24:479–492. doi: 10.1097/00000478-200004000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Weiss SW, Goldblum JR. Enzinger and Weiss’s Soft Tissue Tumors. Mosby; St. Louis, MO: 2001. [Google Scholar]
- 4.Myers BW, Masi AT. Pigmented villonodular synovitis and tenosynovitis: a clinical epidemiologic study of 166 cases and literature review. Medicine. 1980;59:223–238. [PubMed] [Google Scholar]
- 5.Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F. WHO Classification of Tumours of Soft Tissue and Bone. (Fourth edition) 2013 [Google Scholar]
- 6.Chassaignac CME. Cancer de la gaine des tendons. Gaz Hosp Civ Milit. 1852;47:185–186. [Google Scholar]
- 7.Singh R, Grewal DS, Chakravarti RN. Experimental production of pigmented villonodular synovitis in the knee and ankle joints of rhesus monkeys. J Pathol. 1969;98:137–142. doi: 10.1002/path.1710980207. [DOI] [PubMed] [Google Scholar]
- 8.Sakkers RJ, de Jong D, van der Heul RO. X-chromosome inactivation in patients who have pigmented villonodular synovitis. J Bone Joint Surg Am. 1991;73:1532–1536. [PubMed] [Google Scholar]
- 9.Vogrincic GS, O’Connell JX, Gilks CB. Giant cell tumor of tendon sheath is a polyclonal cellular proliferation. Hum Pathol. 1997;28:815–819. doi: 10.1016/s0046-8177(97)90155-6. [DOI] [PubMed] [Google Scholar]
- 10.Choong PF, Willen H, Nilbert M, et al. Pigmented villonodular synovitis. Monoclonality and metastasis - a case for neoplastic origin? Acta Orthop Scand. 1995;66:64–68. doi: 10.3109/17453679508994643. [DOI] [PubMed] [Google Scholar]
- 11.Brandal P, Bjerkehagen B, Heim S. Molecular cytogenetic characterization of tenosynovial giant cell tumors. Neoplasia. 2004;6:578–583. doi: 10.1593/neo.04202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fletcher JA, Henkle C, Atkins L, Rosenberg AE, Morton CC. Trisomy 5 and trisomy 7 are nonrandom aberrations in pigmented villonodular synovitis: confirmation of trisomy 7 in uncultured cells. Genes Chromosomes Cancer. 1992;4:264–266. doi: 10.1002/gcc.2870040312. [DOI] [PubMed] [Google Scholar]
- 13.Mertens F, Orndal C, Mandahl N, et al. Chromosome aberrations in tenosynovial giant cell tumors and nontumorous synovial tissue. Genes Chromosomes Cancer. 1993;6:212–217. doi: 10.1002/gcc.2870060404. [DOI] [PubMed] [Google Scholar]
- 14.Nilsson M, Hoglund M, Panagopoulos I, et al. Molecular cytogenetic mapping of recurrent chromosomal breakpoints in tenosynovial giant cell tumors. Virchows Arch. 2002;441:475–480. doi: 10.1007/s00428-002-0640-y. [DOI] [PubMed] [Google Scholar]
- 15.Sciot R, Rosai J, Dal Cin P, et al. Analysis of 35 cases of localized and diffuse tenosynovial giant cell tumor: a report from the Chromosomes and Morphology (CHAMP) study group. Mod Pathol. 1999;12:576–579. [PubMed] [Google Scholar]
- 16.Bertoni F, Unni KK, Beabout JW, Sim FH. Malignant giant cell tumor of the tendon sheaths and joints (malignant pigmented villonodular synovitis) Am J Surg Pathol. 1997;21:153–163. doi: 10.1097/00000478-199702000-00004. [DOI] [PubMed] [Google Scholar]
- 17.Layfield LJ, Meloni-Ehrig A, Liu K, Shepard R, Harrelson JM. Malignant giant cell tumor of synovium (malignant pigmented villonodular synovitis) Arch Pathol Lab Med. 2000;124:1636–1641. doi: 10.5858/2000-124-1636-MGCTOS. [DOI] [PubMed] [Google Scholar]
- 18.Ushijima M, Hashimoto H, Tsuneyoshi M, Enjoji M, Miyamoto Y, Okue A. Malignant giant cell tumor of tendon sheath. Report of a case. Acta Pathol Jpn. 1985;35:699–709. doi: 10.1111/j.1440-1827.1985.tb00611.x. [DOI] [PubMed] [Google Scholar]
- 19.Mitelman F, Mertens F, Johansson B. Prevalence estimates of recurrent balanced cytogenetic aberrations and gene fusions in unselected patients with neoplastic disorders. Genes Chromosomes Cancer. 2005;43:350–366. doi: 10.1002/gcc.20212. [DOI] [PubMed] [Google Scholar]
- 20.West RB, Rubin BP, Miller MA, et al. A landscape effect in tenosynovial giant-cell tumor from activation of CSF1 expression by a translocation in a minority of tumor cells. Proc Natl Acad Sci USA. 2006;103:690–695. doi: 10.1073/pnas.0507321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Möller E, Mandahl N, Mertens F, Panagopoulos I. Molecular identification of COL6A3-CSF1 fusion transcripts in tenosynovial giant cell tumors. Genes Chromosomes Cancer. 2008;47:21–25. doi: 10.1002/gcc.20501. [DOI] [PubMed] [Google Scholar]
- 22.Kawasaki ES, Ladner MB. Molecular biology of macrophage colony.stimulating factor. In: Dexter TM, Garland JM, Testa NG, editors. Colony Stimulating Factors: Molecular and Cellular Biology. Marcel Dekker; New York, NY: 1990. pp. 155–176. [PubMed] [Google Scholar]
- 23.Cupp JS, Miller MA, Montgomery KD, et al. Translocation and expression of CSF1 in pigmented villonodular synovitis, tenosynovial giant cell tumor, rheumatoid arthritis and other reactive synovitides. Am J Surg Pathol. 2007;31:970–976. doi: 10.1097/PAS.0b013e31802b86f8. [DOI] [PubMed] [Google Scholar]
- 24.Ge H, Liu K, Juan T, Fang F, Newman M, Hoeck W. Fusion Map: detecting fusion genes from next-generation sequencing data at base-pair resolution. Bioinformatics. 2011;27:1922–1928. doi: 10.1093/bioinformatics/btr310. [DOI] [PubMed] [Google Scholar]
- 25.Cleynen I, Van de Ven WJ. The HMGA proteins: a myriad of functions (Review) Int J Oncol. 2008;32:289–305. [PubMed] [Google Scholar]
- 26.Asmann YW, Hossain A, Necela BM, et al. A novel bioinformatics pipeline for identification and characterization of fusion transcripts in breast cancer and normal cell lines. Nucleic Acids Res. 2011;39:e100. doi: 10.1093/nar/gkr362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madureira PA, O’Connell PA, Surette AP, Miller VA, Waisman DM. The biochemistry and regulation of S100A10: a multifunctional plasminogen receptor involved in oncogenesis. J Biomed Biotechnol. 2012;2012:353687. doi: 10.1155/2012/353687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonafé N, Gilmore-Hebert M, Folk NL, Azodi M, Zhou Y, Chambers SK. Glyceraldehyde-3-phosphate dehydrogenase binds to the AU-Rich 3′ untranslated region of colony-stimulating factor-1 (CSF-1) messenger RNA in human ovarian cancer cells: possible role in CSF-1 posttranscriptional regulation and tumor phenotype. Cancer Res. 2005;65:3762–3771. doi: 10.1158/0008-5472.CAN-04-3954. [DOI] [PubMed] [Google Scholar]
- 29.Woo HH, Baker T, Laszlo C, Chambers SK. Nucleolin mediates microRNA-directed CSF-1 mRNA deadenylation but increases translation of CSF-1 mRNA. Mol Cell Proteomics. 2013;12:1661–1677. doi: 10.1074/mcp.M112.025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woo HH, Laszlo CF, Greco S, Chambers SK. Regulation of colony stimulating factor-1 expression and ovarian cancer cell behavior in vitro by miR-128 and miR-152. Mol Cancer. 2012;11:58. doi: 10.1186/1476-4598-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Yi X, Stoffer JB, et al. The multifunctional protein glyceraldehyde-3-phosphate dehydrogenase is both regulated and controls colony-stimulating factor-1 messenger RNA stability in ovarian cancer. Mol Cancer Res. 2008;6:1375–1384. doi: 10.1158/1541-7786.MCR-07-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakheet T, Frevel M, Williams BR, Greer W, Khabar KS. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dewar AL, Cambareri AC, Zannettino AC, et al. Macrophage colony-stimulating factor receptor c-fms is a novel target of imatinib. Blood. 2005;105:3127–3132. doi: 10.1182/blood-2004-10-3967. [DOI] [PubMed] [Google Scholar]
- 34.Blay JY, El Sayadi H, Thiesse P, Garret J, Ray-Coquard I. Complete response to imatinib in relapsing pigmented villonodular synovitis/tenosynovial giant cell tumor (PVNS/TGCT) Ann Oncol. 2008;19:821–822. doi: 10.1093/annonc/mdn033. [DOI] [PubMed] [Google Scholar]
- 35.Cassier PA, Gelderblom H, Stacchiotti S, et al. Efficacy of imatinib mesylate for the treatment of locally advanced and/or metastatic tenosynovial giant cell tumor/pigmented villonodular synovitis. Cancer. 2012;118:1649–1655. doi: 10.1002/cncr.26409. [DOI] [PubMed] [Google Scholar]
- 36.Stacchiotti S, Crippa F, Messina A, et al. Response to imatinib in villonodular pigmented synovitis (PVNS) resistant to nilotinib. Clin Sarcoma Res. 2013;3:8. doi: 10.1186/2045-3329-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]