Abstract
Objective
To examine genetic factors responsible for metabolic syndrome and atherosclerosis in a setting of LDL receptor deficiency in a cross between C57BL/6J (B6) and PERA/Ei (PERA) inbred mouse strains.
Methods and results
Comparison of metabolic phenotypes in B6 and PERA strains revealed the PERA genetic background to be dramatically more susceptible to hyperleptinemia, hyperglycemia, hypertriglyceridemia, elevated insulin levels and body fat increase than the B6 background. To facilitate genetic analysis, metabolic syndrome-related traits and atherosclerotic lesion area were measured in 167 [(PERA × B6.129S7-Ldlrtm1Her) × B6.129S7-Ldlrtm1Her]N2 male and female backcross mice, homozygous for the Ldlr null allele. Quantitative trait locus analysis was performed using 153 polymorphic microsatellite markers spanning the genome. On chromosome 4, we identified a locus influencing plasma triglyceride, insulin, and leptin concentrations, body weight, and atherosclerosis. Several other genetic loci were identified with separate effects on plasma insulin, body weight, HDL-C and atherosclerosis.
Conclusions
The PERA strain is highly susceptible to the development of metabolic syndrome after feeding a WTD. This susceptibility is due, in part, to a locus on murine chromosome 4 in which PERA alleles predispose to adiposity, increased insulin and accelerated atherogenesis in the absence of marked hyperlipidemia.
Introduction
The metabolic syndrome, also known as syndrome X, was reintroduced by Reaven in the late 1980s to describe the clustering of cardiovascular risk factors including insulin resistance, impaired glucose tolerance, high triglyceride (TG) and low high density lipoprotein cholesterol (HDL-C) concentrations, obesity, and hypertension [1]. With increasing prevalence of obesity in the United States, the metabolic syndrome poses a major public health threat, now affecting 22% of the adult population [2]. Atherosclerotic cardiovascular disease is a major clinical complication of the metabolic syndrome; patients with the syndrome have at least a 2-fold increase in risk for atherosclerosis compared to controls [3]. However, the mechanism(s) for the association between metabolic risk factors and atherogenesis are poorly understood. Several studies have attempted to map and identify genetic factors contributing to metabolic syndrome in humans [4–7], but have met with limited success. Due to the inherent difficulties in carrying out linkage analysis in humans, many geneticists have successfully turned to animal models.
Wild-type mice are resistant to atherosclerosis, developing only very small lesions when fed a diet high in fat and cholic acid [8]. Genetically-engineered models, such as the LDL receptor knock-out (Ldlr−/−) and apolipoprotein E knock-out (Apoe−/−) mouse strains, develop larger, more complex lesions with structural and histological features similar to those of humans [9–11]. These knock-out mouse models have been successfully utilized in our laboratory, as well as others, in the mapping of atherosclerosis modifiers [12, 13]. Previously, we identified two atherosclerosis modifier loci in an interspecific cross between MOLF/Ei and B6.129S7-Ldlrtm1Her (B6-Ldlr−/−) that did not affect traditional risk factors for atherogenesis. The goal of the present study was to identify additional atherosclerosis modifier loci in a cross between two genetically distinct mouse strains: PERA/Ei (PERA) and B6-Ldlr−/−. The PERA strain was originally captured in 1961 in the Rimac Valley of Peru and is estimated to have diverged from the B6 strain 1 million years ago [14]. In the process of carrying out this study, we discovered that PERA mice are extremely susceptible to diet-induced obesity and exhibit many features resembling metabolic syndrome in humans. Thus we used quantitative trait locus (QTL) analysis to identify genetic loci influencing metabolic syndrome-related phenotypes in the PERA strain and to study their relationship to atherosclerosis.
Methods
Mice
PERA/Ei (PERA) and B6.129S7-Ldlrtm1Her (B6-Ldlr−/−) mice were purchased from The Jackson Laboratory. PERA females were mated with B6-Ldlr−/− males to produce F1 mice. Female F1s were backcrossed to B6-Ldlr−/− males to produce 167 N2 male and female mice homozygous for the Ldlr knockout allele. N2 mice were weaned onto standard laboratory chow (PicoLab Rodent 20, #5053) at 21 days of age and switched to a Western-type diet (WTD) at 8–12 weeks of age. The WTD contained 21% (wt/wt) butterfat and 0.15% (wt/wt) cholesterol and was approximately 4.53 Kcal/g (42% of calories from fat, 15% from protein, and 43% from carbohydrate) (Harlan Teklad Adjusted Calories TD 88137, Madison, WI). Mice were bled, following a 5 hour fast, after 2 weeks and 3 months of WTD feeding and killed at the 3-month time point. The breeding colony was produced and maintained in a specific pathogen-free environment. All mice were given ad libitum access to food and water and maintained on a standard 12-h light-dark cycle throughout the study. Food intake was measured five times a week for parental strains, which were housed no more than two per cage. All experimental protocols were approved by the Institutional Animal Care and Research Advisory Committee.
Atherosclerotic Lesion Measurements
Mice were anesthetized using Forane (Baxter, Deerfield, IL) and killed by cervical dislocation. The hearts were perfused with 0.9% NaCl and, then, the heart and aortic root were dissected and fixed in 10% formalin. The aortic root was sectioned and stained with oil red O, and lesion areas were quantified as described [15].
Plasma Lipoprotein, Insulin, and Leptin Measurements
Mice were retro-orbitally bled, using Forane anesthesia in the middle of the light cycle after a 5- to 6-h fast. Blood was collected directly into heparinized capillary tubes (Becton Dickson). Plasma was separated from cells by centrifugation and stored at −70 °C. Isolation of HDL-C by chemical precipitation (HDL reagent, Sigma), as well as enzymatic measurements of cholesterol and triglycerides (Wako Pure Chemical, Osaka), were carried out according to the manufacturer’s instructions. Non-HDL-C was calculated by subtracting HDL-C from total cholesterol. To evaluate cholesterol concentrations within lipoprotein sub-fractions from mice grouped by genotype at D4Mit143, 100 μl of plasma was pooled from 10 males per group and separated by fast protein liquid chromatography (FPLC). Plasma was injected onto two Superose 6 columns and eluted at a constant flow rate of 0.5 ml/min with 0.1 M Tris·HCl and 0.4% NaN3. Fractions of 0.5 ml were collected and enzymatic measurements of cholesterol were assayed, as described above. Insulin and leptin were measured by using a commercially available ELISA kit (Crystal Chem, Chicago).
Glucose measurement
Blood samples for glucose analysis were taken from the cut tail and measured using a Blood Glucose Meter (Asencia ELITE, Bayer Corp, NY).
Body Fat Analysis
Dual Energy X-Ray Absorptiometry (DEXA) measurements were used to determine body fat composition. Animals were anesthetized as previously described, placed in the prone position and scanned on a Mouse Densitometer (PIXImus™: GE Medical Systems, WS). Measurements included total mass, total fat mass, total lean mass and body fat percentage.
DNA Extraction and Ldlr Genotyping
DNA was extracted from tail tips by a quick alkaline lysis protocol as previously described [16]. Ldlr genotyping was carried out as previously described [12].
DNA Genotyping and Genome Scan
Initially, 350 fluorescently labeled primers were tested in order to identify polymorphic markers between the B6-Ldlr−/− and PERA/Ei parental strains. Of those tested, 153 polymorphic markers spaced approximately ten centiMorgans (cM) apart throughout the genome were selected for PCR and the genome scan was performed using DNA samples from the N2 generation. Parental and F1 DNA served as controls for each marker. Markers were PCR amplified and products were analyzed by capillary electrophoresis using an Applied Biosystems 3700 DNA sequencer. PCR reactions and electrophoresis were carried out by the Starr Center Genotyping Core Facility at The Rockefeller University using automated technology: Tecan (Durham, NC), Genesis RSP 100, and Robbins Scientific (Mountain View, CA) Hydra 384 robots. Genotypes were determined using Applied Biosystems GENOTYPE 3.6 NT software.
Localization of QTLs
Chromosomal linkage maps were constructed and linkage analysis was performed to localize quantitative trait loci by using MAP MANAGER QTXb19 (http://www.mapmanager.org/) for a backcross. All analyses were performed separately for males and females due to the strong effect of sex on atherosclerosis and lipoprotein phenotypes. Permutation analysis was performed to determine the threshold for “suggestive,” “significant,” and “highly significant” linkage using the genome-wide significance thresholds of P = 0.63, P = 0.05, and P = 0.001, respectively. One thousand permutations were carried out for each trait under investigation.
Statistical Analysis
ANOVA was performed using STATVIEW 5.0 (Abacus Concepts, Berkeley, CA).
Quantification of Lepr, isoform B gene expression
Hypothalami were dissected from each animal and placed in an RNA stabilization buffer (RNAlater™; 1017980: Qiagen GmbH, Germany). RNA was extracted using a guanidine thiocyanate method (RNeasy™ Mini Kit; 74104: Qiagen GmbH, Germany). RNA recovery was quantified by absorbance spectrophotometry (Ultrospec 2000 UV/Visible Spectrophotometer: Pharmacia Biotech, Piscataway, NJ). RNA was reverse transcribed using random hexamers and commercially available reverse transcriptase (SuperScript™ III First-Strand Synthesis System for RT-PCR; 18080-051: Invitrogen, Carlsbad, CA). Quantitative PCR was performed using DyNAmo™ Hot Start SYBR Green qPCR kit (Finnzymes ™: Finland) with gene specific primers. Amplification and fluorescence detection were performed on an Opticon2™ (MJ Research, Waltham, MA). The PCR conditions were 40 cycles of 95 °C for 10s, 55 °C for 10s, and 72 °C for 10s. The forward primer, reverse primer and product size for each amplicon were: Lepr-B, cagggctgtatgtcattg and tgcttggtaaaaagatgct, 187bp; Hprt, agcagtacagccccaaaa and tttggcttttccagtttca, 195bp. Melting curve analysis was performed to verify specificity of amplification. The data were corrected for amplification efficiency and normalized to Hprt mRNA content [17].
Sequence Analysis
Direct bi-directional sequencing was performed on PCR amplified cDNA using an ABI Genetic Analyzer 3100 (Applied Biosystems). Sequence was analyzed using Sequencer version 3.1 and compared to published sequence (Ensembl, ENSMUSG00000035212).
Results
Characterization of metabolic syndrome-related phenotypes in parental and F1 mice
Plasma parameters and body weights of B6, PERA, and F1 strains fed a WTD for two weeks are summarized in Table 1. PERA mice displayed higher levels of plasma total cholesterol than B6 mice (p<0.05), reflecting differences in plasma non-HDL-C levels (p<0.05). Plasma glucose concentrations were increased in the PERA strain relative to the B6 strain (p<0.002). Although not significant, PERA mice also exhibited slightly higher plasma TG and insulin concentrations than the B6 strain.
Table 1.
Assessment of body weights and plasma parameters in C57BL/6J (B6), PERA/Ei (PERA), (B6xB6-Ldlr−/−)F1 and (PERAxB6-Ldlr−/−)F1 male mice fed a Western-type diet (two weeks). Data are expressed as mean ± SD. B6 and PERA mice were bled and weighed at approximately 3 months of age; F1 mice at approximately 6 months of age. Note that B6 and PERA strains are Ldlr+/+; F1 mice are Ldlr+/−.
| Trait | B6 | PERA | (B6xB6-Ldlr−/−)F1 | (PERAxB6-Ldlr−/−)F1 |
|---|---|---|---|---|
| Tot Chol (mg/dl) | 125±11 (n=10) | 143±15* (n=6) | 238±37 (n=11) | 215±23 (n=11) |
| HDL-C (mg/dl) | 47±19 (n=10) | 49±16 (n=7) | 97±25 (n=11) | 88±20 (n=11) |
| Non-HDL-C (mg/dl) | 78±12 (n=10) | 91±9* (n=6) | 140±22 (n=11) | 127±14 (n=11) |
| TG (mg/dl) | 84±22 (n=10) | 104±32 (n=7) | 113±35 (n=11) | 191±58‡ (n=11) |
| BWT (g) | 31±2 (n=6) | 31±3 (n=6) | 39±7 (n=10) | 51±6‡ (n=11) |
| Glucose (mg/dl) | 91±14 (n=6) | 129±17† (n=6) | --- | --- |
| Insulin (ng/ml) | 0.74±0.7 (n=9) | 1.29±0.9 (n=5) | 2.8±1.7 (n=5) | 13±10§ (n=10) |
p < 0.05 vs. B6
p < 0.002 vs. B6
p ≤ 0.001 vs. (B6xB6-Ldlr−/−)F1
p ≤ 0.05 vs. (B6xB6-Ldlr−/−)F1
In order to identify strain background effects on metabolic syndrome-related phenotypes, (PERAxB6-Ldlr−/−)F1 mice were generated and compared to (B6xB6-Ldlr−/−)F1 mice which served as controls (Table 1). After feeding a WTD, plasma TG and insulin concentrations were elevated in (PERAxB6-Ldlr−/−)F1 mice relative to (B6xB6-Ldlr−/−)F1 controls (p ≤ 0.001 and p<0.02, respectively). Since plasma concentration of insulin and TG were also higher in the PERA, relative to the B6, parental strain, this indicated that dominant PERA alleles may contribute to these phenotypes. Further, insulin concentrations of (PERAxB6-Ldlr−/−)F1 mice were dramatically higher than either of the parental strains as well as (B6xB6-Ldlr−/−)F1 controls indicating that this trait may be displaying overdominance. Strikingly, (PERAxB6-Ldlr−/−) F1 mice showed much higher body weights after feeding the WTD compared to controls (p ≤ 0.001).
QTL analysis of phenotypes of metabolic syndrome and atherosclerosis in [(PERAxB6-Ldlr−/−)F1 x B6-Ldlr−/−]N2 mice
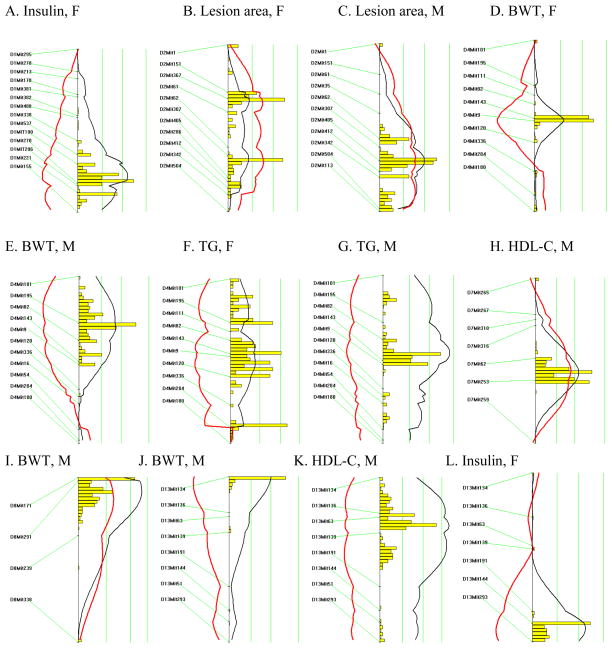
A backcross design was chosen due to the suggestion of dominant alleles affecting metabolic syndrome-related phenotypes in the PERA genetic background and to facilitate Ldlr−/− homozygosity, required for atherosclerotic lesion development. A cross was set up between (PERAxB6-Ldlr−/−)F1 female and B6-Ldlr−/− male mice to produce 167 [(PERAxB6-Ldlr−/−)F1 × B6-Ldlr−/−]N2 mice homozygous for the Ldlr null allele. Animals heterozygous for the Ldlr null allele were not used in this study. In order to detect QTLs for metabolic syndrome phenotypes and aortic lesion area, we performed a genomic scan using 153 polymorphic microsatellite markers distributed throughout the genome, resulting in an average marker spacing of approximately ten cM. Significant QTLs, as determined by permutation analysis, are presented in Table 2 and Figure 1. QTLs influencing plasma insulin concentrations were detected on chromosomes 1 (females; LOD=2.9) and 13 (females and males; LOD=4.3). Body weight QTLs were found on chromosomes 8 (males; LOD=3.4) and 13 (males; LOD=2.9). An atherosclerosis locus was detected on chromosome 2 (females and males; LOD=3.9) and two QTLs influencing HDL-C levels were detected on chromosomes 7 (males; LOD=3.1) and 13 (males; LOD=4.6).
Table 2.
Quantitative trait locus analysis of metabolic syndrome-related phenotypes and atherosclerotic lesion area in [(PERAxB6-Ldlr−/−)F1 × B6-Ldlr−/−]N2 mice. Only significant loci with genome-wide Pemp ≤ 0.05 are presented.
| Linked marker | Chr Position (cM)* | Trait | LOD (%VAR)† males (n=75) | LOD (%VAR) females (n=92) | LOD (%VAR) combined (n=167) | Pemp‡ | Genotype associated with higher value (% increase)§ |
|---|---|---|---|---|---|---|---|
| D1Mit270 | Chr 1 (92) | Insulin|| | --¶ | 2.9 (16%) | -- | ≤0.02 | BP (127%) |
| D2Mit405 | Chr 2 (69) | Lesion area|| | 2.8 (16%) | 1.1 (6%) | 3.9 (11%) | ≤0.04 | BB (27%) |
| D4Mit143 | Chr 4 (43) | TG†† | 3.6 (20%) | 1.2 (6%) | 4.8 (13%) | ≤0.004 | BP (52%) |
| BWT** | 2.0 (11%) | 2.0 (10%) | 4.0 (11%) | ≤0.05 | BP (10%) | ||
| D7Mit253 | Chr 7 (53) | HDL-C†† | 3.1 (18%) | --¶ | -- | ≤0.04 | BB (26%) |
| D8Mit291 | Chr 8 (16) | BWT** | 3.4 (19%) | --¶ | -- | ≤0.02 | BB (15%) |
| D13Mit134 | Chr 13 (6) | BWT†† | 2.9 (16%) | --¶ | -- | ≤0.04 | BP (14%) |
| D13Mit139 | Chr 13 (32) | HDL-C†† | 4.6 (26%) | --¶ | -- | <0.001 | BP (30%) |
| D13Mit144 | Chr 13 (48) | Insulin†† | 1.1 (7%) | 3.2 (15%) | 4.3 (11%) | ≤0.02 | BP (53%) |
BB, homozygous for B6 alleles; BP, heterozygous for B6 and PERA/Ei alleles.
Chr position indicates the distance of the peak marker from the centromere in centiMorgans (cM) as listed in the Mouse Genome Database (http://informatics.jax.org/)
LOD, logarithm of odds ratio score; %VAR, percent variance explained by the locus
Pemp, empirically-determined probability for observing the corresponding LOD score by chance (calculated using the permutation test function of Map Manager QTXb19)
for traits linked in both sexes, the average percent increase for males and females is reported
after feeding a Western-type diet for 3 months; mice were 20 weeks of age
indicates LOD < 1
after feeding a Western-type diet for 2 weeks; mice were 10 weeks of age
after feeding a laboratory chow diet; mice were 8 weeks of age
Figure 1.
Likelihood plots for metabolic syndrome and atherosclerosis quantitative trait loci (QTLs) in [(PERAxB6-Ldlr−/−)F1 × B6-Ldlr−/−]N2 mice. Plots were created using the interval mapping function of Map Manager QTXb19, including a bootstrap test shown as a histogram estimating the confidence interval for the major QTL on each chromosome. Three straight vertical lines on each plot represent significance thresholds for the likelihood ratio statistic (LRS) indicating “suggestive”, “significant” or “highly significant” peaks as calculated by permutation analysis (the genome-wide significance thresholds of P = 0.63, P = 0.05, and P = 0.001, respectively are shown from left to right). Black plots reflect the LRS calculated at 1-cM intervals. Red plots represent the additive regression coefficient indicating the effect of the B6 allele: if B6 represents the high allele then the red plot will be to the right of the graph; if it represents the low allele then the red plot will be to the left.
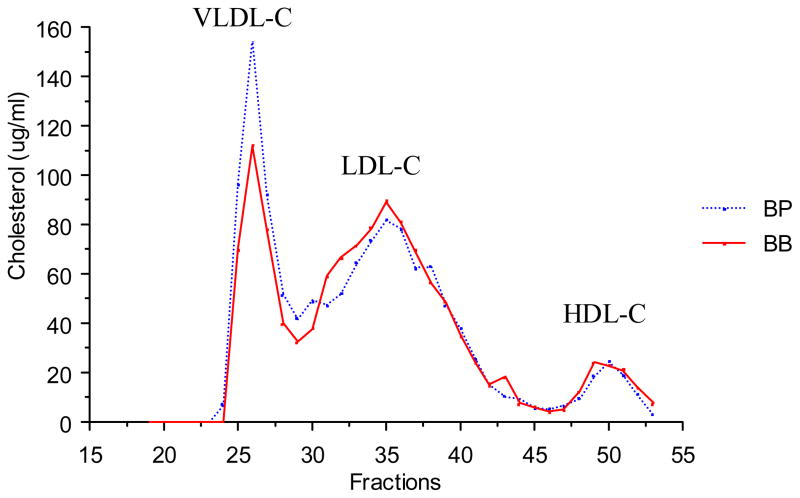
Notably, a QTL on chromosome 4 was associated with multiple traits related to metabolic syndrome. The QTL displayed peak linkage to the microsatellite marker D4Mit143, located 43 cM from the centromere (22–50 cM, +/− 1 LOD support interval) near the leptin receptor (Lepr; 46.7 cM). Primary linkage was to TG (females and males; LOD=4.8) and body weight (females and males; LOD=4.0). Additionally, ANOVA revealed significant genotypic effects on plasma insulin and leptin levels and atherosclerotic lesion area (Table 3). Plasma non-HDL-C and HDL-C levels were not altered in male mice but non-HDL-C was significantly different in females (Table 3). Fast protein liquid chromatography (FPLC) confirmed that total and HDL-C levels were similar in males grouped by genotype at D4Mit143 (Fig. 2), although VLDL levels were slightly elevated in mice carrying the PERA allele, as expected since plasma TG levels were elevated.
Table 3.
Genotypic effect of the chromosome 4 QTL contributing to variations in metabolic syndrome-related phenotypes and atherosclerotic lesion area in [(PERAxB6-Ldlr−/−)F1 × B6-Ldlr−/−]N2 mice grouped by genotype at the peak linked marker, D4Mit143. Data are expressed as mean ± SD.
| Genotype | Lesion area (μm2/section) | TG (mg/dl) | BWTch(g) | BWTWTD(g) | Insulin (ng/ml) | Leptin (ng/ml) | Non HDL-C (mg/dl) | HDL-C (mg/dl) |
|---|---|---|---|---|---|---|---|---|
| Males | ||||||||
| BB (n=45) | (21.7±10)×104 | 408±141 | 25.4±4 | 29.9±5 | 3.18±2 | 11.8±11 | 356±96 | 69±18 |
| BP (n=30) | (28.3±9)×104* | 639±317† | 28.0±4* | 33.3±5* | 4.86±3‡ | 19.3±18§ | 381±111 | 67±20 |
| Females | ||||||||
| BB (n=41) | (32.7±11)×104 | 242±112 | 19.0±2 | 21.4±3 | 1.17±0.8 | 18.3±12 | 286±72 | 57±17 |
| BP (n=51) | (31.6±12)×104 | 348±266§ | 20.5±2.8* | 23.6±3.9* | 1.03±0.9 | 26.1±15§ | 351±128‡ | 53±17 |
BWTch, body weight after feeding a chow diet; BWTWTD, body weight after feeding a Western-type diet. All other traits were measured from mice fed a high fat Western-type diet. BB, homozygous for B6 alleles; BP, heterozygous for B6 and PERA/Ei alleles.
p ≤ 0.005 vs. BB
p <0.0001 vs. BB
p <0.009 vs. BB
p ≤ 0.03 vs. BB
Figure 2.
Distribution of plasma total cholesterol in fractions separated by fast protein liquid chromatography (FPLC) in [(PERAxB6-Ldlr−/−)F1 × B6-Ldlr−/−]N2 male mice fed a Western-type diet (two weeks) grouped by genotype at D4Mit143. Plasma was pooled from 10 mice per group.
BB, homozygous for B6 alleles; BP, heterozygous for B6 and PERA/Ei alleles.
Further characterization of metabolic syndrome-related phenotypes in parental strains
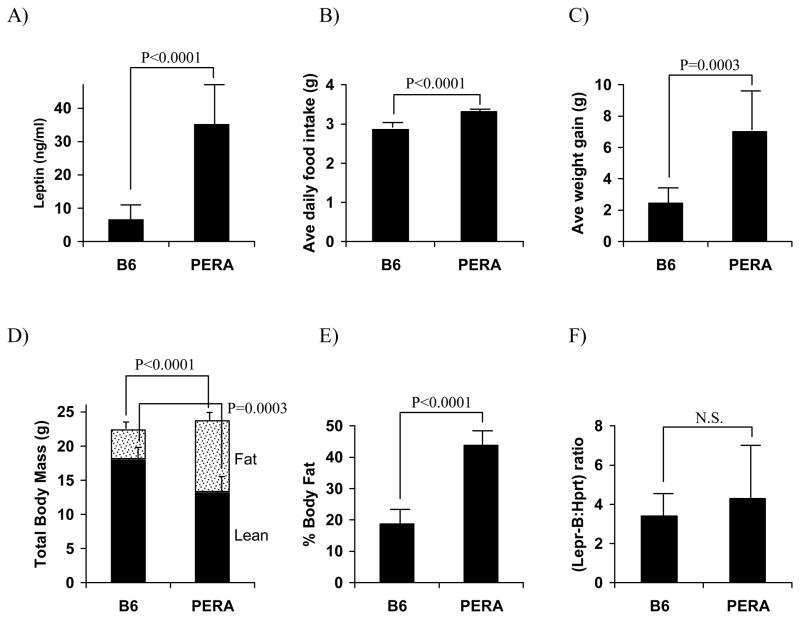
To further characterize the B6 and PERA parental strains, and to test Lepr as a candidate for the chromosome 4 metabolic syndrome locus, we studied the homeostatic response to a two-week, high-fat WTD challenge in the two strains. PERA mice had more than 5-fold higher plasma concentrations of leptin relative to B6 mice (p<0.0001, Fig. 3a). Average daily food intake was also measured. PERA mice consumed approximately 16% more calories per day than B6 mice (p<0.0001, Fig. 3b) and had an average weight gain of 7.1 g, while B6 mice gained an average of 2.5 g (p<0.001, Fig. 3c). These differences reflected greater feed efficiency (g weight gained per g food consumed) in PERA mice compared to B6 mice (0.154 ± 0.057 and 0.061 ± 0.024, respectively, p=0.0008). Dual-energy X-ray absorptiometry (DEXA) analysis showed that while the PERA strain had only slightly higher average total body mass compared to B6 mice, their lean body mass was lower (13.4 g versus 18.2 g) and fat mass was much higher (10.3 g versus 4.2 g) compared to the B6 strain (p<0.0002 and p<0.0001 respectively, Fig. 3d). Overall, average percent body fat was 2.3 fold higher in PERA mice relative to B6 (p<0.0001, Fig. 3e).
Figure 3.
Characterization of plasma leptin levels, food intake, body composition, and the major signal-transducing B isoform of the leptin receptor (Lepr-B) mRNA levels in C57BL/6J (B6) and PERA/Ei (PERA) male mice fed a Western-type diet (two weeks). Values are expressed as mean ± SD of 9–10 B6 and 4–6 PERA mice.
To directly test Lepr as a positional candidate for the chromosome 4 metabolic syndrome locus, we sequenced the coding region of the PERA allele but found no sequence variants between the strains (data not shown). To eliminate the possibility of a promoter or enhancer variant altering expression between PERA and B6, Lepr mRNA abundance was measured in the hypothalamus of WTD fed mice with primers specific for the B isoform, Lepr-B, the major signaling form. Relative levels of Lepr-B mRNA were not statistically different between the strains (Fig. 3f), eliminating Lepr as a candidate for this locus.
Discussion
This study introduces the PERA strain as a mouse model to aid in the elucidation of genetic and molecular aspects of the metabolic syndrome. While the B6 mouse strain itself has been studied as an animal model of diet-induced obesity, insulin resistance, and the metabolic syndrome [18], our studies indicate that the PERA genetic background is strikingly more susceptible to these phenotypes than B6. The mapping population utilized in this study carried Ldlr null alleles. Although Ldlr deficiency has been suggested to contribute to murine obesity in at least one study [19], observation of the metabolic syndrome-related phenotypes in PERA mice carrying wild-type Ldlr alleles suggests that the loci mapped in this study likely represent unrelated genes present in the parental backgrounds.
The PERA mouse serves as a model of the human metabolic syndrome as it develops obesity, hyperleptinemia, hyperglycemia, hypertriglyceridemia and elevated insulin levels on a high-fat, WTD as compared to the B6 strain. While PERA mice fed the high fat diet ingested more calories than B6 mice, they also demonstrated increased feed efficiency suggesting that the extra weight gain in the PERA strain may not be a result of hyperphagia alone. However, strain differences in total energy expenditure, basal metabolic rate, and physical activity were not assessed in this study and warrant further investigation. Several mouse strains carrying genetic variants of the leptin receptor on permissive genetic backgrounds are known to develop obesity or diabetes disorders [20, 21]. Therefore, the leptin receptor was tested as a candidate for the metabolic disorder in PERA mice and eliminated based on sequence analysis and relative hypothalamic mRNA expression.
While atherosclerosis is a major clinical result of the metabolic syndrome, the underlying mechanisms of increased atherosclerosis risk are poorly understood. In humans, part of the increased risk of atherosclerosis is derived from the low HDL-C and increased non-HDL-C levels common in metabolic syndrome patients. But altered lipoprotein levels do not explain all of the increased risk for atherosclerosis. In previous studies of atherosclerosis and metabolic syndrome in the mouse, such as the ob/ob Ldlr-deficient mouse, plasma cholesterol levels were so dramatically different that it is difficult to dissect out the other contributions to increased atherosclerosis susceptibility [22]. Other studies have identified QTLs contributing to obesity, lipoprotein metabolism, and insulin levels but have not assessed effects on atherosclerosis [23–26]. In the current study, we have identified a locus on mouse chromosome 4 that predisposes to both metabolic syndrome phenotypes and atherosclerosis in the absence of marked hyperlipidemia. Since the PERA backcross males, which inherited a PERA allele at the chromosome 4 metabolic syndrome locus, showed increased levels of plasma insulin and also increased atherosclerosis, insulin resistance may be responsible for their increased atherosclerosis.
Insulin resistance is frequently present in individuals with metabolic syndrome and some studies have suggested that it is an important cause of atherosclerosis. Many epidemiological studies have established an association between decreased insulin sensitivity and atherosclerosis susceptibility [27–29] which is not completely accounted for by traditional risk factors. It has been proposed that insulin resistance may elicit a proatherogenic response within the arterial wall. Decreased insulin signaling in vascular endothelial cells has been shown to lower levels of the athero-protective molecule, endothelial nitric oxide synthase (eNOS) [30]. In a separate study, defective insulin signaling in macrophages resulted in increased binding and uptake of oxidized LDL which was partly mediated by elevated CD36 protein levels, predisposing to foam cell formation and presumably, atherosclerosis [31]. It is possible that insulin resistance at the arterial cellular level promotes atherosclerosis in mice carrying the PERA interval of the chromosome 4 locus, explaining why the more insulin resistant male mice develop more atherosclerosis while the females do not.
While environmental factors, common to a “Western” lifestyle such as physical inactivity and a high fat diet, contribute to development of the metabolic syndrome, the syndrome also has a strong genetic component. Some genetic studies have focused on the individual components of the metabolic syndrome, establishing familial aggregation of plasma lipids [32–34], blood pressure [35–37], and non-insulin-dependent diabetes mellitus [38, 39]. But is the metabolic syndrome just a clustering of overlapping phenotypes brought about by environmental factors interacting with independent genetic proclivities, or is there a common, major genetic factor? In a study of 2508 male twins, multivariate genetic modeling suggested the presence of a common factor responsible for the clustering of hypertension, diabetes, and obesity which was influenced by both genetic and environmental effects (59% genetic and 41% environmental) [40]. Evidence for a common genetic component of the syndrome was displayed by the probandwise concordance rate for the clustering of hypertension, diabetes, and obesity which was 5-fold higher in monozygotic versus dizygotic twins. A separate twin study suggested that triglycerides, insulin resistance, HDL-C, body-mass index, and blood pressure are affected by a single genetic factor [41]. Our study indicates the complexity of the genetics of metabolic syndrome. The linkage study revealed many genes contributing to the overall metabolic syndrome phenotype in PERA mice, with most of the loci affecting individual risk factors. However, the chromosome 4 locus may contain a single gene, having primary effects on body weight and TG, but also affecting many features of metabolic syndrome.
Acknowledgments
The assistance of Mikhail Bezouevski, Anna Gorelik, Nashat Latib, and Nick Pleskac are gratefully acknowledged. This study was supported by NIH grant HL54591.
References
- 1.Reaven G. Banting lecture 1988. Role of insulin resistance in human disease. Diabetes. 1988;37:1595–1607. doi: 10.2337/diab.37.12.1595. [DOI] [PubMed] [Google Scholar]
- 2.Ford ES, Giles WH, Dietz WH. Prevalence of the Metabolic Syndrome Among US Adults: Findings From the Third National Health and Nutrition Examination Survey. JAMA. 2002;287:356–359. doi: 10.1001/jama.287.3.356. [DOI] [PubMed] [Google Scholar]
- 3.Grundy SM. Obesity, Metabolic Syndrome, and Cardiovascular Disease. J Clin Endocrinol Metab. 2004;89:2595–2600. doi: 10.1210/jc.2004-0372. [DOI] [PubMed] [Google Scholar]
- 4.Kissebah AH, Sonnenberg GE, Myklebust J, Goldstein M, Broman K, James RG, Marks JA, Krakower GR, Jacob HJ, Weber J, Martin L, Blangero J, Comuzzie AG. Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. PNAS. 2000;97:14478–14483. doi: 10.1073/pnas.97.26.14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loos RJF, Katzmarzyk PT, Rao DC, Rice T, Leon AS, Skinner JS, Wilmore JH, Rankinen T, Bouchard C. Genome-Wide Linkage Scan for the Metabolic Syndrome in the HERITAGE Family Study. J Clin Endocrinol Metab. 2003;88:5935–5943. doi: 10.1210/jc.2003-030553. [DOI] [PubMed] [Google Scholar]
- 6.Olswold C, de Andrade M. Localization of genes involved in the metabolic syndrome using multivariate linkage analysis. BMC Genetics. 2003;4:S57. doi: 10.1186/1471-2156-4-S1-S57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McQueen M, Bertram L, Rimm E, Blacker D, Santangelo S. A QTL genome scan of the metabolic syndrome and its component traits. BMC Genetics. 2003;4:S96. doi: 10.1186/1471-2156-4-S1-S96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishina P, Lowe S, Verstuyft J, Naggert J, Kuypers F, Paigen B. Effects of dietary fats from animal and plant sources on diet-induced fatty streak lesions in C57BL/6J mice. J Lipid Res. 1993;34:1413–1422. [PubMed] [Google Scholar]
- 9.Ishibashi SBM, Goldstein JL, Gerard RD, Hammer RE, Herz J. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J Clin Invest. 1993;92:883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plump AS, SJ, Hayek T, Aalto-Setala K, Walsh A, Verstuyft JG, Rubin EM, Breslow JL. Severe hypercholesterolemia and atherosclerosis in apolipoprotein E-deficient mice created by homologous recombination in ES cells. Cell. 1992;71:343–353. doi: 10.1016/0092-8674(92)90362-g. [DOI] [PubMed] [Google Scholar]
- 11.Zhang SH, RR, Piedrahita JA, Maeda N. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science. 192;258:468–471. doi: 10.1126/science.1411543. [DOI] [PubMed] [Google Scholar]
- 12.Welch CL, Bretschger S, Latib N, Bezouevski M, Guo Y, Pleskac N, Liang CP, Barlow C, Dansky H, Breslow JL, Tall AR. Localization of atherosclerosis susceptibility loci to chromosomes 4 and 6 using the Ldlr knockout mouse model. PNAS. 2001;98:7946–7951. doi: 10.1073/pnas.141239098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dansky HM, Shu P, Donavan M, Montagno J, Nagle DL, Smutko JS, Roy N, Whiteing S, Barrios J, McBride TJ, Smith JD, Duyk G, Breslow JL, Moore KJ. A Phenotype-Sensitizing Apoe-Deficient Genetic Background Reveals Novel Atherosclerosis Predisposition Loci in the Mouse. Genetics. 2002;160:1599–1608. doi: 10.1093/genetics/160.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silver L. Mouse Genetics. New York: Oxford University Press, Inc; 1995. [Google Scholar]
- 15.Plump A, Scott C, Breslow J. Human Apolipoprotein A-I Gene Expression Increases High Density Lipoprotein and Suppresses Atherosclerosis in the Apolipoprotein E-Deficient Mouse. PNAS. 1994;91:9607–9611. doi: 10.1073/pnas.91.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Truett G, Heeger P, Mynatt R, Truett A, Walker J, Warman M. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) Biotechniques. 2000;29:52–54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- 17.Liu W, Saint DA. Validation of a quantitative method for real time PCR kinetics. Biochemical and Biophysical Research Communications. 2002;294:347–353. doi: 10.1016/S0006-291X(02)00478-3. [DOI] [PubMed] [Google Scholar]
- 18.Collins S, Martin TL, Surwit RS, Robidoux J. Genetic vulnerability to diet-induced obesity in the C57BL/6J mouse: physiological and molecular characteristics. Physiology & Behavior. 2004;81:243–248. doi: 10.1016/j.physbeh.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Schreyer SA, Vick C, Lystig TC, Mystkowski P, LeBoeuf RC. LDL receptor but not apolipoprotein E deficiency increases diet-induced obesity and diabetes in mice. Am J Physiol Endocrinol Metab. 2002;282:E207–214. doi: 10.1152/ajpendo.2002.282.1.E207. [DOI] [PubMed] [Google Scholar]
- 20.Kim JH, Taylor PN, Young D, Karst SY, Nishina PM, Naggert JK. New Leptin Receptor Mutations in Mice: Leprdb-rtnd, Leprdb-dmpg and Leprdb-rlpy. J Nutr. 2003;133:1265–1271. doi: 10.1093/jn/133.5.1265. [DOI] [PubMed] [Google Scholar]
- 21.Brown JA, Chua SC, Jr, Liu SM, Andrews MT, Vandenbergh JG. Spontaneous mutation in the db gene results in obesity and diabetes in CD-1 outbred mice. Am J Physiol Regul Integr Comp Physiol. 2000;278:R320–330. doi: 10.1152/ajpregu.2000.278.2.R320. [DOI] [PubMed] [Google Scholar]
- 22.Hasty AH, Shimano H, Osuga J-i, Namatame I, Takahashi A, Yahagi N, Perrey S, Iizuka Y, Tamura Y, Amemiya-Kudo M, Yoshikawa T, Okazaki H, Ohashi K, Harada K, Matsuzaka T, Sone H, Gotoda T, Nagai R, Ishibashi S, Yamada N. Severe Hypercholesterolemia, Hypertriglyceridemia, and Atherosclerosis in Mice Lacking Both Leptin and the Low Density Lipoprotein Receptor. J Biol Chem. 2001;276:37402–37408. doi: 10.1074/jbc.M010176200. [DOI] [PubMed] [Google Scholar]
- 23.Singer JB, Hill AE, Burrage L, Olszens KR, Song JH, Justice M, O’Brien WE, Conti DV, Witte JS, Lander ES, Nadeau JH. Genetic dissection of complex traits with chromosome substitution strains of mice. Science. 2004;304:445–448. doi: 10.1126/science.1093139. [DOI] [PubMed] [Google Scholar]
- 24.Mehrabian M, Wen PZ, Fisler J, Davis RC, Lusis AJ. Genetic loci controlling body fat, lipoprotein metabolism, and insulin levels in a multifactorial mouse model. Journal of Clinical Investigation. 1998;101:2485–2496. doi: 10.1172/JCI1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor BA, Phillips SJ. Detection of obesity QTLs on mouse chromosomes 1 and 7 by selective DNA pooling. Genomics. 1996;34:389–398. doi: 10.1006/geno.1996.0302. [DOI] [PubMed] [Google Scholar]
- 26.Warden CH, Fisler JS, Shoemaker SM, Wen PZ, Svenson KL, Pace MJ, Lusis AJ. Identification of 4 Chromosomal Loci Determining Obesity in a Multifactorial Mouse Model. Journal of Clinical Investigation. 1995;95:1545–1552. doi: 10.1172/JCI117827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard G, O’Leary DH, Zaccaro D, Haffner S, Rewers M, Hamman R, Selby JV, Saad MF, Savage P, Bergman R. Insulin Sensitivity and Atherosclerosis. Circulation. 1996;93:1809–1817. doi: 10.1161/01.cir.93.10.1809. [DOI] [PubMed] [Google Scholar]
- 28.Lempiainen P, Mykkanen L, Pyorala K, Laakso M, Kuusisto J. Insulin Resistance Syndrome Predicts Coronary Heart Disease Events in Elderly Nondiabetic Men. Circulation. 1999;100:123–128. doi: 10.1161/01.cir.100.2.123. [DOI] [PubMed] [Google Scholar]
- 29.Rewers M, Zaccaro D, D’Agostino R, Haffner S, Saad MF, Selby JV, Bergman R, Savage P. Insulin Sensitivity, Insulinemia, and Coronary Artery Disease: The Insulin Resistance Atherosclerosis Study. Diabetes Care. 2004;27:781–787. doi: 10.2337/diacare.27.3.781. [DOI] [PubMed] [Google Scholar]
- 30.Vicent D, Ilany J, Kondo T, Naruse K, Fisher SJ, Kisanuki YY, Bursell S, Yanagisawa M, King GL, Kahn CR. The role of endothelial insulin signaling in the regulation of vascular tone and insulin resistance. J Clin Invest. 2003;111:1373–1380. doi: 10.1172/JCI15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang CP, Han S, Okamoto H, Carnemolla R, Tabas I, Accili D, Tall AR. Increased CD36 protein as a response to defective insulin signaling in macrophages. J Clin Invest. 2004;113:764–773. doi: 10.1172/JCI19528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perusse L, Rice T, Despres JP, Rao DC, Bouchard C. Cross-Trait Familial Resemblance for Body Fat and Blood Lipids: Familial Correlations in the Quebec Family Study. Arterioscler Thromb Vasc Biol. 1997;17:3270–3277. doi: 10.1161/01.atv.17.11.3270. [DOI] [PubMed] [Google Scholar]
- 33.Perusse L, Despres J, Tremblay A, Leblanc C, Talbot J, Allard C, Bouchard C. Genetic and environmental determinants of serum lipids and lipoproteins in French Canadian families. Arteriosclerosis. 1989;9:308–318. doi: 10.1161/01.atv.9.3.308. [DOI] [PubMed] [Google Scholar]
- 34.Fuentes RM, Notkola I-L, Shemeikka S, Tuomilehto J, Nissinen A. Familial Aggregation of Serum Total Cholesterol: A Population-Based Family Study in Eastern Finland. Preventive Medicine. 2000;31:603–607. doi: 10.1006/pmed.2000.0743. [DOI] [PubMed] [Google Scholar]
- 35.Katzmarzyk P, Perusse L, Rice T, Rao D, Bouchard C. Familial aggregation of seven-year changes in blood pressure in Canada. Can J Cardiol. 2001;17:1267–1274. [PubMed] [Google Scholar]
- 36.An P, Rice T, Gagnon J, Borecki IB, Perusse L, Leon AS, Skinner JS, Wilmore JH, Bouchard C, Rao DC. Familial aggregation of resting blood pressure and heart rate in a sedentary population: The heritage family study. American Journal of Hypertension. 1999;12:264–270. doi: 10.1016/s0895-7061(98)00261-1. [DOI] [PubMed] [Google Scholar]
- 37.Adeyemo A, Omotade OO, Rotimi CN, Luke AH, Tayo BO, Cooper RS. Heritability of blood pressure in Nigerian families. J Hypertens. 2002;20:859–863. doi: 10.1097/00004872-200205000-00019. [DOI] [PubMed] [Google Scholar]
- 38.Viswanathan M, McCarthy M, Snehalatha C, Hitman G, Ramachandran A. Familial aggregation of type 2 (non-insulin-dependent) diabetes mellitus in south India; absence of excess maternal transmission. Diabet Med. 1996;13:232–237. doi: 10.1002/(SICI)1096-9136(199603)13:3<232::AID-DIA27>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 39.Thomas F, Balkau B, Vauzelle-Kervroedan F, Papoz L. Maternal effect and familial aggregation in NIDDM. The CODIAB Study. CODIAB-INSERM-ZENECA Study Group. Diabetes. 1994;43:63–67. doi: 10.2337/diab.43.1.63. [DOI] [PubMed] [Google Scholar]
- 40.Carmelli D, Cardon L, Fabsitz R. Clustering of hypertension, diabetes, and obesity in adult male twins: same genes or same environments? Am J Hum Genet. 1994;55:566–573. [PMC free article] [PubMed] [Google Scholar]
- 41.Hong Y, Pedersen N, Brismar K, de Faire U. Genetic and environmental architecture of the features of the insulin-resistance syndrome. Am J Hum Genet. 1997;60:143–152. [PMC free article] [PubMed] [Google Scholar]