Abstract
Over the past decade, considerable advances have been made in the discovery of gene targets in metabolic diseases. However, in vivo studies based on molecular biological technologies such as the generation of knockout mice and the construction of short hairpin RNA vectors require considerable effort and time, which is a major limitation for in vivo functional analysis. Here, we introduce a liver-specific nonviral small interfering RNA (siRNA) delivery system into rapid and efficient characterization of hepatic gene targets in metabolic disease mice. The comparative transcriptome analysis in liver between KKAy diabetic and normal control mice demonstrated that the expression of monoacylglycerol O-acyltransferase 1 (Mogat1), an enzyme involved in triglyceride synthesis and storage, was highly elevated during the disease progression. The upregulation of Mogat1 expression in liver was also found in other genetic (db/db) and diet-induced obese mice. The silencing of hepatic Mogat1 via a liver-specific siRNA delivery system resulted in a dramatic improvement in blood glucose levels and hepatic steatosis as well as overweight with no apparent overall toxicities, indicating that hepatic Mogat1 is a promising therapeutic target for metabolic diseases. The integrated approach with transcriptomics and nonviral siRNA delivery system provides a blueprint for rapid drug discovery and development.
Keywords: in vivo siRNA delivery, liver, microarray, monoacylglycerol O-acyltransferase 1 (Mogat1), type 2 diabetes
Introduction
Type 2 diabetes mellitus is associated with a wide array of dysfunctions resulting from complex interactions between genetic and environmental risk factors.1,2 Numerous attempts have been made to identify the candidate genes by genome-wide association studies and using DNA microarray analysis.3,4,5 Understanding the identity of potential drug targets in the early stages of biomedical research is of great importance for the development of drugs with a high efficacy.6,7,8 The advent of promising biotechnologies such as microarray, proteomics, next-generation sequencing, and other platforms made it possible to identify candidate genes that show marked differences in disease states.4,9,10,11 Furthermore, system biology is an emerging approach, which permits us to understand some of the unknown biological links among them12,13 and is more advantageous for selecting candidate genes. However, a major obstacle is clearly distinguishing between “candidate” and “therapeutic relevant” genes, which arguably remains as a major challenge in drug discovery research.7
A nonviral in vivo nucleic acid delivery technology that can effectively silence a specific gene would be highly desirable, in that it would represent a new molecular biological technology alternative to knockout mice and short hairpin RNA expression adenovirus vectors. Since the appearance of RNA interference in mammalian tissue,14 a great deal of research has been done to deliver chemically synthesized small interfering RNA (siRNA) into various tissues.15,16,17,18,19 Most of these studies focused on a generation of new therapeutic applications, namely, Nanomedicine; however, the siRNA delivery technology would be more beneficial, in particular, in the early stage of drug discovery, in that it can essentially speed up the target selection via a combination with various omics technologies as described in the above.
Here, we propose an interdisciplinary approach with a DNA microarray and a nonviral siRNA delivery system, which has an impact on target selection in the early stage of drug discovery. The first is the DNA microarray-based extraction of a candidate gene that plays essential roles in the progression of type 2 diabetes. The second is an in vivo phenotypic assessment via a liver-specific siRNA delivery system, recently developed in our laboratory,20 for evaluating the responsibility of candidate gene on the onset of diabetes. Efficacy can be evaluated by measuring some important serum biochemical parameters such as glucose and some lipids. Such an approach would be particularly useful in dealing with complex and multifactorial diseases, such as type 2 diabetes, a disease that can be readily evaluated based on elevated serum parameters such as glucose levels. Using the advantage of this approach, we found that the increase of monoacylglycerol O-acyltransferase 1 (Mogat1) expression in liver is responsible for the early onset of type 2 diabetes associated with hepatic steatosis and obesity.
Results
DNA microarray-based selection of a potential target responsible for type 2 diabetes
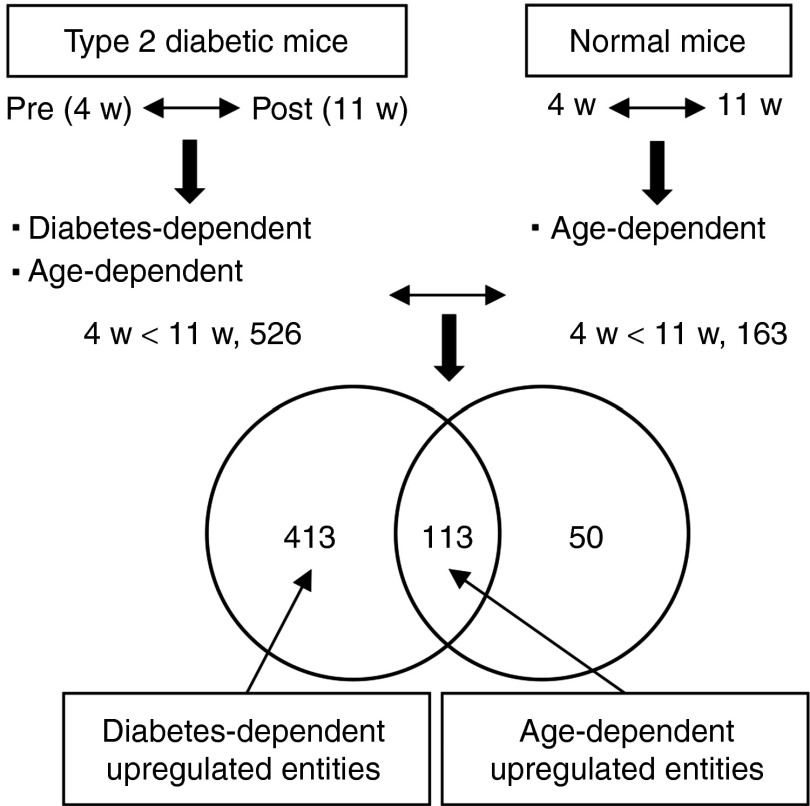
DNA microarray analyses were performed in order to enrich candidate genes in the progression of type 2 diabetes. The scheme shown in Figure 1, outlines the two steps involved in candidate gene enrichment. The first step is to enrich entities that are markedly different between pre-(4w) and post-(11w) diabetic (KKAy) mice. The same procedure was then used to extract entities in response to aging in normal (C57BL/6) mice. The second step involves comparing the two entities' set, a process that results in a substantial enrichment in gene candidates that are exclusively involved in the progression of type 2 diabetes, but not of aging. As shown in Supplementary Figure S1a, 526 and 759 entities were up- and downregulated in post- compared to prediabetic mice. On the other hand, 163 and 732 entities were up- and downregulated in 11w compared to 4w normal control mice. When we compared the 526 and 163 entities that were upregulated in diabetic and normal mice, 113 entities appeared in both, indicating that these overlapped entities were upregulated in response to aging. Therefore, 413 (78.5%) of 526 were determined to be diabetes-dependent upregulated entities (Figure 1). In the same way, 453 (59.7%) out of 759 were diabetes-dependent downregulated entities (Supplementary Figure S1b). Gene ontology analyses revealed that entities associated with lipid metabolic process were functionally enriched in both diabetes-dependent up- and downregulated entities (Supplementary Tables S1 and S2). These results clearly indicate that dysfunctional alteration of lipid metabolism in liver may be responsible for the onset of type 2 diabetes. Further analysis showed that, of 959 entities annotated with the Gene Ontology (GO) term lipid metabolic process (GO: 0006629), 46 and 62 were overlapped with diabetes-dependent up- and downregulated entities, respectively (Supplementary Figure S1c). The diabetes-dependent up- and downregulated entities annotated with lipid metabolic process were listed in Supplementary Tables S3 and S4, respectively.
Figure 1.
A workflow of DNA microarray-based approach for screening candidate genes for type 2 diabetes. Scheme showing the steps involved in candidate gene enrichment. First, differentially expressed genes (DEGs) between pre- and postdiabetic mice are extracted. Diabetes-dependent genes are then extracted by excluding age-dependent DEGs in diabetic mice. DEGs in response to aging in normal mice are defined as age-dependent genes. Venn diagram showing overlaps in upregulated entities between diabetic and normal mice. Of the 526 entities, which were upregulated in the postdiabetic stage, 413 and 113 were determined to be diabetes-dependent and age-dependent entities, respectively.
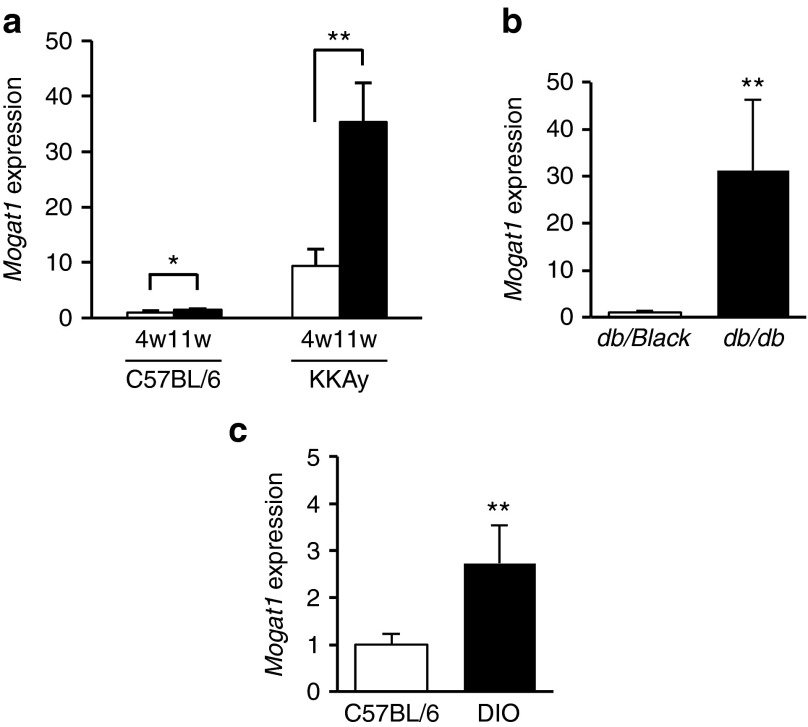
Among the diabetes-dependent upregulated entities associated with lipid metabolism (Supplementary Table S3), we focused on monoacylglycerol O-acyltransferase 1 (Mogat1) because it was reported to be involved in triglyceride biosynthesis and hepatic steatosis,21,22,23 however, there are few reports regarding the role of the gene in the progression of type 2 diabetes. The expression of Mogat1 mRNA in the liver of postdiabetic KKAy mice (11w) was 3.8-fold higher than that of prediabetic KKAy mice (4w), whereas only marginal age-dependent elevation (1.4-fold) was appeared in the normal control mice (C57BL/6; Figure 2a). Hepatic Mogat1 expression was also increased in two other insulin-resistant mouse models, namely, db/db (31-fold increase compared with db/black) and diet-induced obesity (2.7-fold increase compared with normal control C57BL/6; Figure 2b,c). Since acyl-CoA:monoacylglycerol acyltransferase (MGAT) activity is best known for its role in intestinal fat absorption,24,25,26,27 we examined the tissue distribution patterns of Mogat1 and Mogat2, a subtype of Mogat1, in prediabetic KKAy mice. High levels of Mogat1 were expressed in the kidney, stomach, adipose tissue rather than the liver, while Mogat2 was expressed at high levels in the small intestine and kidney (Supplementary Figure S2), indicating that the significant elevation of Mogat1 expression in liver accompanies the progression of diabetes, although its expression in normal liver was maintained at a low level.
Figure 2.
Elevated expression of the hepatic Mogat1 in mouse models of disease. (a) Significant elevation of the hepatic Mogat1 expression accompanied with the progression of diabetes in KKAy mice. Data are shown as relative expression levels against 4w of control mice. (b,c) Higher expression of the hepatic Mogat1 in (b) db/db and (c) DIO mice. The expression of Mogat1 mRNA in liver was analyzed by quantitative reverse transcription PCR (n = 4–5). Mean ± SD are shown for all panels. *P < 0.05, **P < 0.01, Student's t-test. DIO, diet-induced obesity; Mogat1, monoacylglycerol O-acyltransferase 1.
In vivo functional analysis of hepatic Mogat1 via a single injection of nonviral siRNA delivery system
We performed in vivo phenotypic analyses via a liver-specific nonviral siRNA delivery system to determine whether the silencing of hepatic Mogat1 would result in changes in serum parameters related to diabetes. A new pH-sensitive cationic lipid-based nanoparticle, which was designed in our laboratory,20 was used to deliver siRNA against Mogat1 (siMogat1) to the livers of prediabetic mice. A single injection of siMogat1 nanoparticles via the tail vain (3 mg/kg) resulted in a significant knockdown of the hepatic Mogat1 gene for a period of up to 9 days (Figure 3a). We noted that significant knockdown was still observed on Day 12 (43% knockdown compared with nontreatment; Supplementary Figure S3). The siRNA delivery system was highly liver specific, since almost 90% of the total injected dose had accumulated within 2 hours after the injection (Figure 3b). A significant knockdown of Mogat1 was observed only in liver; however, slightly decreased Mogat1 expressions were also detected in adipose tissue and the small intestine at 1 day after the nanoparticle treatment (Figure 3c). Blood glucose monitoring demonstrated an improvement in glucose levels in prediabetic (Figure 3d), but not control C57BL/6 mice (Figure 3e), suggesting that hepatic Mogat1 expression is involved in the elevated blood glucose levels of KKAy mice; however, the improved blood glucose levels by a single injection of siMogat1 were transient.
Figure 3.
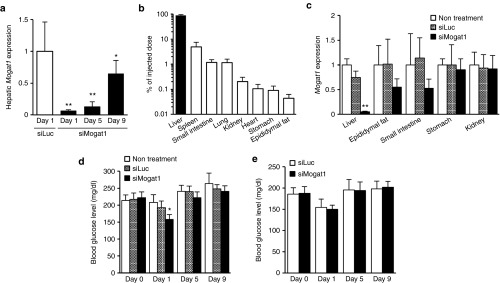
Pharmacological effect of hepatic Mogat1 knockdown as the result of a single injection of siRNA-loaded nanoparticles (3 mg/kg) in prediabetic KKAy mice. (a) Mogat1 mRNA levels in the liver after the intravenous injection of nanoparticles loaded with siMogat1 or siLuc (n = 5). (b) Tissue biodistribution of 3H-CHE-labeled nanoparticles in mice. Each mouse received a single injection of nanoparticles loaded with siMogat1 via the tail vein. Tissue biodistribution was assayed 2 hours after treatment (n = 4). (c) Mogat1 mRNA level in other tissues 1 day after the nanoparticle treatment (n = 5). Monitoring of normal glucose levels of (d) prediabetic KKAy (n = 5) and (e) C57BL/6 (n = 7) mice after the nanoparticle treatment. Mean ± SD are shown for all panels. *P < 0.05, **P < 0.01 versus siLuc (a) and nontreatment (c,d; One-way analysis of variance followed by Dunnett's test). siLuc, siRNA against luciferase; siMogat1, siRNA against monoacylglycerol O-acyltransferase 1.
Preventive effects of hepatic Mogat1 silencing on type 2 diabetes by repeated injection
We next, examined the preventive effects of long-term hepatic Mogat1 silencing in order to estimate future therapeutic opportunities for treating type 2 diabetes (Figure 4a). For the pharmacological study, we treated KKAy mice at 5 days intervals for a total of 21 days by the intravenous injections of siMogat1- and siLuc-loaded nanoparticles at a dose of 2 mg/kg in order to decrease the possibility of undesired toxicity due to the repeated overdose administration. We noted that the blood glucose levels in the fed state of siMogat1 and the siRNA against luciferase (siLuc)-treated groups before the nanoparticle treatment (Day 0) were 234 ± 36 and 240 ± 34 mg/dl, respectively. Hepatic Mogat1 silencing resulted in significantly improved blood glucose levels for at least 5 days after the last injection (Figure 4b). The blood glucose level was considered to be better controlled by insulin in the siMogat1-treated mice, as evidenced by lower serum insulin levels 6 days after the last injection of siMogat1 (Figure 4c). On the other hand, serum adiponectin, which enhances overall insulin sensitivity,28 was significantly increased (Figure 4d), indicating that hepatic Mogat1 silencing has an essential effect in preventing type 2 diabetes. Furthermore, we examined ectopic fat deposition in livers, since of MOGAT enzymes affect triglyceride production, which is the final product of the monoacylglycerol pathway.24,25 A confocal imaging study showed that fat accumulation in liver was substantially reduced in siMogat1-treated mice (Figure 4e). This result is entirely consistent with the quantification of hepatic triglyceride contents (Figure 4f). In addition, the serum triglyceride and cholesterol levels were significantly decreased after the siMogat1 treatment (Figure 4g). Furthermore, the increase in body weight and epididymal fat mass in the siMogat1-treated mice was significantly lower than those in control mice (Supplementary Figure S4), indicating that the long-term silencing of hepatic Mogat1 produced a better pharmacological effect in terms of not only type 2 diabetes, but also improvement in fatty liver and obesity as well.
Figure 4.
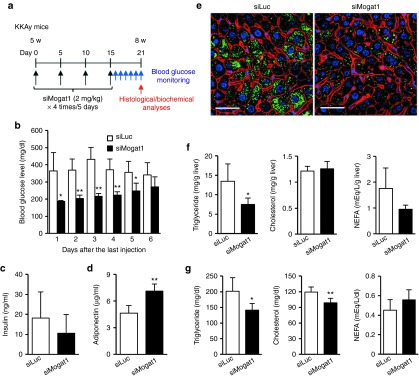
In vivo validation of a candidate Mogat1 gene via long-term silencing. (a) Schedule of siMogat1 treatment in prediabetic KKAy mice. Serum glucose was monitored for 6 days in KKAy mice treated with four injections of siMogat1- or siLuc-loaded nanoparticles. Other histological and biochemical analyses were performed on the sixth day after the last injections (n = 4 and 7 for siMogat1 and siLuc group, respectively). (b) Improved serum glucose levels of siMogat1-treated mice. Normal blood glucose was monitored for 6 consecutive days at the same time points. (c) Serum insulin and (d) adiponectin levels. (e) Representative fluorescent image of decreased ectopic fat deposition in the liver. Lipid droplets, F-actin, and nuclei were stained with boron-dipyrromethene (green), rhodamine-phalloidin (red), and Hoechst 33342 (blue), respectively. Bar = 50 μm. (f) Hepatic and (g) serum lipid contents after the treatment of siMogat1 (n = 4–7). Mean ± SD are shown for all panels. *P < 0.05, **P < 0.01, Student's t-test. siLuc, siRNA against luciferase; siMogat1, siRNA against monoacylglycerol O-acyltransferase 1.
Toxicological study of single or repeated injection of nonviral siRNA-loaded nanoparticles
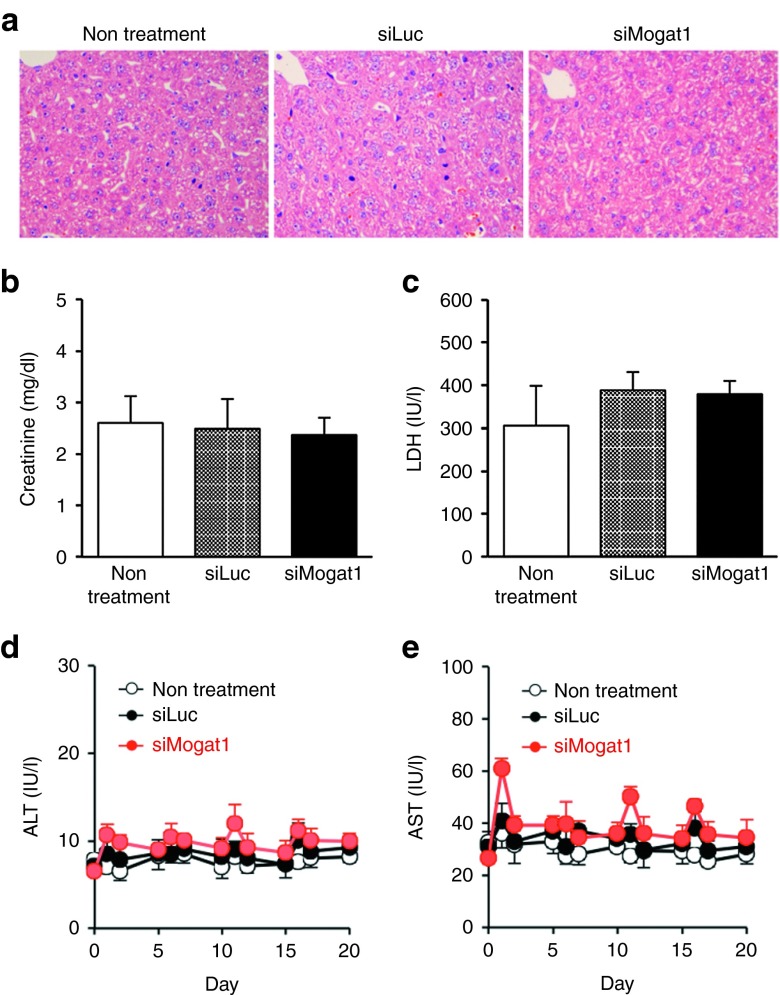
Finally, we evaluated some toxicities of siRNA-loaded nanoparticles used in this study. Pathological examination in livers showed that no apparent increase in the number of mitosis was found (Figure 5a), indicating that prominent liver damage was pathologically undetectable at 1 day after the injection of siRNA-loaded nanoparticles. In addition, both of serum creatinine and lactic dehydrogenase levels, an indicator of kidney and overall organ toxicity, respectively, were not significantly different (Figure 5b,c). Furthermore, the monitoring of serum aspartate aminotransferase and alanine aminotransferase levels showed that cumulative and acute toxicity was not observed throughout the examination period (Figure 5d,e), indicating that the toxicity of the siRNA delivery system was negligible in a single or repeated injection.
Figure 5.
Toxicicological studies of the siRNA delivery system. (a) Representative liver pictures 1 day after the treatment of siRNA-loaded nanoparticles (2 mg/kg). (b) Serum creatinine and (c) LDH levels 1 day after the treatment (n = 6). Monitoring on serum (d) ALT and (e) AST levels in the repeated injections of the siRNA nanoparticles. Each injection dose was 2 mg/kg, every 5 days (n = 5–6). Mean ± SD are shown for all panels. ALT, alanine aminotransferase; AST, aspartate aminotransferase; LDH, lactic dehydrogenase.
Discussion
A great deal of effort has been expended in attempts to discover novel targets in both academic and pharmaceutical settings with many kinds of innovative technology. A recent report demonstrated that 435 drug targets in the human genome have been identified and approved by the FDA until 2009.7 However, this number represents only about 2% of all human genes,29 and no apparent increase in the number of drugs with novel target gene has been reported over the last 30 years,7 suggesting that we need to design a new strategy to break the existing problem. Actually, a tremendous number of efforts have been made to find a novel therapeutic target in various diseases such as type 2 diabetes and obesity,9,30,31 and cancer32 by “genome-wide” gene expression analyses and associated studies. Some of them used a transgenic and knockout technology to test the efficacy of candidate targets in an in vivo setting9,30; however, the generation of these mice is time consuming, very expensive, and labor-intensive, and sometimes leads to the production of lethal embryonic phenotypes, making it difficult to simultaneously identify and functionally validate gene candidates. In addition, Kodama et al. 31 used a specific antibody to target a protein; however, such functional analyses are strictly limited by its cellular localization. On the other hand, adenovirus-mediated gene transfer or small hairpin RNA is a powerful tool to perform gain or loss of function studies in an in vivo setting, however, the generation of these vectors in each target gene appears to be time consuming and labor intensive. Thus, a new strategy will be required to analyze any candidate target in an in vivo setting with a relatively short term and low cost.
In this study, we found that numerous genes (>1,200 entities) were differentially expressed in the liver between pre- (4w) and postdiabetic (11w) KKAy mice (Supplementary Figure S1a). It has been reported that the gene expression profiles in livers of normal rats were considerably altered in an age and sex dependent manner.33 Therefore, the differential gene expression was attributed to both disease and age. In order to identify the candidate genes responsible for the onset of diabetes, we also examined the liver gene expression profiles in 4w and 11w of normal control (C57BL/6) mice, and then, removed the overlapped entities (113 and 306 of up- and downregulated entities, respectively) associated with age (Figure 1 and Supplementary Figure S1b). Furthermore, GO analysis revealed that the genes annotated with lipid metabolic process were functionally enriched in both diabetes-dependent up- and downregulated entities (Supplementary Tables S1 and S2). From this result, we focused on the genes associated with lipid metabolic process and identified Mogat1 as a potential target gene. In these microarray data analyses, the subtraction of age-dependent alteration of gene expression profiles is a crucial step for the extraction of an appropriate candidate gene, since the pathological interpretation of microarray data might be complicated by the genes relevant to age. Actually, GO analysis using the entities selected by comparison between 4w and 11w KKAy mice without the subtraction of age-dependent genes revealed that the genes annotated with cell cycle and mitosis were functionally enriched in the downregulated entities, although the genes associated with lipid metabolic process were representative in the upregulated entities (Supplementary Table S5). Even though there is still limited information of the microarray data on the biological and pathological importance of the candidate genes due to one set of microarray analysis, the approach surely has potential benefits as a method for high throughput screening of the candidates of therapeutic target.
Mogat1 was originally identified in mice as a microsomal enzyme that catalyzes the synthesis of diacylglycerol and triacylglycerol.21 MGAT activity was the highest in the small intestine and lower in the liver in normal mice. However, hepatic Mogat1 expression was increased depending on the progression of type 2 diabetes (Figure 2a), and the expression was also higher in other model mice (Figure 2b,c). It was known that MOGAT1 is involved in an alternative pathway for triglyceride biosynthesis and contributes to, at least partially, hepatic steatosis and insulin resistance in 1-acylglycerol-3-phosphate-O-acyltransferase 2 (Agpat2) deficient mice.34,35 A recent study also demonstrated that the hepatic MGAT pathway was active in nonalcoholic fatty livers of human subjects.22 In addition, Lee et al.23 recently reported that the MGAT pathway induced by hepatic peroxisome proliferator-activated receptor-γ plays an important role in the development of hepatic steatosis during diet-induced obesity. These reports and our findings indicate that Mogat1 might be a potential therapeutic target gene in type 2 diabetes as well as hepatic steatosis and obesity.
In order to evaluate the responsibility of Mogat1 expression in liver on the onset of diabetes, we utilized the liver-specific in vivo siRNA delivery system.20 We found that the systemic administration of siMogat1 via a lipid nanoparticle resulted in a significant reduction of Mogat1 expression in liver and transiently, but significantly, improved blood glucose levels in the fed state in prediabetic KKAy, but not in C57BL/6 mice (Figure 3d,e). The lower effect in normal control mice may be due to the low level of Mogat1 expression in the normal liver. The effect was more prominent with a long silencing via repeated injections of siMogat1 (Figure 4b). Interestingly, silencing of hepatic Mogat1 also resulted in decrease in the amount of triglyceride in the liver (Figure 4e,f) and serum (Figure 4g), even though MGAT pathway is considered to be an alternative pathway to produce diacylglycerol and triacylglycerol. Furthermore, the knockdown of hepatic Mogat1 inhibited in increase of body weight and fat mass (Supplementary Figure S4). These results are in agreement with the recent report.23 Lee et al. demonstrated that the knockdown of hepatic Mogat1 via an adenoviral shMogat1 improved high fat diet-induced hepatic steatosis and glucose tolerance as well as reduced body weight. Based on the different viewpoints and approaches, they (and we) focused on hepatic Mogat1 expression in the disease mouse model and showed the pathological responsibility of hepatic Mogat1 using the viral or nonviral gene silencing techniques. Further investigations will be necessary to completely understand how hepatic Mogat1 is associated with the progression of diseases such as type 2 diabetes and obesity. We strongly believe that the strength of the interdisciplinary strategy is quick and direct evaluation of therapeutic relevance by monitoring serum parameters and phenotypic differences in the early stage of target selection. Although it is also necessary to examine the pharmacodynamics and pharmacokinetic properties of the nanoparticles including clearance or elimination half-life in more detail, these in vivo data will surely drive to minute mechanistic analysis both in vitro and in vivo for deciding the mechanism of action on hepatic Mogat1.
Furthermore, the strategy provides a light for innovative nanomedicine for type 2 diabetes and obesity if the safety issue is guaranteed in preclinical settings. The siRNA delivery system used in this study permits repeated injections without severe toxicity (Figure 5a–e), demonstrating a positive light for preclinical studies.
In conclusion, hepatic Mogat1 expression is responsible for the early onset of type 2 diabetes as well as hepatic steatosis and obesity, and the inhibition of MGAT pathway for alternative triglyceride biosynthesis should be an innovative drug target for these metabolic disorders. The interdisciplinary approach involving the use of nanobiotechnologies and nonviral in vivo siRNA delivery systems is having a deep impact on target selection in the early stage of drug discovery. In particular, the change in use of nonviral siRNA delivery system from therapeutic purposes to target selection process with combination of nanobiotechnologies, allows us to have great benefits in the early stage of drug discovery in a relatively short term and low cost, if the synthesized siRNAs of interest can be obtained. The paradigm shift in the use of siRNA delivery system will be more frequent in the future for speeding up target discovery with high therapeutic opportunities and the subsequent drug development process.
Materials and methods
Animals. KKAy, C57BL/6J, db/db, and diet-induced obesity mice were obtained from CLEA (Tokyo, Japan). All mice used in this study were males and were maintained on a 12-hour light–dark cycle and fed a standard rodent chow. Diet-induced obesity mice were fed a diet containing 60% fat (Research Diets, 58Y1, PMI Nutrition International (Richmond, IN)) for 14 weeks. The experimental protocols were reviewed and approved by the Hokkaido University Animal Care Committee in accordance with the guidelines for the care and use of laboratory animals.
DNA microarray analysis. RNA was extracted from livers of 4- and 11-week-old KKAy and C57BL/6J mice using the RNeasy Mini Kit (QIAGEN, Venlo, Netherlands). Mice were fasted overnight (13.5 hours) before sampling. Four RNA samples from each group were pooled into a single sample. The integrity of the pooled RNA samples was evaluated using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA). Preparation of cRNA and hybridization of whole mouse genome arrays (G4122F) were performed duplicate according to the manufacturer's instructions (Agilent). Gene expression data were analyzed using the GeneSpring GX 12 software program (Agilent) and an entity that was considered to be expressed in a particular experimental condition was analyzed. The results have been deposited in the Gene Expression Omnibus MIAME-compliant database (http://www.ncbi.nlm.nih.gov/geo/. Accession number: GSE36412). GO analyses were conducted using the Database for Annotation, Visualization and Integrated Discovery (http://david.abcc.ncifcrf.gov).
RNA extraction and reverse transcription PCR analysis. RNA was extracted from mouse tissues using the TRIzol Reagent (Life Technologies, Carlsbad, CA). cDNA was prepared from 1 µg of RNA using the high-capacity RNA-to-cDNA Kit (ABI, Foster City, CA), according to the manufacturer's instructions. The resulting cDNA was diluted, and a 5 µl aliquot was used in a 20 µl PCR reaction (SYBR Green; TOYOBO, Osaka, Japan) containing specific primer sets at a concentration of 250 nmol/l each. Primers for the experiment were as follows: Mogat1 forward, 5′-TGCCCTATCGGAAGCTGATCTACA; reverse, 5′- AGGTCGGGTTCAGAGTCTGCTGA; Actb forward, 5′-GAAGGAGATTACTGCTCTGG; reverse, 5′-ACACAGAGTACTTGCGCTCA; Rn18s forward, 5′-GGTAACCCGTTGAACCCCAT, reverse 5′- CAACGCAAGCTTATGACCCG. PCR reactions were run in duplicate and quantified using the Mx3005P real-time QPCR system (Agilent). Cycle threshold (Ct) values were normalized to β-actin expression or 18S ribosomal RNA expression (only used in Supplementary Figure S3) and the results are expressed as the fold change in mRNA compared with control mice.
Lipid nanoparticle formulations. The sequences for the sense and antisense strands of the siMogat1 and siLuc are as follows: siMogat1 sense, 5′-CCGGGUCACAAUUAUAUAUUUdTdT; antisense, 5′-AAAUAUAUAAUUGUGACCCGGdTdT; siLuc sense, 5′-GCGCUGCUGGUGCCAACCCdTdT; antisense, 5′-GGGUUGGCACCAGCAGCGCdTdT. The siMogat1 and siLuc were obtained from Hokkaido System Science (Sapporo, Japan). The gene silencing effect of siMogat1 was confirmed in primary hepatocyte of KKAy mice (Supplementary Figure S5). Primary hepatocytes were prepared as described previously.36 Lipid nanoparticle formulations of siRNA were prepared using YSK05, novel pH responsive cationic lipid.20 The lipid nanoparticles were composed of YSK05, cholesterol, and mPEG-DMG, in molar ratios 70:30:3. siRNAs were formulated in lipid nanoparticles at a total lipid-to-siRNA weight ratio of ~12:1. The siRNA concentration and encapsulation efficiency was determined using the RiboGreen Assay. In brief, formulated lipid nanoparticles were diluted in 10 mmol/l hepes buffer at pH 7.4 and RiboGreen in the presence or absence of 0.1 w/v% TritonX-100. Fluoresence was measured on a spectrofluorometer (Enspire 2300 multilabel Reader, PerkinElmer, Waltham, MA) with excitation and emission wavelengths of 495 and 525 nm, respectively. Particle size and surface charge density measurements were performed using a Zetasizer Nano ZS instrument (Malvern, Worcestershire, UK). The mean particle sizes were 80–90 nm and 80–90% of the siRNA was encapsulated within the lipid nanoparticles.
Lipid nanoparticle-mediated gene silencing. Prediabetic (4–5-week-old) KKAy and C57BL/6J mice were injected via the tail vein with siMogat1 or siLuc at a dose of 2 or 3 mg/kg body weight, respectively. In the case of preventive studies, prediabetic KKAy mice were treated with either siMogat1 or siLuc (2 mg/kg) on every 5 days. Histological and biochemical analyses were performed on the sixth day after the last injection.
Biodistribution experiment in mice. Radiolabled nanoparticles incorporated with 3H-CHE lipid were administered at a siRNA dose of 3 mg/kg via tail vein injection in prediabetic KKAy mice. At 2 hours after the injection, mice were sacrificed and various tissues were collected and solubilized in 2 ml of Soluene-350 overnight at 55 °C. All samples were decolorized by treatment with H2O2. The radioactivity was determined by liquid scintillation counting (LSC-6100; ALOKA, Tokyo, Japan) after adding 10 ml of scintillation fluid (Hionic fluor; PerkinElmer) to the scintillation vials. Tissue accumulation is represented as the percent of the initially injected dose (%ID/tissue).
Metabolic studies. Monitoring of normal blood glucose levels was performed at the same time point on each day. Blood glucose values were determined using an Accu-Check Compact Plus (Roche, Indianapolis, IN).
Confocal imaging studies of livers. Liver tissues were cut into thin sections (around 200 μm) using a microslicer (DSK-1000, DOSAKA-EM, Kyoto, Japan) and these sections were then stained with boron-dipyrromethene (Life Technologies), Rhodamine-labeled phalloidin F-actin (Life Technologies) and Hoechst 33342 (Dojindo, Kumamoto, Japan) for an hour. After mounting the samples on glass slides, they were viewed under a confocal laser scanning microscopy with a water immersion objective lens Plan-Apo x 60/NA.
Measurement of lipid contents in the liver. Lipid contents in liver tissue were measured by the lipid extraction method.37 Briefly, liver tissues (about 0.1 g wet weight) were homogenized at 4 °C in 5 ml of a mixture of CHCl3-MeOH (2:1). After adding 2.5 ml dilute sulfuric acid (0.05%), the resulting suspension was vigorously vortexed and centrifuged at 10,000 rpm at 4 °C for 10 minutes. The chloroform layer containing total lipid was dried and the pellet was dissolved in EtOH solution. The triglyceride, cholesterol, and nonesterified fatty acid concentrations were determined as the same way in plasma biochemical analyses.
Measurement of serum biochemical values. Blood samples were centrifuged at 10,000 rpm at 4 °C for 10 minutes to separate serum. Serum insulin and adiponectin levels were determined with enzyme-linked immunosorbent assay kits (Shibayagi, Gunma, Japan; R&D systems, Minneapolis, MN, respectively). Serum cholesterol, triglyceride, and nonesterified fatty acid, aspartate aminotransferase and alanine aminotransferase values were measured using each detection kit (Wako Pure Chemicals, Osaka, Japan). Serum creatinine and lactic dehydrogenase levels were determined with a Creatinine Assay Kit (BioVision, Milpitas, CA), and a lactic dehydrogenase color endpoint assay kit (BIOO Scientific, Austin, TX), respectively. All experiments were in accordance with manufacturer's instructions.
Liver histopathology. At 20 days after first injection of siMogat1- or siLuc-loaded lipid nanoparticles, mice were killed and perfused with saline via a portal vein to remove the blood. Then, liver tissue was corrected and fixed with 10% formalin in phosphate-buffered saline. The preparation of paraffin-embedded tissue section, hematoxylin and eosin staining and histopathological analyses were conducted by Sapporo General Pathology Laboratory (Sapporo, Japan).
Statistics. All results are presented as mean ± SD. Statistical significance between the multiple groups was determined by analysis of variance, followed by Dunnett's test. Significance between the two groups was calculated using two-tailed t-tests. We assign statistical significance at P < 0.05.
SUPPLEMENTARY MATERIAL Figure S1. Microarray data analyses for the extraction of potential candidate genes. Figure S2. Expression of Mogat1 or Mogat2 mRNA in several tissues of pre-diabetic KKAy mice. Figure S3. In vivo silencing of hepatic Mogat1 expression via a systemic siRNA delivery system. Figure S4. Changes in body weight after siMogat1 treatment. Figure S5. siRNA transfection study of mouse primary hepatocytes. Table S1. Gene Ontology analysis of 413 entities that were up-regulated in diabetic mice Table S2. Gene Ontology analysis of 453 entities that were down-regulated in diabetic mice. Table S3. 46 entities with lipid metabolic process out of the diabetes-dependent up-regulated entities. Table S4. 62 entities with lipid metabolic process out of the diabetes-dependent down-regulated entities. Table S5. Gene Ontology analysis of the differentially expressed genes between pre- and post-diabetic mice.
Acknowledgments
This work was supported by the Special Education and Research Expenses of the Ministry of Education, Culture, Sports, Science and Technology of Japan, and a Grant-in-Aid for Young Scientists (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japanese government. We also thank Milton Feather for editing this manuscript. The authors declare no conflict of interest.
Supplementary material
References
- Staiger H, Machicao F, Fritsche A, Häring HU. Pathomechanisms of type 2 diabetes genes. Endocr Rev. 2009;30:557–585. doi: 10.1210/er.2009-0017. [DOI] [PubMed] [Google Scholar]
- Chen L, Magliano DJ, Zimmet PZ. The worldwide epidemiology of type 2 diabetes mellitus–present and future perspectives. Nat Rev Endocrinol. 2012;8:228–236. doi: 10.1038/nrendo.2011.183. [DOI] [PubMed] [Google Scholar]
- Buechler C, Schäffler A. Does global gene expression analysis in type 2 diabetes provide an opportunity to identify highly promising drug targets. Endocr Metab Immune Disord Drug Targets. 2007;7:250–258. doi: 10.2174/187153007782794353. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Kajimoto K, Iida S, Sato Y, Mizufune S, Kaji N, et al. DNA microarray analysis of whole blood cells and insulin-sensitive tissues reveals the usefulness of blood RNA profiling as a source of markers for predicting type 2 diabetes. Biol Pharm Bull. 2010;33:1033–1042. doi: 10.1248/bpb.33.1033. [DOI] [PubMed] [Google Scholar]
- Keller MP, Attie AD. Physiological insights gained from gene expression analysis in obesity and diabetes. Annu Rev Nutr. 2010;30:341–364. doi: 10.1146/annurev.nutr.012809.104747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinney DC, Anthony J. How were new medicines discovered. Nat Rev Drug Discov. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- Rask-Andersen M, Almén MS, Schiöth HB. Trends in the exploitation of novel drug targets. Nat Rev Drug Discov. 2011;10:579–590. doi: 10.1038/nrd3478. [DOI] [PubMed] [Google Scholar]
- Eckhardt BL, Francis PA, Parker BS, Anderson RL. Strategies for the discovery and development of therapies for metastatic breast cancer. Nat Rev Drug Discov. 2012;11:479–497. doi: 10.1038/nrd2372. [DOI] [PubMed] [Google Scholar]
- Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436:356–362. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- Sugihara Y, Taniguchi H, Kushima R, Tsuda H, Kubota D, Ichikawa H, et al. Proteomic-based identification of the APC-binding protein EB1 as a candidate of novel tissue biomarker and therapeutic target for colorectal cancer. J Proteomics. 2012;75:5342–5355. doi: 10.1016/j.jprot.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- Hwang D, Lee IY, Yoo H, Gehlenborg N, Cho JH, Petritis B, et al. A systems approach to prion disease. Mol Syst Biol. 2009;5:252. doi: 10.1038/msb.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori MA, Liu M, Bezy O, Almind K, Shapiro H, Kasif S, et al. A systems biology approach identifies inflammatory abnormalities between mouse strains prior to development of metabolic disease. Diabetes. 2010;59:2960–2971. doi: 10.2337/db10-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- Judge AD, Robbins M, Tavakoli I, Levi J, Hu L, Fronda A, et al. Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice. J Clin Invest. 2009;119:661–673. doi: 10.1172/JCI37515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer D, Park EJ, Morishita Y, Carman CV, Shimaoka M. Systemic leukocyte-directed siRNA delivery revealing cyclin D1 as an anti-inflammatory target. Science. 2008;319:627–630. doi: 10.1126/science.1149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Yamauchi J, Khalil IA, Kajimoto K, Akita H, Harashima H. Cell penetrating peptide-mediated systemic siRNA delivery to the liver. Int J Pharm. 2011;419:308–313. doi: 10.1016/j.ijpharm.2011.07.038. [DOI] [PubMed] [Google Scholar]
- Hatakeyama H, Akita H, Ito E, Hayashi Y, Oishi M, Nagasaki Y, et al. Systemic delivery of siRNA to tumors using a lipid nanoparticle containing a tumor-specific cleavable PEG-lipid. Biomaterials. 2011;32:4306–4316. doi: 10.1016/j.biomaterials.2011.02.045. [DOI] [PubMed] [Google Scholar]
- Sato Y, Hatakeyama H, Sakurai Y, Hyodo M, Akita H, Harashima H. A pH-sensitive cationic lipid facilitates the delivery of liposomal siRNA and gene silencing activity in vitro and in vivo. J Control Release. 2012;163:267–276. doi: 10.1016/j.jconrel.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Yen CL, Stone SJ, Cases S, Zhou P, Farese RV., Jr Identification of a gene encoding MGAT1, a monoacylglycerol acyltransferase. Proc Natl Acad Sci USA. 2002;99:8512–8517. doi: 10.1073/pnas.132274899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AM, Kou K, Chen Z, Pietka TA, Kumar M, Korenblat KM, et al. Evidence for regulated monoacylglycerol acyltransferase expression and activity in human liver. J Lipid Res. 2012;53:990–999. doi: 10.1194/jlr.P025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Ko EH, Kim JE, Kim E, Lee H, Choi H, et al. Nuclear receptor PPAR?-regulated monoacylglycerol O-acyltransferase 1 (MGAT1) expression is responsible for the lipid accumulation in diet-induced hepatic steatosis. Proc Natl Acad Sci USA. 2012;109:13656–13661. doi: 10.1073/pnas.1203218109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Burn P. Lipid metabolic enzymes: emerging drug targets for the treatment of obesity. Nat Rev Drug Discov. 2004;3:695–710. doi: 10.1038/nrd1469. [DOI] [PubMed] [Google Scholar]
- Cao J, Lockwood J, Burn P, Shi Y. Cloning and functional characterization of a mouse intestinal acyl-CoA:monoacylglycerol acyltransferase, MGAT2. J Biol Chem. 2003;278:13860–13866. doi: 10.1074/jbc.M300139200. [DOI] [PubMed] [Google Scholar]
- Yen CL, Farese RV., Jr MGAT2, a monoacylglycerol acyltransferase expressed in the small intestine. J Biol Chem. 2003;278:18532–18537. doi: 10.1074/jbc.M301633200. [DOI] [PubMed] [Google Scholar]
- Yen CL, Cheong ML, Grueter C, Zhou P, Moriwaki J, Wong JS, et al. Deficiency of the intestinal enzyme acyl CoA:monoacylglycerol acyltransferase-2 protects mice from metabolic disorders induced by high-fat feeding. Nat Med. 2009;15:442–446. doi: 10.1038/nm.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Kamon J, Waki H, Terauchi Y, Kubota N, Hara K, et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Sato C, Shikata K, Hirota D, Sasaki M, Nishishita S, Miyamoto S, et al. P-selectin glycoprotein ligand-1 deficiency is protective against obesity-related insulin resistance. Diabetes. 2011;60:189–199. doi: 10.2337/db09-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama K, Horikoshi M, Toda K, Yamada S, Hara K, Irie J, et al. Expression-based genome-wide association study links the receptor CD44 in adipose tissue with type 2 diabetes. Proc Natl Acad Sci USA. 2012;109:7049–7054. doi: 10.1073/pnas.1114513109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DQ, Hou YF, Wu J, Chen Y, Lu JS, Di GH, et al. Gene expression profile analysis of an isogenic tumour metastasis model reveals a functional role for oncogene AF1Q in breast cancer metastasis. Eur J Cancer. 2006;42:3274–3286. doi: 10.1016/j.ejca.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Kwekel JC, Desai VG, Moland CL, Branham WS, Fuscoe JC. Age and sex dependent changes in liver gene expression during the life cycle of the rat. BMC Genomics. 2010;11:675. doi: 10.1186/1471-2164-11-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West DB, Boozer CN, Moody DL, Atkinson RL. Dietary obesity in nine inbred mouse strains. Am J Physiol. 1992;262 6 Pt 2:R1025–R1032. doi: 10.1152/ajpregu.1992.262.6.R1025. [DOI] [PubMed] [Google Scholar]
- Cortés VA, Curtis DE, Sukumaran S, Shao X, Parameswara V, Rashid S, et al. Molecular mechanisms of hepatic steatosis and insulin resistance in the AGPAT2-deficient mouse model of congenital generalized lipodystrophy. Cell Metab. 2009;9:165–176. doi: 10.1016/j.cmet.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukawa M, Akita H, Masuda T, Hayashi Y, Konno T, Ishihara K, et al. 2-Methacryloyloxyethyl phosphorylcholine polymer (MPC)-coating improves the transfection activity of GALA-modified lipid nanoparticles by assisting the cellular uptake and intracellular dissociation of plasmid DNA in primary hepatocytes. Biomaterials. 2010;31:6355–6362. doi: 10.1016/j.biomaterials.2010.04.031. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Akasaka D, Ogasawara H, Sato K, Miyake M, Saito K, et al. Peripheral serotonin enhances lipid metabolism by accelerating bile acid turnover. Endocrinology. 2010;151:4776–4786. doi: 10.1210/en.2009-1349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.