Abstract
Here, we combined classical biochemistry with novel biophysical approaches to study with high spatial and temporal resolution the organization of GPI-anchored proteins (GPI-APs) at the plasma membrane of polarized epithelial cells. We show that in polarized MDCK cells, following sorting in the Golgi, each GPI-AP reaches the apical surface in homo-clusters. Golgi-derived homo-clusters are required for their subsequent plasma membrane organization into cholesterol-dependent hetero-clusters. By contrast, in non-polarized MDCK cells GPI-APs are delivered to the surface as monomers in an unpolarized manner and are not able to form hetero-clusters. We further demonstrate that this GPI-AP organization is regulated by the content of cholesterol in the Golgi apparatus and is required to maintain the functional state of the protein at the apical membrane. Thus, different from fibroblasts, in polarized epithelial cells a selective cholesterol-dependent sorting mechanism in the Golgi regulates both the organization and the function of GPI-APs at the apical surface.
Introduction
In fibroblasts GPI-APs are organized into a mixture of monomers and of cholesterol dependent clusters (20–40% of total GPI-APs) composed of 3 to 4 GPI-APs of different species1. The mechanisms regulating the formation of these GPI-AP assemblies appear to rely both on the lipid environment, the actin cytoskeleton and the protein itself2-5. The organization of membrane domains is affected by the oligomeric state of proteins6-10, and dimers and oligomers of membrane proteins have been regarded recently as specific domains because they participate to the creation/exclusion of the different sub-compartments4,10 as exemplified in the case of receptor activation6,9,10. Thus one important question is the physiological significance of GPI-APs spatial organization in membranes and whether and how it regulates specific GPI-AP function. This has a particular relevance in polarized epithelial cells where GPI-APs are preferentially sorted to the apical domain of the plasma membrane. Of interest, clustering in the Golgi is a mechanism required for assuring the apical sorting of GPI-APs11,12. The fact that GPIAPs arrive in oligomeric clusters at the apical membrane of polarized cells13, while they are delivered as monomers to the plasma membrane of fibroblasts and non-polarized cells can have major consequences for the formation and hierarchical organization of membrane domains and consequently can affect both the dynamics and the functional state of GPI-APs in polarized vs non-polarized cells. We found that at the apical membrane of polarized MDCK cells GPI-APs are organized in small clusters of single GPI-AP species (referred to as homo-clusters), which are independent of cholesterol. These complexes, but not monomeric GPI-APs, coalesced into larger cholesterol-dependent clusters formed by multiple GPI-APs species (named hetero-clusters). Biochemical and live microscopy data demonstrated that the pre-organization in homo-clusters occurring in the Golgi of polarized epithelial cells is required for hetero-cluster formation at the plasma membrane and regulates the functional state of the protein at the cell surface.
Results
Apical GPI-APs are organized in homo- and hetero-clusters
At the plasma membrane of fibroblasts 20-40% of GPI-APs are organized in cholesterol-dependent nanoclusters containing different species1. To understand whether this was the case in polarized epithelial cells we produced MDCK clones stably transfected with PLAP (Placental Alkaline Phosphatase) and GFP-FR (Folate Receptor)1,11. The relative organization of these model apical GPIAPs (which associated to DRMs and formed HMW complexes; Supplementary Fig. 1a-c) was biochemically assayed by addition to the cell surface of the impermeable cross-linking agent, BS3 [Bis(sulfosuccinimidyl)suberate]14 (Supplementary Fig. 1d-f) followed by a co-immunoprecipitation assay. In agreement with previous findings14 both GFP-FR and PLAP were in HMW complexes (here referred to as homo-clusters) on the apical surface of MDCK cells (Supplementary Fig. 1d-f). Inaddition they could be specifically co-immunoprecipitated in hetero-clusters (Supplementary Fig. 2a,b), which excluded GP114, a transmembrane apical protein, (Supplementary Fig. 2c).
To understand whether homo- and hetero-clusters formation depended on cholesterol we repeated the cross-linking experiments applying the same pharmacological conditions used to perturb the clustered organization of GPI-APs in CHO cells1,2. Addition of saponin did not alter the cross-linking of homo-clusters, as shown by the presence of the HMW bands corresponding to the cross-linked proteins (Supplementary Fig. 1e,f). On the contrary, upon cholesterol depletion homo-clusters were no longer cross-linkable in hetero-clusters (Supplementary Fig. 2a). Importantly, homo-clusters of GFP-FR and PLAP were insensitive to harsh cholesterol depletion conditions (Supplementary Fig. 3a,b), ruling out different threshold sensitivities of homo- and hetero-clusters.
Although confocal microscopy does not provide the spatial resolution to discriminate single GPI-APs clusters we could quantify the effect of saponin on GFP-FR and PLAP co-localization (Supplementary Fig. 3c). Saponin drastically reduced the co-presence between GFP-FR and PLAP resulting in the increase of exclusion areas (50-55%, p< 0.00001) and in a decrease of co-localization (Supplementary Fig. 3d), supporting hetero-clusters dependency of cholesterol.
Apical GPI-AP homo-clusters are cholesterol independent
The above data suggested that differently from fibroblasts1,15 at the apical membrane of polarized cells cholesterol does not influence GPI-AP homo-cluster organization, but regulates the formation of hetero-clusters. To assess the spatio-temporal organization of GPI-APs in live polarized cells we established different biophysical techniques with high spatio-temporal resolution.
In order to visualize single homo-clusters of GFP-FR we used Number and Brightness (N&B) analysis, which provides a pixel resolution map of molecular numbers and aggregation state of fluorescent proteins in living cells16 (see Methods). The aggregation state (or brightness) of fluctuating particles in a specific pixel is given by the variance of their intensity in a pixel/time, which means that larger particles (e.g., dimers, trimers, etc.) would result in higher intensity variance (brightness). Consequently the ratio between Brightness and Intensity (B/I) provides a very good approximation and a high spatial and temporal resolution16 of the number of molecules in a fluctuating particle in living cells (see Methods)16. We observed that the particle brightness, expressed as average B value, of GFPFR (1.25) was higher than the B values corresponding to the monomeric forms of the cytosolic (mGFP, 1.08) and membrane-linked (mGFP-FR, 1.07) GFP (Fig. 1a), indicating that it was in an aggregated state at the apical surface of living MDCK cells. Because photobleaching could influence the brightness of these molecules, we measured the bleaching rate of mGFP-FR and GFP-FR, which were comparable (7-18%; Supplementary Fig. 4a), suggesting that the difference in B was specific.Furthermore, the B map showed that B values were independent of the fluorescence intensity values (Fig. 1b), ruling out the possibility that large B is a consequence of the relatively high local density (compare maps of B and I and scattergram in Supplementary Fig. 4b). The experimental B values ranging from 0.9 to 1.8 indicated heterogeneity in the aggregation state of GFP-FR. Using the K-means function of Matlab software we partitioned the observed B values in groups (with 0.5 intervals), of which we measured the occurrence (% of cells in each group) (Fig. 1c; see Methods). Furthermore, to determine the correspondence between the experimental B values and the aggregation state of GFP-FR (one, two, three, four, etc. molecules) we constructed a standard curve by plotting the experimental B values obtained for monomer, dimer and trimer of soluble mGFP against the number of units/molecule(Supplementary Fig. 4c). Statistical analysis revealed that the difference between the B values for monomeric, dimeric and trimeric GFP were significant (monomer vs dimer p<0.00003; dimer vs trimer p<0.0006; Supplementary Fig. 4d) and B increased linearly with the number of molecules (Supplementary Fig. 4c). Furthermore the perfect overlap with a curve obtained with the experimental B values of another model GPI-AP in which one or two mGFP moieties were linked in tandem to the GPI anchor of the uPAR (Supplementary Fig. 4c), indicated that the B values of a soluble and a membrane-linked GFP of defined molecular mass are in the same range (see Methods).
Figure 1. GFP-FR is in homo-clusters at the apical surface of living MDCK cells.
After 4 days in culture, cells were imaged live by confocal microscopy (scanning ROIs of 256×64 pixels over time) and were analyzed for N&B (see Methods). (a) Quantification of the brightness of GFP-FR in MDCK cells expressing GFP-FR (left) or co-expressing GFP-FR and PLAP (right) from 5 independent experiments is plotted together with the mean B value of mGFP (6 experiments) and mGFP-FR (4 experiments). Error bars, ± SD. *, p<0,000001; (b) Brightness (B) and fluorescence intensity (I) maps of a representative cell are shown in MDCK cells expressing GFP-FR alone (left panels) or co-expressing GFP-FR and PLAP (right panels) showing that brightness and intensity are independent from each other. Bars, 0.9 μm. (c) The percentage of pixels in groups (with a value interval of 0.5) of observed B values of a representative experiment is shown together with their occurrence (inset), expressed as percentage of cells falling in each group (n=30 cells). (d) Graphical representation of the percentage of pixels falling in the different classes of B values (from monomer to hexamer) on the basis of the calibration curve shown in Supplementary Fig. 4g. Values are expressed as mean of 5 independent experiments. Error bars, ± SD. Note that the average B values derived from different area of the plasma membrane and from various clones expressing GFP-FR at different levels did not change (see Methods).
Because the B values for each molecular species (e.g., monomer to trimer) were distributed along a Gaussian curve (Supplementary Fig. 4e), we could ascribe the range of B values corresponding, respectively, to monomer (1.05-1.14), dimer (1.15-1.24), and trimer (1.25-1.29) (Supplementary Fig. 4f). Hence, by interpolating the experimental B values of GFP-FR on the standard curve (Supplementary Fig. 4g) we could extrapolate the range of B values corresponding to tetramer (1.30-1.39), pentamer (1.40-1.44) and hexamer (1.45-1.54) (Fig. 1d).
Overall this analysis revealed that besides monomers and dimers and a few large aggregates, about 30-35% of GFP-FR has a B value within the range of 1.25 to 1.39, corresponding to an organization in aggregates containing three-four molecules (Fig. 1d). As a control for the specificity and accuracy of these calculations we measured the brightness of p75-GFP, a well characterized apical transmembrane protein, which is known to dimerize in the presence of growth factors or serum17,18. Consistently more than 80% of p75-GFP showed B values ranging from 1 to 1.24 (corresponding to monomers and dimers; Supplementary Fig. 4h,i).
Of interest, differently from CHO cells1, the aggregation state of GFP-FR was not affected by the co-expression of another GPI-AP (mCherry-PLAP) indicating that in polarized MDCK cells GPI-AP homo-clusters were independent from each other (Fig. 1a-d). These results were obtained using several clones expressing different levels of mCherry-PLAP suggesting that the GFP-FR clusters are self autonomous and are independent of the number of molecules of another GPI-AP. Furthermore, upon saponin (Fig. 2a,b and Supplementary Fig. 5a,b) or mevinolin (Supplementary Fig. 5c-e) treatments there was no change in the average brightness of GFP-FR nor in the distribution in the specific classes of B values (Fig. 2a,b). As saponin and mevinolin used before to deplete cholesterol from the cells1,2,19,20 should not alter other lipid biosynthesis nor organization21,22, these data demonstrated that cholesterol depletion does not affect the aggregation state of GFP-FR and supported our hypothesis that apical GPI-APs homo-clusters in living MDCK cells are independent of cholesterol.
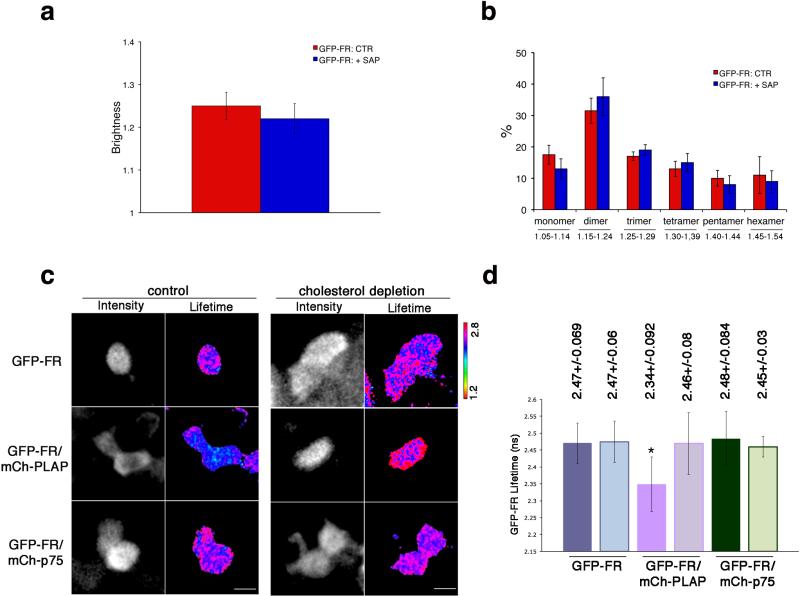
Figure 2. Apical GPI-AP homo- and hetero-clusters have different sensitivity to cholesterol depletion.
After 4 days in culture, polarized MDCK expressing GFP-FR were treated with saponin and imaged in vivo for N&B (a,b) or FLIM (c,d). (a) Quantification of the brightness of GFP-FR from 3 independent experiments is plotted. Error bars, ± SD. (b) Graphical representation of the percentage of pixels falling in the different classes of B values (from monomer to hexamer) on the basis of the calibration curve shown in Supplementary Fig. 4g. Values are expressed as mean of 3 independent experiments. Error bars, ± SD. (c) Intensity and mean fluorescence lifetime maps of GFP-FR alone or in presence of either mCherry-PLAP or mCherry-p75 in control or cholesterol depleted cells. The lifetime scale is from 1.2 ns to 2.8 ns. Cells were imaged live by scanning ROIs of 140×140 pixels corresponding to 4-6 cells of the confluent polarized monolayer. Dark areas correspond to cells either not expressing or out of focus (see methods and comparison between N&B and FLIM images in Supplementary Fig. 6a). Bars, 9 μm. (d) Histograms of GFP-FR lifetime (ns) alone (blue bars) or in combination with mCherry-PLAP (pink bars) or mCherry-p75 (green bars) in control conditions (colored bars) or upon cholesterol depletion (pale colored bars). Experiments were performed 3×, n>35 cells. Error bars, ± SD. *, p<0,0001.
Apical GPI-AP hetero-cluster organization is cholesterol dependent
Next, to study the hetero-cluster organization of GPI-APs in living cells we set up a time domain Fluorescence Lifetime Imaging Microscopy (FLIM) approach23,24, which measures the fluorescence decays (by mean lifetime) of the donor in the absence or presence of a putative acceptor. A decrease of the lifetime of the donor in the presence of an acceptor reveals fluorescence energy transfer (FRET) between the two molecules, indicating close proximity (maximum FRET distance 10 nm). In control cells expressing GFP-FR (donor) alone the mean GFP-FR lifetime was 2.47 ns ± 0.069 (n= 152 cells) and it was significantly decreased to 2.34 ns ± 0.09 (n= 64 cells) in cells co-expressing mCherry-PLAP (as acceptor), revealing FRET occurrence between the two apical GPI-APs (Fig. 2c,d). In contrast the mean GFP-FR lifetime did not change (2.48 ns ± 0.08; n= 40) in cells co-expressing as acceptor mCherry-p75 (an apical transmembrane protein; Fig. 2c,d). This demonstrated that at the apical membrane of polarized MDCK cells two GPI-APs were in close proximity and suggested that they were organized in hetero-clusters, which excluded other apical proteins. To quantitatively interpret these data we determined the quantity of interacting donor (%) using the minimal fraction donor value (mfD) that reflects the proportion of donor proteins engaged in FRET23,24 (see Methods). This was independent of the amount of the proteins expressed at the plasma membrane (as shown by using cells expressing the two proteins at different levels, Supplementary Fig. 6b,c) and therefore independent of both donor and acceptor fluorescence intensities (Supplementary Fig. 6d,e).
Upon cholesterol depletion (saponin or mevilonin) the hetero-FRET between GFP-FR and mCherry-PLAP was lost (Fig. 2c,d) supporting the biochemical findings (Supplementary Fig. 2a) that in polarized MDCK cells GPI-APs hetero-clusters were cholesterol-dependent.
Only cholesterol-dependent GPI-AP clusters exist in CHO cells
Although independent from the molecular concentration as in fibroblasts1,15, the apical organization of GPI-APs in MDCK cells appeared to be different from CHO cells, where 20-40% of GPI-APs are found in cholesterol-dependent nanoclusters containing different GPI-AP species (3 to 4 molecules)1,2. To understand whether this was due to the different approaches used or was dependent on the cell type, we investigated the aggregation state of GFP-FR in CHO cells applying N&B (Supplementary Fig. 7a-d). The average B value in CHO was 1.36 corresponding to an aggregated state of GFP-FR, and the class analysis showed that about 50% of GFP-FR had a B value ranging from 1.25 to 1.44 supporting the existence of clusters of 3 to 4 or 5 molecules (Supplementary Fig. 7a-d). However, in contrast to MDCK cells, upon treatment with saponin there was a reduction of the total B value (compare Supplementary Fig. 7a with Fig. 2a) and a shift in the distribution of the different classes of clusters towards monomers and dimers, indicating cluster dissociation (Supplementary Fig. 7c,d). This was consistent with the nanocluster organization previously reported in CHO cells by using anisotropy1,15 and showed that the organization of GPI-APs is different between fibroblasts and polarized epithelial cells, independently of the methodology used. Furthermore, brightness measurements suggested that GFP-FR clusters in CHO cells are affected by the presence of another GPI-AP (Supplementary Fig. 7a) confirming that different GPI-APs species inhabit the same nanocluster1. Consistently in CHO cells expressing GFP-FR and mCherry-PLAP we could detect hetero-FRET between the two GPI-APs, which was loss upon cholesterol depletion (Supplementary Fig. 7e,f). However in a larger population of CHO cells compared to MDCK cells (30% vs 13%) the GFP-FR interacting fraction was less than 20% (Supplementary Fig. 6f,g). This low quantity of interacting donor (%) in CHO cells is consistent with the nanocluster organization of GPI-APs previously described in CHO cells where energy transfer occurs between 3 to 4 molecules within the same cluster1. On the other hand the fact that in a large proportion of cells (87%) the quantity of GFP-FR in interaction is more than 20% (Supplementary Fig. 6f) supports the occurrence of FRET between hetero-clusters in polarized MDCK.
GPI-APs hetero-clusters derive from pre-existing homo-clusters
To understand the mechanisms of hetero-clusters organization in polarized MDCK cells we used distinct biochemical approaches. To assess whether GFP-FR and PLAP hetero-clusters were derived from monomers or oligomers cross-linking we performed co-immunoprecipitation assays, after protein purification on velocity gradients. We observed that PLAP and GFP-FR co-immunoprecipitated only in fraction 9, which contained HMW complexes, and not in the fractions containing the monomers (Supplementary Fig. 8a). Consistently, DTT (Dithiothreitol) treatment to reduce GFP-FR in monomeric or partially denaturated dimeric forms11 (band at 43 kDa, Supplementary Fig. 8b) impaired its co-immunoprecipitation with PLAP (Supplementary Fig. 8b). These combined experiments demonstrated that only homo-HMW complexes and not GPI-AP monomers could be cross-linked in hetero-clusters and suggested that hetero-clusters were derived from pre-existing homo-clusters.
Golgi homo-clustering regulates apical GPI-AP organization
We hypothesized that in polarized cells apical GPI-AP homo-clusters derive directly from the Golgi apparatus where they form in order to be apically sorted11. If homo-clusters are required for hetero-cluster formation it follows that in polarized epithelial cells the mechanism of GPI-AP apical sorting would determine their spatial organization at the plasma membrane.
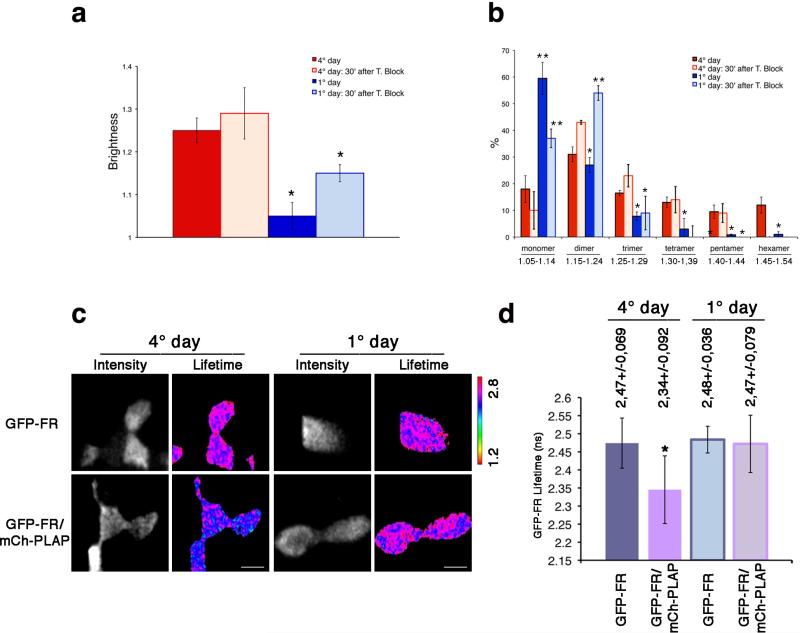
If this hypothesis is correct, GPI-APs, delivered to the surface in their monomeric form25 in nonpolarized MDCK cells, should not be organized in homo/hetero-clusters. In order to test this, we analyzed the organization of the newly arrived proteins at the plasma membrane of polarized and nonpolarized MDCK cells. Cells were subjected to a trypsin treatment to remove resident proteins from the plasma membrane25 and to a temperature shift at 19.5°C to block protein exit from the Golgi. Following release of the block for 30 min at 37°C, we then measured the N&B of the proteins newly arrived at the surface (Fig. 3a,b). Interestingly the average B values (1.29) of newly arrived GFP-FR molecules at the plasma membrane of polarized epithelial cells were higher compared to the average B values of GFP-FR recorded in non-polarized cells (1.15). This showed that in polarized cells newly synthesized GFP-FR molecules reach the apical surface in clusters, while in non-polarized conditions they are delivered as monomers/dimers (Fig. 3a,b). Importantly B values measured at steady state, in non-polarized cells indicated that GFP-FR remained in the form of monomers and dimers (Fig. 3a,b and Supplementary Fig. 9a,b). Consistently, no hetero-FRET was measured between GFP-FR and mCherry-PLAP (Fig. 3c,d). This indicates that monomeric/dimeric GPI-APs at the plasma membrane of non-polarized MDCK cells are not able to form hetero-clusters.
Figure 3. Both homo-cluster and hetero-cluster organization is dependent on the polarity status of MDCK cells.
N&B (a,b) and FLIM (c,d) analysis were performed in MDCK cells after 1 or 4 days in culture. (a) Quantification of the brightness of different populations of GFP-FR molecules at the cell surface for the steady state population (colored bars) and newly arrived population (pale colored bars) from 3 independent experiments is plotted. Error bars, ± SD. *, p<0,000001. (b) Graphical representation of the percentage of pixels falling in the different classes of B values (from monomer to hexamer) on the basis of the calibration curve shown in Supplementary Fig. 4g. Values are expressed as mean of 3 independent experiments. Error bars, ± SD. *, p<0.05; **, p<0.008; ***, p<0.00005. (c) Intensity and mean fluorescence lifetime maps of GFP-FR alone or in combination with mCherry-PLAP. The lifetime scale is from 1.2 ns to 2.8 ns. Bars, 9 μm. (d) Histograms of GFP-FR lifetime alone (blue bars) or in combination with mCherry-PLAP (pink bars) in polarized MDCK cells (4 days of culture, colored bars) or in non-polarized MDCK cells (1 day of culture, pale colored bars). Experiments were performed 2×, n>15 cells. The error bars are the mean ± SD. *, p<0,0001.
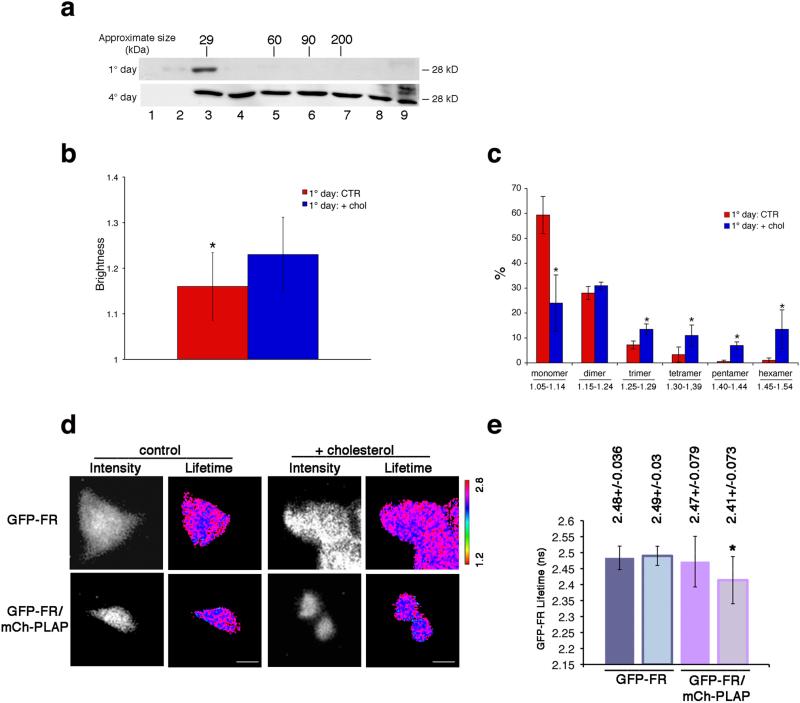
These data suggested that the different GPI-APs organization at the cell surface of polarized vs nonpolarized MDCK cells (e.g., homo- and hetero-clusters vs monomers/dimers) depended directly on the mechanism of Golgi sorting, which requires protein oligomerization11,26. To directly verify this hypothesis we analyzed biochemically the oligomerization state of GFP-FR in the Golgi of polarized and non-polarized MDCK cells. Strikingly, in the Golgi of polarized MDCK cells GFP-FR is in oligomers (30%) while it is almost exclusively monomeric in non-polarized MDCK cells (Fig. 4a) as well as in fibroblasts (Supplementary Fig. 7g).
Figure 4. Cholesterol addition is necessary and sufficient to induce homo- and hetero-cluster of GPI-APs in non-polarized MDCK cells.
(a) MDCK cells expressing GFP-FR grown for 4 days (polarized) or 1 day (non-polarized) were treated with trypsin (to remove surface proteins) and then subjected to temperature block (at 19.5°C in the Golgi) in order to analyze exclusively the Golgi pool of proteins. Cell lysates were run on velocity gradient (see Methods). Fractions were collected from top (Fraction 1) to bottom (Fraction 9), TCA-precipitated, run on SDS-PAGE and GFP-FR was revealed by western blot. Molecular weight markers are indicated on top of the panels. (b-e) 1 day MDCK cells (control or upon cholesterol addition) were used for N&B and FLIM analysis. (b) Quantification of the brightness of GFP-FR from 3 independent experiments is plotted. Error bars, ± SD. *, p<0,0001. (c) Graphical representation of the percentage of pixels falling in the different classes of B values (from monomer to hexamer; see Supplementary Fig. 4g). Values are the mean of 3 independent experiments. Error bars, ± SD. *, p<0.05. (d) Intensity and mean fluorescence lifetime maps of GFP-FR alone or in combination with mCherry-PLAP. The lifetime scale is from 1.2 ns to 2.8 ns. Bars, 9 μm. (e) Histograms of GFP-FR lifetime alone (blue bars) or in combination with mCherry-PLAP (pink bars) in control conditions (colored bars) or upon cholesterol addition (pale colored bars). Experiments were performed 3×, n>15 cells. The error bars are the mean ± SD. *, p<0,005.
Cholesterol addition induces GPI-AP re-organization
Because in polarized MDCK cells cholesterol is a key regulator of the Golgi homo-clustering of apical GPI-APs11,26,27 we asked whether differences in the cellular content of cholesterol between polarized and non-polarized cells could explain the different organization of GPI-APs at the cell surface. To address this we analyzed the B of GFP-FR in non-polarized MDCK cells following exogenous addition of cholesterol (Fig. 4b,c and Supplementary Fig. 9c,d). In these conditions we observed an increase in the average B value of GFP-FR (Fig. 4b) and a shift in the class analysis of the B values from monomer towards higher-order aggregates (Fig. 4c) compared to control cells. Importantly, this was specific for GPI-APs because cholesterol addition did not change the brightness of a control transmembrane protein (p75-GFP; Supplementary Fig. 9e-g). Furthermore, in the presence of mCherryPLAP we recorded a decrease in the lifetime of GFP-FR showing hetero-FRET between the two molecules and supporting the occurrence of hetero-clusters between the two GPI-APs at the cell surface of non-polarized MDCK cells (Fig. 4d,e) upon cholesterol addition.
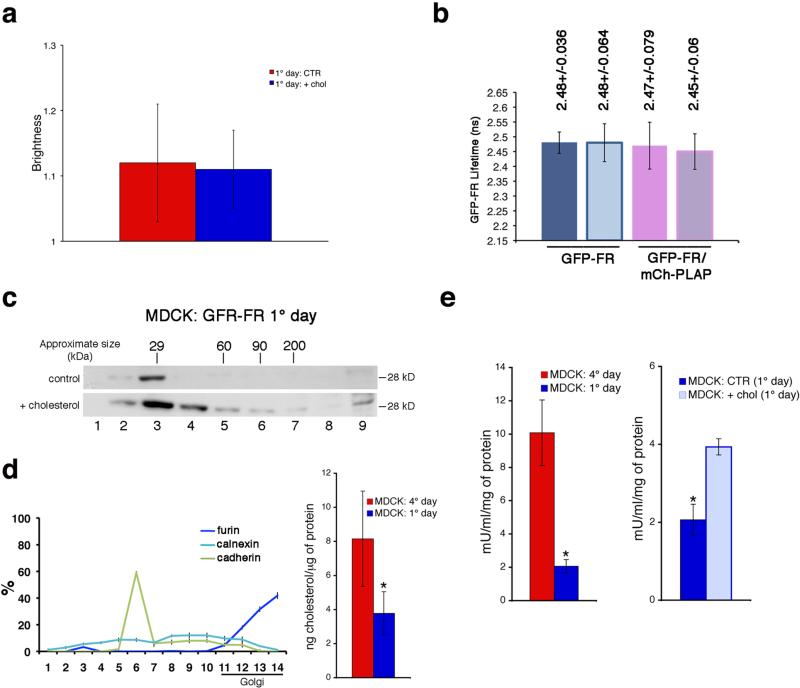
These data indicated that addition of cholesterol was sufficient to induce a whole re-organization of GPI-APs at the cell surface of non-polarized MDCK cells. However they did not show whether this was occurring at the cell surface of non-polarized MDCK cells (by coalescence of monomers, as previously shown in CHO cells) or was the consequence of the re-organization of GPI-APs in the Golgi complex (e.g., inducing homo-clustering, like in polarized condition). In order to discriminate between these two possibilities, we performed N&B (Fig. 5a and Supplementary 9h-j) and FLIM (Fig. 5b and Supplementary 9k) analysis considering the pool of GFP-FR proteins exclusively at the cell surface of non-polarized MDCK cells either in control conditions or upon cholesterol addition. Cells were pre-treated with cycloheximide for two hours (time sufficient to empty the Golgi of GFP-FR) and then incubated with exogenous cholesterol (40 min) still in the presence of cycloheximide in order to exclude from the analysis newly synthesized proteins. Interestingly, in these conditions no homo-clusters (Fig. 5a) or hetero-clusters (Fig. 5b) were detected indicating that cholesterol addition did not affect the organization of pre-existing GPI-APs at the plasma membrane. On the other hand, analysis of the pool of proteins present in the Golgi (by velocity purification on sucrose gradients upon temperature block) revealed that cholesterol addition induced homo-clustering of GFP-FR molecules present in the Golgi, as suggested by the shift towards the heavier fractions of the gradients compared to control conditions (Fig. 5c). Overall these data indicate that cholesterol addition was sufficient to induce homo-clustering of GPI-APs in the Golgi and that this was required and sufficient to promote their organization into homo- and hetero-clusters at the plasma membrane of non-polarized MDCK cells. If this is the case it follows that the amount of cholesterol in the Golgi of polarized vs nonpolarized cells should be different, and tightly regulated. Consistently, measurement of the cholesterol content by using cellular fractionation28 revealed that the Golgi of polarized MDCK cells contains double the amounts of cholesterol compared to the Golgi of non-polarized MDCK cells (Fig. 5d).
Figure 5. Cholesterol addition promotes homo-clustering of GPI-AP in the Golgi and regulates hetero-cluster formation and GPI-AP activity at the cell surface.
N&B (a) and FLIM (b) analysis were performed in 1-day MDCK cells pre-treated (2hrs) with cycloheximide in order to consider exclusively the cell surface pool of GFP-FR, in control conditions or upon cholesterol addition (in presence of cycloheximide). (c) 1 day MDCK cells treated with trypsin were subjected to temperature block, and incubated or not with cholesterol (during the last hour of temperature block) in order to analyse the GFP-FR Golgi pool and purified on velocity gradient. Molecular weight markers are indicated on top of the panels. (d) Cholesterol quantification after subcellular fractionation of MDCK cells. (Left) Quantification of the distribution of ER (calnexin), plasma membrane (cadherin), and trans-Golgi (furin) markers along the sucrose density gradient expressed as percentage of total proteins. (Right) Cholesterol amount in the Golgi-enriched fractions (11–14 fractions) quantified and normalized per microgram of protein in 4-days (red bar) and 1-day (blue bar) culture cells. Experiments were performed 2×. The error bars are the mean ± SD. *, p<0,005. (e) The alkaline phosphatase activity of PLAP in MDCK cells was measured (see Methods) upon different conditions: MDCK cells after 1 or 4 days in culture (left); 1-day MDCK cells in control or upon cholesterol addition (right). In all graphs the ALP activity of PLAP is expressed mU/ml and is normalized for total amount of protein. The error bars are the mean ± SD. *, p< 0,004.
Golgi sorting regulates GPI-APs activity at plasma membrane
By linking the sorting of GPI-APs in the Golgi to their organization at the plasma membrane this mechanism does not allow the missorted proteins (in non-polarized cells) to accomplish the same organization they would have in polarized conditions. Therefore the question arises as to whether this has a functional relevance in regulating proteins activity at the plasma membrane. Thus, we measured the catalytic activity of native PLAP in polarized versus non-polarized MDCK cells. We found that PLAP exhibited an activity three folds higher in polarized MDCK compared to non-polarized MDCK cells (Fig. 5e). Importantly, after addition of cholesterol in non-polarized MDCK cells there was a statistically significant increase (of about two folds) in PLAP activity (Fig. 5e).
Overall these data showed that the cholesterol-dependent mechanism of GPI-AP sorting in the Golgi of polarized cells regulates the organization and modulates the protein function (in the specific case, the enzymatic activity of PLAP) at the plasma membrane (Fig. 6).
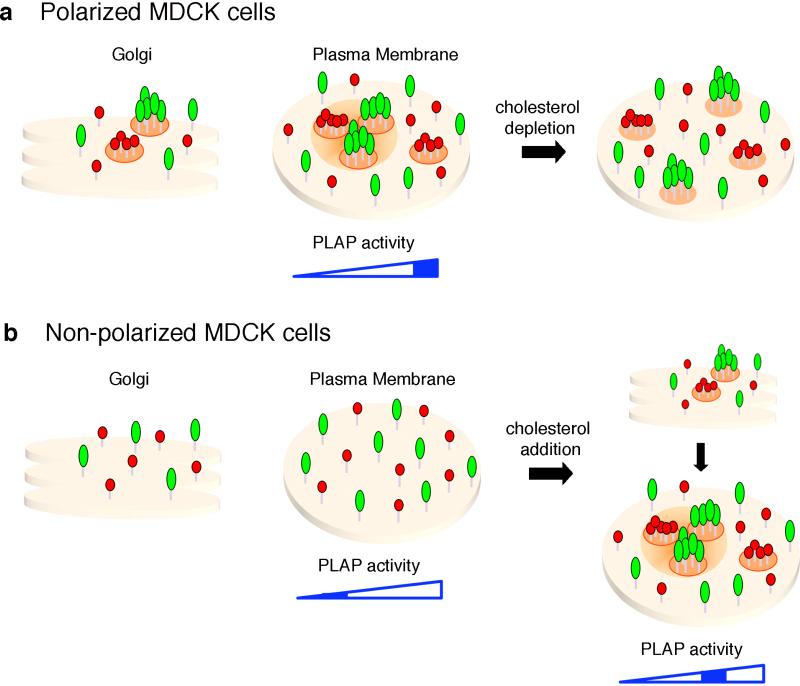
Figure 6. Model of GPI-APs organization and activity in MDCK cells.
(a) In polarized MDCK cells, apical GPI-APs form homo-cluster in the Golgi complex and at the plasma membrane are organized into homo-clusters of the same GPI-AP species (red and green aggregates), and hetero-clusters derived from homo-clusters coalescence. Upon cholesterol depletion homo-clusters are unaffected, while the hetero-cluster organization is lost. We postulate that because of their high affinity for cholesterol and saturated sphingolipids, homo-clusters may generate a local enrichment of raft lipids (including cholesterol) within a confined zone that facilitates their coalescence into hetero-clusters, while excluding GPI-AP monomers and other apical components which have less affinity for lipid domains. (b) In non-polarized MDCK cells GPI-APs do not cluster in the Golgi complex and at the plasma membrane remain in the form of monomer and dimer. Upon cholesterol addition GPI-APs cluster in the Golgi and consequently at the plasma membrane assume the same organization found in polarized cells in homo-and hetero-cluster. Note that clustering organization regulates the activity of PLAP, and in polarized cells this depends on clustering/sorting in the Golgi. In this cartoon, two different GPI-APs are represented as monomer, although it is conceivable that dimers would be the basic unit required for supramolecular organization of GPI-APs (see discussion).
Discussion
GPI-APs homo- and hetero-clusters apical organization
Membrane organization of GPI-APs is crucial for regulating their cellular fates and biological functions7,29. This is particularly relevant in the case of polarized epithelial cells where a selective localization of these proteins at the apical membrane has been reported30,31. In polarized MDCK cells GPI-APs cluster in HMW complexes at the level of the Golgi in order to be apically sorted11,12, while they are in monomers and dimers in non-polarized conditions and in fibroblasts. We postulated that the arrival at the plasma membrane of clustered GPI-APs11,13 would affect their plasma membrane organization and consequently their function. The cholesterol-dependent clustered organization of GPIAPs has been reported previously in non-polarized cells1,2,32, however only scarce data are available in polarized conditions14,33 where the analysis is more complex. Here we used N&B and FLIM techniques to analyse the relative organization of two GPI-APs in their native states in living cells with high spatial-temporal resolution.
Our data showed that at the apical surface of polarized MDCK cells 20-35% of GFP-FR was organized in cholesterol independent homo-clusters containing 3 to 5 molecules of single species of GPI-AP, and hetero-clusters with two or more different species of GPI-APs, which were sensitive to cholesterol depletion (Fig. 6). GPI-APs hetero-clustering was independent on the protein concentration, thus supporting a model of molecular clustering driven by an active mechanism, as previously suggested in fibroblasts1,4,15,34. The apparent discrepancy with TOCCSL data describing only homo-dimers5 might be due to differences in the technique. Because TOCCSL measures only the brightness of molecules diffusing in a bleached area it plausible that monomers and dimers diffuse faster than oligomers35 and thereby would be predominantly detected.
Homo-clusters are a prerequisite for hetero-cluster formation
Hetero-clusters could be formed by the coalescence of homo-clusters or by the collision of monomeric GPI-APs. Our FLIM and biochemical data (Fig. 3 and Supplementary Fig. 8) concurred to show that in polarized MDCK cells hetero-clusters derive from coalescence at the plasma membrane of homo-clusters and not from monomeric GPI-APs. This is fundamentally different from the nanocluster described in CHO cells1 that does not require pre-homo-clustering (Supplementary Fig. 7g). Importantly, we demonstrated that in epithelial cells GPI-AP apical organization is directly linked to the mechanism of their apical sorting, which requires oligomerization in the Golgi and is dependent on cholesterol11. Consistently we found that the cholesterol levels in the Golgi of polarized cells are higher compared to non-polarized MDCK cells (Fig. 5d).
The fact that GPI-APs are organized in size-defined assemblies (homo-clusters of 3-5 molecules) is likely due to the co-occurrence of protein-protein and protein-lipid interactions1,9,10. Indeed based on protein-protein interactions, proteins tend to form infinite linear assemblies (except for the case of circular oligomers36), while in the case of GPI-AP oligomerization both a favourable lipid environment and a specific protein ectodomain are necessary for the process11, 26.
Interestingly a recent study shows that the basic unit required for the supramolecular organization of GPI-APs at the plasma membrane is the dimer, whose formation is dependent on protein-protein interactions and stabilized by interaction with raft-lipids4. Consistent with this hypothesis, we had previously shown that a double point mutation in the GFP sequence (S49/71) that prevents GFP dimerization also impairs oligomerization in the Golgi and the consequent apical sorting of the protein11. Taken together these data indicate that also in the Golgi of epithelial cells the capacity of dimerization is required for the construction of cholesterol-dependent higher order molecular organization of GPI-APs26,27.
Organization of GPI-APs into clusters controls their function
Overall, our data demonstrate that the mechanism of GPI-AP organization at the plasma membrane is different between epithelial and non-epithelial cells. The question therefore arises as to whether it has any physiological significance. GPI-AP clustering (and its partitioning into specific membrane domains) is a way to regulate their functional state4,6,7,10. Differently from fibroblasts in polarized epithelia GPIAPs are selectively delivered to the apical membrane where they perform specialized functions that should not be allowed at the basolateral membrane. Thus while in fibroblasts GPI-AP function needs to be controlled locally at the surface, in epithelial cells it may be important to switch off these functions in case of polarity loss (e.g., cancer, development, infection, etc...) and protein missorting to the basolateral side. The data presented here on the enzymatic activity of PLAP (Fig. 5e) indicate that in polarized epithelial cells, the cluster organization of GPI-APs is achieved only after proper sorting in the Golgi. Thus coupling of the plasma membrane organization to their Golgi sorting allows fine-tuning of GPI-AP activity by maintaining the protein in a sub-optimal state at the plasma membrane until cells reach a fully polarized phenotype.
The discovery of this novel mechanism of regulation of plasma membrane organization of GPI-APs will allow addressing other open questions within a new perspective. For example, what is the possible identity of the cholesterol-sensitive hetero-clusters? We could speculate that the cholesterol-dependent hetero-clusters fulfill the biological and functional definition of cholesterol-dependent microdomains9. Besides working as signalling platforms6,9,37, organization of GPI-APs in nanoclusters is necessary for their targeting into the endocytic GEEC (GPI-AP enriched endosomal compartment) pathway38,39. Furthermore nanoclusters may be induced to form large-scale lipid domains that can enter cells via tubular invaginations and form CLICs38-40. Based on this evidence the presence of hetero-clusters compared to monomers/dimers could define specific internalization pathways in polarized vs nonpolarized cells. Thus it will be interesting to study the dynamics and the pathways of GPI-AP internalization in polarized versus non-polarized MDCK cells and to understand whether and how induced clustering would affect them.
Finally our data also highlight the existence of various mechanisms that control clustering between different cell types. The specific difference between epithelial cells and CHO cells could be due to the fact that these cells originate from different embryonic layers. We can speculate that fibroblasts such as CHO cells lack the selective molecular machineries that have been generated in response to the external cues directing the polarization process in epithelial cells41,42. Thus one needs to consider physiological context when addressing basic cellular mechanisms.
Online methods
Cell culture, transfections and antibodies
MDCK cells were grown in DMEM (Sigma-Aldrich) containing 5% FBS. MDCK cells were cotransfected with sequences encoding for GFP-FR and PLAP or with mCherry-PLAP or mCherry-p75 (the latter two were cloned by PCR in pcDNA3.1 vector carrying hygromicin resistance). In selected experiments we used MDCK cells stably transfected with p75-GFP (kind gift of Dr. A. Le Bivic, IBDML, Marseille, France) or with mGFP-FR (cloned by PCR in PJB20 vector carrying neomicin resistance). Monomeric GFP (mGFP) was derived by site-directed mutagenesis replacing the hydrophobic amino acids (in the external part of the β-barrel) with positively charged amino acids43. One, two or three mGFP in tandem were engineered and cloned by PCR in pcDNA3.1 vector. CHO cells stably expressing GFP-FR (kind gift of Dr. S. Mayor, NCBS Bangalore, India) were grown in HAM's F12 medium containing 10% FBS and were transiently co-transfected with mCherry-PLAP.
The cDNAs encoding for mGFP-GPI (uPAR) and mGFP-mGFP-GPI (uPAR), in which one or two mGFP, respectively, are fused to the GPI attachment signal of uPAR44, were a kind gift of Dr. N. Sidenius (IFOM, Milan, Italy) and were transiently transfected in MDCK cells.
We used the following antibodies: polyclonal and mouse monoclonal anti-PLAP (Rockland and Sigma-Aldrich, respectively), polyclonal anti-GFP (Clontech).
Velocity gradients
Cells, grown to confluence in 100 mm dishes, were lysed in 20 mM Tris, pH 7.4, 100 mM NaCl, 1% TX-100 (with or without 0.4% SDS). Lysates were scraped from dishes, sheared through a 26-g needle and layered on top of a discontinuous sucrose gradient (30-5%) in the same buffer containing 0.1% TX-100. After centrifugation at 45,000 rpm for 16 h in a Beckman SW50 ultracentrifuge, fractions of 500 μl were harvested from the top of the gradient.
Cross-linking
BS3 (Pierce; 0.5 mM) was added to the cells grown on dishes for different incubation times depending on the experiment and quenched for 15 min with 20 mM Tris pH 7.5.
Cell fractionation
MDCK cells were homogenized by 10 strokes in an Isobiotec cell homogenizer with a tungsten carbide ball in 500 μl of 20 mM HEPES/KOH, pH 7.3 and 120 mM sucrose28. A post-nuclear supernatant was obtained by centrifugation at 600 g for 5 min and was loaded on the top of a discontinuous sucrose gradient (45-15%) made up in the same buffer. After centrifugation at 45,000 rpm for 1 h in a Beckman SW50 ultracentrifuge, 14 fractions of 350 μl were collected from the top of the tube.
Temperature block
To achieve an almost complete protein block in the TGN, we used cells were incubated at 19.5°C for 2 hrs in CO2 independent medium (F12 Coon's modified medium without NaHCO3 and with 0.2% BSA and 20 mM Hepes, pH 7.4)25. In the last hour at 19.5°C, they were treated with 150 μg/ml cycloheximide.
Modification of cholesterol content
To deplete cellular cholesterol content we used two specific compounds: saponin, which sequesters cholesterol in situ to form multimeric globular deposits in the membranes45 or mevinolin, which inhibits the production of mevalonate (an intermediate of the metabolic pathway leading to cholesterol synthesis)19. Cells were incubated 30 min on ice with saponin (0.2%) in CO2 independent medium1 or 48 hrs with mevinolin (5 μM) in DMEM supplemented with delipidated serum11.
Loading the cells with cholesterol (10 mM) was carried out using cholesterol-saturated β-cyclodextrin (Sigma-Aldrich), which was added to CO2 independent medium at 37°C for 40 min. To determine the rate of cholesterol depletion or addition we measured cholesterol cellular levels by a colorimetric assay (Calbiochem).
Alkaline Phosphatase assays
Alkaline Phosphatase activity was measured by a colorimetric assay (abcam) used according the manufacturer's instructions. Briefly, cells were washed in PBS containing CaCl2 and MgCl2, homogenized in the assay buffer and centrifuged at 13,000 g for 3 minutes to remove insoluble material. Samples were mixed with pNPP as substrate into 96-well plate and at the end of reaction (60 minutes) O.D. was measured at 405 nm. A standard curve was obtained measuring the O.D. generated by the conversion of defined nmol of pNPP in the colored pNP.
N&B experiments
Number & Molecular Brightness method is a technique based on moment-analysis for the measurements of the average number of molecules and brightness in each pixel in fluorescence microscopy images16. From the average intensity (first moment) in each pixel and the variance of the intensity distribution (second moment), the number and brightness (aggregation) of fluctuating particles is determined, thereby providing the state of aggregation of molecules in living cells with high spatial and temporal resolution16,44.
Microscopy and image analysis
50 frame time-series were acquired with a Zeiss LSM 510 META equipped with a plan apo 63× oil-immersion (NA 1.4) objective lens by using the following settings: 488 nm Argon laser, 0.05 mW of output power, 505-550 nm emission, gain less or equal to 900, offset 0.1, digital gain 1. Scanning parameters were: 512×512 frame window, 25.61 μs/pixel dwell time, no average, zoom 6×, ROI (x, y) 256×64, pinhole corresponding to 1 μm optical slice. Images were collected with resolutions of 70 nm/pixel. All measurements were performed in cells displaying comparable levels of fluorescence intensity. Data from each cell were analyzed by SimFCS software (Globals Software, East Villa Grove, IL 61956, USA) following a described procedure16. Correction was applied for taking into account the analog detection of fluorescence by the photomultiplier tubes of the confocal microscope in order to express the molecular brightness (ε) in terms of photons/s/molecule46. Briefly, the correction parameters S (the conversion factor between one photon detected and the number of digital levels produced by the electronics), offset and sigma0 were determined, for each experiment, plotting the measured average intensity (<I>) vs average variance (<Var>) of 50 frame time-series acquired using same settings as above except 4 different values of laser transmission percentages and filters and beam splitters configured to get reflection images, in order to detect the defined amount of light originating directly from the laser. The obtained plots were linearly interpolated and the equation of straight line (R ≥ 0.99) was used to extract the parameters S and offset based on the following equation: <Var> = S * <I> + q (parameter related to readout noise). The parameter sigma0 was estimated from time-series acquired with laser off, as the half maximum width of the histogram peak of the dark-counts. Its value was constantly lower than 0.1, and consequently was approximated to zero in all the calculations.
By calibrating the percentage of laser power versus the fluorescence intensity we have identified, in our microscope system, the linear range and we chose the minimum intensity to obtain a good signal/noise ratio with low photobleaching and photoxicity. We always verified that the laser power is constant and found that the value of the correction parameter S is relatively constant over the different experiments. As we previously described46, using a laser-scanning microscope we found that a minimum of 10–20 frames is necessary to determine the brightness of particles. Because a larger the number of frames could increase the resolving power of the method, we determined that 50 frames provide the optimal time resolution to determine protein aggregation in the conditions of our experiments without increasing the risk of cell movements.
In analog system brightness was calculated pixel by pixel from the following equation B= V(x,y)/(S*Ix,y) and the relationship with molecular brightness is described by the following equation: B/S= ε+1. Here we indicate with the term brightness the ratio B/S. Hence, the measured brightness (B/S) is > 1 from the pixels with mobile components, while B/S= 1 from the pixels with immobile features.
While possible blinking of GFP has no effect on the brightness as previously described16, photobleaching correction (photobleaching rate measured from the experimental data) has been included in the algorithms used to analyze N&B data16. Specifically we used a high-pass filter to the intensity as a function of time of each pixel, which we experimentally verified to be able to remove slowly varying signals. After removal of the trend, we added a constant equal to the average intensity at that pixel. Therefore, the variance of the “immobile” part is unaffected by bleaching after correction and we can recover the variance of the mobile part16.
The presence of membrane ruffles, microvilli or other membrane protusions do not influence the brightness of molecules as previously demonstrated47. Furthermore, in order to account for the possible influence of microvilli on the GFP-FR distribution we measured B in membrane areas with various amounts of microvilli. In all experiments a detrend function (the same used for bleaching correction) was applied to image stacks before determining the B in order to avoid that slow changes of the intensity due to the cell movement or protrusion/retraction events could interfere with our measurements16, 46. Finally all acquisitions where we monitored aberrant movements (e.g., microvilli movement or fluctuations of the apical membrane) were discarded.
Data Analysis
We implemented a method to analyze the experimental data by using a mathematical analysis that allows to partition observations in groups (clusters of observations)52. Specifically, by Matlab software (The Mathworks Inc. Natick, MA) using the K-means function52 we partitioned, with an interval of 0.5, the observed brightness values upon different experimental conditions into N exclusive groups with statistical reliability. In particular, for each experiment (number of cells > 15) we obtained the percentage of pixels in each group (calculated as average of single cell values from an experiment) and its occurrence, defined as percentage of cells falling in each group.
In order to determine the correspondence of the B values to the aggregation state of GFP-FR (two, three, four molecules, etc.) we determined a standard curve plotting the experimental B values for monomeric, dimeric, trimeric GFP vs number of units per aggregate (see Supplementary Fig. 4c) by expressing the different GFP chimeric proteins both in MDCK and HeLa cells. We found that B is directly proportional to the number of GFP subunits (correlation degree 0.99). Thus we can extrapolate the values of B corresponding to tetramer, pentamer etc with high accuracy. Furthermore, because the B values experimentally obtained for monomeric, dimeric, trimeric GFP are statistically different (Supplementary Fig. 4d) and are distributed along a Gaussian curve (Supplementary Fig. 4e) we could ascribe the range of B values corresponding, respectively, to monomer (1.05-1.14), dimer (1.15-1.24), and trimer (1.25-1.29) (Supplementary Fig. 4f) and extrapolate from the standard curve the range of B values associated with tetramer (1.30-1.39), pentamer (1.40-1.44), hexamer (1.45-1.54). Hence, on the basis of the percentage of pixels falling in each range we could estimate the amount of GFP-FR present as monomer, dimer, trimer...etc.
The accurateness of the standard curve made with soluble GFP is guaranteed by the fact that the brightness of soluble and membrane proteins is comparable because the maximum possible difference between 2D and 3D depends on the value of the gamma factor (0.5 for a plane intersecting the PSF and 0.36 for 3D)16,47, which multiplies the measurement of the variance from which the brightness is derived. As a consequence of the fact that the membrane is not perfectly flat and not exactly in the focus of the laser beam (random position of the membrane in the focus is equivalent of molecules randomly distributed in 3D), the difference between 2D and 3D tends to be cancelled out as we experimentally tested47. Consistently, we found that the B values for the soluble (mGFP) and two different membrane-linked (mGFP-FR; mGFP-GPI uPAR) forms of monomeric GFP are absolutely comparable (see Supplementary Fig. 4d) and a standard curve made with membrane-linked forms (mGFP-GPI uPAR, mGFP-mGFP-GPI uPAR) overlaps perfectly with the curve from soluble GFP (see Supplementary Fig. 4c).
FLIM experiments
The FRET-FLIM system was previously described23. Briefly, the FRET-FLIM apparatus combines multifocal multiphoton excitation (TriMscope, LaVisionBiotec, Bielefeld, Germany) connected to an inverted microscope (IX 71, Olympus, Tokyo, Japan) and a fast-gated CCD camera (Picostar, LaVisionBiotec, Bielefeld, Germany). A mode-locked Ti:Sa laser at 950 nm for the excitation of GFP (Spectra Physics, France) is split into 2 to 64 beams by utilizing a 50/50 beam splitter and mirrors. A line of foci is then created at the focal plane, which can be scanned across the sample. A filter wheel of spectral filters (535AF45 for GFP) is used to select the fluorescence imaged onto a fast-gated light intensifier connected to a CCD camera. The gate of the intensifier (adjusted at 2 ns) is triggered by an electronic signal coming from the laser and a programmable delay box was used to acquire a stack of five time-correlated images of the 10 ns fluorescence decay window. The acquisition time of the CCD camera was adjusted considering the fluorescence signal level (between 3 to 5 s). All instrumentation is controlled by Imspector software developed by LaVision Biotec. Regions of interest (ROIs) of 140×140 pixel (4-6 cells) were acquired.
Analysis of the data was done using image-J (Rasband, W.S., Image J, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/). The methodology used in order to perform quantitative analysis was previously developed23. Briefly, the five images coming from a time-gated stack are first smoothened by a 3×3 mask to decrease the noise and to recover mean lifetime, <τ>, the following equation is applied:
| Eq., 1 |
where Λti= 2i – 1 corresponds to the time delay after the laser pulse of the ith image acquired and Ii to the pixel intensity map in the ith image.
Finally, the following equation is applied on mean lifetime images using fixed lifetime donor values (τD, extracted from cells expressing GFP-fused protein alone) to recover mfD.
| Eq., 2 |
mfD is a method to quantify the minimal percentage of donor engaged in FRET. Our time-gated method is fast and data analysis of mean lifetimes is carried out by a direct calculation without any fitting (to reduce the photon budget which increases the rate of acquisition). Moreover with the mean lifetime, we are able to quantify the mfD without the need of knowing the lifetime of the FRET (or the real FRET efficiency). Indeed differently from fD, mfD is extracted directly from raw data without any fitting procedure. Importantly, in a large case study23, 24, mfD was found not to underestimate by more than 30% the fD value of the fraction of donor engaged in FRET (Figure 7 in23). In addition, we took into account that a large fraction (40 to 60%) of mCherry protein is in a “dark state” that cannot be involved in FRET process with a green donor24,48, we therefore have multiplied by 2 the recovered mfD values and named it quantity of interacting donor (%). Thus, we can quantitatively interpret the FLIM/FRET data.
Both in N&B and FLIM experiments MDCK and CHO cells, grown either on bottom-glass dishes or on filters, were imaged in vivo in CO2 independent medium (150 mM NaCl, 5 mM KCl, 1 mM, CaCl2, 1 mM MgCl2, and 20 mM Hepes, pH 7.4).
Except where indicated, we always use confluent fully polarized cells grown either on plastic dishes or polycarbonate filters for 4 days, after which time they attain complete polarization25. The cells grown in polarized fashion are elongated along the axial axle and it is quite easy to focus on the apical or basal membrane by finding first the focal plane corresponding to the filter (or to the opposite membrane plane which contains easily identifiable microvilli) and then move up or down.
Since CHO cells are very flat we generally image the cells only at the cell periphery in order to detect only signals coming from surface and to avoid intracellular contamination. This is very easy for N&B experiments because cells were imaged with a large zoom. Instead, in FLIM experiments, in which images were acquired with smaller zoom, it is not always possible to avoid collecting intracellular fluorescence (see comparison between N&B and FLIM images in Supplementary Fig. 6a). In these cases, in post-acquisition analysis we always take into account only the signal coming from the cell periphery.
Sucrose Density Gradients
Sucrose gradient analysis was performed using cells grown on 150 mm (20-25 × 106) dishes to confluence. Cells were scraped from dishes in PBS containing CaCl2 and MgCl2, resuspended in 1 ml of lysis buffer containing 1% TX-100, 10 mM Tris buffer, pH 7.5, 150 mM NaCl, 5 mM EDTA and sheared through a 23-g needle. The lysate was mixed with an equal volume of 85% sucrose and run on discontinuous sucrose gradients (40–5%) at 200,000 g for 17 hrs at 4°C49. Fractions were collected starting from the top of the tube and TCA-precipitated. Samples were revealed by western blotting using specific antibodies.
DTT assay
To change the oxidation state of proteins, cells were incubated for 5 min at 37°C with DTT (10 mM) in normal culture medium50 and then were cross-linked with BS3 at 37°C for 10 min still in presence of DTT (5 mM). Cells were quenched for 10 min with 20 mM Tris pH 7.5 in presence of iodacetic acid (20 mM) to prevent oxidation, and then lysed.
Co-localization analysis
Fluorescence measurements were quantified by using the Matlab software (The Mathworks Inc. Natick, MA). 10-20 cells randomly chosen for two different experiments were analyzed. In each image, regions of interest (ROIs) were identified by applying the Otsu algorithm. A homemade matlab application was used to find and quantify areas of the images where the single fluorophores localized, as number of pixels. Then, co-presence (and/or exclusion) of the region occupied by each fluorophore was expressed as percentage of the total area. Similar data were obtained by measuring the fluorescence intensity (data not shown). The degree of co-localization in the co-presence regions was calculated by using the Pearson coefficient51 measured pixel by pixel and showed as correlation maps. The percentage of co-localization was calculated from pixels whose Pearson coefficient was equal to or higher than 0.6 (highly significant threshold; we chose this cut-off on the basis of control experiments where fluorescence from the two fluorophores was fully co-localizing).
Statistical analysis
In all experiments we used two-tailed student test as statistical analysis.
Acknowledgments
We thank P. Casanova (Institut Pasteur) for his technical support, N. Auduge (Institut Jacques Monod, ImagoSeine) for his help in FLIM analysis, P. Riccio (University of Naples Federico II) for her help in statistical analysis and the imaging facility at Pasteur Institute (PFID) and at CEINGE Institute (DIM). This work was supported by ANR (05-BLAN 296-01, ANR-09-BLAN-0122), European Union FP7 (Priority, Grant 222887) to CZ; AIRC grant (MFAG 2007-2009) to SP; ANR PFTV2007, FRM “Grands Equipements” to MC-M; NIH-P41 P41-RRO3155, P50-GM076516 to EG.
Abbreviations
- GPI-APs
GPI-anchored proteins
- HMW
high molecular weight
- N&B
Number and Brightness
- mFD
minimal fraction donor
Footnotes
Author contributions
C.Z. conceived the project; CZ, S.P. and S.L. designed experiments. S.P. planned and performed N&B and biochemical experiments and S.L. planned and performed FLIM experiments; S.T. planned and performed biochemical experiments. E.G. developed the N&B technique, EG and GO helped with N&B data analysis, M.T and M.C developed FLIM technique and helped with data analysis. F.F. performed the mathematical analysis of N&B data. S.P, S.L, and C.Z wrote the manuscript. All authors discussed the results and manuscript text.
Competing financial interest
The authors declare no competing financial interests.
References
- 1.Sharma P, et al. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–589. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- 2.Goswami D, et al. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell. 2008;135:1085–1097. doi: 10.1016/j.cell.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gowrishankar K, et al. Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell. 2012;149:1353–1367. doi: 10.1016/j.cell.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki KG, et al. Transient GPI-anchored protein homodimers are units for raft organization and function. Nat Chem Biol. 2012;8:774–783. doi: 10.1038/nchembio.1028. [DOI] [PubMed] [Google Scholar]
- 5.Brameshuber M, et al. Imaging of Mobile Long-lived Nanoplatforms in the Live Cell Plasma Membrane. J Biol Chem. 2010;285:41765–41771. doi: 10.1074/jbc.M110.182121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harder T. Lipid raft domains and protein networks in T-cell receptor signal transduction. Curr Opin Immunol. 2004;16:353–359. doi: 10.1016/j.coi.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Fivaz M, et al. Differential sorting and fate of endocytosed GPI-anchored proteins. Embo J. 2002;21:3989–4000. doi: 10.1093/emboj/cdf398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuck S, Simons K. Polarized sorting in epithelial cells: raft clustering and the biogenesis of the apical membrane. J Cell Sci. 2004;117:5955–5964. doi: 10.1242/jcs.01596. [DOI] [PubMed] [Google Scholar]
- 9.Simons K, Gerl MJ. Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 10.Kusumi A, Suzuki KG, Kasai RS, Ritchie K, Fujiwara TK. Hierarchical mesoscale domain organization of the plasma membrane. Trends Biochem Sci. 2011;36:604–615. doi: 10.1016/j.tibs.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Paladino S, et al. Protein oligomerization modulates raft partitioning and apical sorting of GPI-anchored proteins. J Cell Biol. 2004;167:699–709. doi: 10.1083/jcb.200407094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paladino S, Sarnataro D, Tivodar S, Zurzolo C. Oligomerization is a specific requirement for apical sorting of glycosyl-phosphatidylinositol-anchored proteins but not for non-raft-associated apical proteins. Traffic. 2007;8:251–258. doi: 10.1111/j.1600-0854.2006.00522.x. [DOI] [PubMed] [Google Scholar]
- 13.Hannan LA, Lisanti MP, Rodriguez-Boulan E, Edidin M. Correctly sorted molecules of a GPI-anchored protein are clustered and immobile when they arrive at the apical surface of MDCK cells. J Cell Biol. 1993;120:353–358. doi: 10.1083/jcb.120.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedrichson T, Kurzchalia TV. Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature. 1998;394:802–805. doi: 10.1038/29570. [DOI] [PubMed] [Google Scholar]
- 15.Varma R, Mayor S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- 16.Digman MA, Dalal R, Horwitz AF, Gratton E. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys J. 2008;94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catino MA, Paladino S, Tivodar S, Pocard T, Zurzolo C. N- and O- glycans are not directly involved in the oligomerization and apical sorting of GPI proteins. Traffic. 2008;9:2141–2150. doi: 10.1111/j.1600-0854.2008.00826.x. [DOI] [PubMed] [Google Scholar]
- 18.Breuza L, Garcia M, Delgrossi MH, Le Bivic A. Role of the membrane-proximal O- glycosylation site in sorting of the human receptor for neurotrophins to the apical membrane of MDCK cells. Exp Cell Res. 2002;273:178–186. doi: 10.1006/excr.2001.5442. [DOI] [PubMed] [Google Scholar]
- 19.Keller P, Simons K. Cholesterol is required for surface transport of influenza virus hemagglutinin. J Cell Biol. 1998;140:1357–1367. doi: 10.1083/jcb.140.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ilangumaran S, Hoessli DC. Effects of cholesterol depletion by cyclodextrin on the sphingolipid microdomains of the plasma membrane. Biochem J. 1998;335:433–440. doi: 10.1042/bj3350433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ridgway ND. Interactions between metabolism and intracellular distribution of cholesterol and sphingomyelin. Biochim Biophys Acta. 2000;1484:129–141. doi: 10.1016/s1388-1981(00)00006-8. [DOI] [PubMed] [Google Scholar]
- 22.Ohvo-Rekila H, Ramstedt B, Leppimaki P, Slotte JP. Cholesterol interactions with phospholipids in membranes. Prog Lipid Res. 2002;41:66–97. doi: 10.1016/s0163-7827(01)00020-0. [DOI] [PubMed] [Google Scholar]
- 23.Padilla-Parra S, Auduge N, Coppey-Moisan M, Tramier M. Quantitative FRET analysis by fast acquisition time domain FLIM at high spatial resolution in living cells. Biophys J. 2008;95:2976–2988. doi: 10.1529/biophysj.108.131276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padilla-Parra S, et al. Quantitative comparison of different fluorescent protein couples for fast FRET-FLIM acquisition. Biophys J. 2009;97:2368–2376. doi: 10.1016/j.bpj.2009.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paladino S, Pocard T, Catino MA, Zurzolo C. GPI-anchored proteins are directly targeted to the apical surface in fully polarized MDCK cells. J Cell Biol. 2006;172:1023–1034. doi: 10.1083/jcb.200507116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paladino S, et al. Different GPI-attachment signals affect the oligomerisation of GPI-anchored proteins and their apical sorting. J Cell Sci. 2008;121:4001–4007. doi: 10.1242/jcs.036038. [DOI] [PubMed] [Google Scholar]
- 27.Lebreton S, Paladino S, Zurzolo C. Selective roles for cholesterol and actin in compartmentalization of different proteins in the Golgi and plasma membrane of polarized cells. J Biol Chem. 2008;283:29545–29553. doi: 10.1074/jbc.M803819200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imjeti NS, et al. N-Glycosylation Instead of Cholesterol Mediates Oligomerization and Apical Sorting of GPI-APs in FRT Cells. Mol Biol Cell. 2012;22:4621–4634. doi: 10.1091/mbc.E11-04-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cunningham O, et al. Dimerization controls the lipid raft partitioning of uPAR/CD87 and regulates its biological functions. Embo J. 2003;22:5994–6003. doi: 10.1093/emboj/cdg588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown DA, Crise B, Rose JK. Mechanism of membrane anchoring affects polarized expression of two proteins in MDCK cells. Science. 1989;245:1499–1501. doi: 10.1126/science.2571189. [DOI] [PubMed] [Google Scholar]
- 31.Lisanti MP, Caras IW, Davitz MA, Rodriguez-Boulan E. A glycophospholipid membrane anchor acts as an apical targeting signal in polarized epithelial cells. J Cell Biol. 1989;109:2145–2156. doi: 10.1083/jcb.109.5.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sengupta P, et al. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat Methods. 2011;8:969–975. doi: 10.1038/nmeth.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meder D, Moreno MJ, Verkade P, Vaz WL, Simons K. Phase coexistence and connectivity in the apical membrane of polarized epithelial cells. Proc Natl Acad Sci U S A. 2006;103:329–334. doi: 10.1073/pnas.0509885103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kenworthy AK, Petranova N, Edidin M. High-resolution FRET microscopy of cholera toxin B-subunit and GPI-anchored proteins in cell plasma membranes. Mol Biol Cell. 2000;11:1645–1655. doi: 10.1091/mbc.11.5.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 36.Ali MH, Imperiali B. Protein oligomerization: how and why. Bioorg Med Chem. 2005;13:5013–5020. doi: 10.1016/j.bmc.2005.05.037. [DOI] [PubMed] [Google Scholar]
- 37.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 38.Sabharanjak S, Sharma P, Parton RG, Mayor S. GPI-anchored proteins are delivered to recycling endosomes via a distinct cdc42-regulated, clathrin-independent pinocytic pathway. Dev Cell. 2002;2:411–423. doi: 10.1016/s1534-5807(02)00145-4. [DOI] [PubMed] [Google Scholar]
- 39.Kirkham M, Parton RG. Clathrin-independent endocytosis: new insights into caveolae and non-caveolar lipid raft carriers. Biochim Biophys Acta. 2005;1746:349–363. doi: 10.1016/j.bbamcr.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Johannes L, Mayor S. Induced domain formation in endocytic invagination, lipid sorting, and scission. Cell. 142:507–510. doi: 10.1016/j.cell.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 41.Datta A, Bryant DM, Mostov KE. Molecular regulation of lumen morphogenesis. Curr Biol. 2010;21:R126–136. doi: 10.1016/j.cub.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanos B, Rodriguez-Boulan E. The epithelial polarity program: machineries involved and their hijacking by cancer. Oncogene. 2008;27:6939–6957. doi: 10.1038/onc.2008.345. [DOI] [PubMed] [Google Scholar]
- 43.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 44.Hellriegel C, Caiolfa VR, Corti V, Sidenius N, Zamai M. Number and brightness image analysis reveals ATF-induced dimerization kinetics of uPAR in the cell membrane. Faseb J. 2011;25:2883–2897. doi: 10.1096/fj.11-181537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bridgman PC, Nakajima Y. Membrane lipid heterogeneity associated with acetylcholine receptor particle aggregates in Xenopus embryonic muscle cells. Proc Natl Acad Sci U S A. 1981;78:1278–1282. doi: 10.1073/pnas.78.2.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dalal RB, Digman MA, Horwitz AF, Vetri V, Gratton E. Determination of particle number and brightness using a laser scanning confocal microscope operating in the analog mode. Microsc Res Tech. 2008;71:69–81. doi: 10.1002/jemt.20526. [DOI] [PubMed] [Google Scholar]
- 47.Malengo G, et al. Fluorescence correlation spectroscopy and photon counting histogram on membrane proteins: functional dynamics of the glycosylphosphatidylinositol-anchored urokinase plasminogen activator receptor. J Biomed Opt. 2008;13:031215. doi: 10.1117/1.2940570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hillesheim LN, Chen Y, Muller JD. Dual-color photon counting histogram analysis of mRFP1 and EGFP in living cells. Biophys J. 2006;91:4273–4284. doi: 10.1529/biophysj.106.085845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tivodar S, et al. Analysis of detergent-resistant membranes associated with apical and basolateral GPI-anchored proteins in polarized epithelial cells. FEBS Lett. 2006;580:5705–5712. doi: 10.1016/j.febslet.2006.09.022. [DOI] [PubMed] [Google Scholar]
- 50.Mezghrani A, et al. Manipulation of oxidative protein folding and PDI redox state in mammalian cells. Embo J. 2001;20:6288–6296. doi: 10.1093/emboj/20.22.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manders EMM, Verbeek FJ, Aten JA. Measurement of co-localization of objects in dualcolor confocal images. Journal of Microscopy. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 52.Seber GAF. Multivariate Observations. J. Wiley & Sons; New York, USA: 1984. [Google Scholar]